Abstract
Purpose
Gluten proteins, the culprits in celiac disease (CD), show striking similarities in primary structure with human salivary proline-rich proteins (PRPs). Both are enriched in pro-line and glutamine residues that often occur consecutively in their sequences. We investigated potential differences in the spectrum of salivary PRPs in health and CD.
Experimental design
Stimulated salivary secretions were collected from CD patients, patients with refractory CD, patients with gastrointestinal complaints but no CD, and healthy controls. PRP isoforms/peptides were characterized by anionic and SDS-PAGE, PCR, and LC-ESI-MS.
Results
The gene frequencies of the acidic PRP isoforms PIF, Db, Pa, PRP1, and PRP2 did not differ between groups. At the protein level, PRPs peptides showed minor group differences, but these could not differentiate the CD and/or refractory CDs groups from the controls.
Conclusions and clinical relevance
This extensive study established that salivary PRPs, despite similarity to gluten proteins, show no apparent correlation with CD and thus will not serve as diagnostic markers for the disease. The structural basis for the tolerance to the gluten-like PRP proteins in CD is worthy of further exploration and may lead to the development of gluten-like analogs lacking immunogenicity that could be used therapeutically.
Keywords: Celiac disease, Gliadin, Proline-rich protein, Saliva
1 Introduction
Human saliva contains a high concentration of proline-rich proteins (PRPs) secreted by the major and minor salivary glands [1–4]. The PRPs in saliva are divided into acidic and basic PRP protein families [5]. Functionally, the phosphorylated domains within the acidic PRPs have a high affinity for hydroxyapatite and participate in the formation of the acquired enamel pellicle, a protective proteinaceous layer covering the tooth surface [6–8]. A clear function for the basic PRPs within the oral cavity, other than tannin binding, and lubrication by the glycosylated PRB3 variant [9, 10] has not yet been established. Therefore, discovery of additional functional roles for PRPs can be expected. Such functions could extend well beyond the oral cavity since significant volumes of saliva (about 0.8 L) are being swallowed each day, thus reaching the gastrointestinal tract.
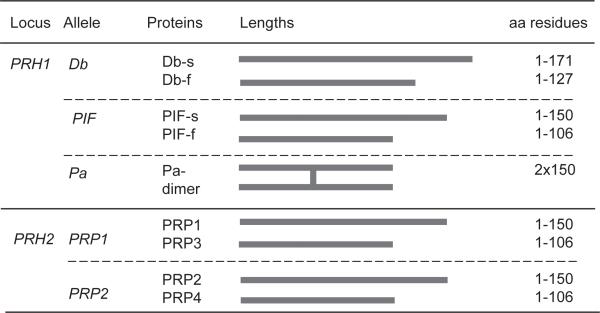
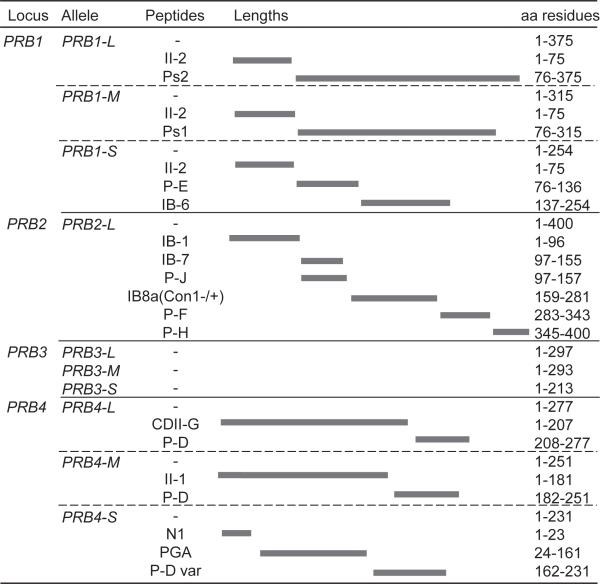
Acidic PRPs are encoded by five different genes: Db, PIF, and Pa at the PRH1 locus, and PRP1 and PRP2 at the PRH2 locus [11]. They are proteins of approximately 150 residues in length, with the exception of the Db protein which is 171 amino acid residues long. Pa protein contains a cysteine residue, and in human saliva appears in a dimeric form of 300 residues (Fig. 1). Native acidic PRP gene products as well as posttranslationally cleaved isoforms can be detected in human saliva. The major basic PRPs in saliva are encoded by four genes: PRB1, PRB2, PRB3, and PRB4. Three of the four basic PRP alleles show polymorphisms, resulting in variable length isoforms due to different numbers of repeat domains in exon 3 [12]. In contrast to acidic PRPs, basic PRPs are completely degraded within the glandular secretory vesicles, with the exception of the glycosylated PRB3 (Fig. 2) [13]. Due to the various alleles and posttranslational processing of PRPs within the gland and after secretion, human saliva contains a rich mixture of acidic and basic PRP-derived proteins and peptides [14–21].
Figure 1.
Most common acidic PRP isoforms in human saliva.
Figure 2.
Most common basic PRP isoforms in human saliva.
Interestingly, salivary PRPs show an unusual structural similarity to the dietary gluten proteins found in wheat, rye, and barley [22]. Upon ingestion, gluten peptides can cause a celiac disease (CD) in genetically predisposed individuals carrying the human lymphocyte antigens HLA-DQ2 or HLADQ8 (where HLA is human leukocyte antigen) on professional antigen presenting cells, leading to T-cell activation and small intestinal inflammation, villous atrophy, crypt hyperplasia, and a broad spectrum of intestinal and extraintestinal symptoms [23–25]. In most patients, CD can be treated by a strict gluten-free diet (GFD), except for patients with refractory CD (RCD) whose disease does not respond to dietary gluten exclusion [26–28]. Like PRPs, gluten proteins, which in wheat encompasses the monomeric gliadins and the cross-linked glutenins, are rich in proline and glutamine residues that comprise approximately 50% of their amino acid composition [29] in repetitive XPQ sequences (X = variable amino acid). Importantly, these sequences are found in gluten peptides that are presented by HLA-DQ2 or HLA-DQ8 and that are recognized by inflammatory T cells from the duodenum of CD patients [30].
Despite the structural similarities between gluten proteins and salivary PRPs, salivary PRPs are unlikely to cause CD, since gluten exclusion from the diet usually resolves CD-associated symptoms in most cases, while salivary PRPs are continually “ingested.” However, PRP might play a role in either promoting or mitigating CD or its remission on a GFD. In this vein, apart from the central predisposition (HLA-DQ2 or HLA-DQ8) and minor known genetic associations [25,31, 32], PRP variants could predispose to or protect patients from CD. Furthermore, PRP variants might contribute to the small fraction of patients with RCD. Given these considerations, the salivary gluten-like PRP gene family clearly deserves further exploration in relation to CD.
The present study was undertaken to test the hypothesis that CD and/or RCD patients differ in the type and quantity of PRP isoforms that can be found in their saliva, and to decipher and compare the highly complex PRP isoform patterns in patients and controls. The goal was to elucidate differences in health and CD that could potentially be useful diagnostically or therapeutically.
