Summary
It is widely accepted that D1 dopamine receptor-expressing striatal neurons convey their information directly to the output nuclei of the basal ganglia while D2-expressing neurons do so indirectly via pallidal neurons. Combining optogenetics and electrophysiology we show that this architecture does not apply to mouse nucleus accumbens projections to the ventral pallidum. Thus, current thinking attributing D1/D2 selectivity to accumbens projections akin to dorsal striatal pathways needs to be reconsidered.
Motivated behavior is thought to be encoded through the ventral striatum in two parallel pathways: direct projections from the nucleus accumbens to the output nuclei of the basal ganglia, namely the ventral mesencephalon (VM), and projections conveying information to the VM indirectly through synapses in the ventral pallidum (VP)1–3. Canonical understanding is that the striatomesencephalic pathway consists of D1-expressing medium spiny neurons (D1-MSNs) while the striatopallidal pathway consists of D2-MSNs, akin to the direct and indirect pathways of the dorsal striatum4. This widely-held assumption has gained momentum with the development of transgenic mice that allow investigators to disentangle the role of D1- and D2-MSNs in regulating motivated behaviors. While recent studies in the dorsal striatum question the complete segregation of the direct/indirect pathways to D1- and D2-MSNs, respectively5, 6, studies in the ventral striatum conclude that manipulations of D1-MSNs in the accumbens are relevant to the striatomesencephalic pathway, while regulating D2-MSNs applies only to the striatopallidal pathway1, 3, 7. However, earlier anatomical data cast doubt on whether the known segregation of the dorsal direct and indirect pathways to D1- and D2-MSNs can be applied to accumbens-VP projections8. First, in contrast to the globus pallidus external (GPe) that sends projections within the basal ganglia, the VP serves also as an output nucleus sending projections outside of the basal ganglia to the mediodorsal thalamus (MDT)9, 10. This poses the possibility that innervation of the VP by D2-MSNs, which are thought to inhibit the thalamus through the striatopallidal pathway, may unexpectedly lead to disinhibition of the thalamus, similar to the activation of the striatomesencephalic pathway. Second, retrograde tracer into the VP co-localizes with both D1- and D2-receptor mRNA in the accumbens11, raising questions regarding the canonical understanding that only accumbens D2-MSNs innervate the VP.
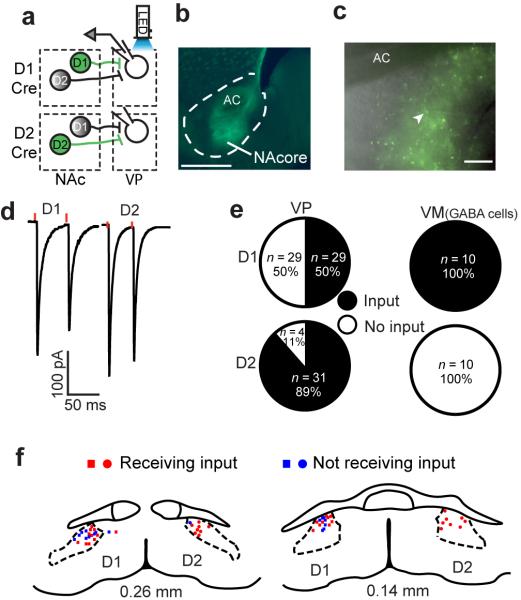
In order to definitively determine the circuitry architecture of D1- and D2-MSN coding of the striatomesencephalic and striatopallidal pathways from accumbens we microinjected a Cre-dependent (floxed) channelrhodopsin (ChR2) viral vector (AAV) largely into the core subcompartment (NAcore) of D1-Cre and D2-Cre transgenic mice (Fig. 1b), with minimal infiltration of the adjacent shell subcompartment (Supplementary Fig. 1a). This allowed selective optical activation of D1- or D2-MSN GABAergic synapses in the VP or the VM (Fig. 1a). Adult mice (P60–P80) were sacrificed 2–3 weeks following bilateral AAV microinjections and neurons were patched in the dorsal VP compartment (dVP; Supplementary Figs. 1b,c) corresponding to the location of GFP expression in MSN afferents (Fig. 1c). Optical stimulation of D1- or D2-MSN terminals in dVP was used to evoke GABAergic IPSCs (eIPSCs). In contrast to