Abstract
Divergent gene pairs (DGPs) are abundant in eukaryotic genomes. Since two genes in a DGP potentially share the same regulatory sequence, one might expect that they should be co-regulated. However, an inspection of yeast DGPs containing cell-cycle or stress response genes revealed that most DGPs are differentially-regulated. The mechanism underlying DGP differential regulation is not understood. Here, we showed that co- versus differential regulation cannot be explained by genetic features including promoter length, binding site orientation, TATA elements, nucleosome distribution, or presence of non-coding RNAs. Using time-lapse fluorescence microscopy, we carried out an in-depth study of a differentially regulated DGP, PFK26-MOB1. We found that their differential regulation is mainly achieved through two DNA-binding factors, Tbf1 and Mcm1. Similar to ‘enhancer-blocking insulators’ in higher eukaryotes, these factors shield the proximal promoter from the action of more distant transcription regulators. We confirmed the blockage function of Tbf1 using synthetic promoters. We further presented evidence that the blockage mechanism is widely used among genome-wide DGPs. Besides elucidating the DGP regulatory mechanism, our work revealed a novel class of insulators in yeast.
INTRODUCTION
In eukaryotes, co-regulated genes can be clustered or paired, suggesting that co-regulation might result from proximity. In one of these cases, two ‘head-to-head’, closely-spaced genes are divergently transcribed on opposite DNA strands, forming a ‘divergent gene pair’ (DGP). DGPs are highly abundant in eukaryotic genomes. With an upper limit of 1 kb between two divergent open reading frames (ORFs), about 46%, 32%, and 10% of all genes in budding yeast, fruit fly and human genomes respectively are present as DGPs (1,2). Elucidating the regulatory mechanism of DGP is thus an essential component in understanding eukaryotic gene regulation.
Two genes in a DGP potentially share the same upstream regulatory sequences. It is thus commonly thought that DGPs tend to be co-regulated, and the co-regulated divergent genes tend to be functionally related. Consistently, in budding yeast, histone genes (H2A/H2B and H3/H4) and galactose metabolic genes (GAL1/GAL10) are organized into co-regulated DGPs (3,4). Similar examples are also present in higher eukaryotes: human collagen genes (COL4A1/COl4A2) and mitochondrial chaperonin genes (HSP60/HSP10) form co-regulated DGPs (5,6). Since most transcription factors were shown to regulate in an orientation insensitive manner (7), co-regulation can occur naturally through bidirectional recruitment of transcription machinery. Recent genome-wide studies in both yeast and mammalian cells detected divergent transcription at a large fraction of promoters with positively correlated activities in the two orientations (8–11). Although many of these bidirectional transcriptions generate sense/antisense RNAs instead of two protein-coding mRNAs, these results nevertheless indicate that co-regulated divergent transcription is an intrinsic property of promoters (12).
However, not all DGPs are co-regulated. In yeast, this is best exemplified by the PET56-HIS3 gene pair: transcription factor Gcn4 selectively activates HIS3 but not PET56, despite the short distance between the two genes (<200 bp between transcription start sites (TSSs)) (13). Differentially regulated DGPs with very short intergenic regions can also be found in higher eukaryotes—TK-KF in mouse and COL4A1–COL4A2 in human (14,15). At the genome-wide level, there are many cases where deletion of a transcription factor, or change in growth condition, only alter the expression of one gene in a DGP, indicating that differential regulation is a wide-spread phenomenon (16).
How are two genes differentially regulated, if both of them are immediately downstream from the same regulatory elements? In principle, the differential regulation can be determined at multiple steps of transcription, including initiation, elongation, and RNA processing. At the initiation step, certain promoter features (e.g. TATA sequences) may bias the regulation towards one gene over the other (13). Alternatively, the transcript sequences may signal for different rates of elongation, termination or degradation, leading to apparent differential regulation. The latter scenario contributes to the differential regulation of sense/antisense RNA pairs in mammalian cells: antisense RNA tends to be rapidly terminated and degraded because it is polyA-enriched and U1 snRNP signal depleted (17,18). Importantly, the DGPs studied here involve two mRNAs, and their differential regulation can be recapitulated by the divergent promoter driving identical mRNAs in two orientations (see ‘Results’ section), indicating that the regulatory pattern is at least partially encoded in the promoter sequence.
Several promoter features have been proposed to direct gene regulation. In the case of PET56-HIS3 DGP, it was proposed that HIS3 and PET56 respond differently to the Gcn4 signaling due to their distinct TATA elements (13). How general this mechanism is among genome-wide DGPs is not clear. Nucleosomes may also play a role in coordinating divergent transcription. Many divergent transcripts originate from a shared nucleosome depleted region (NDR) (11). A few nucleosome-related factors, including CAF-I and Isw2, repress antisense transcription while having little effect on the sense mRNA (19,20). Since these factors are thought to be sequence-independent, how they selectively function in the non-coding direction remains elusive. Finally, the regulation of divergent genes can be decoupled by insulators. Studies in Drosophila, Arabidopsis and human indicate that insulators preferentially bind to the intergenic regions of differentially expressed DGPs (21–23). However, to the best of our knowledge, this type of ‘enhancer-blocking insulators’ has not been reported in budding yeast.
Here, we investigated the promoter features that affect co- versus differential regulation in yeast. We first established that most DGPs containing cell cycle regulated or stress-responding genes are differentially regulated. We then showed that the differential regulation is not predominantly determined by several promoter features. Using differentially-regulated PFK26-MOB1 DGP as a model, we discovered that certain sequence-specific DNA binding factors, including Tbf1 and Mcm1, can ‘block’ regulation from a distal upstream activating sequence (UAS) and thus prevent crosstalk between the divergent genes. These factors represent a novel class of insulators in yeast. The blockage function of Tbf1 was confirmed by synthetic promoters with different UASs, and under different promoter contexts. We also presented evidence that Tbf1 is widely used to decouple genome-wide DGPs. The molecular origin of the blockage function will be further discussed.
MATERIALS AND METHODS
Plasmids and synthetic promoter construction
All plasmids used in this study (detailed below) were derived from pRS yeast shuttle vectors (24). The reporter genes were inserted into the multiple cloning sites. (i) pCY01 containing wt or mutated PFK26-MOB1pr driving destabilized VENUS from either orientations; (ii) pLB19 was identical to pCY01 except that a nuclear localization sequence (NLS) was inserted upstream of VENUS gene; (iii) CLN2-PFK26 hybrid promoter in Figure 4A was constructed by ligating the CLN2 UAS (−413 to −295 relative to CLN2 TSS) to the PFK26 promoter (starting from −170 relative to PFK26 TSS) containing wt or mutated Tbf1 binding site; (iv) CLN2 promoter variant in Figure 4B was constructed by inserting two copies of wt (TTAGGGG) or mutated (gcAGGGG) Tbf1 binding sites at −282 (downstream of UAS) or −552 (upstream of UAS) relative to CLN2 TSS; (v) GAL1 promoter variant in Figure 4C was constructed by inserting wt or mutated Tbf1 binding site at −100 relative GAL1 TSS. The TSS information was retrieved from Rhee et al. (25).
Figure 4.
