Abstract
Toxoplasma rhoptries, an unusual set of apical organelles that are associated with Toxoplasma infection may cause subversion of the host cell functions. Parasite rhoptry protein 16 (ROP16) is a regulator of host cell transcription during cell invasion in which it migrates into the host cell cytoplasm and subsequently localizes to the nucleus. In the present study, we found that overexpression of ROP16 could partially mediate human neuroblastoma SH-SY5Y apoptosis (12.47%) and cell cycle arrest in G1 phase (60.77%) in a p53 dependent manner by influencing the expression of Bax/Bcl-2 and p21/CDKs. ROP16 was identified to co-localize with p53, a novel direct interaction partner in the nucleus of SH-SY5Y. Furthermore, SH-SY5Y apoptosis via the mitochondria-dependent p53 pathway and cell cycle arrest caused by ROP16 dealt with direct serine 15/37 phosphorylation of p53. Our studies provide a new mechanism by which ROP16 interacts with the nucleus proteins which subsequently subverts the host cells functions.
Keywords: Toxoplasma rhoptries, Parasite rhoptry protein 16
Introduction
Toxoplasma gondii(T.gondii) is an opportunistic protozoan pathogen belongs to the phylum Apicomplexa, which can be defined by the presence of an apical complex which includes secretory organelles 1, 2. T. gondii is capable of invading and replicating in all nucleated cells as an obligate intracellular parasite. The rhoptries are a type of apical secretory organelle of T. gondii that have shown close relationship with the parasites' pathogenesis, host cell invasion and host cell interaction 3. There are more than 30 proven rhoptry proteins that most of which have shown clear homology to protein kinases 1. Recent studies had found that many of rhoptry proteins were involved in the invasive process and played an important role for growth and survival in the host cell. ROP16, a key virulence determinant, is a member of the ROP2 family and can invade into the host cell nucleus quickly after the parasites infection 4. ROP16 has serine - threonine kinase activity with a molecular weight of 96KD constituted by 707 amino acids. This protein invades host cell and accumulates in the host cell nucleus via the nucleus localized sequence (NLS) 5. The evidences showed that ROP16 unique to the apicomplexa was crucial in the host-pathogen interaction 6. ROP16 of type I or III strains of T. gondii is a regulator of host cell transcription that subverts the host functions by direct tyrosine phosphorylation of STAT pathways. It affected the activation of STAT3/6 signaling pathways and influenced the consequent downstream host cytokine, interleukin-12 7, 8. In addition, ROP16 also induced the phosphorylation and nuclear translocation of STAT5 to generate protective immunity 9, 10. In order to gain a better understanding of the molecular functions of ROP16 in the host cell nucleus as well as the roles of ROP16 in changing the functions of human neural cell, we carried out tests to identify novel interacting host's nuclear protein with ROP16 and interplay each other in the response of human neuroblastoma SH-SY5Y cell line to ROP16.
Materials and methods
Cell culture, plasmids construction and transfection
The SH-SY5Y cell lines obtained from American Type Culture Collection (ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM, Hyclone) which was supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco ). NE-4C cell lines(from ATCC) that lacks functional p53 protein were maintained on poly-L-lysine-coated dishes in Eagle MEM(Gibco) supplemented with 10% FBS, 1% Glutamax(Invitrogen) and 1% Non-essential Amino Acids. Cells were incubated in a humidified atmosphere containing 5% CO2 at 37°C and were passaged every 2-4 days by trypsinization. The coding region of T. gonndii ROP16 was amplified using ROP16 forward primer containing EcoRI: 5'-GAGAATTCCATGAAAGTGACCACGAAAGG3-3'; and reverse primer containing Flag-tag gene sequence EcoRv: 5'-GCGATATCCTTGTCATCGTCGTCCTTGTAGTCCATCCGATGTGAAGAAAGTTC-3'.
All constructs were verified by sequencing. SH-SY5Y cell lines transfected with a total of 4.0 μg of either empty vector or the indicated plasmids (4 μg Flag-tagged ROP16) via Lipofectamine 2000 as specified by the manufacturer (Invitrogen) were cultured in atmosphere containing 5% CO2 at 37°C for 48h before harvest.
RNA extraction and cDNA synthesis
RNA from T.gondii and SH-SY5Y cells were isolated using TRIzol reagent (Invitrogen). The process of cDNA synthesis used a template that was reverse-transcribed via SuperScript Ⅱ RNase H-reverse transcriptase and oligo(dT)25 as the primer (Invitrogen). PCR was completed under the following conditions after cDNA synthesis: a denaturation cycle at 94°C for 5 min, 94°C for 30 s, annealing at 55°C for 30 s and elongation at 68°C for 150 s, and a final extension at 68°C for 5 min.
DNA fragmentation
SH-SY5Y cells were grown in a 10-cm dish when cells were 70-80% confluent. Cells were harvested by scraping and centrifuging and later lysed with lysis buffer (5 mM Tris-HCl, pH 8.0, 20 mM EDTA, 0.5% Triton X-100) on ice for 15min. Fragmented DNA in the supernatant after centrifugation at 12,000 rpm was extracted twice with phenol/chloroform/isopropanol (25/24/1, v/v) and once with chloroform and then were precipitated with ethanol and 5 M NaCl. The DNA pellet was washed once with 70% ethanol and resuspended in Tris-EDTA buffer (pH 8.0) with 100g/ml RNase at 37°C for 2 h. The DNA fragments were separated by 1.5% agarose gel electrophoresis.
Flow cytometric analysis for cell apoptosis
The extent of apoptosis was determined by flow cytometry via Annexin V-FITC-PI apoptosis detection kit (Biovision). Briefly, SH-SY5Y cells and SH-SY5Y-ROP16 cells were harvested and rinsed twice with cold PBS (pH7.4) respectively before resuspended in 1×binding buffer at a concentration of 1×106 cells/ml. 5 µl of Annexin V-FITC and 5 µl of propidium iodide were added to 500 µl of cell suspension and then were incubated for 15 mins at room temperature in darkness. The stained samples were analyzed by Flow cytometer system (FACS Calibur BD Flow Cytometer) and Cell Quest software (BD) within 1 hour.
