Abstract
Enhancer-dependent transcription involving the promoter specificity factor σ54 is widely distributed amongst bacteria and commonly associated with cell envelope function. For transcription initiation, σ54-RNA polymerase yields open promoter complexes through its remodelling by cognate AAA+ ATPase activators. Since activators can be bypassed in vitro, bypass transcription in vivo could be a source of emergent gene expression along evolutionary pathways yielding new control networks and transcription patterns. At a single test promoter in vivo bypass transcription was not observed. We now use genome-wide transcription profiling, genome-wide mutagenesis and gene over-expression strategies in Escherichia coli, to (i) scope the range of bypass transcription in vivo and (ii) identify genes which might alter bypass transcription in vivo. We find little evidence for pervasive bypass transcription in vivo with only a small subset of σ54 promoters functioning without activators. Results also suggest no one gene limits bypass transcription in vivo, arguing bypass transcription is strongly kept in check. Promoter sequences subject to repression by σ54 were evident, indicating loss of rpoN (encoding σ54) rather than creating rpoN bypass alleles would be one evolutionary route for new gene expression patterns. Finally, cold-shock promoters showed unusual σ54-dependence in vivo not readily correlated with conventional σ54 binding-sites.
INTRODUCTION
Amongst the strategies used to regulate transcription initiation in bacteria, the emergence of the enhancer-dependent system that uses the σ54 RNA polymerase is of special interest because it deviates from the transcription factor recruitment mechanisms commonly seen (1,2). Rather, binding and hydrolysis of aenosine triphosphate (ATP) by cognate AAA+ ATPase activator proteins remodel σ54 containing closed promoter complexes to yield open promoter complexes (3,4). Potentially, σ54 initially acted as a global repressor of RNA polymerase (RNAP) activity since it effectively road blocks the DNA entry path used for delivering melted DNA into the DNA binding cleft of RNAP. This silenced state of RNAP may then have become a target for regulation. Intriguingly, to date no bacteria have been found where the gene encoding σ54 (rpoN) is present in the absence of any cognate AAA+ activator (4,5). This suggests forms of σ54 functioning without activators are uncommon. Yet variants of σ54 bypassing activator requirements for making open promoter complexes exist and have been characterized extensively in vitro (6,7). Single amino acid substitutions in σ54 can yield activator bypass forms. Quite why σ54 has seemingly not (or rarely) evolved to activator independence is rather unclear. Here we have investigated the in vivo global actions of wild-type σ54 in Escherichia coli bacteria and one bypass variant to explore potential restrictions on evolving bypass gene regulation systems.
We found that only a very limited number of knock-outs (KOs) of non-essential genes in E. coli increased activator bypass transcription suggesting multifactorial global constraints prevent unregulated transcription by the σ54 containing RNAP holoenzyme (σ54RNAP). Intriguingly, some genes with σ54 binding sites were found to be repressed in a σ54-dependent manner. In some cases the putative σ54 binding site overlapped that of a putative σ70 binding site suggesting a competition for promoter sites, which we confirmed by in vitro experiments. In other cases, σ54 and σ70 RNAP appear to act antagonistically through e.g. convergent transcription events.
MATERIALS AND METHODS
Bacterial strains and growth conditions
For the construction of the glnAp2 strains (Supplementary Table S1) P1vir transduction was used to insert rpoN208::Tn10 (K1471 donor strain) into the native chromosomal rpoN locus of BW25113 (recipient). Subsequently, transformation was used to transfer the pSB4A3 plasmid harbouring ΦPglnAp2-gfp (a gift from Baojun Wang, University of Edinburgh, UK) and pBAD18 cm (containing Klebsiella pneumonia rpoN, rpoNΔRI or vector only). For the construction of the pspA strains (Supplementary Table S1) P1vir transduction was used to insert rpoN208::Tn10 (K1471 donor strain) into the native chromosomal rpoN locus of BW25113 (recipient). Next the ΦPpspA-lacZ (MVA4 donor strain; 8) was inserted into the att site of the BW25113 rpoN208::Tn10 chromosome, using P1vir transduction. Finally, transformation was used to transfer pBAD18cm (containing Klebsiella pneumonia rpoN, rpoNΔRI or vector only) into the BW25113 rpoN208::Tn10 ΦPpspA-lacZ. The F3K3 plasmid containing the filamentous phage pIV secretin was also transformed into this strain. To transfer the single-gene KOs from the KEIO collection (9) and small peptide/ small RNA library (10), P1vir lysates were produced from the pooled strains. These were then used as a donor for P1vir transduction of the KOs into the pspA-SABRS recipient strain (Supplementary Table S1).
For gene expression assays in vivo, strains were grown on LB indicator (XGal and MacConkey) plates and in minimal Gutnick liquid media supplemented with L-glutamine (11) as the nitrogen source to allow good growth in the absence of σ54 function (since the nitrogen assimilation pathways in E. coli rely on σ54 for expressing appropriate levels of glutamine synthetase). The antibiotic concentrations used were 30 μg/ml kanamycin, 50 μg/ml chloramphenicol and 50–100 μg/ml ampicillin.
Gene expression assays invivo
To measure activity of ΦPpspA-lacZ in vivo, β-Galactosidase assays were carried out as described (12). To determine gene expression levels of the ΦPglnAp2-gfp reporter in vivo, cells were grown in black 96-well clear-bottom tissue culture plates. Optical density at 600 nm (OD600) and green fluorescence (excitation: 485 nm; emission: 520 ± 10 nm, gain: 1000) were measured simultaneously using a BMG FLUOstar Omega microplate reader. Promoter activity was expressed as fluorescence emission at 520 nm per OD600 measured in triplicate.
Inverse PCR
Inverse polymerase chain reaction (PCR) was used to identify selected KO mutants (Supplementary Table S2). Chromosomal DNA from strains that were positive for β-galactosidase activity on indicator plates and in liquid culture (Miller assay) was purified and digested with NcoI, a 6-base cutter. Re-ligation of the resulting fragments into circular DNA was done in a volume of 100 μl. This DNA was used as a template for PCR, using kanamycin cassette-specific forward and reverse primers (9). Products were run on a gel, extracted and sequenced. NCBI Blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to align sequenced regions to the E. coli chromosome and to determine the identity of the deleted gene.