expectation, we found that 50% of VP cells received input from D1-MSNs, while 89% showed eIPSCs generated by D2-MSN input (Fig. 1d,e). These findings indicate that up to 50% of the dVP neurons are innervated by both D1- and D2-MSN afferents. Accordingly, the location of D1- and D2-responsive neurons overlapped in the dVP where we focused our recordings (Fig. 1f). The maximum eIPSC amplitudes were the same for D1- and D2-MSN inputs to dVP neurons (392±90 pA and 537±103 pA, respectively) (Fig. 1d, Supplementary Table 1). Interestingly, some parameters of dVP neurons that should not depend on efficacy of ChR2 transfection differed depending on whether they were D1- or D2-MSN receptive, such as D1-MSN receptive neurons having higher capacitance, lower input resistance and greater action potential amplitude (Supplementary Table 1). Finally, Cre-dependent ChR2 expression was specific to Cre-expressing neurons since WT mice injected with Cre-dependent ChR2 in the NAcore showed no GFP expression or eIPSCs in the dVP (Supplementary Figure 2).
Figure 1. NAcore D1-MSNs send input to the VP.
(a) Schematic of the experimental protocol. Cre-dependent ChR2 was injected into the NAcore of D1-Cre (top) or D2-Cre (bottom) mice. Terminals of the infected MSNs (green) were optogenetically activated and responses were recorded in VP neurons. (b) Representative image of injection sites (see Supplementary Figure 1a for composite of injection sites) of Cre-dependent ChR2 into the NAcore (D1-Cre mouse). Calibration bar - 500 μm. AC - anterior commissure. (c) Representative micrograph of Cre-dependent ChR2 expression surrounding the recording pipette (arrowhead) in the dVP. ChR2 expression in dVP was used to define recording site for all experiments. Calibration bar - 150 μm. (d) Representative responses to optogenetic stimulation (red dots) of D1-MSN or D2-MSN input to the dVP, obtained in all experiments involving dVP recordings. (e) Half of the neurons in the dVP received input from NAcore D1-MSNs. In the VM only D1-MSN input from the NAcore was detected. n represents number of cells. n in figure is cells recorded, number of animals was: D1-VP n= 12, D2-VP n= 10, D1-VM n= 3, D2-VM n=3. (f) Location of patched cells exhibiting (red) or not (blue) eIPSCs after stimulation of D1-MSN (squares) or D2-MSN (circles) terminals.
To determine whether the direct projection from accumbens to VM is also comprised of a mixture of D1-MSN and D2-MSN afferents we recorded from cells in the VM surrounded by anterogradely transported GFP. As previously indicated12, only D1-MSN stimulation elicited eIPSCs in the VM (Figure 1e), preferentially innervating electrophysiologically-characterized GABA-like neurons (Supplementary Figure 3)13.
Recently, it was suggested that dorsal striatal innervation of GP is a mixed D1-MSN and D2-MSN projection5, 6. To evaluate this possibility, we injected Cre-ChR2 into the dorsal striatum (Supplementary Figure 4a) and recorded from GP neurons. In contrast to the VP, only 2 out of 11 cells responded to selective D1-MSN optical stimulation, while nearly all GP cells responded to D2-MSN input (Supplementary Figure 4b). The average D1-MSN input detected in GP cells was weaker than accumbens D1-MSN input to the VP (Supplementary Figure 4c,e), while other parameters did not differ (Supplementary Figure 4d,f; Supplementary Table 1). Thus, under the present recording conditions, a contribution by dorsal striatal D1-MSN innervation to GP was confirmed.