Cell cycle analysis
The cells were collected by trypsinization, washed with cold PBS, and resuspended in cold PBS. Afterwards, the cells were fixed in ice-cold 70% methanol for 24 hrs at 4°C. The cells were then centrifuged, and washed again with cold PBS, suspended in 500 µl PBS and incubated with RNase (20 µg/ml, final concentration) for 30 min at 37°C. The cells were then chilled over ice for 10 min, stained with propidium iodide (50 µg/ml, final concentration) for 30min and then were analyzed by flow cytometry (Calibur BD Flow Cytometer). Cell cycle analysis was analyzed with ModFit LT (Verity Software House, USA).
cDNA microarray
Microarray experiments were performed by using Agilent human Genome 8× 60K oligonucleotide arrays. The sample preparation and microarray hybridization were performed according to the manufacturer's standard protocols with minor modifications. Briefly, mRNA from SH-SY5Y cells and SH-SY5Y-ROP16 cells were purified from total RNA after removal of rRNA (mRNA-ONLYTM Eukaryotic mRNA Isolation Kit, Epicentre). Each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts. The labeled cRNAs were hybridized onto the Human Array at 45℃ overnight. The arrays were scanned by the Agilent Scanner G2565CA after having washed the slides. Feature Extraction Software was used to read out and translate the microarray image files into digital, further nomalized using the percentile shift gene expression data analysis system (GeneSpring GX Software). Gene transcripts were considered to be differentially expressed when the absolute fold change≥2 (up or down) from the controls with the P value set at <0.05.
Immunoprecipitation
Cells were collected and disrupted in lysis buffer supplemented with protease inhibitors. Cell lysates was precleared with protein G-agarose (Roche) for 1 h at 4°C in a rotation wheel. Samples were immunoprecipitated using 2 μg anti-Flag antibody (Cell Signaling Technology), anti-p53 antibody (Cell Signaling Technology), or normal rabbit IgG (Santa Cruz) with 30 μl of protein G-agarose overnight at 4°C respectively. Beads were washed five times with cold lysis buffer, resuspended in 2× SDS loading buffer and analyzed by SDS-PAGE. The antibodies used were monoclonal anti-Flag and anti-p53 (Cell Signaling Technology).
MALDI-TOF/TOF mass spectrometer assay
The immunoprecipitation complex was loaded on a 10% SDS-PAGE gel and visualized by coomassie staining. The bands on the SDS-PAGE gel were excised and digested with trypsin at 37 ℃ overnight. The digest production was incubated with 50% acetonitrile and 0.1% methanoic acid before concentrated and subjected to mass spectrometer. Protein candidates were chosen by Swiss-Prot and NCBInr with MASCOT search engine. Proteins identified by MALDI-TOF/TOF tandem mass spectrometer assay were considered as candidates when scores were 2 fold changed (up or down) between non-transfected controls and ROP16 expression cells with the P value set at <0.05.
Immunoblot analysis
Cells were harvested after 48hrs of transfection. Proteins of cell extracts were denatured and heated in 5× protein loading buffer before separated by SDS-PAGE. Proteins on 10% polyacrylamide-SDS gels were transferred to nitrocellulose membranes. The membranes were next immunoblotted with primary antibodies at 4℃ overnight detecting Flag-tagged ROP16 (Abmart), p53, Bax, p21 and p53-Ser 6/9/15/20/33/37/46/392 and T81 (Cell Signal Technology) followed by incubation with appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signal Technology). Blots were exposed to ECL.
Confocal microscopy
SH-SY5Y expressing ROP16 were seeded onto 13-mm circular glass coverslips with regard to the immunofluorescence studies,. The next day, cells were washed with PBS, warmed to 37°C, and fixed with 4% paraformaldehyde for 20 min at room temperature. Cells were washed three times with PBS and permeabilized with 0.2% Triton X-100 in PBS for 5 mins. Nonspecific staining was reduced by blocking with 5% goat serum overnight at 4°C and then followed by incubation with anti-rabbit IgG-FITC and anti-mouse IgG-Cy3 (Santa Cruz) as the secondary antibodies.
Quantitative real-time PCR
Total RNA was extracted from SH-SY5Y and SH-SY5Y-ROP16 cells with TRIzol reagent (invitrogen) and followed by cDNA synthesis performance (TAKARA). Real-time PCR amplification was used via ABI Prism 7300 sequence detector (Applied Biosystems) and SYBR Green reagent. The mRNA levels of the indicated genes were measured by qRT-PCR with gene-specific primers (Table 1). All amplification reactions were carried out under the following conditions: an initial stage of 95 °C for 30 sec, then a two-step program of 95 °C for 5 sec, 60°C for 31 sec over 40 cycles and were performed in triplicate. The relative target mRNA levels were analyzed with ABI Prism 7300 software and normalized to that of internal control GAPDH.
TABLE 1.
Sequences of primers used in RT-qPCR analyses
| Gene | Sequence | |
|---|---|---|
| Forward primer | Reverse primer | |
| Bax | 5'-ATCAGAACCATCATGGGCTGGACA-3' | 5'-AGCCCATCTTCTTCCAGATGGTGA-3' |
| Bcl-2 | 5'-TTGTGGCCTTCTTTGAGTTCGGTG-3' | 5'-ACTCACATCACCAAGTGCACCTAC-3' |
| Caspase 9 | 5'-TGGTGCTCAGACCAGAGATT-3' | 5'-ACGGGGTGGCATCTGGCTCG-3' |
| p21 | 5'-GTCACTGTCTTGTACCCTTGTG-3' | 5'-GGCGTTTGGAGTGGTAGAAA-3' |
| CDK6 | 5'-TGCACAGTGTCACGAACAGACAGA-3' | 5'-TTAGATCGCGATGCACTACTCGGT-3' |
| Cyclin D1 | 5'-CAAGCTCAAGTGGAACCTGG-3' | 5'-GCGGATGATCTGTTTGTTCT-3' |
| GAPDH | 5'-TCGACAGTCAGCCGCATCTTCTTT-3' | 5'-GCCCAATACGACCAAATCCGTTGA-3' |
In Vitro Kinase Assay
Recombinant His-p53 protein was purified in BL-21 E.coli transformed with the PROEX-HTb-His-p53 plasmid in 200 mL LB for 3 h's induction of expression with 1 mM IPTG. Cells were harvested and lysed with PBS and 2% (vol/vol) Triton-X 100 plus 1%PMSF and sonicated for 15 min. Cell lysates were incubated with 200μL Ni beads (GE Healthcare) overnight at 4 ℃ and washed four times in washing buffer (20 mM sodium phosphate, 0.5 M NaCl, 30-100 mM imidazole, pH 7.4) followed by a single wash in elution buffer (20 mM sodium phosphate, 0.5 M NaCl, 500 mM imidazole, pH 7.4). Recombinant ROP16-GST was produced in 293T cells transfected with the ROP16-GST plasmid and then collected with GST beads (GE Healthcare) before incubated in 1×kinase buffer (25mM Tris-HCl, ph 7.5, 10mM MgCl2,2mM DTT, 5mM β-Glycerolyphosphate, 0.1mM Na3VO4, 2mM EGTA and 1%PRI) with His-p53 and ATP at 30℃ for 30mins. Kinase reactions were analyzed by SDS-PAGE as described before.