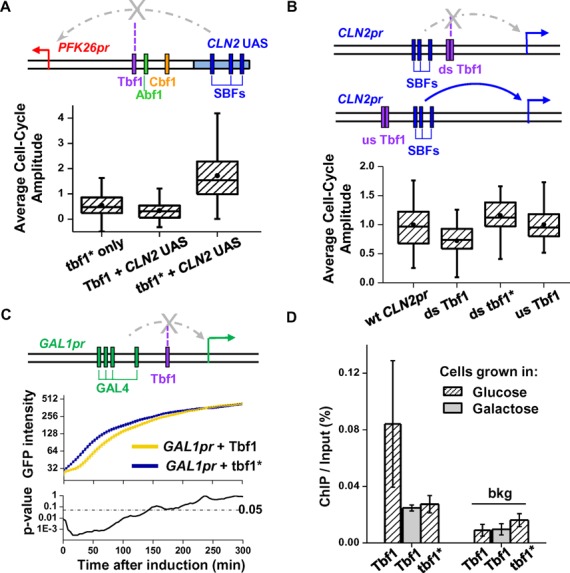
Testing the blockage activity of Tbf1 using synthetic promoters. (A) Tbf1 in PFK26 UAS1 blocks the activation by CLN2 UAS. In this hybrid promoter, we replaced UAS2 of the PFK26-MOB1 promoter with the CLN2pr UAS, which contains three SBF (cell-cycle dependent activator) binding sites. The arrow represents TSS (same as in B and C). The box plot shows the distribution of the cell cycle amplitude of VENUS expression driven by this hybrid promoter and its variants. tbf1* only: PFK26 promoter with mutated Tbf1 site (no CLN2 UAS); Tbf1/tbf1* + CLN2 UAS: hybrid promoter with wt/mutated Tbf1 site. (B) Downstream Tbf1 sites, but not upstream ones, reduce the activation by CLN2 UAS in native CLN2pr background. In CLN2pr, we inserted two adjacent Tbf1 binding sites (wt or mutated) either downstream or upstream the CLN2 UAS. The box plot shows the cell cycle activity of GFP driven by these artificial promoters, relative to that of the wt CLN2pr. ds: downstream; us: upstream. (C) Tbf1 attenuates the initial GAL1pr induction. A Tbf1 binding site (wt or mutated) was inserted into GAL1pr downstream the Gal4 binding sites. The intensity of GFP driven by these promoters was measured as a function of time during galactose induction (middle panel). Based on the P-value calculated by t-test (lower panel), the two curves are significantly different during the initial stage of the induction, but not afterwards. (D) ChIP measurements of Tbf1 binding to the promoters in C. For the promoter with the wt Tbf1 site, ChIP was performed in glucose (repressive) and galactose (activating) media. bkg: background region in YER129W ORF.
Strain construction
We used GC46 to construct all the yeast strains for single cell measurement. GC46 is the same as W303 except it is ADE2+ and contains mCherry labeled MYO1 gene. We integrated the plasmids into chromosome at the CLN2 locus. These strains were then subjected to gene expression analysis using time-lapse fluorescence microscopy.
Strains used for chromatin immunoprecipitation (ChIP) measurements are derived from S288C, and each strain has an ORF (Tbf1, Abf1, Reb1, Cbf1, Mcm1 and Fkh2) with tandem affinity purification (TAP) tag at its C-terminal (26). In the ChIP strains containing PFK26-MOB1 promoter derivatives or CLN2-PFK26 hybrid promoter, the endogenous PFK26-MOB1 promoter was deleted. This step was to keep these promoter sequences as a single copy in the genome to avoid complication for the subsequent qPCR analysis. Since the MOB1 gene is essential, we inserted an ACT1 promoter driving MOB1 into the genome to maintain the viability of the strains. For the strains in Figure 4B and C, the plasmids containing the synthetic promoters were first integrated into the endogenous CLN2 or GAL1 loci. We then grew the strains in non-selective medium during which a small fraction of cells will ‘popout’ the wt copies of CLN2 or GAL1 promoters through natural recombination, resulting in the loss of URA3 marker. These ‘popout’ colonies were then selected on FOA plates and confirmed by sequencing.
Identifying DGPs containing cell-cycle-regulated or stress-responding genes
Three sets of cell-cycle-regulated genes were previously identified based on different experimental results and computational algorithms (27–29). Seven hundred and twenty-one genes appeared in at least two of the three sets, and they were considered as cell-cycle-regulated; 620 genes appeared in only one of the three sets, and they were marked as ambiguous; the rest of the genome was considered as non-cell-cycle-regulated. All DGPs containing ambiguous genes were excluded from future analysis. DGPs containing at least one cell-cycle genes were further selected based on the following criteria. (i) We only included DGPs with distance between the two TSSs less than 800 bp. (ii) We discarded DGPs containing ‘dubious genes’. (iii) We discarded DGPs with incorrect annotation, e.g. mapped transcription start site is downstream the translational start site. About 45% of DGPs were eliminated based on these criteria. The remaining DGPs were used for the statistical analysis in Table 1 and Figure 1. DGPs containing two cell cycle genes were defined as co- (anti-) regulated if their mRNA peak times differ by less (more) than 0.2 cell cycle. We obtained the list of stress responding genes from Gasch et al. (30). DGPs containing stress-response genes were identified and categorized with similar procedures. DGPs containing two stress genes are co-regulated if two genes have the same response (activated or repressed) in the presence of stress. See Supplementary Tables S1 and S2 for detailed information on these DGPs.
Table 1. Number of DGPs containing cell-cycle or stress response genes in different categories.
| Category | Cell-cyclea | Stress-responsea | ||||
|---|---|---|---|---|---|---|
| # | Null 1 | Null 2 | # | Null 1 | Null 2 | |
| Co-regulated | 33 | 16 | 6 | 31 | 21 | 11 |
| Anti-regulated | 5 | 22 | 9 | 3 | 13 | 6 |
| Differential-regulated | 106 | 211 | 195 | 228 | ||
aOut of the 144 cell-cycle DGPs and 229 stress-response DGPs, there are 29 in common.
Figure 1.
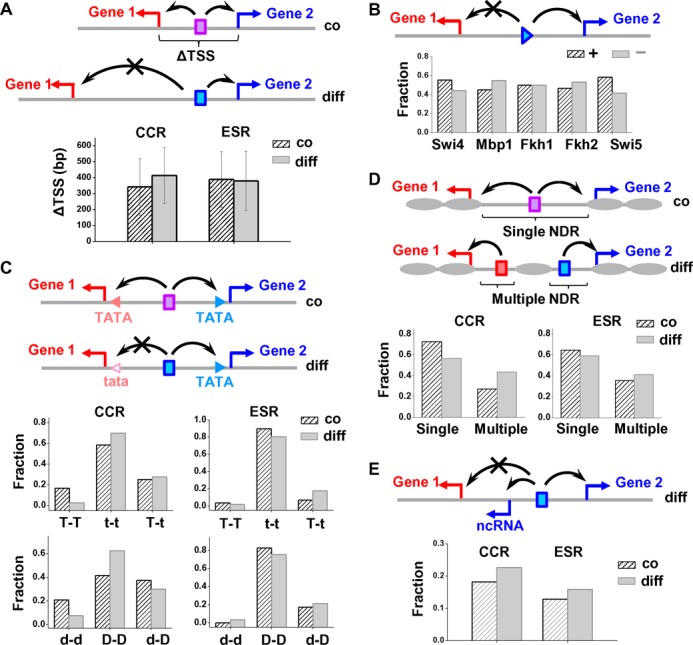
Co/differential regulation of DGP cannot be explained by the length of the divergent promoter, orientation of the transcription factor, TATA box, nucleosome configuration or non-coding RNA. We generated five hypotheses of promoter features that may contribute to co/differential regulation of DGPs, and tested their function by analyzing their occurrence frequency in different DGP groups. The notations in the diagrams are: triangles in B and all rectangles: transcription factors; red and blue arrows: TSS; black arrows: direction of activation; gray ovals: nucleosomes; triangles in C: TATA elements. (A) Hypothesis 1: differentially regulated DGPs may have longer divergent promoters. The plot shows the distance between TSSs of co- versus differentially regulated DGPs containing cell-cycle regulated (CCR) or environmental stress response (ESR) genes. The error bars represent standard deviation. (B) Hypothesis 2: orientation of transcription factors may lead to directional regulation. We determined the binding sites of five cell-cycle regulators on differentially-regulated divergent promoters, and plotted the probability of these factors facing toward (+; arbitrarily defined) or against (−) the genes they regulate. (C) Hypothesis 3: asymmetry in the TATA elements may contribute to DGP differential regulation. Using consensus and non-consensus TATA elements (TATA/tata) mapped in previous ChIP-Exo measurements (25), we plotted the fraction of divergent promoters that contain two TATAs (T–T), two tatas (t–t), or one TATA and one tata (T–t). Since TATA-containing promoters tend to be depleted of the TFIID complex, we also plotted the fraction of divergent promoters that are TFIID-depleted or TFIID-enriched on both sides (d–d/D–D), or TFIID depleted on one side (d–D). (D) Hypothesis 4: co-/differential regulation may be determined by the number of NDRs on the divergent promoter. The plot shows the fraction of divergent promoters that contain either single or multiple NDRs. (E) Hypothesis 5: the presence of non-coding RNA (ncRNA) may decouple the transcription of the two mRNAs. The plot shows the fraction of divergent promoters that overlaps with ncRNA. All of these features are about equally represented in the co- versus differentially regulated divergent promoters, indicating that they are not the dominant factors in determining the DGP regulation mode.