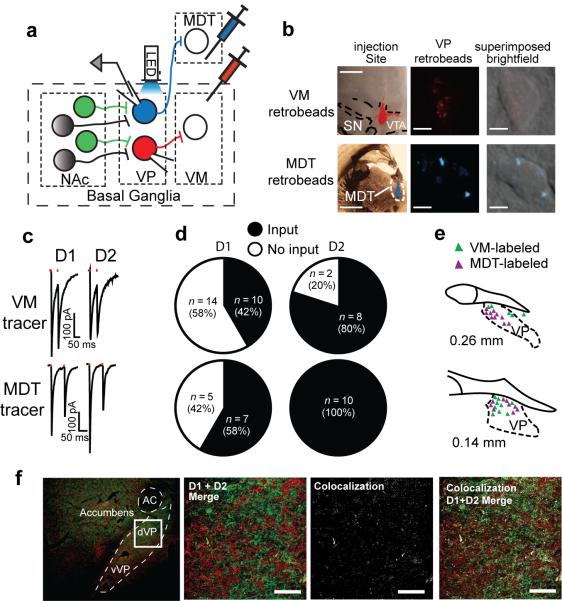
The dVP serves as the relay point between the NAcore and the VM in the indirect pathway. However, the dVP has substantial projections to brain regions outside of the basal ganglia, mainly the MDT9, 10. Thus, although D1-MSNs synapse robustly in the dVP, if they preferentially innervate MDT-projecting dVP neurons their activation will still disinhibit the thalamus, akin to the striatomesencephalic pathway. In order to specifically test the connectivity between D1- and D2-MSNs and VP neurons with mesencephalic or thalamic projections, we microinjected retrograde-tracing microspheres (retrobeads) into the MDT or VM of D1-Cre and D2-Cre mice, and Cre-dependent ChR2 into the NAcore (Figure 2a,b). We then recorded from identified VP neurons projecting to VM or MDT (Figure 2b) and tested whether they received D1-MSN and D2-MSN input. We found that 42% of the VM-projecting dVP neurons received D1-MSN input (Figure 2c,d), thereby definitively showing that D1-MSNs comprise a significant portion of the indirect projections from the NAcore through the VP to the VM. In addition, as expected, most (80%) of the VM-projecting dVP neurons responded to D2-MSN stimulation (Figure 2c,d).
Figure 2. NAcore D1-MSNs form a striatopallidal pathway to the VM.
(a) Schematic of experimental protocol. Cre-dependent ChR2 was injected into the NAcore of D1-Cre or D2-Cre mice while retrobeads were injected into the VM or MDT. Infected terminals (green) were optogenetically activated and retrogradely-labeled VP projection neurons were recorded from. (b) Representative images of retrobeads in the injection sites and in retrogradely-labeled VP neurons. Red MDT retrobeads are shown in cyan pseudo-color. Injection site scale bars - 1 mm. Retrobeads scalebars - 5 μm. (c) Representative traces of optogenetically-generated IPSCs recorded from retrogradely-labeled VP neurons in D1-Cre and D2-Cre mice. (d) Approximately half of the VM-projecting VP neurons received D1-MSN input from the NAcore while 80% received D2-MSN input. All MDT-projecting VP neurons responded to D2-MSN activation while 58% received D1-MSN input from the NAcore. n in figure represents number of cells, number mice was: D1-VM n= 6, D2-VM n= 3, D1-MDT n= 3, D2-MDT n= 3. (e) In the more rostral VP VM-projecting (red) neurons tended to be dorsolateral to the MDT-projecting (blue) neurons while at the caudal VP both projection cells were intermingled. (f) 2.4% (n= 3) co-localization of D1- and D2-expression in fibers in the dVP was quantified in D1-tomato X D2-GFP BAC transgenic mice. The left panel shows low magnification of the location in the dVP where we quantified fiber co-expression shown in the next 3 panels. Bar= 50 μm.