Statistical analysis
Statistical analysis of all the data was performed by Student's t-test. The P values were <0.05.
Results
ROP16 mediated partially cell apoptosis and induces cell-cycle arrest in G1 phase
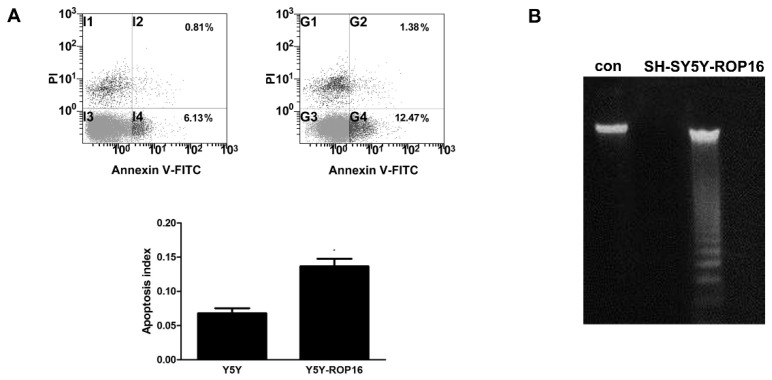
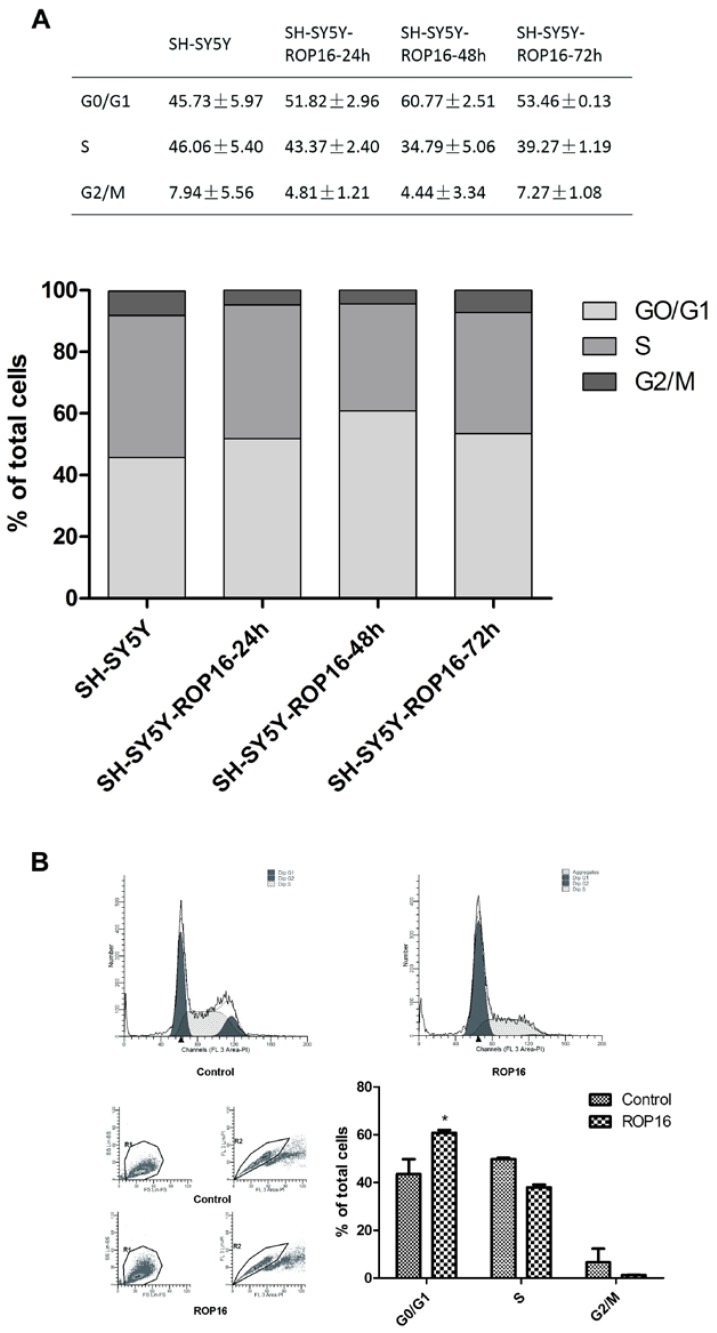
We first examined the cell apoptosis and cell cycle arrest by flow cytometry analysis in order to determine the role of ROP16 in altering host cell function after ROP16 invasion into the nucleus. Figure 1A revealed a marked increase in the percentage of apoptotic cells in cells transfected with ROP16 (approximately 12.47%) compared with the control group (6.13%).ROP16 could result in the fragmented DNA ladder, suggesting that ROP16 partially mediated cell apoptosis (Figure 1B). Furthermore, the population of cells in G1 phase increased and reached a peak at 48hrs after being transfected. The G1 phase cells were much more in ROP16-expressing cells (60.77%) than control (45.73%) (Figure 2). It was concluded that ROP16 induced G1 arrest in SH-SY5Y cells.
Figure 1.
ROP16 mediated partially human SH-SY5Y apoptosis. (A) Cell apoptosis assessed by double staining of FITC-AnnexinV and propidium iodide with flow cytometry. Error bars showed mean ±SEM of at least 3 replicates (p<0.05). (B) Cells were harvested and DNA was prepared and examined using agarose gel electrophoresis for the presence of 180 bp-fragmentation.
Figure 2.
Overexpression of ROP16 induced SH-SY5Y cell cycle arrest in G1 phase. (A) SH-SY5Y cells stained with PI were analyzed by FACS after transfected with pPEXPR-IBA-105IP-ROP16 for 24hs, 48hs and 72hs. The mean values were used in histogram drawn. (B) Cell cycle analysis was performed 48hrs later after SH-SY5Y transfected with pPEXPR-ROP16 or pPEXPR by FACS. Error bars showed mean ±SEM of at least 3 replicates (p<0.05).