The null model 1 in Table 1 calculated the random chance of co-regulation among DGPs containing two cell-cycle regulated genes. The null model 2 calculated the expected number of co-, anti-, and differentially regulated DGPs if all the cell cycle genes were randomly distributed in the genome. The same analysis was applied to the stress genes.
Bioinformatics analysis of promoter features
We used the following genome-wide datasets for the bioinformatics analysis: TSS annotation (11), factor binding (ChIP-chip) (31), TATA consensus (ChIP-exo) (25), TFIID enrichment (32), nucleosome occupancy (33), ncRNA (11) and locations of ‘orphan’ Pre-Initiation Complexes (25).
To compute the orientation of the cell-cycle regulators in Figure 1B, we searched for DPGs that (i) are bound by one of the factors, (ii) contain at least one gene whose regulation is consistent with the factor's activity. For instance, G1/S activator Mbp1 binds to the HTA2-HTB2 and EMW1-RFA2 divergent promoters, among which HTA2, HTB2, and RFA2 are G1/S activated, but not EMW1. We assume that the first three genes are regulated by Mbp1. The orientation of Mbp1 binding sites relative to the TSSs of HTA2, HTB2 and RFA2 versus EMW1 were computed and pooled into two different groups.
We analyzed the NDR distribution on divergent promoters (Figure 1D) based on the data in Lee et al. (33). We first localized regions where nucleosome density was lower than –1 (log2 scale), and joined the neighboring low density regions when the distance in between was smaller than 100 bp. The low-density region was considered an NDR if its length was longer than 50 bp (otherwise the region is likely to be a regular linker region between nucleosomes). We then examined each divergent promoter to see whether it contains a single or multiple NDRs.
Time-lapse fluorescence microscopy and data analysis
Sample preparation for the time-lapse assay was performed using similar methods as previously described (34,35). To collect the heat shock data, we first incubated the cells under an agarose pad at room temperature for 4 h, then placed them on a heated microscope objective and immediately started imaging. We adjusted the objective temperature so that the sample surface reached 37°C (measured by Omega surface thermocouple). For the galactose induction experiment, cells were first cultured overnight in 3% raffinose before transferred to agarose pads containing 3% galactose. The time of this transfer was recorded as time 0 of induction. We started time-lapse measurements of these cells ∼5 min afterward.
The instrumentation of the time-lapse microscopy and the MATLAB software for data acquisition and analysis have been described in Charvin et al. (34). Images were acquired every 4 min for 8 h (cell cycle data) or 6 h (heat shock data). MOB1pr has weak cell cycle regulation. To enhance signal to noise ratio, we used NLS-VENUS (VENUS containing nuclear localization signal) so that the real VENUS signal will be concentrated in the nucleus, allowing us to differentiate it from uniformly distributed auto-fluorescence background. Accordingly, for the cell cycle data, the VENUS signal was quantified as the variance of intensity for all the pixels within the cell boundary. The curves shown in Figure 2C were smoothed and corrected by subtracting a baseline connecting flanking troughs. We calculated the peak height for each cell cycle and used them as an indicator for the cell cycle intensity. For the heat shock data in Figure 3C, we quantified the VENUS signal using the average VENUS intensity in single cells. We calculated the slope of each trace during the arrest phase as the heat shock responsiveness.
Figure 2.
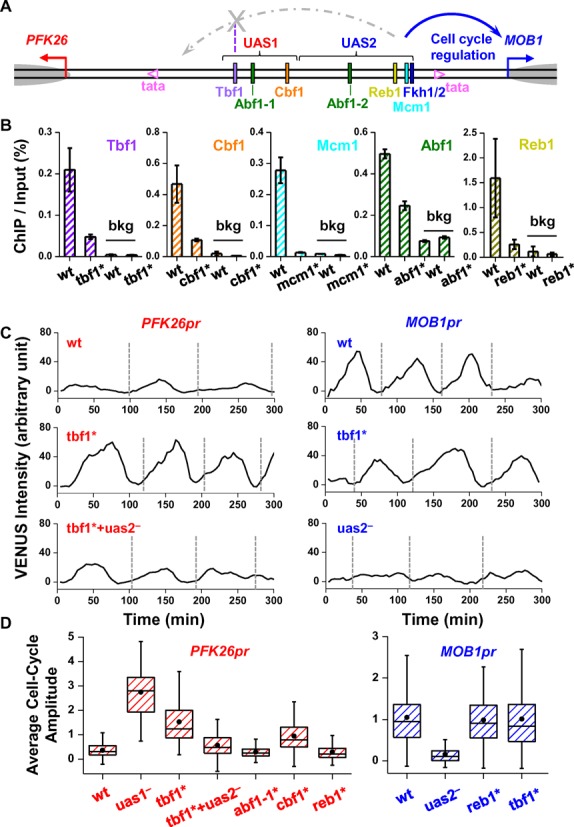
Tbf1 ‘blocks’ the cell cycle signal from UAS2 to PFK26. (A) Genomic structure of PFK26-MOB1 divergent promoter. The regulatory region was divided into UAS1 and UAS2, with colored rectangles representing factor binding sites. The two non-consensus TATA elements (mapped in (25)) were shown as pink hollow triangles, TSSs as red and blue arrows, and +1 nucleosomes as grey ovals. The diagram also shows a model we concluded from the data below: factors bound in UAS2 lead to cell cycle regulation of MOB1pr, but on the PFK26 side, the signal is blocked by Tbf1. (B) ChIP measurements of factor binding on PFK26-MOB1 promoter and a background region (bkg; YER129W ORF). The ChIP assay was carried out with either the wild type PFK26-MOB1 promoter (wt), or the ones with mutations in the putative factor binding sites, tbf1*, cbf1*, mcm1*, reb1* and abf1* (abf1* represents mutations on both Abf1 binding sites). The error bars represent the standard errors in the ChIP signal among three biological replicates. (C) Typical traces of VENUS expression driven by PFK26pr (left panels) or MOB1pr (right panels) during vegetative growth. Each trace represents the VENUS intensity as a function of time in a single yeast cell over multiple cell cycles. The vertical dashed lines indicate the cell division time. The promoters are either wt (upper panels), or mutated. uas2−: UAS2 deletion; tbf1* + uas2−: combined Tbf1 site mutation and UAS2 deletion. Similar notations apply to all text below. (D) Box plots of the cell-cycle amplitude of VENUS expression driven by wt or mutant PFK26-MOB1 promoter. See Supplementary Table S7 for the number of measurements (same in Figures 3 and 4). In the left / right panel, VENUS was driven from the PFK26 / MOB1 orientation, respectively. The amplitude is calculated as the peak-to-trough difference in the VENUS signal per cell cycle. All data in this plot were normalized to that of the wt MOB1pr.
Figure 3.
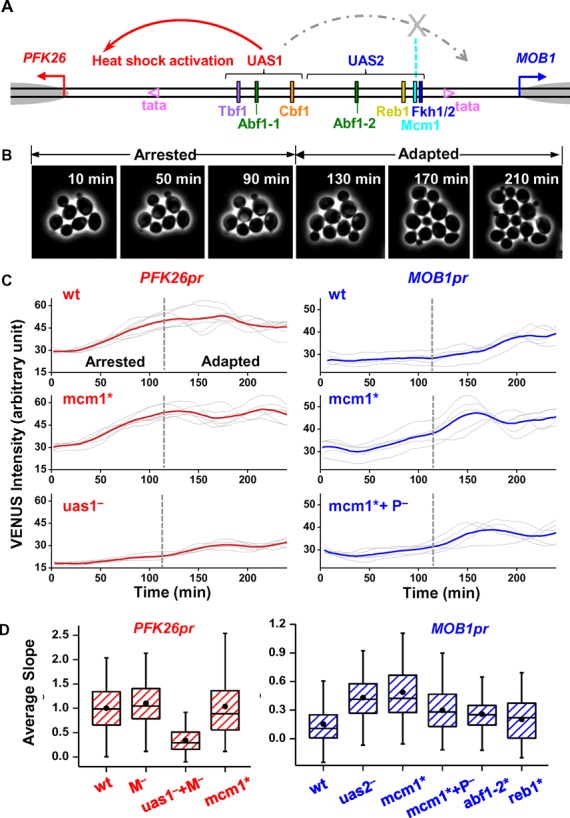
Mcm1 ‘blocks’ the heat shock signal from UAS1 to MOB1. (A) The same as Figure 2A except showing the decoupling of heat shock activation: UAS1 induces heat shock activation of PFK26, but its effect on MOB1 is blocked by Mcm1. (B) Time-lapse images showing two stages of yeast growth during heat shock. During the first ∼120 min after heat shock, yeast experiences cell-cycle arrest (arrested phase). The cells then adapted to high temperature and resumed cell-cycle (adapted phase). (C) Typical traces of VENUS expression driven by PFK26pr (left panels) or MOB1pr (right panels) under heat shock condition. Each gray trace represents the VENUS intensity in a single cell during the first 4 h of heat shock, and the red / blue curves are the average of the gray traces. Vertical dashed lines are the boundaries between the arrested and adapted phases. The promoters are either wt (upper panels), or mutated. P−/M−: deletion of the PFK26/MOB1 side of the promoter up to the boundary between UAS1 and UAS2. (D) Box plots of the heat shock intensity driven by wt PFK26-MOB1 promoter and its variants. This intensity is calculated based on the slope of the heat shock traces during the arrested phase (‘Materials and Methods’ section). All data in this plot were normalized to that of the wt PFK26pr.