RNA sequencing (RNAseq)
A total of 100 ml cell cultures were grown to exponential phase at 42 or 37°C, in minimal Gutnick media supplemented with 5 mM L-Glutamine and 0.4% glucose. Cell pellets were sent to Vertis Biotechnologie AG (Germany) for further processing. Total RNA was isolated from the cell pellets using a bead mill and the mirVana RNA isolation kit (Ambion) including DNase treatment. Ribosomal RNA molecules were depleted from the total RNA preparations using the RiboZero rRNA Removal Kit (Bacteria) (Epicentre). The rRNA depleted RNAs were fragmented with RNase III. Then, the RNA fragments were poly(A)-tailed using poly(A) polymerase and the 5′ PPP structures were removed using RNA 5′ polyphosphatase (Epicentre). An RNA adapter was ligated to the 5′-phosphate of the RNA fragments. First-strand cDNA synthesis was performed using an oligo(dT)-adapter primer and M-MLV reverse transcriptase. The resulting cDNA was PCR-amplified using a high fidelity DNA polymerase. The cDNA was purified using the Agencourt AMPure XP kit (Beckman Coulter Genomics). The cDNA pools were size fractionated in the size range of 200–450 bp using a differential clean-up with the Agencourt AMPure kit. The primers used for PCR amplification were designed for TruSeq sequencing according to the instructions of Illumina and sequenced on a Illumina HiSeq 2000 system using 50 bp read length. Near equimolar amounts of cDNA were used for sequencing. The cDNA reads were analysed via the RNA-seq workflow within Partek® Genomics suite 6.6, including a QA/QC step to gauge the sequencing quality. Each sample yielded close to equivalent total reads with 100% of reads aligned to the E. coli K-12 reference genome (NC_000913) (Supplementary Table S4). For each gene, reads were normalized for both the number of reads and length of RNA and expressed as RPKM (reads per kilobase per million) (13; Supplementary Table S4). A one-way ANOVA statistical analysis was performed to test for significant differences in genome-wide gene expression between all 3 strains (Supplementary Table S4).
In vitro spRNA assays
Small primed RNAs (spRNAs) were synthesized in 10 μl volumes containing: 0.125 μM σ70/σ54RNAP holoenzyme, 20 nM linear promoters, 0.5 mM dinucleotide primers, 0.2 mg/ml heparin and 0.2 μCi/μl 32P-NTP in STA buffer (2.5 mM Tris-acetate pH8, 8 mM Mg-acetate, 10 mM KCl, 1 mM DTT, 3.5% (w/v) PEG 8000) at 37°C for 10 min. For the σ70/σ54 competition assays, 0.4 μM σ54 were either added before or with σ70 for binding competition. The reactions were quenched by formamide stop dye and run on a 20% sequencing gel. Signals were detected by Fuji PhosphorImager and quantified using the AIDA Image Analyzer software (raytest).
RESULTS
Genome-wide screens for increased bypass transcription suggest constraints exist
A form of σ54 lacking the repressive Region I, σ54ΔRI, can bypass activator requirement for transcription in vitro (Figure 1;14,15). Using a lacZ fusion to the pspA promoter (PpspAlacZ) in single copy integrated into the att site of the E. coli chromosome we sought bypass activity from the σ54-dependent pspA promoter in vivo. Reporter strains lacked rpoN and were complemented with either the wild-type or the ΔRI form of σ54 expressed from pBAD18 cm (Supplementary Table S1, Figure S1). While rpoN and rpoNΔRI were under control of the araBAD arabinose inducible promoter, leaky expression (without arabinose) yielded sufficient levels of σ54 to enable pIV-dependent transcription from the pspA promoter (Supplementary Figure S2). Thus unless stated otherwise (Supplementary Table S2) the experiments were performed in the absence of arabinose. Under standard growth conditions, no clear bypass transcription was observable with the σ54ΔRI form (Figure 2). Secretin pIV-dependent activation of the pspA promoter (16) via its cognate activator PspF acting on the σ54RNAP holoenzyme was clearly observed in vivo (Figure 2).
Figure 1.
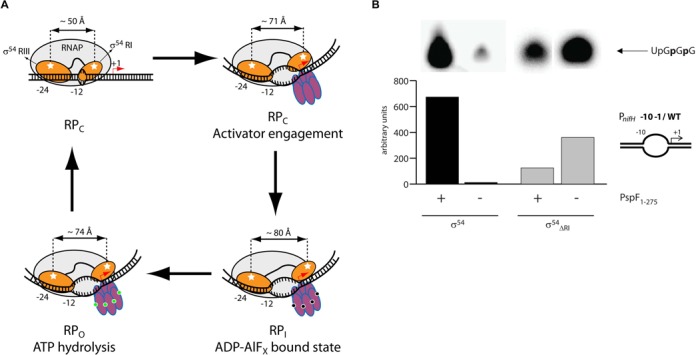
(A) The Region I of σ54 acts to inhibit RNA polymerase activity by blocking promoter DNA entry into the major DNA binding cleft. (B) Removal of Region I enables in vitro transcription from the late-melted nifH promoter (−10–1/WT) without the AAA+ activator PspF1–275.
Figure 2.
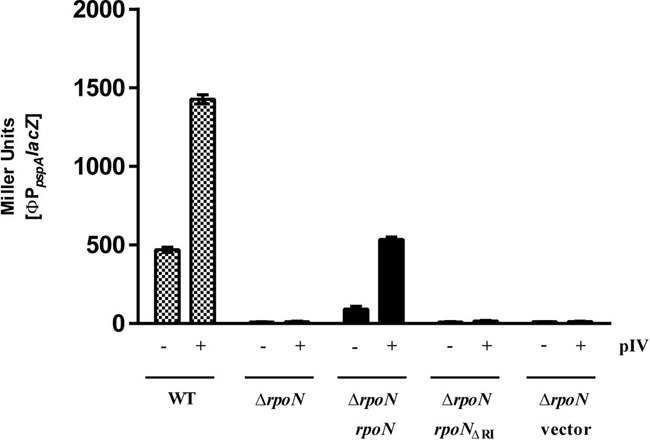
The σ54 dependent pspA promoter is active in vivo in the presence of stress inducing signals (here pIV secretin) and non-bypass wild-type σ54, but is inactive with the ΔRI σ54 form. Experiments were performed with three biological replicates.
It was reported that increased temperature enhances σ54 activator bypass transcription in vitro at the glnAp2 promoter (6). We therefore cultured cells at 30, 37 and 42°C to favour detecting bypass transcription in vivo. Indeed, we observed a four-fold increase in σ54ΔRI-dependent glnAp2 promoter activity at 42°C compared to 30 or 37°C. This bypass activity accounts for almost 50% of wild-type σ54-dependent transcription (Figure 4).
Figure 4.