2 Materials and methods
2.1 Subjects and inclusion/exclusion criteria
Parotid secretion (PS) and whole saliva (WS) were collected from four groups of subjects: (i) healthy subjects (healthy controls [HC]) having no signs (genetic, serological, or histological) or symptoms of CD or gluten sensitivity and presenting in overall good health (n = 19), (ii) nonceliac patients (gastrointestinal [GI]) suffering from nonimmune-mediated GI symptoms and in whom CD was excluded by serological and histological testing (n = 11), (iii) CD patients: with positive anti-deamidated gliadin peptide and/or anti-TG2 (tissue transglutaminase) antibodies [33], duodenal villous atrophy at diagnosis and who were clinically responsive to a GFD (n = 20), and (iv) RCD patients (nonmalignant type I) who were previously diagnosed with CD and met criteria for RCD after a minimum of 6 months on a GFD [34] (n = 8). CD, RCD, and nonceliac GI subjects were recruited at Beth Israel Deaconess Medical Center, Boston, and HC subjects were recruited at Boston University Goldman School of Dental Medicine. All subjects enrolled were at least 18 years old and able to comply with study requirements. Exclusion criteria were illicit drug or excessive alcohol use, unstable or uncontrolled heart, kidney or liver disease, a clinically defined mental illness, sicca syndrome, or overt signs of severe dental or periodontal health issues. The study was approved by the Committee for Clinical Investigations (CCI) at the Beth Israel Deaconess Medical Center and the Institutional Review Board at Boston University, and all subjects provided their informed consent prior to participation in the study.
2.2 Collection of saliva samples
Donors of saliva samples refrained from eating, smoking, drinking (except water), mouth wash, and tooth brushing for at least 1 h. Donors were asked to rinse their mouth with water three times and were then given a 20 × cm 20 cm piece of Parafilmtm (Sigma, St. Louis, MO) for masticatory stimulation of saliva secretion. WS was expectorated into a graded 50 mL centrifuge tube placed on ice and the time for collection of a 10 mL volume was recorded. After vortexing, the collected WS was centrifuged at 16 000 × g for 10 min at 4°C to separate the WS supernatant from the WS pellet.
PS was collected by placing a Curby cup over the orifice of the Stensen's duct [35]. Sour candies were provided to donors for stimulation of PS secretion. Volumes of 10 mL PS were collected on ice and the collection time was recorded. Immediately after collection and processing all saliva samples were aliquoted and stored at −80°C until analysis.
2.3 SDS-PAGE
Aliquots of 35 μL PS were boiled for 5 min in diluted 4 × 2-[Bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)-1,3-propanediol (Bis-Tris) sample buffer and applied to NuPAGE ® Novex ® 12% Bis-Tris Gels (Invitrogen, Carlsbad, CA, USA). Electrophoresis was carried out at a constant voltage of 120 V for about approximately 1.5 h at room temperature. The gels were stained with 0.1% CBB R-250 in 40% ethanol and 10% acetic acid for 16 h, and destained in 10% acetic acid for 2–3 days as described [36].
2.4 Anionic gel electrophoresis
Anionic gel electrophoresis was conducted as previously described [4, 37, 38] using the mini-gel system (Bio-Rad, Hercules, CA, USA). The separating gel contained 7.5% acrylamide, 0.375 M Tris (pH 8.9), and 0.07% ammonium persulfate. The stacking gel contained 6.7% acrylamide, 41.7 mM Tris (pH 6.7), 0.3% riboflavin, and 26.7% sucrose. For qualitative assessment of the PRP patterns, aliquots of 50 μL PS were dried using a speedvac (Savant, ThermoFisher Scientific, Waltham, MA) and resolved in sample buffer containing 0.06 M Tris-HCl (pH 6.7), 19.2% sucrose, and 11.9 μM bromophenol blue. The running buffer contained 24.9 mM Tris and 191.8 mM glycine (pH 8.3). Electrophoresis was performed at 120 V for approximately 2 h. The gels were stained with 0.5% Amido Black in 7% acetic acid for 16 h, and destained with 7% acetic acid.
2.5 DNA isolation from WS pellet
Bacterial DNA was isolated from WS pellet derived from 1 mL WS using the MasterPure-Gram Positive DNA Purification Kit (Epicentre, Madison, USA) according to the manufacturer's instructions. Briefly, to each aliquot of WS pellet 150 μL TE buffer containing Ready-Lyse Lysozyme was added and samples were incubated for 16 h at 37°C. Subsequently, 150 μL of proteinase K/Gram positive lysis solution was added followed by incubation at 65–70°C for 15 min. After cooling to room temperature, 175 μL of protein precipitation reagent was added to the 300 μL of lysed sample. The supernatant was separated from the debris by centrifugation at 16 000 × g at 4°C for 10 min, to which an aliquot of 1 μL of RNase A (5 μg/μL) was added, followed by incubation at 37°C for 30 min. Thereafter, 500 μL of isopropanol was added, and the DNA was pelleted by centrifugation at 4°C for 10 min at 16 000 × g. The supernatant was removed and the pellet was washed once with 70% ethanol, and suspended in 50 μL of 10 mM Tris (pH 8.0). The DNA was kept at −20°C until analysis.
2.6 PRH1 gene amplification with PCR
Primers for the PRH1 gene were designed for amplification of exon 3 by Zakhary et al. [39]. The forward primer was 5′-GTGATGGGAACCAGGATGATGG-3′, and the reverse primer was 5′-AAACTGGAATCGTACCTGTCATT-3′. PCR was performed on DNA isolated from WSP. The PCR reaction (total volume 25 μL) contained 0.3 μL WS pellet DNA (containing 20.7–195.3 μg DNA, subject variation), 12.5 μL TopTaq Mater Mix containing 0.92 units of TopTaq DNA Polymerase (Qiagen, Hilden, Germany), 0.5 μL forward and reverse primer mix (both final concentrations are 200 nM), and 11.7 μL RNAse free water (Qiagen). The PCR was performed in Bio-Rad T100 Thermal Cycler PCR system (Bio-Rad) under the following conditions (38 cycles): DNA denaturation at 94°C for 30 s, annealing at 62°C for 1 min, and extension at 72°C for 1 min. The amplification products were separated on a 2.5% agarose gel and visualized with SYBR Safe DNA gel stain (Invitrogen).
2.7 RP-HPLC-ESI-MS for acidic and basic PRP's characterization and quantification
For PRP peptide analysis by LC-ESI-MS, saliva samples were immediately mixed 1:1 (v/v) with 0.2% TFA in ice bath and the solution centrifuged at 8000 × g at 4°C for 5 min. The acidic supernatant was separated from the precipitate and 100 μL (corresponding to 50 μL of saliva) analyzed by LC-ESI-MS. The HPLC-ESI-IT-MS apparatus was a surveyor HPLC system connected by a T splitter to a photo diode-array detector and to an LCQ Advantage mass spectrometer (ThermoFisher, San Jose, CA, USA). The MS apparatus was equipped with an ESI source. The chromatographic column was a 150 × 2.1 mm Vydac (Hesperia, CA, USA) C8 column, with 5-μm particle diameter. The following solutions were utilized for RP-HPLC separation: (eluent A) 0.056% aqueous TFA and (eluent B) 0.05% TFA in ACN–water 80/20. The gradient applied for the analysis of saliva was linear from 0 to 55% of B in 40 min, and from 55 to 100% of B in 10 min, at a flow rate of 0.30 mL/min. The T splitter permitted 0.20 mL/min to flow toward the diode-array detector and 0.10 mL/min to flow toward the ESI source. The diode-array detector was set at 214 and 276 nm. Mass spectra were collected every 3 ms in the positive ion mode. The MS spray voltage was 5.0 kV, and the capillary temperature was 260°C. Deconvolution of the averaged ESI-MS spectra was automatically performed by using MagTran 1.0 software [40].