ChIP assay
ChIP was performed following a previously described protocol with some modifications (36). Briefly, TAP-tagged yeast strains (26) were cultured in YPD overnight to an OD ∼0.4. Then we took 100 ml culture and cross-linked them with formaldehyde (final concentration of 1%) for 15 min at room temperature. The cross-linking was then quenched by the addition of glycine to a final concentration of 125 mM. Cells were disrupted by vortexing with glass beads, and the chromatin was fragmented using Qsonica Sonicator until major fragments are in a 300–500 bp range. The resulting whole cell extract were divided into two equal portions: one was subjected to DNA extraction and will be used as input DNA; the other one was incubated with an anti-TAP tag antibody (Thermo Scientific, CAB1001) and Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, sc-2003) sequentially. Immunoprecipitated DNA was then washed off the agarose beads and subjected to DNA extraction. Immunoprecipitated DNA and input DNA were quantified by real-time PCR. When the ChIP efficiency from multiple strains needed to be directly compared, we performed their ChIP measurements in parallel to minimize the batch-to-batch variations in antibody quality, the extent of crosslinking, pull-down, and washing.
For the PFK26-MOB1 promoter, primers used here were 5′-GCTATTGTCAGTAATGGCGC-3′ and 5′-TCAAACGGGCTGAGCTAAG-3′. We chose YER129W ORF as the background with primers 5′-CTTCATCCAACGGAAACCAC-3′ and 5′-TGTCCAAGGATGCAAATGAG-3′ (labeled as ‘bkg’ in the corresponding figures). The following are the primers for ChIP experiments on the synthetic promoters: CLN2-PFK26 hybrid promoters were detected with 5′-CTCAAAACTGCGTGTTCTAGTC-3′ and 5′-GCTATTGTCAGTAATGGCGC-3′; CLN2pr variants with 5′-ACCTCAAAACTGCGTGTTCTAG-3′ and 5′-GTAGGCCTGTCAGATCAACATT-3′ (downstream), and 5′-GGAGTGCGGTAACTTTTATTG-3′ and 5′-AAGCGCGCATGAATTGATAAC-3′ (upstream). GAL1pr variants with 5′-CGCACTGCTCCGAACAATAAAG-3′ and 5′-CGCATTATCATCCTATGGTTG-3′. For each promoter, at least three biological replicates were performed.
In vivo nucleosome mapping
We used the method described by Bai et al. (36), where nucleosomes were mapped by micrococcal nuclease digestion followed by stacking real-time PCR (37,38). PCR primers were designed across the whole PFK26-MOB1 promoter region. In the nucleosome map (Figure 5), the x-axis represents the mid-point of the PCR product relative to the MOB1 TSS. We used the nucleosome −1 on the PHO5pr as the standard to scale the nucleosome occupancy from 0 to 1. The error bars on the nucleosome occupancy represent the standard errors from three independent biological replicates.
Figure 5.
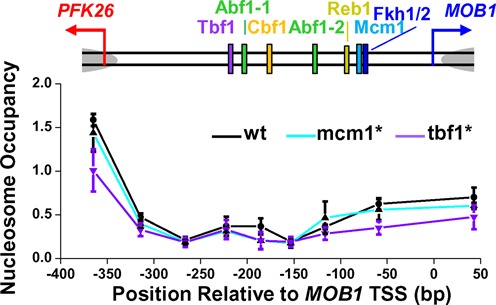
Mutations in the Tbf1 and Mcm1 binding sites do not alter nucleosome positioning on the PFK26-MOB1 promoter. The plot shows the measured nucleosome occupancy at different positions on the wt PFK26-MOB1 promoter (black), or with Mcm1 binding site mutation (cyan) or Tbf1 binding site mutation (purple). The error bars represent the standard errors in three biological replicates.
Bioinformatics analysis of factor association on a genome-wide scale
Due to a lack of TSS information for every gene, we defined ‘DGPs’ as head-to-head gene pairs with distance between the two ORFs less than 1000 bp. The expression correlation of each DGP was retrieved from serial pattern of expression levels locator (SPELL) (39). DGPs without expression correlation data in SPELL were discarded and the rest were ranked from high to low by the expression correlation value (Supplementary Table S5). We assigned the top 20% DGPs (most correlated) as co-regulated, and the bottom 20% (least correlated) as differentially regulated (228 DGPs in each category).
We then analyzed the binding of 163 factors in co- versus differentially regulated divergent promoters. The binding data was retrieved from Swiss Regulon Database (40). For each factor, two parameters were calculated: (i) fraction of divergent promoters bound by this factor; (ii) binding site density of this factor (total number of binding sites divided by the total intergenic sequence length) (Supplementary Table S6). The statistical difference between the two groups was evaluated using a Chi-squared test.
Furthermore, we focused on a few factors studied in this manuscript (Tbf1, Cbf1, Mcm1, Abf1 and Reb1). For each factor, we divided the genome-wide DGPs into ‘bound’ and ‘unbound’ groups based on the Swiss Regulon Database (40). We then extracted the correlation score of the DGPs in each group from SPELL (39). P-values were calculated using student t test. For blockage factors, we expect the average correlation score in the bound group to be significantly lower than the unbound group.
RESULTS
Only a small fraction of divergent gene pairs are co-regulated
DGPs containing cell-cycle regulated genes represent a dataset that can be clearly scored for co- vs differential regulation. Using previously published genome-wide microarray data (27–29), we identified 144 DGPs in yeast containing at least one cell-cycle regulated gene (‘Materials and Methods’ section; Supplementary Table S1). We categorized these DGPs into three groups: (i) co-regulated: two genes are both cell-cycle regulated and express at the same phase of cell-cycle; (ii) anti-regulated: two genes are both cell-cycle regulated but express at different phases; (iii) differentially regulated: only one of the two genes is cell-cycle regulated (Table 1). The number of co-regulated DGPs is significantly larger than that predicted by the null models, consistent with the function of DGP in supporting co-regulation. But overall, only a small fraction of DGPs (23%) are co-regulated. Analysis of DGPs containing general stress response genes yielded similar statistics (Table 1; Supplementary Table S2) (30). These results show that close proximity and divergent arrangement of two genes do not guarantee co-regulation.
Co/Differential regulation is not predominantly determined by promoter length, transcription factor binding orientation, TATA elements, nucleosome distribution or the presence of ncRNAs
We investigated the mechanisms of co-/differential regulation using bioinformatics analysis. We generated five hypotheses of promoter features that may contribute to co-/differential regulation: (1) length of divergent promoter. The regulatory elements in yeast tend to function over relatively short distances (41). Differentially regulated DGPs may have longer intergenic region so that one gene simply falls out of the functional range of transcription factor. (ii) Orientation of transcription factors. Many transcription factors recognize non-palindromic sequences so that their binding orientation potentially influences the direction of regulation. We consider this mechanism unlikely because out of 75 transcription factors tested, 69 were shown to regulate with equal strength in both orientations (7). (iii) TATA elements. A small fraction of yeast promoters contain the consensus TATA sequence, and the others lack the consensus (25). TATA-containing promoters tend to use SAGA to recruit transcription initiation complex and associate with stress-response genes; the ones without the consensus tend to use TFIID and associate with constitutive (house-keeping) genes (32,42). Therefore, asymmetric TATA configuration may be responsible for differential regulation (13). (iv) Nucleosome configuration. Genome-wide data indicated that NDRs may support co-regulated divergent transcription (12). Since the vast majority of divergent promoters in yeast contain either one or two NDRs, it is possible that single/multiple NDRs lead to co-/differential regulation. (v) Non-coding RNAs (ncRNA). 90% of ncRNAs in yeast initiate from the NDRs in the promoters of protein-coding genes (11). If an ncRNA is initiated within a divergent promoter, it will disrupt the divergent pattern of the two genes and potentially decouple their transcription regulation.