A parallel experiment examining D1- and D2-MSN input to MDT-projecting dVP neurons revealed substantial D2-MSN input onto this population of dVP neurons serving as output neurons of the basal ganglia (Figure 2c,d). Over half of the MDT-projecting neurons received D1-MSN input and 100% of the neurons responded to D2-MSN stimulation. This experiment definitively shows that accumbens D2-MSNs innervate output neurons of the basal ganglia. Thus, activation of these D2-MSNs is expected to disinhibit thalamic activity, akin to the activation of the striatomesencephalic pathway. Previous studies estimate 5–15% of neurons across the VP send collaterals to both the VM and MDT9, 14, indicating that the D2-MSN projection to MDT-projecting dVP neurons is largely a selective projection out of the basal ganglia. Moreover, low collateralization is consistent with the MDT-projecting neurons in rostral dVP being located more medial and ventral than the VM-projecting neurons, although there was no difference in the topographical organization of recorded VM- and MDT-projecting neurons in the caudal VP (Fig. 2e). Only minor differences in passive membrane properties (Supplementary Table 2) between VM-projecting or MDT-projecting VP neurons were observed.
It has been estimated that up to 6% of NAcore MSNs co-express D1 and D2 receptors (D1/D2-MSNs)15, posing the possibility that dual innervation of dVP may arise from coexpressing MSNs. This is improbable given that ~30% of NAcore neurons retrogradely labeled from dVP express D1 or preprotachykinin mRNA11. Moreover, previous estimates of co-localization were indirect15, while the literature16 and our own cell counts of neurons double-labeled for D1/D2 in adult BAC transgenic mice indicate <2% of the neurons are D1/D2-MSNs (Supplementary Fig. 5, Supplementary Table 3). Finally, quantitative analysis of D1- and D2-expressing fibers in dVP revealed 2.37±0.22% (n=3) overlap between reporter proteins (Fig. 2f), strongly indicating separate D1- and D2-MSN projections to dVP.
Our data provide definitive evidence that the classic NAcore-dVP-VM projection includes major contributions from both D1- and D2-MSNs. Similarly, the D1- and D2-MSNs directly innervate output neurons of the basal ganglia located in the VP by synapsing on dVP neurons projecting to the MDT. Thus, both D1- and D2-MSNs may inhibit or disinhibit thalamic activity depending on their projection pattern and not on their genetic characteristics. Therefore, the `unanchoring' of D1- and D2-MSNs from their coding of the classic striatal pathways indicates that genetic targeting of D1- or D2-MSNs does not confer specificity for regulating NAcore direct or indirect projections to output nuclei of the basal ganglia1, 17, 18. The importance of the NAcore projection to the dVP in regulating motivated behavior has been known for decades19, and recently validated using genetic strategies20. The breakdown of selective D1- and D2-MSN synaptic modulation of the different projections from the NAcore indicates that the regulation of behavior by NAcore MSNs synapsing in the dVP may be substantially more nuanced than has been assumed based on extensive study of largely non-overlapping projections from dorsal striatal D1- and D2-MSNs.
Supplemental Experimental Procedures
Animals
Mice were C57BL/6J (wild type, P60-P80) or heterozygous bacterial artificial chromosome (BAC) transgenic mice in which cyclic recombinase (Cre) was expressed under the control of the D1R promoter (Drd1a (D1)-Cre, line FK150) or the D2R promoter (Drd2 (D2)-Cre, line ER44). Mice were obtained from N. Heintz and P. Greengard (Rockefeller), C. Gerfen (NIMH) and NINDS/GENSAT (www.gensat.org)21, 22. Males and females were used. Mice were fed ad libitum and maintained at ~22 °C in a humidity-controlled environment on a reverse 12-hr light/dark cycle (lights off at 9:00 AM). Mice were single housed after surgery. All experiments were conducted in accordance with the National Institute of Health Guidelines for the Care and Use of Laboratory Animals and all procedures were approved by the Institutional Animal Care and Use Committee at the Medical University of South Carolina.