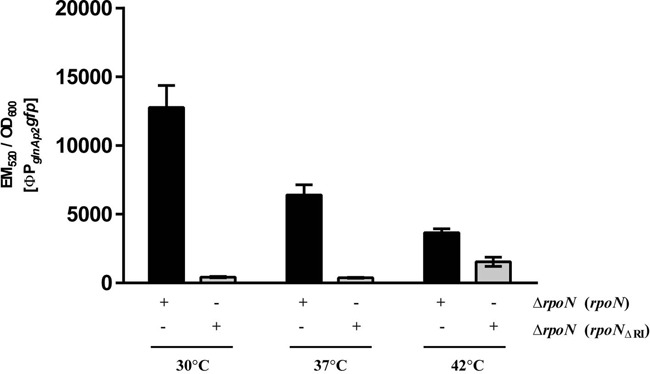
Elevated temperature increases bypass transcription from the glnAp2 promoter in vivo (grey), by over 40% activity of the wild-type rpoN control at the same temperature (black). Experiments were performed with three biological replicates.
To seek genetic determinants potentially restricting in vivo bypass transcription from the pspA promoter we introduced non-essential gene KOs into the reporter strains from the KEIO collection of E. coli gene KOs (9; a gift from the Carol Gross laboratory, UCSF, USA) and the small RNA/small peptide KO collection (10; a gift from the Gigi Storz laboratory, NIH, USA). Gene KOs were introduced into the reporter strains by phage P1vir transduction following growth of the phage on pooled KOs. Transductants were screened on indicator media (X-gal, MacConkey) or on lactose as a carbon source for increased expression of PpspAlacZ. Screens were carried out at 37 and 42°C, and in the absence and presence of 0.001% arabinose (Supplementary Table S2). Colony PCR was used to confirm all necessary genetic elements (rpoN KO, pBAD18 cm, rpoNΔRI allele, PpspAlacZ fusion) were intact in candidate upregulated clones. Coverage was three to four-fold of the chromosome assuming all KOs were represented in the library.
From the above screens, we found 26 KOs which led to apparent bypass transcription from the pspA promoter based on an increased β-galactosidase activity on indicator plates (Supplementary Table S2). Of these, six KOs (asnA, greA, hldE, nanT, ttdR, yhbX) which were found in a screen at 37°C also show increased β-galactosidase activity in liquid culture. These KOs were directly identified by inverse PCR and sequencing. Five KOs were discounted as being direct restrictive determinants of in vivo levels of bypass transcription on the basis of (i) just causing changes to the cell's envelope which led to enhanced β-galactosidase activity, potentially a permeability issue with the ONPG substrate or (ii) reconstitution of the native chromosomal rpoN locus. The ttdR gene however encodes a nucleoid binding protein and its loss seems to genuinely cause increased bypass transcription (Figure 3). The basis of the effect may lie in changes to DNA superhelicity, which is known to impact on bypass transcription in vitro (6). We conclude that in E. coli very few non-essential genes act to restrict bypass transcription in vivo. Those that do may act globally on e.g. DNA superhelicity and so may also impact on many additional gene expression events which themselves are not directly σ54 dependent.
Figure 3.
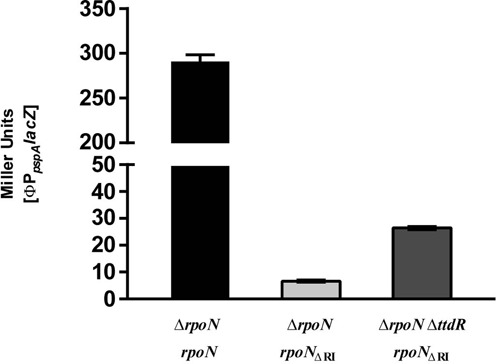
Single-gene knock-out screening at 37°C yielded ttdR (encoding a nucleoid DNA-binding protein) as negatively impacting σ54 bypass transcription from the pspA promoter in vivo. In addition to increased bypass transcription from the ΦPpspAlacZ on XGal and MacConkey screens (Supplementary Table S2), the ΔttdR mutant also increased bypass transcription as measured in Miller Units. Experiments were performed with three biological replicates.
Overexpressed genes do not yield increased bypass transcription
We considered that the amount of a gene product might be limiting, rather than inhibitory, for supporting bypass transcription by σ54. We therefore introduced a library of genes from the closely related Salmonella typhimurium bacteria on a multi-copy plasmid (a gift from a gift from Diarmaid Hughes; Uppsala University, Sweden; 17) into the E. coli reporter strain and screened for increased expression of PpspAlacZ. None was observed, suggesting no single gene product limits bypass transcription in E. coli (Supplementary Table S2).
RNAseq analysis of the σ54 bypass allele ΔRI activity reveals activator independent promoter sites
We conducted RNAseq of cells containing the ΔRI bypass form of σ54 and compared outcomes to those from cells with wild-type σ54 and to cells lacking σ54. Cells were cultured at 42°C, which enhances bypass in vitro (6), to favour detecting bypass transcription in vivo. We confirmed using the glnAp2 promoter this was so in vivo (Figure 4). We chose minimal media supplemented with L-glutamine as the nitrogen source to allow good growth in the absence of σ54 function (since the nitrogen assimilation pathways in E. coli rely on σ54 for expressing appropriate levels of glutamine synthetase).