To test the hypotheses above, we analyzed the occurrence frequency of the five features in co- versus differentially regulated divergent promoters (‘Materials and Methods’ section). Anti-regulated DGPs were not included in the analysis due to the small sample size. If some of the hypotheses were correct, we expected the enrichment of the corresponding feature(s) to be significantly different between co- versus differentially regulated groups. Unexpectedly, all five features were about equally represented (Figure 1). Due to the noisy nature of the genome-wide data, we could not completely rule out these mechanisms, especially if they only function in a small fraction of DGPs. However, the results in Figure 1 strongly indicate that none of these mechanisms play a dominant role in DGP decoupling. Interestingly, many differentially regulated DGPs (33/106 among the cell cycle set) have short intergenic regions, symmetric TATA distribution, single NDR, and no ncRNA association. This observation raises the possibility that there are additional mechanisms that can lead to DGP differential regulation.
Tbf1 and Mcm1 contribute to the differential regulation of PFK26-MOB1 gene pair
We carried out a targeted and in-depth analysis to investigate additional mechanisms that lead to differential regulation. Our strategy was to choose a differentially-regulated DGP that cannot be explained by any of the mechanisms above, introduce mutations to its promoter, and examine which mutations can lead to symmetric firing of the two genes. To quantify the promoter activity, we constructed yeast strains carrying destabilized VENUS (a yellow fluorescent protein) driven by a wild type (wt) or a mutated promoter with either orientation, then monitored fluorescent intensity in single cells using time-lapse fluorescence microscopy (Materials and Methods; see Supplementary Table S3 for strain list). Note that in this setup, we are probing the divergent transcription separately, one direction at a time.
We selected a DGP, PFK26-MOB1, to be our model. This DGP has an intriguing expression pattern: MOB1 is cell cycle regulated but not heat-shock activated; conversely, PFK26 is heat-shock activated but not cell cycle regulated (28–30). The promoter of this DGP is short (distance between two TSSs < 400 bp) and nucleosome-free (Figure 2A) (33). In both orientations, there are non-consensus TATA-elements that were predicted to function as transcription initiation sites (25). The ChIP-Exo experiment on this promoter did not detect any RNA polymerase II (pol II) binding between the two TATA elements (25), indicating that this promoter is not associated with non-coding transcription (Supplementary Figure S1).
We first investigated the decoupling of cell cycle regulation between PFK26 and MOB1. We constructed strains containing either PFK26pr or MOB1pr (two orientations of the same promoter) driving VENUS and monitored fluorescence in individual cells during vegetative growth at 30°C. Consistent with the literature, VENUS driven by MOB1pr, but not PFK26pr, showed significant cell-cycle dependent oscillation in its intensity (Figure 2C). MOB1pr is regulated by the cell cycle transcription factor Fkh1/2. First, MOB1 belongs to the G2/M regulon, of which Fkh1/2 is the major activator (43). Second, a segment on the PFK26-MOB1 promoter, defined as ‘UAS2’, contains a consensus site of Fkh1/2 (Figure 2A). We confirmed the binding of Fkh2 to the UAS2 using ChIP assay (Supplementary Figure S2). Third, deletion of the UAS2 segment (Figure 2C), or depletion of the Fkh2 factor (43), largely eliminates MOB1pr cell cycle activity. Finally, deletion of the UAS2 upstream sequences did not significantly affect MOB1pr activity (data not shown).
Besides Fkh1/2, there are several highly-conserved transcription factor binding sites on the PFK26-MOB1 promoter, including those of Tbf1, Cbf1, Abf1, Reb1 and Mcm1 (Figure 2A; see Supplementary Table S4 for detailed promoter annotation). These factors are considered as ‘general regulatory factors’ and involved in many cellular processes, including transcriptional regulation, chromatin remodeling and DNA replication (44–46). Previous genome-wide ChIP experiments found enrichment of Cbf1, Reb1 and Mcm1 on this promoter (47–49), but not of Tbf1 or Abf1 (50,51). Since both false positives and negatives are present in genome-wide ChIP measurements, we performed small-scale ChIP experiments to specifically look for these factors on the PFK26-MOB1 promoter with YER129W ORF as our background (bkg). All five factors were shown to be highly enriched on the promoter compared to the background (Figure 2B). More importantly, mutating each site significantly reduced the ChIP signal of the corresponding factor (Figure 2B). Overall, these results confirmed the binding of these factors to the identified sites, and showed that our mutations were effective in depleting these factors from the promoter.
Strikingly, a 2 bp mutation in the Tbf1 binding site (tbf1*) strongly enhanced the cell-cycle activity of PFK26pr (single traces in Figure 2C and statistical summary in Figure 2D). This is not due to an accidentally generated cell-cycle regulator binding site, since deletion of the Tbf1 site also increased PFK26pr cell-cycle regulation. tbf1* did not alter MOB1pr expression, therefore this effect is directional (Figure 2CD). When we introduced tbf1* and UAS2 deletion simultaneously, the cell-cycle activity of PFK26pr became very weak again. These results suggest that Fkh1/2 activation is intrinsically bidirectional, but Tbf1 ‘shields’ PFK26 from this activation. An increase in the PFK26pr cell-cycle regulation was also observed upon mutation of the Cbf1 site, but not the Abf1 or Reb1 site (Figure 2D).
We next studied the directionality in the heat shock activation (Figure 3A). Previous work showed that PFK26, but not MOB1, can be activated under heat shock or other stress conditions (30). This asymmetric regulation could be recapitulated in our single cell gene expression assay. In this experiment, we placed cells on a heated objective (37°C) and monitored the response in real time (‘Materials and Methods’ section). The heat shock led to a transient cell cycle arrest for ∼2 h, and then cells adapted to the higher temperature and recommitted to growth (Figure 3B). Consistent with PFK26 being a heat shock gene, the intensity of PFK26pr-VENUS increased linearly during the arrest phase before reaching a plateau (Figure 3C). In contrast, MOB1pr-VENUS showed little activation during the arrest phase, but started to oscillate when cells resumed growth (Figure 3C). We used the slopes of the fluorescence traces during the arrest phase to quantify the heat-shock activity (Figure 3D).
A general stress-responsive transcription factor, Msn2/4, is at least partially responsible for PFK26 induction (Supplementary Figure S3). Msn2/4 mutation significantly reduced the heat shock response of PFK26 (30). Also, following the ectopic expression of Msn2, PFK26 was induced similarly as some well-known Msn2/4 target genes (52). A segment on the PFK26-MOB1 promoter, ‘UAS1,’ contains a few potential binding sites of Msn2/4 (Supplementary Table S4), and deletion of UAS1 significantly reduced the PFK26 heat shock activity (Figure 3CD). The key factor that prevents Msn2/4 from activating MOB1 turned out to be Mcm1 in UAS2. Mutation in the Mcm1 binding site (mcm1*) did not affect the PFK26pr induction, but significantly increased the heat shock activity of MOB1pr (Figure 3CD). This enhanced activity was reduced back down when we deleted UAS1 on top of mcm1* (Figure 3CD). In contrast to Mcm1, mutations in Abf1 or Reb1 binding sites in UAS2 had little effect on MOB1pr activation.
To summarize, the data in Figures 2 and 3 indicate that the differentially regulation of PFK26 and MOB1 is achieved through ‘blockage factors’: on PFK26 side, Tbf1, and to a less extent Cbf1, block the cell cycle regulation from UAS2; on MOB1 side, Mcm1 blocks the heat shock activation from UAS1. Since Reb1 and Abf1 do not have the same function, this blockage activity is specific to certain DNA-binding factors.
Tbf1 blockage activity is confirmed by synthetic promoters
To understand the generality of the observations above, we measured the blockage of gene regulation using synthetic promoters. The advantage of this approach is that we can selectively perturb the promoter configuration in a controlled manner, allowing us to uncover the causal relationship between factor binding and the blockage effect. We focused on Tbf1 since it showed the highest blockage strength on the PFK26-MOB1 promoter.