Microinjections
Mice were anesthetized with ketamine (150 mg/kg) and xylazine (12 mg/kg) and placed in a stereotaxic instrument (Kopf, Tujunga, CA). Thirty-three gauge needles were used for microinjecting either AAV2-EF1a-DIO-hChR2(H134R)-EYFP (Cre-dependent ChR2) (University of North Carolina Vector Core Facility) or red retrobeads (Lumafluor, Naples, FL). ChR2 was bilaterally infused (0.3 μl) at 0° angle into the NAcore (AP +1.55; ML ±1.00, DV −4.80) or the dorsal striatum (AP +0.32; M/L ±2.0; D/V −3.1). The red retrobeads were microinjected (0.3 μl) at 0° angle either into the ventral mesencephalon (VM) (AP −2.80; ML ±0.70; DV −4.55) or the mediodorsal thalamus (MDT) (AP −1.50; ML ±0.5; DV −3.7). Injections were performed using a 2.5 μl syringe (Hamilton Company, Reno, NV). After the injection the needle was left in place for 5 min to allow diffusion of the virus/retrobeads. Microinjected mice were allowed at least 2 weeks to allow for virus expression before slices were prepared.
Histology
For verification of microinjection accuracy, fresh slices (100 μm) were obtained from brains used for electrophysiological recordings in the VP. Slices were then fixed in 4% (w/v) paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 1−2 h and mounted on slides for visualization. Slices were visualized using an Olympus BX40 fluorescent microscope. The EYFP tag expressed together with ChR2 was examined with an Olympus U-MWIB filter (excitation at 460–490 nm) while the red retrobeads were visualized using an Olympus U-MWIG filter (520–500 nm).
Immunohistochemistry and cell counting
D1-Tomato X D2-GFP BAC transgenic mice were perfused with 4% (w/v) PFA in phosphate-buffered saline (PBS) followed by cryoprotection in 30% sucrose. Brains were sectioned on a cryostat (Leica Biosystems) in the sagittal plane at 40 μm. Sections were blocked in 3% normal donkey serum with 0.01% Triton-X in PBS for 1 h on the shaker at room temperature and then incubated in primary antibodies in block overnight on the shaker at room temperature. Antibodies used were the following: mouse anti-NeuN (1:1000, catalog #MAB377, Millipore), chicken anti-GFP (1:5000, catalog #10–20, Aves), and rabbit anti-dsRed (1:2000, catalog #632496, Clontech). The next day, sections were rinsed in PBS followed by a 1 h incubation in secondary antibodies: donkey anti-rabbit Cy3, donkey anti-mouse Alexa-647, and donkey anti-chicken Alexa-488 (Jackson ImmunoResearch Laboratories). Cell counting was performed with ImageJ Cell Counter software (ImageJ, NIH). Cells were counted using 200 μm × 200 μm nucleus accumbens (core and shell) images from three D1-tomato X D2-GFP mice. NeuN cells were counted per brain region per mouse, and then the number of GFP+, tomato+, and GFP+:tomato+ cells were counted in each region.
Imaging and co-localization analysis
Image stacks were taken at 63x using a Leica SP6 confocal (Leica Microsystems, Germany) with a step size of 0.5 micrometer in the z-plane and 1024 × 1024 pixel resolution in the xy-plane (0.24 μm per pixel). Series were deconvolved using AutoQuant X2 (MediaCybernetics, Rockville MD) and 3D co-localization analysis was performed using ImarisColoc (Bitplane, Concord MA). Co-localization was calculated as percentage overlap within a masked region of interest for each channel, and then averaged. Where possible co-localization threshold values were determined automatically using the Costes and Lockett algorithm. In total 3 to 5 stacks were taken of the subcommissural dVP for each animal.