Salivary peptide semiquantitative analysis was based on the area of the RP-HPLC-ESI-MS extracted ion current (XIC) peaks, measured when the S/N was at least 5. The XIC analysis reveals the peak associated with the protein of interest by searching the specific multiple charged ions generated at the source by the protein. The ions used to quantify the proteins/peptides were carefully selected to exclude values in common with other coeluting proteins, and were the same as those reported previously [41]. The area of the ion current peak is proportional to the concentration, and under constant analytical conditions can be used for a semiquantitative analysis and to compare levels of the same analyte in different samples [42]. When samples were of insufficient quality, for example, when there was evidence of extensive degradation, they were excluded from analysis.
2.8 Statistical analysis
SPSS 15.0 software was used for statistical analysis. Comparisons of PS flow rate and protein concentrations were made with the Mann–Whitney test. Significant differences between the expected and observed frequencies in the PRH1 allele frequencies between groups were tested with the chi-square test. The ANOVA (GraphPad Prism, version 4.0, one-way ANOVA) was used to evidence differences between groups in levels of acidic and basic PRPs determined by MS, and the post hoc analysis was performed with the Tukey's test when the overall p values were less than 0.05.
3 Results
3.1 Unusual structural similarity between PRPs and gliadin proteins
The primary amino acid sequences of four gliadins from wheat and two basic PRP proteins from human saliva are shown in Fig. 3. The shared high content of P and Q (highlighted in red), and the consecutive occurrence of these residues in both gliadins and PRPs is evident. Differences can also be noted. For instance, basic PRPs contain repeat domains, typically approximately 21 amino acids in length, which cannot be readily discerned in the gliadin proteins. Given the high overall similarities though, and the association of gluten (gliadins) with CD, we next investigated if salivary PRPs patterns would differ in health and CD.
Figure 3.
Primary structures of four wheat gliadins (top four proteins) and two human salivary basic PRPs (bottom two proteins).
3.2 Demographic information and saliva sample characteristics
PSs were collected from the four groups of subjects, HC, nonceliac GI patients, CD patients, and RCD patients. The patient demographics and PS sample characteristics (stimulated flow rate and protein concentration) are summarized in Supporting Information Table 1 and Table 1. The subjects with diet-responsive CD were following a GFD (mean 43.1 months, SD 56.1 months). The subjects with RCD were also following a GFD (mean 85 months, SD 57.5 months). All CD patients in the study had abnormal small bowel biopsies diagnostic for CD. Repeat biopsies were not obtained as part of this study but subjects with RCD had all shown persisting villous atrophy (Marsh III lesions) on repeat biopsy, despite long-term treatment with a strictly GFD. In subjects with responsive CD average IgA anti tissue transglutaminase (from serum drawn on average 2 wk from the day of saliva collection) were 36.3 units (SD 33.5). The corresponding IgA anti tissue transglutaminase titers in serum, drawn on average less than 1 wk from the day of saliva collection, for subjects with RCD were 23.6 units (SD 25.7). Race and gender distribution of the healthy donors (HC) were matched to the CD groups. The average age of the RCD group was higher than that of the HC and CD subjects, consistent with the occurrence of RCD in older people [43]. The stimulated PS flow rates did not differ between groups, and the average PS protein concentration was slightly higher in the RCD group compared to the CD group (p = 0.037) but no statistical differences were found between any other groups.
Table 1.
Demographic information, PS flow rate, and protein concentration in samples collected from CD patients and controls
| HC n = 19 | GI n = 11 | CD n = 20 | RCD n = 8 | |
|---|---|---|---|---|
| Age (Mean ± SD) | 33.6 + 13.9 | 41.0 + 148 | 35.1 ± 16.6 | 54.1 + 13.5a),b) |
| Gender (%) | M 5 (26.3%) F 14 (73.796) | M 3 (27.3%) F 8 (72.7%) | M 3(15%) F 17(85%) | M 3 (37.5%) F 5 (62.5%) |
| Race | Caucasian | Caucasian | Caucasian | Caucasian |
| PS flow rate (mL/min) | ||||
| Mean ± SD | 0.84 ± 0.47 | 0.82 ± 0 42 | 0.73 ± 0.39 | 1.10 ± 0.96 |
| Minimum/maximum | 0.3–2.08 | 0.19–1.41 | 0 21–1.93 | 0.3–3.05 |
| Median | 0.75 | 0.93 | 0.65 | 0.695 |
| PS protein concentration (mg/mL) | ||||
| Mean ± SD | 1.07 + 0.25 | 1.29 ± 0.52 | 1.03 + 0.27 | 1.21 + 0.13c) |
| Minimum/maximum | 0.57–1.51 | 0.62–2.4 | 047–1.36 | 0.99–1.33 |
| Median | 1.11 | 1.13 | 1.03 | 1 27 |
HC, healthy controls; GI, (unrelated) gastrointestinal disorders; CD, celiac disease; RCD, refractory CD.
Statistically significant difference between HC and RCD group (p = 0.007).
Statistically significant difference between CD and RCD group (p = 0.018).
Statistically significant difference between CD and RCD group (p = 0.037).
All statistical analysis were performed with the Mann-Whitney test.
3.3 PRPs in saliva from HC, CD, RCD, and GI patients assessed by SDS-PAGE
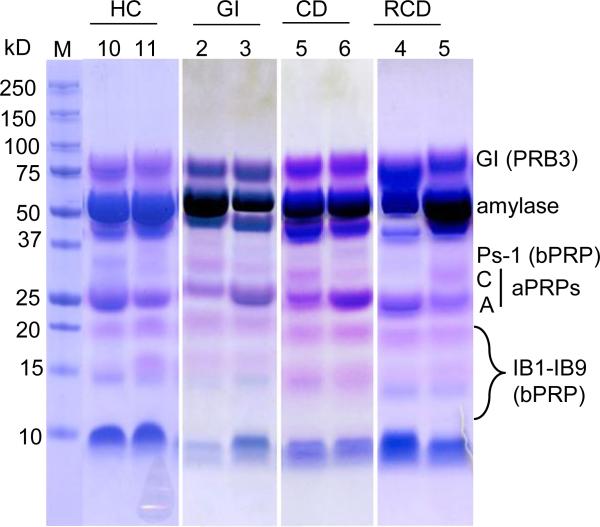
SDS-PAGE and a modified Commassie blue staining method [36] were used to assess the general pattern of acidic and basic PRPs in PS from patient and control groups. The utilized method of destaining takes advantage of the metachromasia of PRPs which, upon destaining, turn pink or violet, while other proteins remain blue-stained. The aim was to determine if a distinct pattern was associated with a particular patient group. A gel with two subjects per patient group is shown in Fig. 4, and gels of all subjects are shown in Supporting Information Fig. 1. Salivary protein patterns, including PRP patterns, show overall similarities, but also noticeable inter-subject variation. This can in part be explained by the large number of PRP polymorphic isoforms in human saliva (Figs. 1 and 2). After careful comparison of patterns in each group, we were unable to elucidate a banding pattern selectively associated with a particular group of subjects. A more detailed analysis of acidic and basic PRP isoform patterns in the four groups was subsequently performed.