We incorporated Tbf1 into three sets of synthetic promoters. In the first set, we replaced UAS2 of the PFK26-MOB1 promoter with the CLN2 promoter UAS (Figure 4A, Materials and Method). Like MOB1, CLN2 is also cell-cycle regulated, but instead of Fkh1/2, it is activated by the Swi4/Swi6 complex at the G1/S transition (53,54). We measured the activity of this hybrid promoter with either wt or mutated Tbf1 (tbf1*) downstream the CLN2pr UAS. The binding of Tbf1 on the wt site was confirmed by ChIP measurements, and the ChIP signal was significantly reduced with tbf1* (Supplementary Figure S4A). Consistent with our previous observation, tbf1* generated a 5-fold increase in the cell-cycle activity of this promoter (Figure 4A). This result shows that Tbf1 can block the regulatory signal from different activators, and the blockage function can tolerate small changes in the promoter structure, e.g. the locations of the activator binding sites.
In the second set, we inserted two Tbf1 binding motifs (wt or mutated) into the CLN2 promoter background, either downstream or upstream the UAS (Figure 4B, ‘Materials and Methods’ section). Tbf1 binding on these promoters was again measured by ChIP, and it was consistent with our design (Supplementary Figure S4B). In the downstream case, the insertion of the wt Tbf1 sites, but not tbf1*, caused a significant decrease (∼30%) in the cell cycle activity. In contrast, wt Tbf1 insertion upstream the UAS had no effect on the activity (Figure 4B). These data confirm the blockage activity of Tbf1 in a completely different promoter.
Finally, we tested the Tbf1 blockage activity in a promoter that is not cell cycle regulated. We integrated a Tbf1 motif (wt or mutated) into GAL1pr, downstream the Gal4 binding sites (Figure 4C, ‘Materials and Methods’ section). We measured the activity of these synthetic promoters during galactose induction. Interestingly, compared to the promoter containing tbf1*, the one with the wt Tbf1 site had significantly attenuated activity at the early stage of induction (<100 min; P-value < 10−2), but this difference gradually disappeared at later time points (Figure 4C). ChIP measurements showed that Tbf1 was enriched on this promoter in glucose but not in galactose (Figure 4D), indicating that Tbf1 was displaced from this promoter when fully activated, causing the loss of the blockage effect.
Overall, the measurements in Figure 4 support the generality of the Tbf1 blockage function against different activators with different promoter background sequences. The key variables that influence blockage activity may include activator strength and location of blocking factor binding sites. Elucidation of more detailed relations among these variables requires further experimentation.
Blockage activity of Tbf1 and Mcm1 is not mediated by nucleosome distribution
Both Tbf1 and Mcm1 were shown to cause nucleosome depletion on a subset of promoters (55). Depletion of these factors on the PFK26-MOB1 promoter may lead to nucleosome repositioning, which can potentially affect gene expression. To investigate the possibility that the blockage effect is mediated by nucleosome configuration, we measured nucleosome positioning on the wt PFK26-MOB1 promoter, and the promoters with tbf1* and mcm1* (‘Materials and Methods’ section). The wt PFK26-MOB1 promoter contains a long stretch of NDR between the two TSSs (Figure 5). To make sure that this is a bona-fide NDR but not unstable nucleosomes, we performed the nucleosome mapping with two lower MNase concentrations (0.2× and 0.4×), and the NDR remained intact in all of these measurements (Supplementary Figure S5). Importantly, this NDR was not altered with tbf1* or mcm1* mutation (Figure 5), consistent with previous genome-wide nucleosome measurements with Tbf1 and Mcm1 protein deletion (55). These results indicate that the blockage effect of Tbf1 or Mcm1 is not related to nucleosome positioning.
Tbf1 preferentially binds to differentially regulated DGPs on a genome-wide scale
Figure 4 demonstrates the generality of the blockage function in synthetic promoters, but it is not clear whether the blockage factors are widely used in the native genome to decouple DGPs. To address this question, we carried out bioinformatics analysis to compute the enrichment of transcription factors in differentially-regulated versus co-regulated divergent promoters on a genome-wide scale (‘Materials and Methods’ section; Supplementary Table S5). Remarkably, among all 163 sequence-specific factors we examined, Tbf1 had the most significant enrichment in differentially regulated promoters, consistent with its proposed role (Figure 6AB; Supplementary Table S6). The reverse is also true: DGPs driven by Tbf1-associated divergent promoters on average have lower correlation in their expression (Figure 6C).
Figure 6.
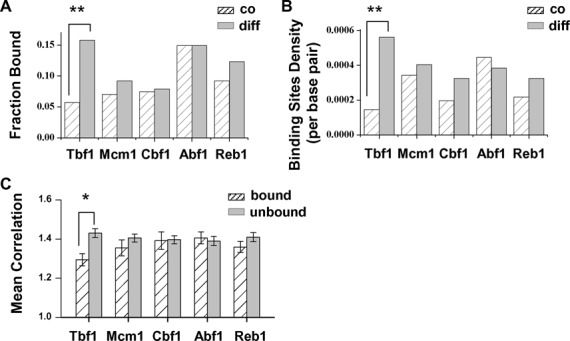
Tbf1 binding sites are highly enriched in the genome-wide differentially regulated promoters. (A) Fraction of co- versus differentially regulated divergent promoters that are associated with Tbf1, Mcm1, Cbf1 and Abf1. **P-value < 10−3 (same as in B). (B) Binding sites density of these factors in the co- versus differentially regulated divergent promoters. (C) Mean correlation score of DGPs that are associated versus not associated with these four factors. A larger score represents a higher level of co-regulation of the two divergent genes. Error bars stand for standard errors. *P-value < 10−2.
Using P-value 0.01 as a cutoff, four other factors besides Tbf1 (Rph1, YER130C, Rox1 and Stp4) showed enrichment in the differentially regulated DGPs (Supplementary Table S6). The blockage function of these factors will be investigated in the future. Notably, Rox1 is known as a repressor for anaerobic genes, e.g. the activation of HEM13 is repressed by the three Rox1 sites downstream the UAS (56–58). In addition, Rox1 binds to anti-regulated DGP, ANB1 and CYC1, and prevents the aerobic activation of ANB1, but not CYC1 (59). These observations are consistent with the possibility that Rox1 acts as a blockage factor.
Four other general regulatory factors, Mcm1, Cbf1, Abf1 and Reb1, are not significantly over-represented in the differentially regulated promoters (P-value > 0.05 in all cases; Figure 6A and B). For Abf1 and Reb1, this result is consistent with our observation that they do not contribute to the differential regulation of PFK26-MOB1. The low enrichment of Mcm1 or Cbf1 may be due to their weak blockage strengths. In addition, the locations of their binding sites on some of these divergent promoters may prevent them from effectively decoupling the two genes.
DISCUSSION
Novel ‘enhancer-blocking insulators’ in S. cerevisiae
It has been over 20 years since the discovery of insulators—DNA elements that protect gene from inappropriate regulation (60). In higher eukaryotes, there are two general categories of insulators, ‘enhancer-blocking elements’ and ‘barrier elements.’ The former prevents more distant enhancers from regulating promoters, and the latter sets the borders at heterochromatin to prevent the spread of silencing. So far, studies of insulators in budding yeast have all been focusing on ‘barrier elements’ (60). The ‘blockage factors’ identified in this study can prevent the activation from distant UASs and therefore are functionally similar to enhancer-blocking elements in higher eukaryotes. To our knowledge, this is the first time such elements are reported in Saccharomyces cerevisiae.
Blockage factors may have functions beyond DGPs. A recent study has shown that mRNAs can be produced from the non-coding direction of a divergent promoter, indicating that the transcription initiation of non-coding RNAs and mRNAs are mechanistically similar (20). Therefore, we speculate that blockage factors may also decouple divergent coding / non-coding RNA pairs. Among the tandem genes across the yeast genome, about 80% have no detectable divergent non-coding RNAs (11). In these cases, blockage factors may serve as one mechanism to attenuate the level of non-coding RNA. Consistent with this idea, it was found that Tbf1 inactivation led to increased cryptic transcription initiated divergently from the 5′ NDR of a subset of protein coding genes (55).