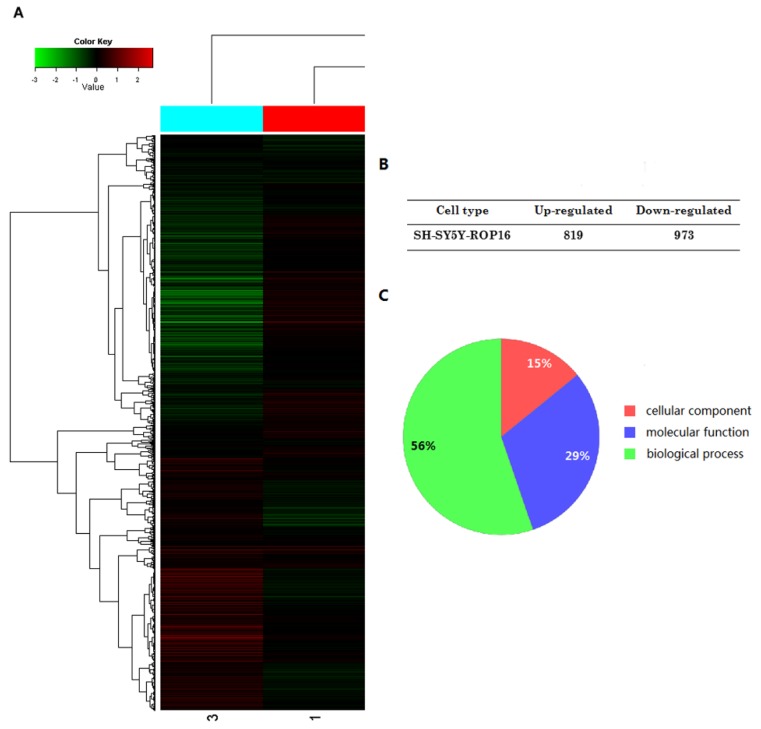
The expressional profiles of SH-SY5Y-ROP16 were examined by cDNA chip analysis showing 819 up-regulated genes and 973 down-regulated genes (Figure 3). mRNA expression level of many genes was changed (Table 2), especially TP53 and its downstream genes acting as important modulators that regulate cell apoptosis and cell-cycle arrest.
Figure 3.
A summary of gene expression changes in SH-SY5Y cells ectopically expressing ROP16. (A) A tree view diagram showing the fold change of genes expression. (B) Summary of changes in genes expression. (C) Gene Ontology (GO) analysis of genes expression profile showed that the genes differently expressed in SH-SY5Y ectopically expressing ROP16 are involved in biological process, molecular function and cellular component.
TABLE 2.
Apoptosis and Cell-cycle Arrest-associated Genes Expression Changes in SH-SY5Y Cells Overexpressing ROP16
| No. | Gene Name | Length | Location | Function | Regulation |
|---|---|---|---|---|---|
| 1 | Casp3 | 277 | Cytoplasm | Promotes apoptosis | Up |
| 2 | Casp8 | 480 | Cytoplasm | Promotes apoptosis | Up |
| 3 | Casp9 | 416 | Cytoplasm | Promotes apoptosis | Up |
| 4 | Bax | 192 | Cytoplasm | Promotes apoptosis | Up |
| 5 | p21 | 164 | Nucleus | Inhibit cellular proliferation | Up |
| 6 | Bcl-2 | 342 | Cytoplasm | Inhibit apoptosis | Down |
| 8 | SIRT | 747 | Nucleus | Participates in cell cycle, response to DNA damage, apoptosis and autophagy | Down |
| 13 | PUMA | 261 | Mitochondrion | Promote apoptosis | Up |
| 14 | SLTM | 1034 | Nucleus | Promote apoptosis | Up |
| 15 | NOXA | 454 | Mitochondrion | Promote apoptosis | Up |
| 16 | FAS | 335 | Cell membrane | Apoptosis | Up |
| 17 | EP300 | 2414 | Nucleus | Cell cycle | Up |
| 18 | E2f1 | 430 | Nucleus | Control cell-cycle progression from G1 to S phase | Down |
| 7 | CDKN1B | 198 | Cytoplasm Nucleus |
Induce G1 arrest | UP |
| 19 | UBE2L3 | 154 | Cytoplasm Nucleus |
Involved in cell cycle progression | Up |
| 20 | RB1 | 928 | Nucleus | Induce G1 arrest | UP |
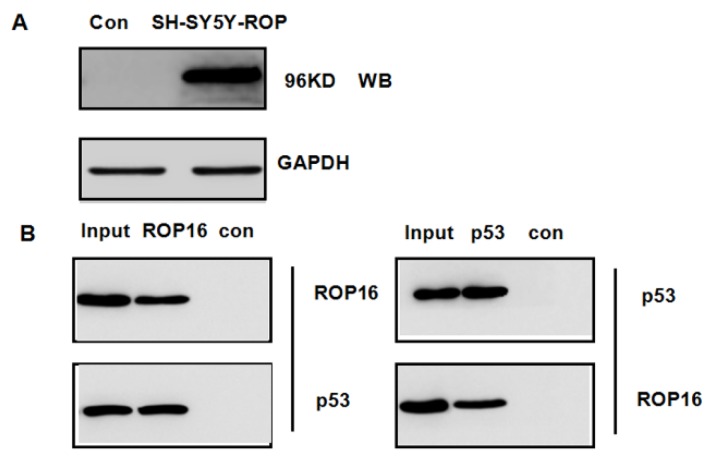
p53, as a novel ROP16-interaction partner isolated in mass spectrum and co-immunoprecipitation
We hypothesized that ROP16 might be involved in p53 signal pathway since it induces cell apoptosis and cell cycle arrest. p53 was one of the probable candidate proteins that interacted with ROP16 by the mass spectrum (Table 3). The interaction between ROP16 and p53 was detected by reciprocal co-immunoprecipitation (co-IP) assays. ROP16 was co-immunoprecipitated with p53 from SH-SY5Y-ROP16 cells. Likewise, p53 protein from SH-SY5Y-ROP16 cells also co-precipitated with ROP16 protein (Figure 4).
TABLE 3.
Apoptosis - associated Candidate Interacting Proteins with ROP16 in SH-SY5Y Cells
| No. | Protein Name | Function |
|---|---|---|
| 1 | Proliferating cell nuclear antigen | Apoptosis |
| 2 | Cell growth-regulating nucleolar protein | Apoptosis |
| 3 | Proliferation-associated protein 2G4 | Involved in growth regulation |
| 4 | Apoptosis inhibitor 5 isoform a | Anti-apoptotic factor |
| 5 | neurabin-2 | Apoptosis |
| 6 | Programmed cell death protein 5 | Apoptosis |
| 7 | Cellular tumor antigen p53 | Induces growth arrest or apoptosis |
| 8 | Apoptosis regulator BAX isoform alpha | Accelerates programmed cell death |
| 9 | Bcl-2-associated transcription factor 1 | Induction of apoptosis |
| 10 | Apoptosis-inducing factor 1 | Apoptotic process |
Figure 4.