Figure 4.
SDS-PAGE (12% gel) of PS from two patients of each group. Aliquots of 35 μL PS were loaded for each subject. The gel was stained with Coomassie Blue R-250 using a modified destaining method [36]. The PRPs stain pink or violet while other proteins stain blue.
3.4 Acidic and basic PRP isoforms in CD patients and healthy controls
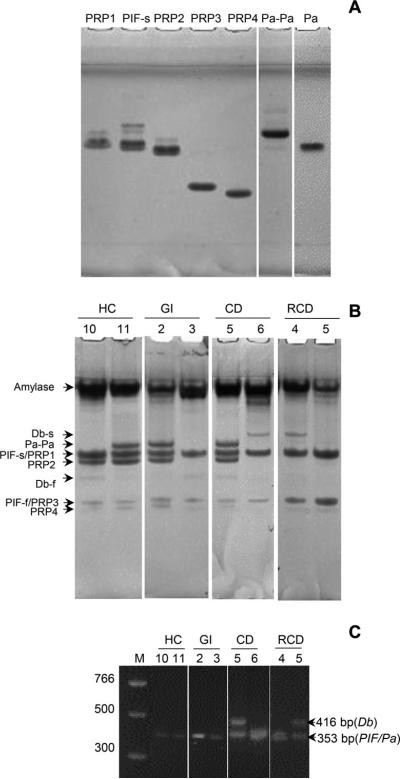
To selectively visualize the acidic PRP isoforms, the PS samples were analyzed by anionic PAGE. Figure 3A shows the position in the gel of acidic PRP isoforms, which had previously been purified in our laboratory [5]. Except for PIF-s and PRP1, all acidic PRP isoforms could be separated based on electrophoretic mobility. The identity of the Pa dimer (Pa-Pa) was confirmed with the dissociation of dimers into monomers upon incubation with DTT (Fig. 5A, far right lane). Figure 5B shows the analysis of two PS samples per patient group, and the gels of all subjects are shown in Supporting Information Fig. 2.
Figure 5.
Analysis of acidic PRP isoforms in human PS. (A) Ornstein Davis gel electrophoresis of PRP isoforms. Lanes 1–6, pure and semipure PRP isoforms (20 μg protein/lane); lane 7, Pa dimer (20 μg) incubated with DTT. (B) Ornstein Davis gel electrophoresis of PS (50 μL/lane) from two subjects per group. (C) Agarose gel (2.5%) of PCR products obtained after amplification with primers specific for the PRH1 locus. Lane 1, bp standard; lane 2–9, PCR products representing three products: Db (416 bp) and PIF/Pa (each 353 bp, indistinguishable).
Since the Db band was faint in some patients and no purified Db protein was available for comparison, PCR was conducted on the PRH1 gene to detect the Db allele, which could be distinguished based on size from the other alleles at this locus (Pa and PIF). Results with two patients from each group are shown in Fig. 5C, clearly showing that patients CD5 and RCD5 contain the Db allele, while the others do not. PCR was conducted on all patient samples (data not shown). Based on the gels and PCR results combined, for 41 of 58 subjects, the alleles encoded at the PRH1 and PRH2 locus could be deciphered. The frequencies of the Pif, Db, Pa, PRP1, and PRP2 alleles in each group were calculated (Table 2). Despite the relatively small sample size, the obtained frequencies matched previously reported frequencies of acidic PRPs in HCs [11, 44]. Statistical analysis revealed no differences in PRH1 or PRH2 allele frequencies between healthy and diseased groups (p > 0.05).
Table 2.
Acidic PRP genotypes in the subjects
| PRH1 |
PRH2 |
n | |||||
|---|---|---|---|---|---|---|---|
| Db | PIF | Pa | PRP1 | PRP2 | |||
| HC | 0.19 | 0.69 | 0.11 | 0.88 | 0.11 | 13 | |
| G | 0.25 | 0.5 | 0.25 | 0.7 | 0.3 | 6 | |
| CD | 0.156 | 0.81 | 0.031 | 0.96 | 0.04 | 16 | |
| RCD | 0.25 | 0.75 | 0 | 1 | 0 | 6 | |
| χ 2a) | 0.769 | 4.387 | 6.786 | 7.041 | |||
| P | 0.857 | 0.223 | 0.079 | 0.071 | |||
| Azen and Maeda [11] | HC | 0.17 | 0.66 | 0.21 | 0.71 | 0.25 | 149 |
| Hay et al. [44] | HC | 0.15 | 0.67 | 0.18 | 0.76 | 0.23 | 125 |
Pearson's chi-squared test (level of significance p < 0.05).
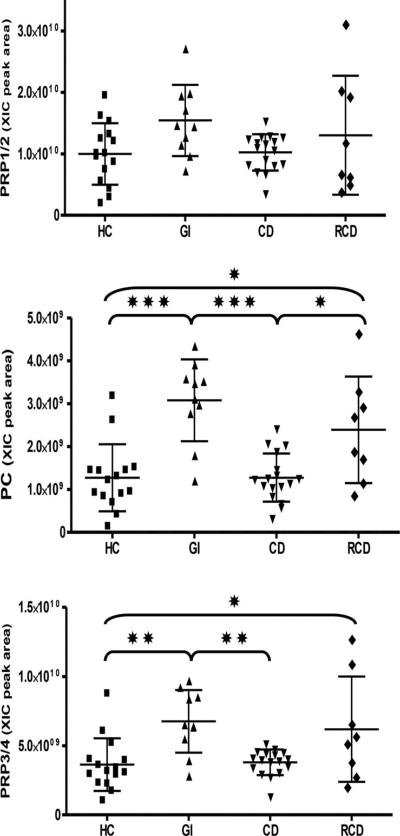
The acidic PRP protein levels were quantitated in WS by RP-HPLC-ESI-MS [45, 46]. The four groups showed no statistically significant differences of intact aPRPs levels (Fig. 6, Table 3). Conversely, PC peptide (the N-terminal 44 residues in the acidic PRPs) and the truncated PRPs (PRP3/4) were found to be more abundant in saliva of GI and RCD subjects compared to HC and CD. The CD group did not differ from the healthy control group.
Figure 6.
Distribution of the XIC peak areas of the aPRPs in the four groups. Means ± SD of the XIC peak areas are reported on each diagram. Significant differences between pairs of the groups (one way ANOVA with Tukey's post hoc multiple comparison test) are represented by asterisks.*p ≤ 0.05, **p ≤ 0.01, ≤ ***p ≤ 0.001.