We propose that the general function of blockage factors is to mark the ‘boundary’ of individual promoters. This boundary decreases the undesired interaction between regulatory elements and off-target promoters, and as a consequence, it may help the regulation to ‘focus’ on the target genes. Consistent with this idea, Tbf1 was shown to demarcate the majority of yeast snoRNA promoters, and elimination of the Tbf1 binding on these promoters decreased the snoRNA expression (50).
It should be noted that only 36 out of 228 differentially regulated divergent promoters bind to Tbf1 (Figure 6). Therefore, Tbf1 can only be partially responsible for DGP differential regulation. The rest of the DGPs could be decoupled by other blockage factors. Alternatively, differential regulation may be caused by multiple mechanisms, including the ones we discussed in Figure 1, e.g. promoter length, TATA distribution, and nucleosome configuration. It is possible that each mechanism works on a small fraction of DGPs, and when averaged among all DGPs, they do not appear to be statistically significant. These ideas will be tested in the future.
Potential mechanism of blockage function
We identified Tbf1 as one of the blockage factors that lead to DGP differential regulation. The molecular mechanism of the blockage function requires further investigation. We first considered the function of Tbf1 as a barrier between the silenced telomeric region and the rest of the chromatin (61). In budding yeast, telomeric silencing is established by the physical spreading of the Sir complex, which depends on H3/H4 deacetylation by Sir2 (62). It was proposed that Tbf1 antagonizes the spreading by nucleosome exclusion and recruitment of histone acetyletransferase (60,63). This mechanism is unlikely to apply to our case since (i) there is no spreading of Sir complex, (ii) depletion of Tbf1 from the PFK26-MOB1 promoter didn't change the nucleosome positioning (Figure 5).
We also considered the mechanism of a well-studied enhancer-blocking insulator, CTCF. CTCF and the blockage factors are functionally related; in particular, a fraction of TSS-proximal CTCFs were proposed to decouple closely positioned DGPs in both Drosophila and human cells (21,23). However, most CTCFs in mammalian cells are positioned far from promoters and block enhancers kilo- to mega-bases away (64). To function at this distance, CTCF was proposed to form large and isolated ‘chromatin loop domains’ through interaction with each other or structural proteins (65). In contrast, regulatory elements in S. cerevisiae tend to be very close to TSSs (typically < a few hundred bases), and it is hard to imagine chromatin domains or loops at this length scale. We thus speculate that CTCF and the blockage factors may use different mechanisms for insulation.
Finally, since we found multiple blockage factors (Tbf1, Mcm1, and Cbf1) on the PFK26-MOB1 divergent promoter, we reasoned that the blockage effect may stem from some common activities of these factors. Tbf1 and Mcm1 both have nucleosome depletion activity, but the blockage effect is not mediated by change in nucleosome occupancy or positioning. Another common feature of all three factors (or their orthologs) is that they can severely bend DNA near their binding sites in vitro (66–68). Rox1, another potential blockage factor we found through the bioinformatics analysis, also bends DNA (69,70). If such bending indeed occurs in vivo, the distortion in DNA may directly interfere with the assembly of transcription initiation complex. Alternatively, the bent DNA may be rigid and thus prevent the interaction between UAS and TSS. It is an interesting future direction to test the role of DNA bending in the blockage function. Since DNA bending is such a fundamental activity and is likely to generate different effects at different genomic locations, it may also explain why factors like Tbf1 carry out many functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors acknowledge Dr B. Franklin Pugh for the TAP-tagged strains; Han Zheng and Kuangyu Yen for the help of MNase assay; Qian Zhang for technical support, and all the members in Bai lab for insightful comments on the manuscript.
Author contribution: C.Y. designed and performed most of the experiments and bioinformatics analysis; D.Z. performed ChIP measurements; J.A.G. and M.M.M. improved the time-lapse image analysis software; L.B. designed the project and performed some initial measurements and bioinformatics analysis; C.Y. and L.B. wrote the manuscript.
FUNDING
Penn State start-up funding from Penn State University (to L.B. and M.M.). Funding for open access charge: Penn State start-up funding from Penn State University (to L.B. and M.M.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Trinklein N.D., Aldred S.F., Hartman S.J., Schroeder D.I., Otillar R.P., Myers R.M. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang L., Yu J. A comparative analysis of divergently-paired genes (DPGs) among Drosophila and vertebrate genomes. BMC Evol. Biol. 2009;9:55. doi: 10.1186/1471-2148-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.West R.W., Jr, Yocum R.R., Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol. Cell. Biol. 1984;4:2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osley M.A. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 5.Burbelo P.D., Martin G.R., Yamada Y. Alpha 1(IV) and alpha 2(IV) collagen genes are regulated by a bidirectional promoter and a shared enhancer. Proc. Natl. Acad. Sci. U.S.A. 1988;85:9679–9682. doi: 10.1073/pnas.85.24.9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen J.J., Bross P., Westergaard M., Nielsen M.N., Eiberg H., Borglum A.D., Mogensen J., Kristiansen K., Bolund L., Gregersen N. Genomic structure of the human mitochondrial chaperonin genes: HSP60 and HSP10 are localised head to head on chromosome 2 separated by a bidirectional promoter. Hum. Genet. 2003;112:71–77. doi: 10.1007/s00439-002-0837-9. [DOI] [PubMed] [Google Scholar]
- 7.Sharon E., Kalma Y., Sharp A., Raveh-Sadka T., Levo M., Zeevi D., Keren L., Yakhini Z., Weinberger A., Segal E. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat. Biotechnol. 2012;30:521–530. doi: 10.1038/nbt.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Core L.J., Waterfall J.J., Lis J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seila A.C., Calabrese J.M., Levine S.S., Yeo G.W., Rahl P.B., Flynn R.A., Young R.A., Sharp P.A. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neil H., Malabat C., d’Aubenton-Carafa Y., Xu Z., Steinmetz L.M., Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 11.Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Munster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L.M. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seila A.C., Core L.J., Lis J.T., Sharp P.A. Divergent transcription: a new feature of active promoters. Cell cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- 13.Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol. Cell. Biol. 1986;6:3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollner R., Schmidt C., Fischer G., Kuhn K., Poschl E. Cooperative and competitive interactions of regulatory elements are involved in the control of divergent transcription of human Col4A1 and Col4A2 genes. FEBS Lett. 1997;405:31–36. doi: 10.1016/s0014-5793(97)00152-x. [DOI] [PubMed] [Google Scholar]
- 15.Schuettengruber B., Doetzlhofer A., Kroboth K., Wintersberger E., Seiser C. Alternate activation of two divergently transcribed mouse genes from a bidirectional promoter is linked to changes in histone modification. J. Biol. Chem. 2003;278:1784–1793. doi: 10.1074/jbc.M204843200. [DOI] [PubMed] [Google Scholar]
- 16.Su C.H., Shih C.H., Chang T.H., Tsai H.K. Genome-wide analysis of the cis-regulatory modules of divergent gene pairs in yeast. Genomics. 2010;96:352–361. doi: 10.1016/j.ygeno.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Almada A.E., Wu X.B., Kriz A.J., Burge C.B., Sharp P.A. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360-U141. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ntini E., Jarvelin A.I., Bornholdt J., Chen Y., Boyd M., Jorgensen M., Andersson R., Hoof I., Schein A., Andersen P.R., et al. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat. Struct. Mol. Biol. 2013;20:923–928. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- 19.Yadon A.N., Van de Mark D., Basom R., Delrow J., Whitehouse I., Tsukiyama T. Chromatin remodeling around nucleosome-free regions leads to repression of noncoding RNA transcription. Mol. Cell. Biol. 2010;30:5110–5122. doi: 10.1128/MCB.00602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marquardt S., Escalante-Chong R., Pho N., Wang J., Churchman L.S., Springer M., Buratowski S. A chromatin-based mechanism for limiting divergent noncoding transcription. Cell. 2014;157:1712–1723. doi: 10.1016/j.cell.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Negre N., Brown C.D., Shah P.K., Kheradpour P., Morrison C.A., Henikoff J.G., Feng X., Ahmad K., Russell S., White R.A., et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X., Winter C., Xia X., Gan S. Genome-wide analysis of the intergenic regions in Arabidopsis thaliana suggests the existence of bidirectional promoters and genetic insulators. Curr. Top. Plant Biol. 2011;12:15–33. [Google Scholar]