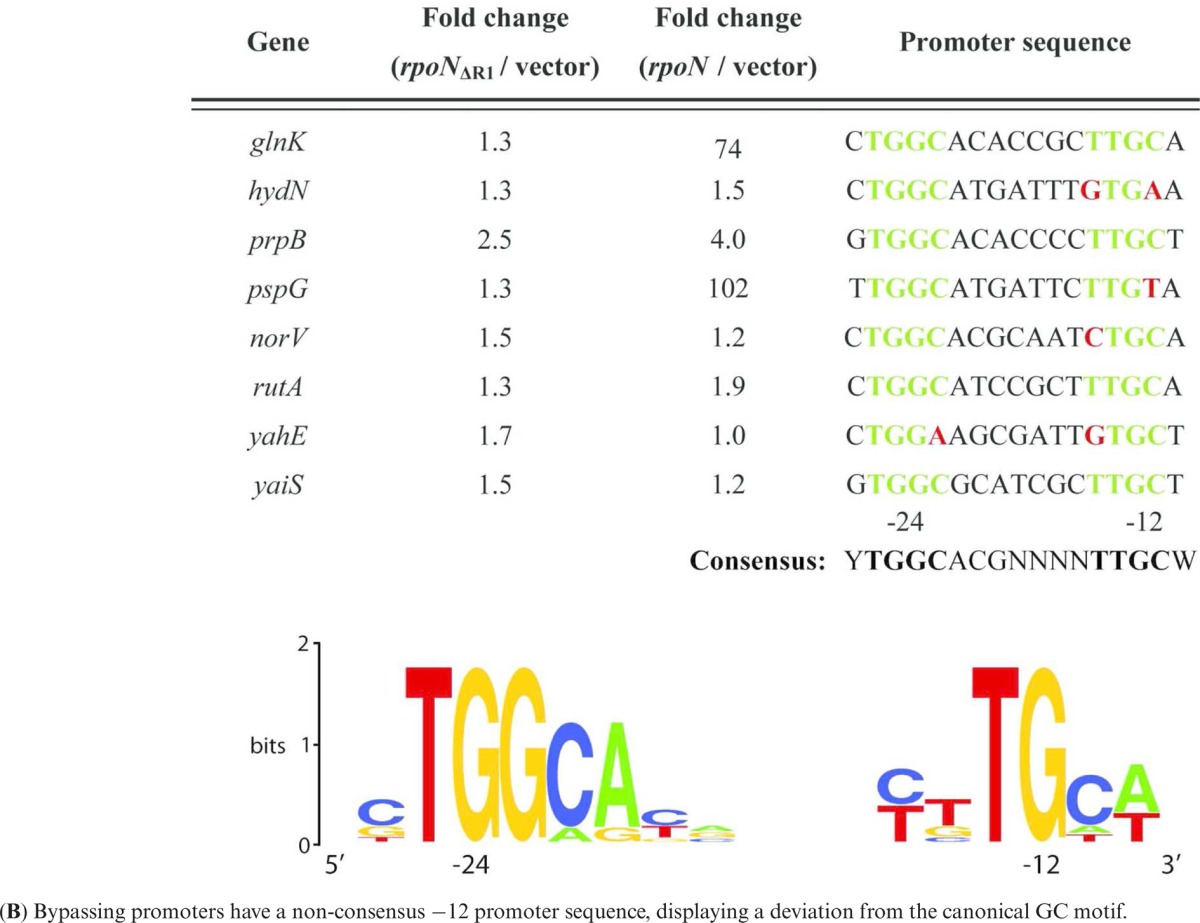
RNAseq profiles of cells with wild-type σ54 compared to cells with σ54ΔRI or no σ54 revealed no major global bypass transcription (Figure 5). By scoring for more RNAseq reads in presence of the bypass σ54ΔRI form compared to the no σ54 we identified eight sites of low-level activator-independent transcription from sequences downstream of a canonical −12/−24 σ54 promoter DNA recognition sites (Table 1). We conclude that a small subset of σ54 binding sites can yield transcripts independently of the conventional AAA+ ATPase driven activation pathway, and that this bypassing is mediated by the loss of Region I of σ54. The C residue in the GC −12 element is conserved in 96% of bacterial σ54 promoters (18). Alignment (19,20) of the bypassing −12/−24 σ54 motifs indicates that the C residue is not strictly conserved at these promoters (Table 1). This finding is in line with in vitro studies showing that the consensus C at −12 is important for keeping basal transcription in check (21).
Figure 5.
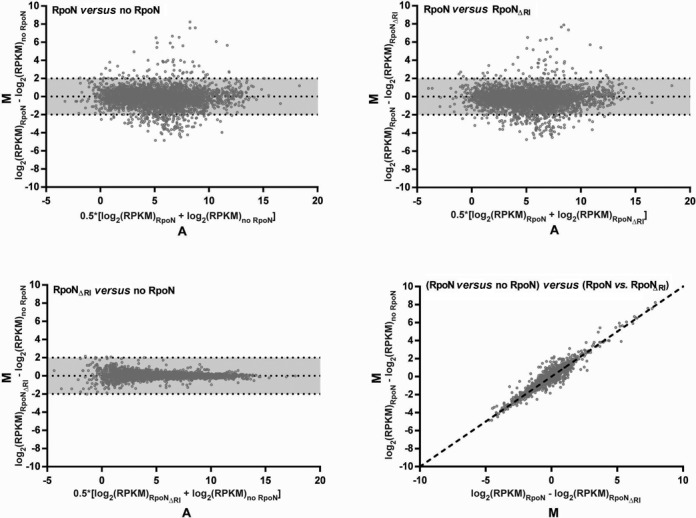
Genome-wide transcriptional profiling by RNAseq revealed no major global bypass transcription by RpoNΔRI. Shown are MA scatterplots comparing cells with wild-type RpoN to cells with no RpoN (Top left) or RpoNΔRI (Top right) as well as cells with RpoNΔRI to cells with no RpoN (Bottom left). M is the difference in number of reads for each gene between a pair of strains expressed as log2(RPKM)Strain 1 – log2(RPKM)Strain 2, i.e. values > 0 indicate higher expression in strain 1 while values < 0 indicate higher expression in strain 2. A is the average number of reads of a gene between both strains expressed as 0.5*[log2(RPKM)Strain 1 + log2(RPKM)Strain 2]. Data points within the grey shaded area (M = log2(RPKM) = +/− 2) depict genes less than equal to four-fold differently expressed between the strains. The scatterplot in the Bottom right corner compares the M-values of the Top left and Top right plot showing that the differences in gene expression between cells with wild-type RpoN and no RpoN are similar to the differences in gene expression between cells with wildtype RpoN and RpoNΔRI (dotted line: y = x). The data is derived from single sample analysis.
Table 1. (A) σ54 promoter sites showing increased bypass transcription at 42°C in vivo when Region I of σ54 is absent.