Identification of p53 as a novel ROP16-interacting protein. (A) Overexpression of ROP16 in SH-SY5Y. (B) Reciprocal co-immunoprecipitation by using SH-SY5Y cells transfected with Flag-ROP16.
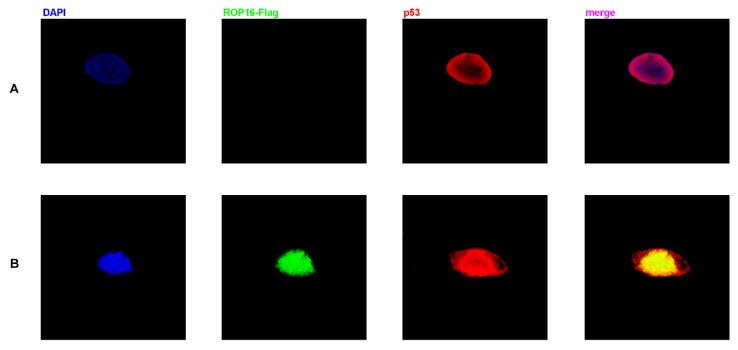
Co-localization of ROP16 and p53 in nucleus of SH-SY5Y cells
Both ROP16 and p53 were localized in the nucleus as shown in Figure 5. The merged images revealed the clear co-localization of the two proteins in SH-SY5Y cells. Overexpression of ROP16 clearly activated cell apoptosis compared with the control in Figure 5B.
Figure 5.
Co-localization of p53 and ROP16 in the nucleus of SH-SY5Y. SH-SY5Y cells were examined by indirect double immunofluorescence staining of the Flag-tagged ROP16 protein and p53 protein using a mixture of monoclonal anti-Flag antibody and anti-p53 antibody. Nucleus were visualized by DAPI staining. The merged images presented co-localization of p53 and ROP16 in SH-SY5Y nucleus.
ROP16 affects apoptosis and cell cycle-associated genes expression of SH-SY5Y
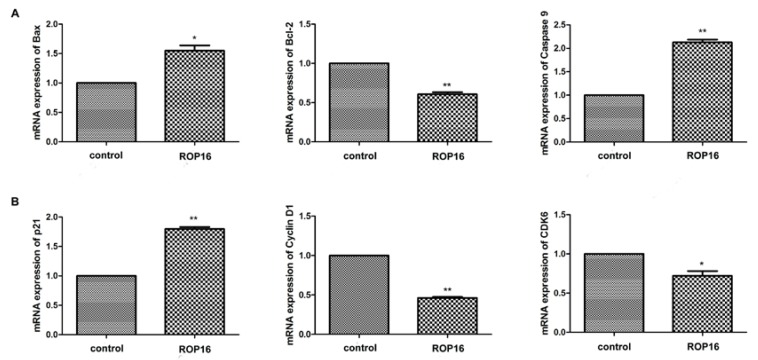
The expressions of Bax and caspase 9 were significantly up-regulated while the Bcl-2 expression was markedly down-regulated (Figure 6A). Afterwards, the cell cycle-associated genes expressions were determined by real time PCR. Our results showed that p21 expression was up-regulated significantly and the expressions of cyclin D1 and CDK6 were decreased (Figure 6B). In addition, the protein expressions of Bax and p21 were increased as well (Figure 7).
Figure 6.
ROP16 affected the apoptosis and cell cycle arrest-associated genes expression in SH-SY5Y. (A) qRT-PCR was applied to measure mRNA expression levels of p53-dependent apoptosis genes, including Bax, Bcl-2 and caspase 9 in SH-SY5Y transfected with ROP16-Flag. (B)The expression of cell cycle-associated gene p21,cyclin D1 and CDK6 were detected by qRT-PCR in SH-SY5Y cells overexpressing ROP16-Flag.Error bars showed mean ±SEM of at least 3 replicates (p<0.05).
Figure 7.
ROP16 mediated the expression of proteins associated with cell apoptosis and cell cycle arrest in SH-SY5Y. Western blot revealed that the protein expressions of Bax and p21 were significantly increased, which mediated partially cell apoptosis and cell cycle arrest (p<0.01).
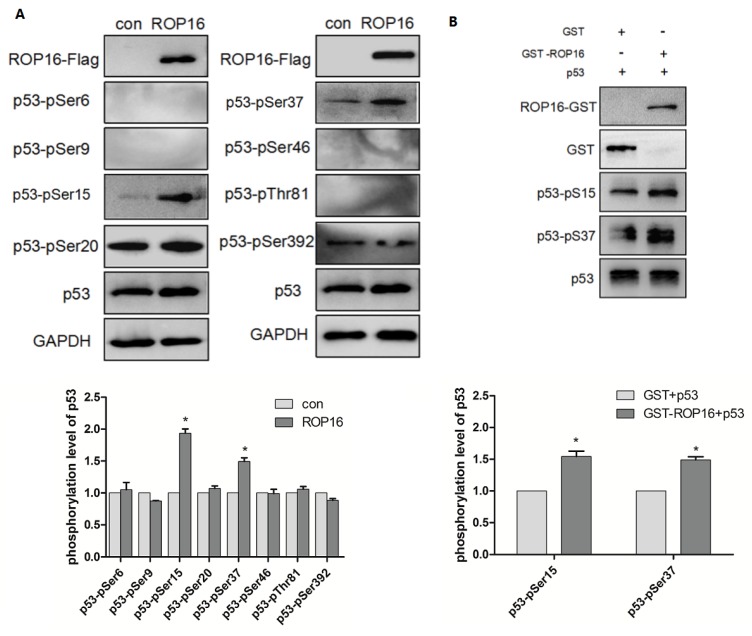
ROP16 produced an effect on p53 stabilization and its phosphorylation levels at Ser 15/37
ROP16 contains serine/threonine kinase activity while p53 protein possesses multiple phosphorylation sites that plays important roles in cell-cycle arrest and apoptosis. Therefore, we hypothesize that ROP16 mediates cell apoptosis and cell-cycle arrest by phosphorylating p53 at different sites. Western blot analysis showed that p53 expression level was significantly increased. Several phosphorylation sites of p53 including: Ser6, Ser9, Ser15, Ser20, Ser37, Ser46, Thr81, and Ser392 were examined by western blot in order to identify the phosphorylation sites of p53. Results showed that only Ser15 and Ser37 phosphorylation levels were increased significantly (Figure 8A). The in vitro kinase assay demonstrated that the recombinant His-p53 was directly phosphorylated at Ser15 and Ser37 by ROP16-GST (Figure 8B).