Slice preparation for electrophysiology
Coronal slices were prepared 2–3 weeks following bilateral injection of the Cre-dependent ChR2. Mice were anesthetized with ketamine (150 mg/kg) and decapitated. Fresh VP slices (190–200 μm; VT1200S Leica vibratome) were collected into a vial containing artificial cerebrospinal fluid (aCSF) (in mM: 126 NaCl, 1.4 NaH2PO4, 25 NaHCO3, 11 glucose, 1.2 MgCl2, 2.4 CaCl2, 2.5 KCl, 2.0 NaPyruvate, 0.4 ascorbic acid, bubbled with 95% O2 and 5% CO2) and a mixture of 5 mM kynurenic acid and 100 μM MK-801 maleate. Slices were kept at room temperature until they were used for recordings. Microinjection placements were verified for every mouse. Mice with missed injections were not further used.
Whole-cell patch-clamp recording
Slices were constantly perfused with oxygenated aCSF heated to 32 °C (TC-344B, Warner Instruments, Hamden CT). Neurons were visualized with an Olympus BX51WI microscope at 60X. For visualization of EYFP or the red retrobeads 460 nm or 530 nm LEDs (Prizmatix, Israel), respectively, were used. Multiclamp 700B (Axon Instruments, Union City, CA, USA) was used to record synaptic currents and cellular parameters. Recordings of synaptic currents were performed in voltage-clamp configuration at a membrane potential of −80 mV. For detection of Ih currents in VM neurons membrane potential was held at −60 mV. Action potentials and membrane potential were recorded immediately after invading the cell in current clamp configuration. Glass microelectrodes (1.1–1.9 MΩ tip resistance) were prepared using a PC-10 vertical puller (Narishige, Tokyo, Japan) and filled with internal solution (in mM: 68 KCl, 65 D-gluconic acid potassium salt, 7.5 HEPES potassium, 1 EGTA, 1.25 MgCl2, 10 NaCl, 2.0 MgATP, and 0.4 NaGTP; pH 7.2–7.3, 275 mOsm). GABAergic projection neurons were selected by virtue of their morphology and passive electrophysiological properties23, 24. Data were acquired at 10 kHz and filtered at 2 kHz using Axograph X software (Axograph Scientific, Sydney, Australia). Inhibitory postsynaptic currents (IPSCs) were evoked optogenetically by a 4 ms long LED pulse that was transmitted on the slice through the microscope objectives. Recordings of synaptic transmission were started 10 min after invading the cell to allow for internal solution diffusion to remote dendrites. Membrane capacitance and input resistance were calculated automatically from a −2 mV pulse by Axograph X. Recordings with unstable series resistance or with a series resistance higher than 15 MΩ were discarded.
Statistics
All statistical analyses were performed using Graphpad Prism 6.0, and data collection and analysis were not performed blind to the conditions of the experiments. No data points were removed from statistical analysis. Two-tailed unpaired t-test was used in most comparisons as data was normally distributed (examined with D'Agostino & Pearson omnibus normality test). Amplitudes of D1- and D2 eIPSC amplitudes did not distribute normally and the Mann-Whitney test was used. Statistical tests are indicated in the main text and each figure's legend. In all experiments no statistical methods were used to predetermine sample sizes, but sample size is similar to sample sizes routinely used in the field for similar experiments24. In analysis of GP neurons sample size was limited by the number of GP neurons responding to D1-MSN activation.
Supplementary Material
Acknowledgements
We thank Dr. T.C. Jhou and R.J. Smith for their assistance with the histology experiments. We thank Dr. E.J. Nestler at Mt Sinai School of Medicine for providing D1-Cre and D2-Cre breeding pairs, and R. Gregory and J. Hyang for genotyping of the mice. This work was supported by USPHS grants DA03906, DA12513, DA015369 and a fellowship from the Neuroscience Institute, Medical University of South Carolina.