Table 3.
XIC peak areas of the salivary proteins /peptides: Mean ± SD (arbitrary units × 108) and frequency in the four groups
| HC n = 15 | GI n = 10 | CD n = 17 | RCD n = 8 | One way ANOVA p Value | |
|---|---|---|---|---|---|
| aPRPs | |||||
| PRP1/2 total | 99.8 ± 50.1 | 154.3 ± 57.8 | 102.4 ± 29.6 | 130.3 ± 96.8 | ns |
| 15 | 10 | 17 | 8 | ||
| PRP3/4 total | 36.4 ± 19.0 | 67.6 ± 22.6 | 37.9 ± 9.2 | 61.9 ± 38.0 | 0.0008*** |
| 15 | 10 | 17 | 8 | ||
| PC | 12.7 ± 7.8 | 30.3 ± 9.5 | 12.7 ± 5.6 | 23.9 ± 12.4 | < 0.0001** |
| 15 | 10 | 17 | 8 | ||
| bPRPs | |||||
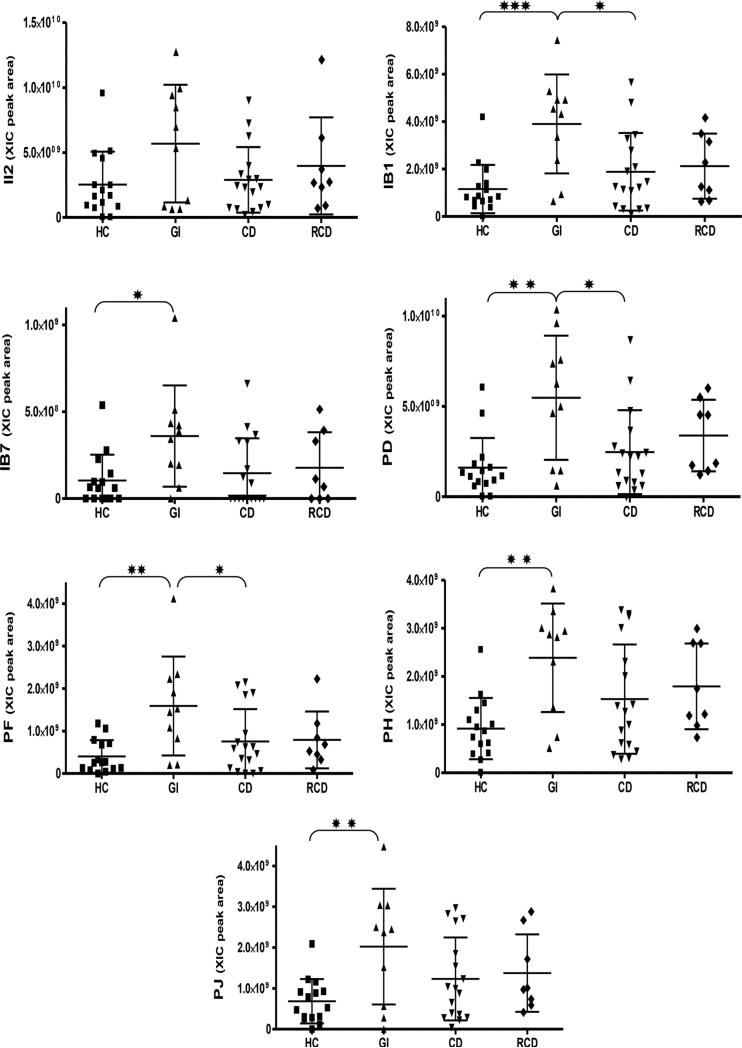
| II2 | 25.3 + 25.5 | 56.9 ± 45.3 | 29.0 ± 25.3 | 39.7 ± 37.3 | ns |
| 13 | 10 | 17 | 8 | ||
| IB1 | 11.6 ± 10.2 | 39.0 ± 20.9 | 18.9 ± 16.4 | 21.2 ± 13.7 | 0.0009*** |
| 14 | 10 | 17 | 8 | ||
| IB7 | 1.0±1.5 | 3.6 ± 2.9 | 1.5 ± 2.0 | 1.8 ± 2.0 | 0.0298* |
| 9 | 9 | 8 | 5 | ||
| PD | 16.0 ± 16.4 | 54.7 ± 34.4 | 24.7 ± 23.3 | 33.9 ± 19.8 | 0.0021** |
| 13 | 10 | 16 | 8 | ||
| PF | 4.0 ± 3.8 | 15.9 ± 11.6 | 7.6 ± 7.6 | 7.9 ± 6.7 | 0.0046** |
| 14 | 10 | 15 | 8 | ||
| PH | 9.2 ± 6.4 | 23.9 + 11.3 | 15.3 ± 11.3 | 17.9 ± 8.9 | 0.0055** |
| 14 | 10 | 15 | 8 | ||
| PJ | 6.8 ± 5.4 | 20.2 ± 14.2 | 12.3 ± 10.2 | 13.7 ± 9.5 | 0.0179* |
| 14 | 9 | 17 | 8 |
ns, not significant. The p values were obtained by the ANOVA test, and refer to the comparison of the four groups globally.
Lastly, basic PRP peptides were quantitated using the same methodology. Basic PRPs show a higher degree of overlap to gliadin proteins than acidic PRPs, with some domains displaying an approximately 50% homology to immunogenic gluten domains. The following seven abundant basic PRP peptides were quantitated: II-2, IB-1, IB-7, PD, PF, PH, and PJ. In WS no differences were found between CD and HC (Fig. 7, Table 3), but GI showed a higher concentration of several bPRPs with respect to HC and CD. Conversely, no differences in aPRPs and bPRPs were found among groups in parotid saliva samples (Supporting Information Table 2), which may point to altered oral proteolytic processing in GI and RCD groups.
Figure 7.
Distribution of the XIC peak areas of the bPRPs in the four groups. Means ± SD of the XIC peak areas are reported on each diagram. Significant differences between pairs of the groups (one way ANOVA with Tukey's post hoc multiple comparison test) are represented by asterisks: *p ≤ 0.05, **p ≤ 0.01, ≤ ***p ≤ 0.001.
4 Discussion
Based on the structural similarities between gluten proteins and salivary PRPs, the apparent involvement of non-HLA genes in CD pathogenesis, and the fact that certain patients (RCD) do not respond to a GFD, we hypothesized that healthy and CD/RCD patients might differ in their composition/isoforms of PRPs, and that these differences may modulate the manifestation or onset of CD, or have a diagnostic potential. The results demonstrated that PRP patterns, isoforms, and amounts showed minor differences between groups and were not specific for the presence of CD or RCD versus controls.