|
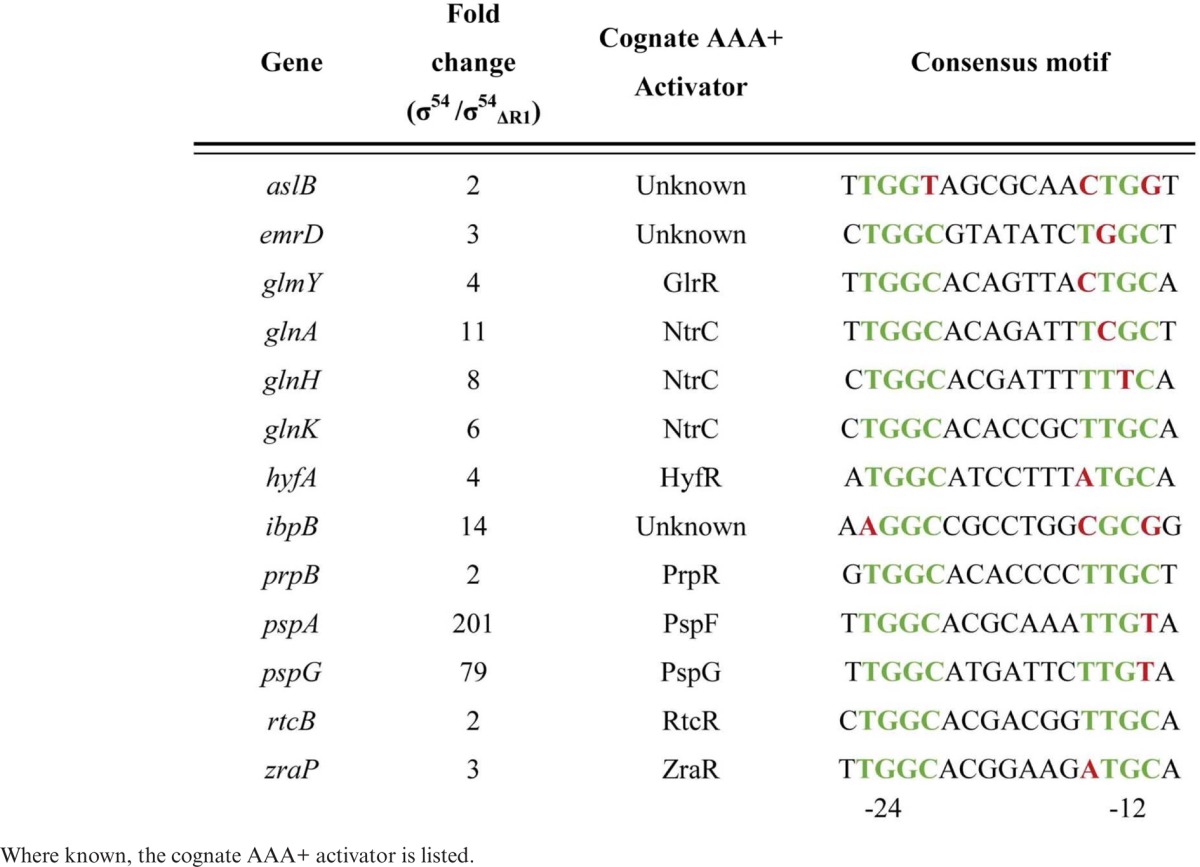
By scoring for more RNAseq reads in presence of wild-type σ54 compared to σ54ΔRI we identified canonical −12/−24 promoters that are likely to be activated in a Region I-dependent manner under the growth conditions of the experiment. Table 2 lists these promoters and the genes under their control. The majority are promoters from several well characterized σ54-dependent systems such as those controlled by the activators such as NtrC, PspF, PrpR and RtcR.
Table 2. σ54 promoter sites active at 42°C in vivo.
|
σ54 is repressive at some promoters
We also compared reads from cells lacking σ54 to cells with the wild-type σ54 or its ΔRI form. Intriguingly, in several cases reads were higher when σ54 was absent, suggesting that the σ54RNAP holoenzyme can be repressive for some genes, either directly or indirectly. We inspected such genes for −12/−24 sequences and grouped them into four different classes based on the location of the σ54 binding motifs relative to the active −10/−35 promoter sequences (Figure 6 and Supplementary Table S3). In several cases (class I and II), we find that the σ54 recognition site is overlapping with the likely footprint of a σ70RNAP holoenzyme on these sites, suggesting a simple binding competition mechanism of repression by the σ54RNAP. Using purified components, we tested four σ70-regulated linear homoduplex promoters (argT, chaC, mdfA and patA) from class I and II for binding repression by σ54 in vitro (Figure 7 and Supplementary Figure S3). The σ70RNAP holoenzyme was allowed to generate small primed RNAs (spRNAs) using initiating dinucleotides (−1/+1 or +1/+2). The experiments were performed under two conditions. In the first, σ54 and σ70 were co-incubated with the RNAP prior to addition of promoter DNA probes. Here σ54 was in three-fold molar excess to σ70 and RNAP. This competition from excess σ54 reduced the amount of spRNA synthesised by 10–60% (Figure 7). In the second, σ54 was pre-incubated with the promoter DNA probes prior to addition of the σ70RNAP holoenzyme. Here, σ70 was in more than three-fold molar excess to σ54 and RNAP. Again, we observed a similar σ54-dependent reduction in spRNA synthesis (Supplementary Figure S3). Since the binding affinity and dissociation constant of σ54 and σ70 for RNAP are very similar (22) and similar results were obtained in both conditions, we suggest that inhibition is observed due to a binding competition from σ54 containing RNAP within the −10/−35 promoter sites rather than a direct competition for RNAP by σ70 and σ54.
Figure 6.
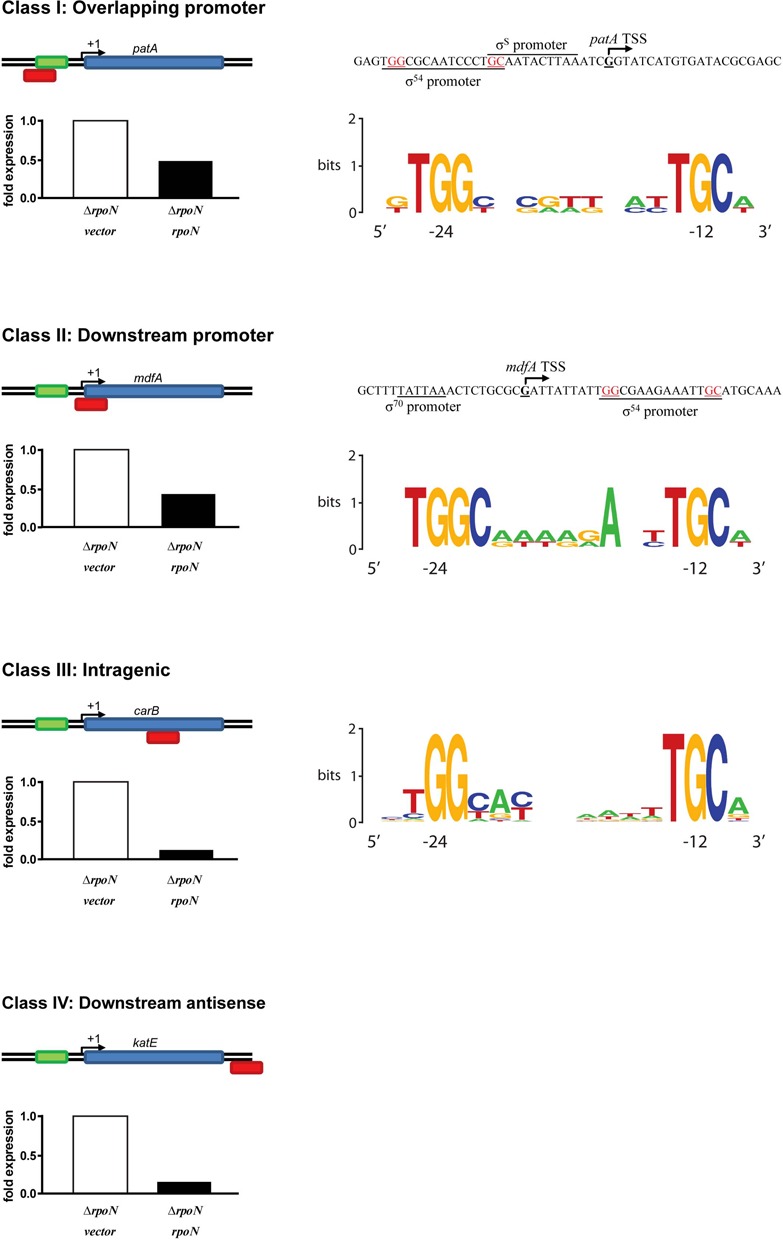
σ54 -dependent repression was observed at multiple genes with σ54 binding sites identified through ChIP Seq (Supplementary Table S3). These genes were grouped in 4 classes based on the location of the σ54 motifs relative to the promoter sequences. Shown are examples of each class and consensus motifs of class I–III. Class IV consists of only one member, katE.