Footnotes
Author contributions Y.M.K. and P.W.K. designed the experiments, Y.M.K., R.M.B. and D.J.S. performed microinjection surgeries, J.A.H. and M.K.L. performed double-labeling studies, Y.M.K. performed electrophysiology and histology experiments and analyzed the data, Y.M.K and P.W.K. wrote the paper; all authors discussed the results and commented on the manuscripts.
References
- 1.Bock R, et al. Nature neuroscience. 2013;16:632–638. doi: 10.1038/nn.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacAskill AF, Cassel JM, Carter AG. Nat Neuroscience. 2014;17:1198–1207. doi: 10.1038/nn.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yawata S, Yamaguchi T, Danjo T, Hikida T, Nakanishi S. PNAS. 2012;109:12764–12769. doi: 10.1073/pnas.1210797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerfen CR, Surmeier DJ. Ann Rev Neuroscience. 2011;34:441–466. doi: 10.1146/annurev-neuro-061010-113641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cazorla M, et al. Neuron. 2014;81:153–164. doi: 10.1016/j.neuron.2013.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saunders A, et al. Nature. 2015 [Google Scholar]
- 7.MacAskill AF, Little JP, Cassel JM, Carter AG. Nat Neuroscience. 2012;15:1624–1626. doi: 10.1038/nn.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith RJ, Lobo MK, Spencer S, Kalivas PW. Curr Opin Neurobiol. 2013;23:546–552. doi: 10.1016/j.conb.2013.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tripathi A, Prensa L, Mengual E. Brain Struct Funct. 2013;218:1133–1157. doi: 10.1007/s00429-012-0451-0. [DOI] [PubMed] [Google Scholar]
- 10.Zahm DS, Zaborszky L, Alheid GF, Heimer L. J Comp Neurol. 1987;255:592–605. doi: 10.1002/cne.902550410. [DOI] [PubMed] [Google Scholar]
- 11.Lu X-Y, Ghasemzadeh MB, Kalivas PW. Neuroscience. 1998;82:767–780. doi: 10.1016/s0306-4522(97)00327-8. [DOI] [PubMed] [Google Scholar]
- 12.Bocklisch C, et al. Science. 2013;341:1521–1525. doi: 10.1126/science.1237059. [DOI] [PubMed] [Google Scholar]
- 13.Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, Uchida N. Neuron. 2012;74:858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Leung BK, Balleine BW. J Neuroscience. 2015;35:4953–4964. doi: 10.1523/JNEUROSCI.4837-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertran-Gonzalez J, et al. J Neuroscience. 2008;28:5671–5685. doi: 10.1523/JNEUROSCI.1039-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thibault D, Loustalot F, Fortin GM, Bourque MJ, Trudeau LE. PLoS One. 2013;8:e67219. doi: 10.1371/journal.pone.0067219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kravitz AV, Tye LD, Kreitzer AC. Nat Neuroscience. 2012;15:816–818. doi: 10.1038/nn.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lobo MK, Nestler EJ. Front Neuroanatomy. 2011;5:41. doi: 10.3389/fnana.2011.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mogenson GJ, Jones DJ, Yim CY. Prog Neurobiology. 1980;14:69–97. doi: 10.1016/0301-0082(80)90018-0. [DOI] [PubMed] [Google Scholar]
- 20.Stefanik MT, Kupchik YM, Brown RM, Kalivas PW. J Neuroscience. 2013;33:13654–13662. doi: 10.1523/JNEUROSCI.1570-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong S, et al. J Neuroscience. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong S, et al. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 23.Bengtson CP, Osborne PB. J Neurophysiology. 2000;83:2649–2660. doi: 10.1152/jn.2000.83.5.2649. [DOI] [PubMed] [Google Scholar]
- 24.Kupchik YM, Kalivas PW. Brain Struct Funct. 2013;218:1487–1500. doi: 10.1007/s00429-012-0471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.