The oral cavity is the entrance to the GI system, and saliva is an integral part of the collection of fluids that are released into the digestive tract. The oral cavity has been probed for the potential to diagnose CD since this anatomical location is more easily accessible for collection of excretions or biopsies than the small intestine. Some studies have thus focused on the oral mucosa, and its ability to reflect an immunological signature or antibody expression profile that would be representative of processes in the small intestine of CD patients. However, lymphocytic infiltration in the oral mucosal epithelium and lamina propria was not higher in untreated compared to treated CD patients [47]. A subcutaneous oral mucosal challenge with gliadins, on the other hand, significantly increased the number of CD4+ and CD8+ T cells in the oral lamina propria of CD patients, but not in nonceliac controls, suggesting an induced oral mucosal immune response to gluten in CD [48]. Cultured oral mucosal and duodenal biopsies from CD patients showed a comparable anti-TG2 and anti-endomysal antibody response in the culture media, with a sensitivity between 57 and 100% [49,50], likely depending on biopsy size [49,51]. Saliva as a diagnostic fluid for CD has also been explored. Anti-TG2 antibodies were detectable in saliva of 31 of 32 children who tested positive in serum [52], suggesting saliva could potentially serve as a diagnostic alternative to a blood drawn for CD screening, although the salivary anti-TG studies in general had much less predictive results. Other, visible, changes in CD in the oral cavity are enamel defects [53–55], and recurrent aphthous stomatitis [55–57]. Dental enamel anomalies are strongly correlated with and predictors of CD [58–60]. Enamel defects are most likely due to impaired absorption of calcium and vitamin D in CD, both of which are required for proper enamelogenesis. It is less likely that enamel protective proteins are implicated, supported by our observation that the acidic PRPs did not differ in health and CD.
The PRH1 and PRH2 gene alleles showed a similar distribution in patients and controls, ruling out the potential link of a specific allele with CD/RCD. More detailed analysis with RP-HPLC-ESI-MS of basic PRP isoforms further confirmed the similarity of salivary PRP peptide characteristics in CD/RCD compared to controls. Using anionic PAGE, in two subjects an isoform was noted with lower electrophoretic mobility compared to normal PRP1/3, which we previously reported and named PRB1/3 RB variants (Roma-Boston Ser22 (Phos) →Phe variant) [61]. The variant was observed in one RCD and one HC, and was thus unlikely to be linked to CD/RCD pathogenesis. Other known single nucleotide substitutions in the PRPs are summarized in Table 4. It can be speculated that some of the substitutions might render PRPs more gluten like, or even immunogenic by creating sequence changes that could favor recognition by TG2 enabling glutamine deamidation, which usually increases the immunogenicity of gluten domains [62]. Our investigations did not yet go as far as to search for these SNPs in our patient populations and this could be explored further.
Table 4.
Known single nucleotide polymorphisms in PPR alleles
| Allele |
|||||
|---|---|---|---|---|---|
| PRP2 | Basic PRB1L | Basic PRB2L | Basic PRB3L | Basic PRB4L | |
| Positiona) (aa change) | 4 (D to N) | 24 (S to P) | 52 (P to S) | 24 (R to P) | 12 (S to P) |
| 22 (S to F) | 112 (K to R) | 72 (K to R) | 35 (G to S) | 21 (E to Q) | |
| 26 (I to L) | 255 (R to Q) | 135 (Q to P) | 44 (R to P) | 50 (N to D) | |
| 50 (D to N) | 258 (Q to R) | 255 (G to D) | 65 (P to Q) | 85 (R to E) | |
| 103 (R to C) | 262 (G to R) | 68 (Q to P) | 113 (H to N) | ||
| 147 (Q to K) | 291 (Q to T) | 103 (K to E) | 176 (N to D) | ||
| 310 (A to P) | 121 (R to H) | 196 (N to D) | |||
| 314 (S to C) | 142 (H to R) | ||||
| 321 (A to S) | 221 (Q to R) | ||||
Position indicated is in the secreted protein (without signal peptide); information derived from www.uniprot.org and lavarone et al. [61].
Importantly, our studies do not rule out an effect of PRPs in general and of PRP variants in particular on innate or adaptive immune responses in CD or RCD. Such effect could either be stimulatory or suppressive. To this aim, further studies using innate immune cell and CD-derived T cell cultures, or biopsy cultures will be required. Such studies are currently under way. The delicate difference between tolerance versus immunogenicity in CD of PRPs and gluten, respectively, is a challenging and interesting topic for further investigation.
Supplementary Material
Clinical Relevance.
CD is a gluten-sensitive enteropathy triggered by the ingestion of gluten proteins. Gluten proteins show an unusual structural similarity to human salivary PRPs. In this study, we applied gel electrophoretic and proteomics approaches to analyze the PRP isoforms pattern and quantities in saliva from CD patients, RCD patients, and control subjects to investigate a potential relation with CD.
Acknowledgments
These studies were supported by NIH grants AI087803 (EJH), AI101067 (EJH), and DK095937 (DAL), Cagliari University (IM), Catholic University of Rome (MC), Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR; IM, MC), Italian National Research Council (MC), and Nando Peretti Foundation (MC).
Abbreviations
- CD
celiac disease
- GFD
gluten-free diet
- GI
gastrointestinal
- HC
healthy controls
- HLA
human leukocyte antigen
- PRPs
proline-rich proteins
- PS
parotid secretion
- RCD
refractory celiac disease
- WS
whole saliva
- XIC
extracted ion current
Footnotes
The authors have declared no conflict of interest.