Figure 8.
ROP16 affects the p53 stablilization and its phosphorylation level at Ser 15/37 directly in vivo and in vitro. (A)Cells were harvested after 48h of transfection and lysates were subjected to immunoblot analysis using antibodies to p53-pSer6/9/15/20/37/46/392 and p53-pThr81. (B)Recombinant ROP16-GST was reacted with His-p53 in vitro. Phosphorylation resulting from ROP16-GST catalyzed His-p53 as the substrate was detected with anti-p53 pS15 and anti-p53 pS37 antibodies (p<0.05).
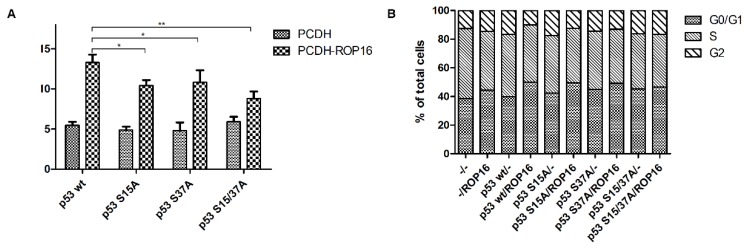
NE-4C-ROP16 cells injected with Lenti-p53-S15A37A survived much more than cells injected with other three types of p53 lentivirus (Lenti-p53 wt, Lenti-p53-S15A, Lenti-p53-S37A) which suggested that p53 Ser15 and Ser37 phosphorylated by ROP16 could induce the apoptosis index of the neural stem cells (Figure 9).
Figure 9.
ROP16 induced apoptosis and G1 arrest by phosphorylating p53 at Ser15/37 directly in NE-4C cells. NE-4C cells were infected with lentiviruce pCDH-p53wt/S15A/S37A/S15A37A and pCDH-ROP16/Flag before harvest. Cell cycle and apoptosis analysis of NE-4C cells were detected by flow cytometry for 3 times. Error bars showed mean ±SEM of at least 3 replicates (p<0.05).
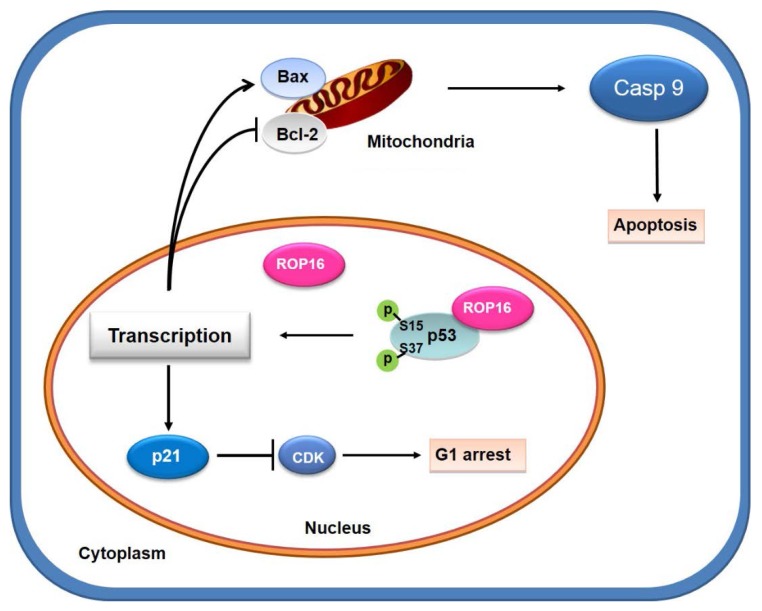
A model for Toxoplasma ROP16 interacting with p53 partially mediated apoptosis and G1 arrest in neuron cells
ROP16, overexpressed in SH-SY5Y cells, co-localizing and interacting with a novel partner p53 in the nucleus, leads to the increase phosphorylation level of Ser15 and Ser37 as well as the transcriptional activity. This interaction facilitates the expression of Bax while inhibits the expression of Bcl-2 thus partially mediates the mitochondria-dependent p53 apoptosis pathway. On the other hand, ROP16 expression in neuron cells results in cell-cycle arrest at G1 phase by promoting the expression of p21 and inhibiting the expression of CDK6 and cyclin D1 (Figure 10).
Figure 10.
A model for Toxoplasma ROP16 directly interacting with p53 partially mediates apoptosis and G1 phase arrest in SH-SY5Y. ROP16 overexpressed in SH-SY5Y cells co-localizing and interacting with p53 in the nucleus, influences p53 phosphorylation levels, triggering partially apoptosis and cell cycle arrest in G1 phase.
Discussion
Apoptosis is the process of programmed cell death that maybe caused by intracellular organisms. Some intracellular protozoans such as T. gondii have developed successful strategies to modulate host cell apoptosis. There still exist some controversies with the regard of the effect of T. gondii on apoptosis. T. gondii can lead to cell apoptosis by disrupting host cell signaling pathways 11-13. Toxoplasma strain virulence plays a major role in the fate of host cells as higher virulence leads to a great degree of host cell apoptosis 14, 15. The toxoplasma rhoptries are known to secrete their contents during the process of invasion into a host cell. ROP16, an essential virulence factor released from the apical organelles can quickly migrate into the host cell nucleus and intersect the host signal transducer and activator of transcription pathways through its kinase activity that result in the suppression of immune responses 1, 7, 9. The mechanisms by which ROP16 interferes with apoptosis signals and host cell functions are poorly understood. In this experiment, we found that ROP16 interacted with p53, as a novel interaction partner in the nucleus of host cell and ROP16 transfected into SH-SY5Y cells could partially mediate cell apoptosis and G1 arrest. The rate of apoptosis that occurred in SH-SY5Y was 12.47%. The protein p53 plays a crucial role in multi-cellular organisms as it served as a regulator of cell apoptosis and cell cycle. When a cell becomes exposed to stimulation, p53 protein will activate or repress various downstream target genes such as Bax, PUMA, Bcl-2 and p21 that are involved in apoptosis and cell-cycle arrest 16. p53 can directly activate the pro-apoptotic protein Bax, a p53 primary-response gene that leads to permeabilizitation mitochondria that induces the apoptotic program. Moreover, the p53 that are accumulated in the cytosol can function to activate the expression of Bax and subsequently trigger cell apoptosis 17. The anti-apoptotic proteins Bcl-2 has been shown to prevent disruption of mitochondrial physiology and block cytochrome c release from mitochondria by serving as a response gene of p53 thus in involved in p53 regulated apoptosis 18. Our data showed that the overexpression of ROP16 in SH-SY5Y led to the up-regulation of Bax, caspase 9 and down-regulation of Bcl-2. It suggested that ROP16 induced SH-SY5Y apoptosis via the mitochondria-dependent p53 pathway by increasing Bax expression while decreasing the Bcl-2 expression.