- 23.Xie X., Mikkelsen T.S., Gnirke A., Lindblad-Toh K., Kellis M., Lander E.S. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhee H.S., Pugh B.F. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 27.de Lichtenberg U., Jensen L.J., Fausboll A., Jensen T.S., Bork P., Brunak S. Comparison of computational methods for the identification of cell cycle-regulated genes. Bioinformatics. 2005;21:1164–1171. doi: 10.1093/bioinformatics/bti093. [DOI] [PubMed] [Google Scholar]
- 28.Pramila T., Wu W., Miles S., Noble W.S., Breeden L.L. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006;20:2266–2278. doi: 10.1101/gad.1450606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spellman P.T., Sherlock G., Zhang M.Q., Iyer V.R., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacIsaac K.D., Wang T., Gordon D.B., Gifford D.K., Stormo G.D., Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huisinga K.L., Pugh B.F. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 33.Lee W., Tillo D., Bray N., Morse R.H., Davis R.W., Hughes T.R., Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 34.Charvin G., Cross F.R., Siggia E.D. A microfluidic device for temporally controlled gene expression and long-term fluorescent imaging in unperturbed dividing yeast cells. PLoS One. 2008;3:e1468. doi: 10.1371/journal.pone.0001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bean J.M., Siggia E.D., Cross F.R. Coherence and timing of cell cycle start examined at single-cell resolution. Mol. Cell. 2006;21:3–14. doi: 10.1016/j.molcel.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 36.Bai L., Ondracka A., Cross F.R. Multiple sequence-specific factors generate the nucleosome-depleted region on CLN2 promoter. Mol. Cell. 2011;42:465–476. doi: 10.1016/j.molcel.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kent N.A., Mellor J. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 1995;23:3786–3787. doi: 10.1093/nar/23.18.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekinger E.A., Moqtaderi Z., Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Hibbs M.A., Hess D.C., Myers C.L., Huttenhower C., Li K., Troyanskaya O.G. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics. 2007;23:2692–2699. doi: 10.1093/bioinformatics/btm403. [DOI] [PubMed] [Google Scholar]
- 40.Pachkov M., Erb I., Molina N., van Nimwegen E. SwissRegulon: a database of genome-wide annotations of regulatory sites. Nucleic Acids Res. 2007;35:D127–131. doi: 10.1093/nar/gkl857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobi K.C., Winston F. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:5575–5586. doi: 10.1128/MCB.00459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basehoar A.D., Zanton S.J., Pugh B.F. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 43.Zhu G., Spellman P.T., Volpe T., Brown P.O., Botstein D., Davis T.N., Futcher B. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature. 2000;406:90–94. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]
- 44.Chasman D.I., Lue N.F., Buchman A.R., LaPointe J.W., Lorch Y., Kornberg R.D. A yeast protein that influences the chromatin structure of UASG and functions as a powerful auxiliary gene activator. Genes Dev. 1990;4:503–514. doi: 10.1101/gad.4.4.503. [DOI] [PubMed] [Google Scholar]
- 45.Miyake T., Loch C.M., Li R. Identification of a multifunctional domain in autonomously replicating sequence-binding factor 1 required for transcriptional activation, DNA replication, and gene silencing. Mol. Cell. Biol. 2002;22:505–516. doi: 10.1128/MCB.22.2.505-516.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 47.Lavoie H., Hogues H., Mallick J., Sellam A., Nantel A., Whiteway M. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 2010;8:e1000329. doi: 10.1371/journal.pbio.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuch B.B., Galgoczy D.J., Hernday A.D., Li H., Johnson A.D. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 2008;6:e38. doi: 10.1371/journal.pbio.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kasinathan S., Orsi G.A., Zentner G.E., Ahmad K., Henikoff S. High-resolution mapping of transcription factor binding sites on native chromatin. Nat. Methods. 2014;11:203–209. doi: 10.1038/nmeth.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Preti M., Ribeyre C., Pascali C., Bosio M.C., Cortelazzi B., Rougemont J., Guarnera E., Naef F., Shore D., Dieci G. The telomere-binding protein Tbf1 demarcates snoRNA gene promoters in Saccharomyces cerevisiae. Mol. Cell. 2010;38:614–620. doi: 10.1016/j.molcel.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 51.Schlecht U., Erb I., Demougin P., Robine N., Borde V., van Nimwegen E., Nicolas A., Primig M. Genome-wide expression profiling, in vivo DNA binding analysis, and probabilistic motif prediction reveal novel Abf1 target genes during fermentation, respiration, and sporulation in yeast. Mol. Biol. Cell. 2008;19:2193–2207. doi: 10.1091/mbc.E07-12-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elfving N., Chereji R.V., Bharatula V., Bjorklund S., Morozov A.V., Broach J.R. A dynamic interplay of nucleosome and Msn2 binding regulates kinetics of gene activation and repression following stress. Nucleic Acids Res. 2014;42:5468–5482. doi: 10.1093/nar/gku176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cross F.R., Hoek M., McKinney J.D., Tinkelenberg A.H. Role of Swi4 in cell cycle regulation of CLN2 expression. Mol. Cell. Biol. 1994;14:4779–4787. doi: 10.1128/mcb.14.7.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stuart D., Wittenberg C. Cell cycle-dependent transcription of Cln2 is conferred by multiple distinct cis-acting regulatory elements. Mol. Cell. Biol. 1994;14:4788–4801. doi: 10.1128/mcb.14.7.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Bakel H., Tsui K., Gebbia M., Mnaimneh S., Hughes T.R., Nislow C. A compendium of nucleosome and transcript profiles reveals determinants of chromatin architecture and transcription. PLoS Genet. 2013;9:e1003479. doi: 10.1371/journal.pgen.1003479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klinkenberg L.G., Mennella T.A., Luetkenhaus K., Zitomer R.S. Combinatorial repression of the hypoxic genes of Saccharomyces cerevisiae by DNA binding proteins Rox1 and Mot3. Eukaryotic Cell. 2005;4:649–660. doi: 10.1128/EC.4.4.649-660.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sertil O., Kapoor R., Cohen B.D., Abramova N., Lowry C.V. Synergistic repression of anaerobic genes by Mot3 and Rox1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:5831–5837. doi: 10.1093/nar/gkg792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amillet J.M., Buisson N., Labbe-Bois R. Characterization of an upstream activation sequence and two Rox1p-responsive sites controlling the induction of the yeast HEM13 gene by oxygen and heme deficiency. J. Biol. Chem. 1996;271:24425–24432. doi: 10.1074/jbc.271.40.24425. [DOI] [PubMed] [Google Scholar]
- 59.Lowry C.V., Zitomer R.S. ROX1 encodes a heme-induced repression factor regulating ANB1 and CYC7 of Saccharomyces cerevisiae. Mol. Cell. Biol. 1988;8:4651–4658. doi: 10.1128/mcb.8.11.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.West A.G., Gaszner M., Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 61.Fourel G., Revardel E., Koering C.E., Gilson E. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 1999;18:2522–2537. doi: 10.1093/emboj/18.9.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu F., Zhang Q., Zhang K., Xie W., Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol. Cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bi X., Broach J.R. Chromosomal boundaries in S. cerevisiae. Curr. Opin. Genet. Dev. 2001;11:199–204. doi: 10.1016/s0959-437x(00)00179-9. [DOI] [PubMed] [Google Scholar]
- 64.Kim T.H., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Phillips J.E., Corces V.G. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niedenthal R.K., Sengupta M., Wilmen A., Hegemann J.H. Cpf1 protein-induced bending of yeast centromere DNA element-1. Nucleic Acids Res. 1993;21:4726–4733. doi: 10.1093/nar/21.20.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bianchi A., Smith S., Chong L., Elias P., deLange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tan S., Richmond T.J. Crystal structure of the yeast MATalpha2/MCM1/DNA ternary complex. Nature. 1998;391:660–666. doi: 10.1038/35563. [DOI] [PubMed] [Google Scholar]
- 69.Deckert J., Khalaf R.A., Hwang S.M., Zitomer R.S. Characterization of the DNA binding and bending HMG domain of the yeast hypoxic repressor Rox1. Nucleic Acids Res. 1999;27:3518–3526. doi: 10.1093/nar/27.17.3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deckert J., Rodriguez Torres A.M., Simon J.T., Zitomer R.S. Mutational analysis of Rox1, a DNA-bending repressor of hypoxic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:6109–6117. doi: 10.1128/mcb.15.11.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