Additional supporting information may be found in the online version of this article at the publisher's web-site
References
- 1.Azen EA, Denniston CL. Genetic polymorphism of human salivary proline-rich proteins: further genetic analysis. Biochem. Genet. 1974;12:109–120. doi: 10.1007/BF00487820. [DOI] [PubMed] [Google Scholar]
- 2.Azen EA, Oppenheim FG. Genetic polymorphism of proline-rich human salivary proteins. Science (New York, NY) 1973;180:1067–1069. doi: 10.1126/science.180.4090.1067. [DOI] [PubMed] [Google Scholar]
- 3.Bennick A. Salivary proline-rich proteins. Mol. Cell. Biochem. 1982;45:83–99. doi: 10.1007/BF00223503. [DOI] [PubMed] [Google Scholar]
- 4.Oppenheim FG, Hay DI, Franzblau C. Proline-rich proteins from human parotid saliva. I. Isolation and partial characterization. Biochemistry. 1971;10:4233–4238. doi: 10.1021/bi00799a013. [DOI] [PubMed] [Google Scholar]
- 5.Oppenheim FG, Salih E, Siqueira WL, Zhang W, Helmerhorst EJ. Salivary proteome and its genetic polymorphisms. Ann. NY Acad. Sci. 2007;1098:22–50. doi: 10.1196/annals.1384.030. [DOI] [PubMed] [Google Scholar]
- 6.Bennick A. The binding of calcium to a salivary phospho-protein, protein A, common to human parotid and submandibular secretions. Biochem. J. 1976;155:163–169. doi: 10.1042/bj1550163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hay DI, Carlson ER, Schluckebier SK, Moreno EC, Schlesinger DH. Inhibition of calcium phosphate precipitation by human salivary acidic proline-rich proteins: structure-activity relationships. Calcif. Tissue Int. 1987;40:126–132. doi: 10.1007/BF02555696. [DOI] [PubMed] [Google Scholar]
- 8.Lamkin MS, Oppenheim FG. Structural features of salivary function. Crit. Rev. Oral Biol. Med. 1993;4:251–259. doi: 10.1177/10454411930040030101. [DOI] [PubMed] [Google Scholar]
- 9.Mehansho H, Clements S, Sheares BT, Smith S, Carlson DM. Induction of proline-rich glycoprotein synthesis in mouse salivary glands by isoproterenol and by tannins. J. Biol. Chem. 1985;260:4418–4423. [PubMed] [Google Scholar]
- 10.Bennick A. Interaction of plant polyphenols with salivary proteins. Crit. Rev. Oral Biol. Med. 2002;13:184–196. doi: 10.1177/154411130201300208. [DOI] [PubMed] [Google Scholar]
- 11.Azen EA, Maeda N. Molecular genetics of human salivary proteins and their polymorphisms. Adv. Hum. Genet. 1988;17:141–199. doi: 10.1007/978-1-4613-0987-1_5. [DOI] [PubMed] [Google Scholar]
- 12.Azen EA, Latreille P, Niece RL. PRBI gene variants coding for length and null polymorphisms among human salivary Ps, PmF, PmS, and Pe proline-rich proteins (PRPs). Am. J. Hum. Genet. 1993;53:264–278. [PMC free article] [PubMed] [Google Scholar]
- 13.Levine MJ, Reddy MS, Tabak LA, Loomis RE, et al. Structural aspects of salivary glycoproteins. J/ Dent. Res. 1987;66:436–441. doi: 10.1177/00220345870660020901. [DOI] [PubMed] [Google Scholar]
- 14.Inzitari R, Cabras T, Onnis G, Olmi C, et al. Different iso-forms and post-translational modifications of human salivary acidic proline-rich proteins. Proteomics. 2005;5:805–815. doi: 10.1002/pmic.200401156. [DOI] [PubMed] [Google Scholar]
- 15.Isemura S, Saitoh E. Molecular cloning and sequence analysis of cDNA coding for the precursor of the human salivary proline-rich peptide P-B. J. Biochem. (Tokyo) 1994;115:1101–1106. doi: 10.1093/oxfordjournals.jbchem.a124464. [DOI] [PubMed] [Google Scholar]
- 16.Isemura S, Saitoh E, Sanada K. Isolation and amino acid sequences of proline-rich peptides of human whole saliva. J. Biochem. (Tokyo) 1979;86:79–86. [PubMed] [Google Scholar]
- 17.Isemura S, Saitoh E, Sanada K. The amino acid sequence of a salivary proline-rich peptide, P-C, and its relation to a salivary proline-rich phosphoprotein, protein C. J. Biochem. (Tokyo) 1980;87:1071–1077. [PubMed] [Google Scholar]
- 18.Kauffman D, Wong R, Bennick A, Keller P. Basic proline-rich proteins from human parotid saliva: complete covalent structure of protein IB-9 and partial structure of protein IB-6, members of a polymorphic pair. Biochemistry. 1982;21:6558–6562. doi: 10.1021/bi00268a036. [DOI] [PubMed] [Google Scholar]
- 19.Kauffman DL, Bennick A, Blum M, Keller PJ. Basic proline-rich proteins from human parotid saliva: relationships of the covalent structures of ten proteins from a single individual. Biochemistry. 1991;30:3351–3356. doi: 10.1021/bi00228a001. [DOI] [PubMed] [Google Scholar]
- 20.Saitoh E, Isemura S, Sanada K. Complete amino acid sequence of a basic proline-rich peptide, P-F, from human parotid saliva. J. Biochem. (Tokyo) 1983;93:883–888. doi: 10.1093/jb/93.3.883. [DOI] [PubMed] [Google Scholar]
- 21.Saitoh E, Isemura S, Sanada K. Complete amino acid sequence of a basic proline-rich peptide, P-D, from human parotid saliva. J. Biochem. (Tokyo) 1983;93:495–502. doi: 10.1093/oxfordjournals.jbchem.a134204. [DOI] [PubMed] [Google Scholar]
- 22.Helmerhorst EJ, Zamakhchari M, Schuppan D, Oppenheim FG. Discovery of a novel and rich source of gluten-degrading microbial enzymes in the oral cavity. PloS One. 2010;5:e13264. doi: 10.1371/journal.pone.0013264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuppan D, Junker Y, Barisani D. Celiac disease: from pathogenesis to novel therapies. Gastroenterology. 2009;137:1912–1933. doi: 10.1053/j.gastro.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Green PH, Cellier C. Celiac disease. N. Engl. J. Med. 2007;357:1731–1743. doi: 10.1056/NEJMra071600. [DOI] [PubMed] [Google Scholar]
- 25.Abadie V, Sollid LM, Barreiro LB, Jabri B. Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu. Rev. Immunol. 2011;29:493–525. doi: 10.1146/annurev-immunol-040210-092915. [DOI] [PubMed] [Google Scholar]
- 26.Roshan B, Leffler DA, Jamma S, Dennis M, et al. The incidence and clinical spectrum of refractory celiac disease in a north American referral center. Am. J. Gastroenterol. 2011;106:923–928. doi: 10.1038/ajg.2011.104. [DOI] [PubMed] [Google Scholar]
- 27.Malamut G, Cellier C. Refractory celiac disease. Exp. Rev. Gastroenterol. Hepatol. 2014;8:323–328. doi: 10.1586/17474124.2014.887438. [DOI] [PubMed] [Google Scholar]
- 28.van Berge-Henegouwen GP, Mulder CJ. Pioneer in the gluten free diet: Willem-Karel Dicke 1905–1962, over 50 years of gluten free diet. Gut. 1993;34:1473–1475. doi: 10.1136/gut.34.11.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wieser H. The precipitating factor in coeliac disease. Bail-liere. Clin. Gastroenterol. 1995;9:191–207. doi: 10.1016/0950-3528(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 30.Sollid LM, Qiao SW, Anderson RP, Gianfrani C, Koning F. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics. 2012;64:455–460. doi: 10.1007/s00251-012-0599-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wijmenga C, Gutierrez-Achury J. Celiac disease genetics: past, present and future challenges. J. Pediatr. Gastroenterol. Nutr. 2014;59(Suppl 1):S4–S7. doi: 10.1097/01.mpg.0000450392.23156.10. [DOI] [PubMed] [Google Scholar]