Figure 7.
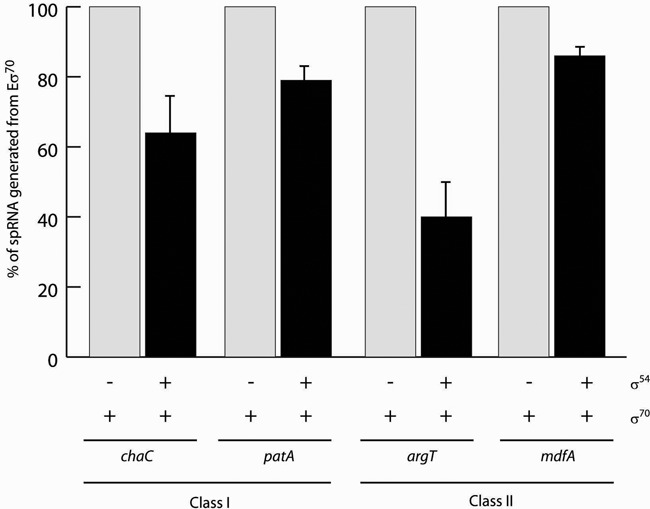
σ54 represses transcription by σ70in vitro. Purified holoenzymes were incubated with linear homoduplexes from four promoters (argT, chaC, mdfA and patA WT/WT) and transcription was measured using dinucleotide primers (−1/+1 or +1/+2) and a radioactive rNTP corresponding to +1 and or +2 to allow initiation of spRNA synthesis at the −10/−35 promoter sequences.
In one other case the putative −12/−24 recognition site is far away from where the upregulated transcription start site is mapped to by RNAseq (Figure 6 class IV). Here, repression may occur not by simple competition for overlapping promoters, but for example through direct or indirect interferences between convergent transcribing RNA polymerases.
Interestingly, in contrast to bypass promoters (Table 1) the C −12 residue is present in all observed σ54 motifs correlating with local repression of transcription, consistent with the aforementioned repressive nature of GC at −12 (21).
Cold-shock promoters have unusual σ54 dependencies
Notably, some of the promoters dependent upon σ54 in vivo were driving the expression of cold-shock genes (Figure 8A). ChIP Seq identified a σ54 binding site in two of these genes (cspA and cspH) despite lacking any clear consensus −12/−24 sequences (Joe Wade, University of Albany, USA; personal communication). However, in vitro transcription from cspA (Figure 8B) and cspH (Figure 8C) promoters was found to be σ70-dependent and largely unresponsive to σ54. Thus, the basis of their σ54 dependence in E. coli remains unclear, however in Bacillus subtilis the σ54 homologue (σL) has been shown to be essential for mounting a cold shock response through the transcription of σ54-dependent cold shock transcriptional regulators BkdR and YplP (23). Intriguingly, both cspA and cspH promoters contain putative heat shock sigma factor (σ32) binding motifs (24), consistent with the transcriptional start site mapped via the RNASeq reads (Figure 8B and C). Moreover, we only observe this σ54-dependent upregulation of cold shock genes at 42°C and not 37°C (Figure 8A). In all RNAseq experiments cells were cold harvested. The expression of cold shock promoters may therefore reflect the difference in the temperature shift from 42°C to ice and from 37°C to ice. We note however that σ32 activity has been suggested to be regulated by σ54 (25), raising the possibility that σ54 may be required for the expression of cold shock genes cspA and cspH by modulating the activity of σ32.
Figure 8.
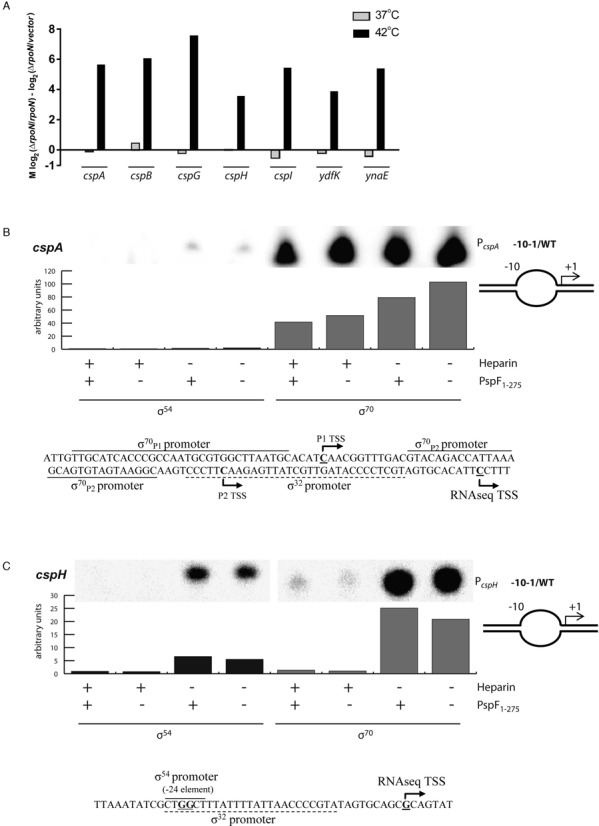
(A) Expression of cold-shock genes in vivo is upregulated in a σ54-dependent manner at 42°C but not at 37°C. In vitro transcription of cspA (B) and cspH (C) with the -10–1/WT heteroduplex promoters is σ70 dependent and largely σ54 independent.