The cell interphase is composed of G1, S, G2 and M phases that defines a series of changes that occur in a newly formed cell and its nucleus. The previous data indicated that T. gondii induced G1/S transition in host cell in the presence of UHRF1 (ubiquitin-like, containing PHD and RING finger domains1), followed by G2 arrest after cyclin B1 down-regulation as probable major cause of the arrest 19. Toxoplasma tachyzoites attachment increased as both the Chinese hamster ovary cells and bovine kidney cells proceeded from the G1 phase to the mid-S phase and later decreased as the cells entered the G2-M boundary 20. The ectopic expression of ROP16 led to the up-regulation of p21 and down-regulation of CDK6 and cyclin D1. Moreover, ROP16 overexpression in mammalian cell could induce cycle-arrest in G1 phase (Figure 2). The protein p21, a direct p53 downstream target gene, is a potent cyclin-dependent kinase inhibitor of CDK6 and plays an important role in the cell cycle G1 phase progression and G1/S transition 21, 22. Evidences show that Toxoplasma GRA16 (dense granule protein) alters p53 levels in an HAUSP-dependent manner and induces nuclear translocation of the PP2A holoenzyme. The effects of GRA16 on p53 protein levels also affect to some extent its transcriptional activity toward p21 23. Our results strongly supported that ROP16 mediated SH-SY5Y cell-cycle arrest in G1 phase by increasing the p21 expression and decreasing the CDK expression.
ROP16's serine/threonine kinase activity has been proven. Many studies have been demonstrated a variety of p53 post-translational modifications that includes phosphorylation, ubiquitination, acetylation and methylation. p53 can be phosphorylated at multiple sites in vivo and by several different protein kinases in vitro 24. The modification at different sites plays different roles in the host cell function. Several serine residues such as Ser6, 9, 15, 37, 46 at N-terminal transcriptional domain have great impact on cell cycle arrest and apoptosis 24-29. The phosphorylation of serine at position 15 could reduce interaction between p53 and its negative regulator, the oncoprotein MDM2. It also induces stabilization of p53 regulating pro-apoptotic genes downstreams transcription and thus mediates cell apoptosis 30-32. Furthermore, activation of p53 results cell-cycle arrest that are reversible under some circumstances while may also lead to apoptotic cell death 33. It was well-known that elevated phosphorylation of p53 Ser37 led to cell cycle arrest in G1 phase 34. In order to define how ROP16 subverts the functions of the host cell at the specific sites phosphorylated of p53, we tested the eight sites in total that are composed of S6, S9, S15, S20, S37, S46, S392 and T81 that were involved in cell apoptosis while assessing the DNA damage as well as the transcriptional activity. The results showed that the S15 and S37 phosphorylation levels were significantly increased in cells expressing ROP16. In vitro kinase assay and the rescue experiment in NE-4C cells lacking the functional TP53 gene were performed with results demonstrating that ROP16 phosphorylated p53 at Ser15 and Ser37 residues (Figure 8B and Figure 9). These results indicated that ROP16 mediated neuron cell apoptosis by direct serine 15/37 phosphorylation of p53.
On the basis of our research, we propose the model of ROP16 triggering apoptosis in SH-SY5Y. We believe that ROP16 triggers apoptosis in SH-SY5Y due to the experimental results obtained from our research. ROP16 interacts and phosphorylates with a novel partner p53 in nucleus that increases stabilization and transcriptional activities of p53. Subsequently, the expression of downstream target genes of p53 such as Bax, Bcl-2 and p21 are affected as well resulting in G1 phase arrest and apoptosis via the mitochondria-dependent p53 pathway. The model provides an explanation of the relationship between T.gondii-induced apoptosis and cell arrest through the ROP16-p53 direct interaction.
Acknowledgments
We thank Huijun Wang for expert technical assistance and Shuhui Sun for advice on preparation and operation of FACS.
References
- 1.Boothroyd JC. Have it your way: how polymorphic, injected kinases and pseudokinases enable toxoplasma to subvert host defenses. 2013. [DOI] [PMC free article] [PubMed]
- 2.Dubremetz JF. Rhoptries are major players in Toxoplasma gondii invasion and host cell interaction. Cellular microbiology. 2007;9:841–8. doi: 10.1111/j.1462-5822.2007.00909.x. [DOI] [PubMed] [Google Scholar]
- 3.Mueller C, Klages N, Jacot D, Santos JM, Cabrera A, Gilberger TW. et al. The Toxoplasma protein ARO mediates the apical positioning of rhoptry organelles, a prerequisite for host cell invasion. Cell host & microbe. 2013;13:289–301. doi: 10.1016/j.chom.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Weilhammer DR, Rasley A. Genetic approaches for understanding virulence in Toxoplasma gondii. Briefings in functional genomics. 2011;10:365–73. doi: 10.1093/bfgp/elr028. [DOI] [PubMed] [Google Scholar]
- 5.Boothroyd JC, Dubremetz J-F. Kiss and spit: the dual roles of Toxoplasma rhoptries. Nature Reviews Microbiology. 2008;6:79–88. doi: 10.1038/nrmicro1800. [DOI] [PubMed] [Google Scholar]
- 6.Taylor S, Barragan A, Su C, Fux B, Fentress S, Tang K. et al. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science. 2006;314:1776–80. doi: 10.1126/science.1133643. [DOI] [PubMed] [Google Scholar]
- 7.Saeij J, Coller S, Boyle J, Jerome M, White M, Boothroyd J. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature. 2007;445:324–7. doi: 10.1038/nature05395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto M, Standley DM, Takashima S, Saiga H, Okuyama M, Kayama H. et al. A single polymorphic amino acid on Toxoplasma gondii kinase ROP16 determines the direct and strain-specific activation of Stat3. The Journal of experimental medicine. 2009;206:2747–60. doi: 10.1084/jem.20091703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosowski EE, Saeij JP. Toxoplasma gondii clonal strains all inhibit STAT1 transcriptional activity but polymorphic effectors differentially modulate IFNγ induced gene expression and STAT1 phosphorylation. 2012. [DOI] [PMC free article] [PubMed]