- 32.Romanos J, Rosen A, Kumar V, Trynka G, et al. Improving coeliac disease risk prediction by testing non-HLA variants additional to HLA variants. Gut. 2014;63:415–422. doi: 10.1136/gutjnl-2012-304110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leffler DA, Schuppan D. Update on serologic testing in celiac disease. Am. J. Gastroenterol. 2010;105:2520–2524. doi: 10.1038/ajg.2010.276. [DOI] [PubMed] [Google Scholar]
- 34.Abdallah H, Leffler D, Dennis M, Kelly CP. Refractory celiac disease. Curr. Gastroenterol. Rep. 2007;9:401–405. doi: 10.1007/s11894-007-0049-5. [DOI] [PubMed] [Google Scholar]
- 35.Curby WA. Device for collection of human parotid saliva. J. Lab. Clin. Med. 1953;41:493–496. [PubMed] [Google Scholar]
- 36.Beeley JA, Sweeney D, Lindsay JC, Buchanan ML, et al. Sodium dodecyl sulphate-polyacrylamide gel electrophoresis of human parotid salivary proteins. Electrophoresis. 1991;12:1032–1041. doi: 10.1002/elps.1150121207. [DOI] [PubMed] [Google Scholar]
- 37.Davis BJ. Disc electrophoresis. II. Method and application to human serum proteins. Ann. NY Acad. Sci. 1964;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- 38.Ornstein L. Disc electrophoresis. I. Background and theory. Ann. NY Acad. Sci. 1964;121:321–349. doi: 10.1111/j.1749-6632.1964.tb14207.x. [DOI] [PubMed] [Google Scholar]
- 39.Zakhary GM, Clark RM, Bidichandani SI, Owen WL, et al. Acidic proline-rich protein Db and caries in young children. J. Dent. Res. 2007;86:1176–1180. doi: 10.1177/154405910708601207. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z, Marshall AG. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 41.Cabras T, Pisano E, Boi R, Olianas A, et al. Age-dependent modifications of the human salivary secretory protein complex. J. Proteome Res. 2009;8:4126–4134. doi: 10.1021/pr900212u. [DOI] [PubMed] [Google Scholar]
- 42.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 43.Rubio-Tapia A, Murray JA. Classification and management of refractory coeliac disease. Gut. 2010;59:547–557. doi: 10.1136/gut.2009.195131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hay DI, Ahern JM, Schluckebier SK, Schlesinger DH. Human salivary acidic proline-rich protein polymorphisms and biosynthesis studied by high-performance liquid chromatography. J. Dent. Res. 1994;73:1717–1726. doi: 10.1177/00220345940730110701. [DOI] [PubMed] [Google Scholar]
- 45.Inzitari R, Vento G, Capoluongo E, Boccacci S, et al. Proteomic analysis of salivary acidic proline-rich proteins in human preterm and At-term newborns. J. Proteome Res. 2007;6:1371–1377. doi: 10.1021/pr060520e. [DOI] [PubMed] [Google Scholar]
- 46.Castagnola M, Cabras T, Iavarone F, Vincenzoni F, et al. Top-down platform for deciphering the human salivary proteome. J. Matern. Fetal Neonat. Med. 2012;25:27–43. doi: 10.3109/14767058.2012.714647. [DOI] [PubMed] [Google Scholar]
- 47.Lahteenoja H, Toivanen A, Viander M, Raiha I, et al. Increase in T-cell subsets of oral mucosa: a late immune response in patients with treated coeliac disease? Scand. J. Immunol. 2000;52:602–608. doi: 10.1046/j.1365-3083.2000.00794.x. [DOI] [PubMed] [Google Scholar]
- 48.Lahteenoja H, Maki M, Viander M, Toivanen A, Syrjanen S. Local challenge of oral mucosa with gliadin in patients with coeliac disease. Clin. Exp. Immunol. 2000;120:38–45. doi: 10.1046/j.1365-2249.2000.01177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vetrano S, Zampaletta U, Anania MC, Di Tola M, et al. Detection of anti-endomysial and anti-tissue transglutaminase autoantibodies in media following culture of oral biopsies from patients with untreated coeliac disease. Dig. Liver Dis. 2007;39:911–916. doi: 10.1016/j.dld.2007.07.158. [DOI] [PubMed] [Google Scholar]
- 50.Carroccio A, Campisi G, Iacono G, Iacono OL, et al. Oral mucosa of coeliac disease patients produces antiendomysial and antitransglutaminase antibodies: the diagnostic usefulness of an in vitro culture system. Aliment. Pharmacol. Ther. 2007;25:1471–1477. doi: 10.1111/j.1365-2036.2007.03335.x. [DOI] [PubMed] [Google Scholar]
- 51.Compilato D, Campisi G, Pastore L, Carroccio A. The production of the oral mucosa of antiendomysial and anti-tissue-transglutaminase antibodies in patients with celiac disease: a review. Sci. World J. 2010;10:2385–2394. doi: 10.1100/tsw.2010.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonamico M, Nenna R, Montuori M, Luparia RP, et al. First salivary screening of celiac disease by detection of anti-transglutaminase autoantibody radioimmunoassay in 5000 Italian primary schoolchildren. J. Pediatr. Gastroenterol. Nutr. 2011;52:17–20. doi: 10.1097/MPG.0b013e3181e6f2d0. [DOI] [PubMed] [Google Scholar]
- 53.Maki M, Aine L, Lipsanen V, Koskimies S. Dental enamel defects in first-degree relatives of coeliac disease patients. Lancet. 1991;337:763–764. doi: 10.1016/0140-6736(91)91375-5. [DOI] [PubMed] [Google Scholar]
- 54.Smith DM, Miller J. Gastro-enteritis, coeliac disease and enamel hypoplasia. Br. Dent. J. 1979;147:91–95. doi: 10.1038/sj.bdj.4804290. [DOI] [PubMed] [Google Scholar]
- 55.Cheng J, Malahias T, Brar P, Minaya MT, Green PH. The association between celiac disease, dental enamel defects, and aphthous ulcers in a United States cohort. J. Clin. Gastroenterol. 2010;44:191–194. doi: 10.1097/MCG.0b013e3181ac9942. [DOI] [PubMed] [Google Scholar]
- 56.Ferguson MM, Wray D, Carmichael HA, Russell RI, Lee FD. Coeliac disease associated with recurrent aphthae. Gut. 1980;21:223–226. doi: 10.1136/gut.21.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferguson R, Basu MJ, Asquith P, Cooke WT. Proceedings: recurrent aphthous ulceration and its association with coeliac disease. Gut. 1975;16:393. [PubMed] [Google Scholar]
- 58.Krzywicka B, Herman K, Kowalczyk-Zajac M, Pytrus T. Celiac disease and its impact on the oral health status—review of the literature. Adv. Clin. Exp. Med. 2014;23:675–681. doi: 10.17219/acem/37212. [DOI] [PubMed] [Google Scholar]
- 59.Trotta L, Biagi F, Bianchi PI, Marchese A, et al. Dental enamel defects in adult coeliac disease: prevalence and correlation with symptoms and age at diagnosis. Eur. J. Intern. Med. 2013;24:832–834. doi: 10.1016/j.ejim.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 60.Rashid M, Zarkadas M, Anca A, Limeback H. Oral manifestations of celiac disease: a clinical guide for dentists. J. Can. Dent. Assoc. 2011;77:b39. [PubMed] [Google Scholar]
- 61.Iavarone F, D'Alessandro A, Tian N, Cabras T, et al. High-resolution high-performance liquid chromatography with electrospray ionization mass spectrometry and tandem mass spectrometry characterization of a new isoform of human salivary acidic proline-rich proteins named Roma-Boston Ser22 (Phos) –>Phe variant. J. Sep. Sci. 2014;37:1896–1902. doi: 10.1002/jssc.201400227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vader LW, de Ru A, van der Wal Y, Kooy YM, et al. Specificity of tissue transglutaminase explains cereal toxicity in celiac disease. J. Exp. Med. 2002;195:643–649. doi: 10.1084/jem.20012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.