DISCUSSION
The σ54-dependent gene expression paradigm is widely distributed amongst bacteria and is responsible for the expression of stress-induced genes important in many bacterial driven processes, especially the nitrogen cycle (26). Bioinformatics suggests the σ54 system is commonly associated with cell envelope function (27). Recent structural studies highlight how σ54 interacts with the core RNA polymerase to yield a holoenzyme unable to manipulate promoter DNA for making open promoter complexes until re-organised by its cognate AAA+ activator proteins (28). Studied here, the loss of the σ54 Region I from the holoenzyme frees up RNAP for promoter DNA delivery within and the subsequent templated synthesis of RNA (6,29–33). Our analysis of the properties of the bypass form of σ54 in vivo suggests such DNA delivery into the RNAP once Region I is missing is rarely occurring in vivo. In vitro the activator bypass phenotype of the ΔRI σ54 form is enhanced by using pre-opened DNA templates and conditions favouring DNA opening such as DNA supercoiling (6,32). The lack of an appropriate state of the promoter DNA available to the ΔRI form of the σ54RNAP holoenzyme may well be limiting for bypass transcription in vivo. The single gene KO enhancing bypass transcription encodes a nucleoid-associated protein (TtdR), and may influence DNA structure to normally repress bypass transcription.
Earlier work from the Gralla lab established that with single promoter studies (the glnAp2 promoter) bypass transcription could be elevated by altering the TGCA sequence around the −12 GC element (21). Where we did observe evidence for bypass transcription in vivo we noted a non-consensus −12 promoter sequence, the place where the Region I of σ54 interacts with DNA (34,35). We propose that the repressive interactions made by Region I may be compensated for by some promoter sequences associated with DNA delivery into RNAP. Indeed destroying the repressive fork junction structure formed near the −12 promoter element yields bypass transcription in vitro (6,21,36). Overall we conclude that to increase bypass transcription in vivo appropriate changes in local DNA structure favoring DNA opening and diminishing the repressive contributions of the −12 promoter sequences would need to occur. This view is congruent with the following observations: (i) correlation between low-level bypass transcription with a less conserved C −12 nucleotide and (ii) an apparent lack of in trans encoded repressors of bypass transcription.
Overlapping promoters and competition between sigma factors are recognized as being important regulatory features of a range of genes (37–41), although rather few cases are documented for σ54 (40). Here, we provide further evidence that, in addition to being a target of activators for regulated gene expression, σ54 can act to repress gene transcription by other forms of RNA polymerase through a binding competition for promoter sites. Our analysis of any repressive capacity is unlikely to be comprehensive, since a single growth condition was studied. Given more σ54 binding sites than conventional activator dependent σ54 promoters have been identified using a range of genomic methods, we propose that the repressive function is pervasive. Indeed, the σ54 factor is designed to inhibit RNAP by penetrating and blocking DNA entry routes (27,42,43), and it may well be that the earliest function of σ54 or some of its domains was to globally repress transcription. The unusual behaviours of the cold-shock promoters with respect to σ54 dependence is intriguing, and suggests some non-canonical functions of σ54 and or its holoenzyme remain to be discovered. These may involve factors outside of the well-recognized AAA+ activators which reorganize σ54 at its classical promoter sites.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank the lab of Dr Joseph Wade (University of Albany, NY, USA) for sharing their data on genomewide σ54 ChIP binding. We also thank Goran Jovanovic, Patricia Burrows, Simone Wiesler and Joerg Schumacher for their advice and comments.
FUNDING
Biotechnology and Biological Sciences Research Council, UK Research Grant [BB/J00717X/1]. Funding for open access charge: Biotechnology and Biological Sciences Research Council, UK Research Grant [BB/J00717X/1].
Conflict of interest statement. None declared.
REFERENCES
- 1.Wigneshweraraj S., Bose D., Burrows P.C., Joly N., Schumacher J., Rappas M., Pape T., Zhang X., Stockley P., Severinov K., et al. Modus operandi of the bacterial RNA polymerase containing the sigma54 promoter-specificity factor. Mol. Microbiol. 2008;68:538–546. doi: 10.1111/j.1365-2958.2008.06181.x. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh T., Bose D., Zhang X. Mechanisms for activating bacterial RNA polymerase. FEMS Microbiol. Rev. 2010;34:611–627. doi: 10.1111/j.1574-6976.2010.00239.x. [DOI] [PubMed] [Google Scholar]
- 3.Rappas M., Bose D., Zhang X. Bacterial enhancer-binding proteins: sigma54-dependent gene transcription. Curr. Opin. Struct. Biol. 2007;17:110–116. doi: 10.1016/j.sbi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Bush M., Dixon R. The role of bacterial enhancer binding proteins as specialized activators of sigma54-dependent transcription. Microbiol. Mol. Biol. Rev. 2012;76:497–529. doi: 10.1128/MMBR.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Studholme D.J., Buck M. The biology of enhancer-dependent transcriptional regulation in bacteria: insights from genome sequences. FEMS Microbiol. Lett. 2000;186:1–9. doi: 10.1111/j.1574-6968.2000.tb09074.x. [DOI] [PubMed] [Google Scholar]
- 6.Wang J.T., Syed A., Gralla J.D. Multiple pathways to bypass the enhancer requirement of sigma54 RNA polymerase: roles for DNA and protein determinants. Proc. Natl. Acad. Sci. U.S.A. 1997;94:9538–9543. doi: 10.1073/pnas.94.18.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaney M., Buck M. The sigma 54 DNA-binding domain includes a determinant of enhancer responsiveness. Mol. Microbiol. 1999;33:1200–1209. doi: 10.1046/j.1365-2958.1999.01566.x. [DOI] [PubMed] [Google Scholar]
- 8.Engl C., Jovanovic G., Lloyd L.J., Murray H., Spitaler M., Ying L., Errington J., Buck M. In vivo localizations of membrane stress controllers PspA and PspG in Escherichia coli. Mol. Microbiol. 2009;73:382–396. doi: 10.1111/j.1365-2958.2009.06776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hobbs E.C., Astarita J.L., Storz G. Small RNAs and small proteins involved in resistance to cell envelope stress and acid shock in Escherichia coli: analysis of a bar-coded mutant collection. J. Bacteriol. 2010;192:59–67. doi: 10.1128/JB.00873-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutnick D., Calvo J.M., Klopotowski T., Ames B.N. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J. Bacteriol. 1969;100:215–219. doi: 10.1128/jb.100.1.215-219.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller J.H. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 13.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 14.Dago A.E., Wigneshweraraj S.R., Buck M., Morett E. A role for the conserved GAFTGA motif of AAA+ transcription activators in sensing promoter DNA conformation. J. Biol. Chem. 2007;282:1087–1097. doi: 10.1074/jbc.M608715200. [DOI] [PubMed] [Google Scholar]