- 10.Jensen KD, Hu K, Whitmarsh RJ, Hassan MA, Julien L, Lu D. et al. Toxoplasma gondii rhoptry 16 kinase promotes host resistance to oral infection and intestinal inflammation only in the context of the dense granule protein GRA15. Infection and immunity. 2013;81:2156–67. doi: 10.1128/IAI.01185-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang T, Zhou J, Gan X, Wang H, Ding X, Chen L. et al. Toxoplasma gondii induce apoptosis of neural stem cells via endoplasmic reticulum stress pathway. Parasitology. 2014;141:988–95. doi: 10.1017/S0031182014000183. [DOI] [PubMed] [Google Scholar]
- 12.Hwang I-Y, Quan JH, Ahn M-H, Ahmed HAH, Cha G-H, Shin D-W. et al. Toxoplasma gondii infection inhibits the mitochondrial apoptosis through induction of Bcl-2 and HSP70. Parasitology research. 2010;107:1313–21. doi: 10.1007/s00436-010-1999-3. [DOI] [PubMed] [Google Scholar]
- 13.Peter ME. Programmed cell death: Apoptosis meets necrosis. Nature. 2011;471:310–2. doi: 10.1038/471310a. [DOI] [PubMed] [Google Scholar]
- 14.Gavrilescu LC, Denkers EY. IFN-γ overproduction and high level apoptosis are associated with high but not low virulence Toxoplasma gondii infection. The Journal of Immunology. 2001;167:902–9. doi: 10.4049/jimmunol.167.2.902. [DOI] [PubMed] [Google Scholar]
- 15.Hunter CA, Sibley LD. Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nature Reviews Microbiology. 2012;10:766–78. doi: 10.1038/nrmicro2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH. et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes & development. 2000;14:981–93. [PMC free article] [PubMed] [Google Scholar]
- 17.Beilharz K, Nováková L, Fadda D, Branny P, Massidda O, Veening J-W. Control of cell division in Streptococcus pneumoniae by the conserved Ser/Thr protein kinase StkP. Proceedings of the National Academy of Sciences. 2012;109:E905–E13. doi: 10.1073/pnas.1119172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunelle JK, Letai A. Control of mitochondrial apoptosis by the Bcl-2 family. Journal of cell science. 2009;122:437–41. doi: 10.1242/jcs.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brunet J, Pfaff AW, Abidi A, Unoki M, Nakamura Y, Guinard M. et al. Toxoplasma gondii exploits UHRF1 and induces host cell cycle arrest at G2 to enable its proliferation. Cellular microbiology. 2008;10:908–20. doi: 10.1111/j.1462-5822.2007.01093.x. [DOI] [PubMed] [Google Scholar]
- 20.Grimwood J, Mineo JR, Kasper LH. Attachment of Toxoplasma gondii to host cells is host cell cycle dependent. Infection and immunity. 1996;64:4099–104. doi: 10.1128/iai.64.10.4099-4104.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zupkovitz G, Grausenburger R, Brunmeir R, Senese S, Tischler J, Jurkin J. et al. The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation. Molecular and cellular biology. 2010;30:1171–81. doi: 10.1128/MCB.01500-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saurin AT, Brownlow N, Parker PJ. Protein kinase C epsilon in cell division: Control of abscission. Cell Cycle. 2009;8:549–55. doi: 10.4161/cc.8.4.7653. [DOI] [PubMed] [Google Scholar]
- 23.Bougdour A, Durandau E, Brenier-Pinchart M-P, Ortet P, Barakat M, Kieffer S. et al. Host cell subversion by Toxoplasma GRA16, an exported dense granule protein that targets the host cell nucleus and alters gene expression. Cell host & microbe. 2013;13:489–500. doi: 10.1016/j.chom.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Kruse J-P, Gu W. Modes of p53 regulation. Cell. 2009;137:609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T. et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–62. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- 26.Du D, Wang L, Shao Y, Wang J, Engelhard MH, Lin Y. Functionalized graphene oxide as a nanocarrier in a multienzyme labeling amplification strategy for ultrasensitive electrochemical immunoassay of phosphorylated p53 (S392) Analytical chemistry. 2011;83:746–52. doi: 10.1021/ac101715s. [DOI] [PubMed] [Google Scholar]
- 27.Friedler A, Veprintsev DB, Freund SM, Karoly I, Fersht AR. Modulation of binding of DNA to the C-terminal domain of p53 by acetylation. Structure. 2005;13:629–36. doi: 10.1016/j.str.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 28.Karni-Schmidt O, Friedler A, Zupnick A, McKinney K, Mattia M, Beckerman R. et al. Energy-dependent nucleolar localization of p53 in vitro requires two discrete regions within the p53 carboxyl terminus. Oncogene. 2007;26:3878–91. doi: 10.1038/sj.onc.1210162. [DOI] [PubMed] [Google Scholar]
- 29.Smeenk L, Van Heeringen SJ, Koeppel M, Gilbert B, Janssen-Megens E, Stunnenberg HG. et al. Role of p53 serine 46 in p53 target gene regulation. PloS one. 2011;6:e17574–e. doi: 10.1371/journal.pone.0017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Can G, Akpinar B, Baran Y, Zhivotovsky B, Olsson M. 5-Fluorouracil signaling through a calcium-calmodulin-dependent pathway is required for p53 activation and apoptosis in colon carcinoma cells. Oncogene. 2013;32:4529–38. doi: 10.1038/onc.2012.467. [DOI] [PubMed] [Google Scholar]
- 31.Amano T, Nakamizo A, Mishra SK, Gumin J, Shinojima N, Sawaya R. et al. Simultaneous phosphorylation of p53 at serine 15 and 20 induces apoptosis in human glioma cells by increasing expression of pro-apoptotic genes. Journal of neuro-oncology. 2009;92:357–71. doi: 10.1007/s11060-009-9844-1. [DOI] [PubMed] [Google Scholar]
- 32.Yamada C, Ozaki T, Ando K, Suenaga Y, Inoue K-i, Ito Y. et al. RUNX3 modulates DNA damage-mediated phosphorylation of tumor suppressor p53 at Ser-15 and acts as a co-activator for p53. Journal of Biological Chemistry. 2010;285:16693–703. doi: 10.1074/jbc.M109.055525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leontieva OV, Gudkov AV, Blagosklonny MV. Weak p53 permits senescence during cell cycle arrest. Cell Cycle. 2010;9:4323–7. doi: 10.4161/cc.9.21.13584. [DOI] [PubMed] [Google Scholar]
- 34.Tawara H, Fujiuchi N, Sironi J, Martin S, Aglipay J, Ouchi M. et al. Loss of p53-regulatory protein IFI16 induces NBS1 leading to activation of p53-mediated checkpoint by phosphorylation of p53 SER37. Frontiers in bioscience: a journal and virtual library. 2007;13:240–8. doi: 10.2741/2674. [DOI] [PubMed] [Google Scholar]