- 15.Zhang N., Joly N., Burrows P.C., Jovanovic M., Wigneshweraraj S.R., Buck M. The role of the conserved phenylalanine in the sigma54-interacting GAFTGA motif of bacterial enhancer binding proteins. Nucleic Acids Res. 2009;37:5981–5992. doi: 10.1093/nar/gkp658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joly N., Engl C., Jovanovic G., Huvet M., Toni T., Sheng X., Stumpf M.P., Buck M. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 2010;34:797–827. doi: 10.1111/j.1574-6976.2010.00240.x. [DOI] [PubMed] [Google Scholar]
- 17.Bergman J.M., Hammarlöf D.L., Hughes D. Reducing ppGpp level rescues an extreme growth defect caused by mutant EF-Tu. PLoS One. 2014;9:e90486. doi: 10.1371/journal.pone.0090486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrios H., Valderrama B., Morett E. Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic Acids Res. 1999;27:4305–4313. doi: 10.1093/nar/27.22.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L., Guo Y., Gralla J.D. Regulation of sigma 54-dependent transcription by core promoter sequences: role of −12 region nucleotides. J. Bacteriol. 1999;181:7558–7565. doi: 10.1128/jb.181.24.7558-7565.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda H., Fujita N., Ishihama A. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 2000;28:3497–3503. doi: 10.1093/nar/28.18.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiegeshoff F., Beckering C.L., Debarbouille M., Marahiel M.A. Sigma L is important for cold shock adaptation of Bacillus subtilis. J. Bacteriol. 2006;188:3130–3133. doi: 10.1128/JB.188.8.3130-3133.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowing D.W., Bardwell J.C., Craig E.A., Woolford C., Hendrix R.W., Gross C.A. Consensus sequence for Escherichia coli heat shock gene promoters. Proc. Natl. Acad. Sci. U.S.A. 1985;82:2679–2683. doi: 10.1073/pnas.82.9.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pallen M. RpoN-dependent transcription of rpoH. Mol. Microbiol. 1999;31:393. doi: 10.1046/j.1365-2958.1999.01148.x. [DOI] [PubMed] [Google Scholar]
- 26.Kustu S., Santero E., Keener J., Popham D., Weiss D. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol. Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francke C., Groot K.T., Hagemeijer Y., Overmars L., Sluijter V., Moezelaar R., Siezen R.J. Comparative analyses imply that the enigmatic Sigma factor 54 is a central controller of the bacterial exterior. BMC Genomics. 2011;12:385. doi: 10.1186/1471-2164-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bose D., Pape T., Burrows P.C., Rappas M., Wigneshweraraj S.R., Buck M., Zhang X. Organization of an activator-bound RNA polymerase holoenzyme. Mol. Cell. 2008;32:337–346. doi: 10.1016/j.molcel.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallegos M.T., Cannon W.V., Buck M. Functions of the sigma(54) region I in trans and implications for transcription activation. J. Biol. Chem. 1999;274:25285–25290. doi: 10.1074/jbc.274.36.25285. [DOI] [PubMed] [Google Scholar]
- 30.Wang J.T., Syed A., Hsieh M., Gralla J.D. Converting Escherichia coli RNA polymerase into an enhancer-responsive enzyme: role of an NH2-terminal leucine patch in sigma 54. Science. 1995;270:992–994. doi: 10.1126/science.270.5238.992. [DOI] [PubMed] [Google Scholar]
- 31.Wang J.T., Gralla J.D. The transcription initiation pathway of sigma 54 mutants that bypass the enhancer protein requirement. Implications for the mechanism of activation. J. Biol. Chem. 1996;271:32707–32713. doi: 10.1074/jbc.271.51.32707. [DOI] [PubMed] [Google Scholar]
- 32.Cannon W., Chaney M., Buck M. Characterisation of holoenzyme lacking sigmaN regions I and II. Nucleic Acids Res. 1999;27:2478–2486. doi: 10.1093/nar/27.12.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Syed A., Gralla J.D. Identification of an N-terminal region of sigma 54 required for enhancer responsiveness. J. Bacteriol. 1998;180:5619–5625. doi: 10.1128/jb.180.21.5619-5625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaney M., Buck M. The sigma 54 DNA-binding domain includes a determinant of enhancer responsiveness. Mol. Microbiol. 1999;33:1200–1209. doi: 10.1046/j.1365-2958.1999.01566.x. [DOI] [PubMed] [Google Scholar]
- 35.Chaney M., Grande R., Wigneshweraraj S.R, Cannon W., Casaz P., Gallegos M.T., Schumacher J., Jones S., Elderkin S., Dago A.E., et al. Binding of transcriptional activators to sigma 54 in the presence of the transition state analog ADP-aluminum fluoride: insights into activator mechanochemical action. Genes Dev. 2001;15:2282–2294. doi: 10.1101/gad.205501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Y., Wang L., Gralla J.D. A fork junction DNA-protein switch that controls promoter melting by the bacterial enhancer-dependent sigma factor. EMBO J. 1999;18:3736–3745. doi: 10.1093/emboj/18.13.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wade J.T., Castro R.D., Grainger D.C., Hurd D., Busby S.J., Struhl K., Nudler E. Extensive functional overlap between sigma factors in Escherichia coli. Nat. Struct. Mol. Biol. 2006;13:806–814. doi: 10.1038/nsmb1130. [DOI] [PubMed] [Google Scholar]
- 38.Grainger D.C., Goldberg M.D., Lee D.J., Busby S.J. Selective repression by Fis and H-NS at the Escherichia coli dps promoter. Mol. Microbiol. 2008;68:1366–1377. doi: 10.1111/j.1365-2958.2008.06253.x. [DOI] [PubMed] [Google Scholar]
- 39.Cho B.K., Kim D., Knight E.M., Zengler K., Palsson B.O. Genome-scale reconstitution of the sigma factor network in Escherichia coli: topology and functional states. BMC Biol. 2014;12:4. doi: 10.1186/1741-7007-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zafar M.A., Carabetta V.J., Mandel M.J., Silhavy T.J. Transcriptional occlusion caused by overlapping promoters. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1557–1561. doi: 10.1073/pnas.1323413111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levi-Meyrueis C., Monteil V., Sismeiro O., Dillies M.A., Kolb A., Monot M., Dupuy B., Duarte S.S., Jagla B., Coppee J.Y., et al. Repressor activity of the RpoS/σS-dependent RNA polymerase requires DNA binding. Nucleic Acids Res. 2015;43:1456–1468. doi: 10.1093/nar/gku1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burrows P.C., Joly N., Buck M. A prehydrolysis state of an AAA +ATPase supports transcription activation of an enhancer-dependent RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9376–9381. doi: 10.1073/pnas.1001188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma A., Leach R.N., Gell C., Zhang N., Burrows P.C., Shepherd D.A., Wigneshweraraj S., Smith D.A., Zhang X., Buck M., et al. Domain movements of the enhancer-dependent sigma factor drive DNA delivery into the RNA polymerase active site: insights from single molecule studies. Nucleic Acids Res. 2014;42:5177–5190. doi: 10.1093/nar/gku146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


