Abstract
MicroRNA-122 (miR-122), which is expressed at high levels in hepatocytes, is selectively stabilized by 3′-adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Here, we report that poly(A)-specific ribonuclease (PARN) is responsible for the deadenylation and destabilization of miR-122. The 3′-oligoadenylated variant of miR-122 was detected in Huh7 cells when PARN was down-regulated. In addition, both the steady-state level and stability of miR-122 were increased in PARN knockdown cells. We also demonstrate that CUG-binding protein 1 (CUGBP1) specifically interacts with miR-122 and other UG-rich miRNAs, and promotes their destabilization. Overexpression of CUGBP1 or PARN in Huh7 cells reduced the steady-state levels of these miRNAs. Because CUGBP1 interacts directly with PARN, we hypothesized that it specifically recruits PARN to miR-122. In fact, CUGBP1 enhanced PARN-mediated deadenylation and degradation of miR-122 in a dose-dependent manner in vitro. These results indicate that the cellular level of miR-122 is determined by the balance between the opposing effects of GLD-2 and PARN/CUGBP1 on the metabolism of its 3′-terminus.
INTRODUCTION
MicroRNAs (miRNAs) are small non-coding RNAs of ∼21 nucleotides (nt) that regulate various biological processes in animals and plants, including development, differentiation, physiology and pathology. RNA-induced silencing complexes that incorporate miRNAs (miRISCs) target the 3′-untranslated region (UTR) of mRNAs and repress their translation and/or induce their degradation (1,2). The steady-state levels of miRNAs and their activities are regulated at the transcriptional and post-transcriptional levels (3). The 3′-termini of miRNAs frequently undergo post-transcriptional uridylation, adenylation and 2′-O-methylation (3,4); these modifications play major roles in regulating the biogenesis, function and stability of miRNAs.
Recent reports demonstrated that Lin-28 recruits the terminal uridyltransferase TUT4/Zcchc11 or TUT7/Zcchc6 to the let-7 precursor RNA (pre-let-7) and induces 3′-uridylation in mammals; this event blocks processing by Dicer and eventually leads to degradation of the RNA by the 3′-to-5′ exonuclease Dis3l2 (5–8). Similarly, miR-26 is also 3′-uridylated by TUT4/Zcchc11, and this modification regulates its ability to repress interleukin-6 expression (9). In myotonic dystrophy, reduced levels of muscleblind-like 1 protein (MBNL1), which binds to the loop of pre-miR-1 and competes with Lin-28 binding, promote the 3′-uridylation of pre-miR-1 by TUT4/Zcchc11, leading to a reduction in the level of miR-1 and dysregulation of cardiac function (10). In plants, the 3′-termini of miRNAs and small interfering RNAs (siRNAs) are modified by 2′-O-methylation during maturation (11). The methyltransferase Hen1 is essential for this modification and prevents 3′-oligouridylation by Hen 1 Suppressor 1 (HESO1) (11–13), and Hen1 mutant plant cells display reduced accumulation of mature miRNAs and abnormal development (14). In mammals and Drosophila melanogaster, the 3′-termini of piwi-interacting RNAs (piRNAs) and siRNAs, rather than those of miRNAs, are 2′-O-methylated by Hen1 homologs (Pimet and DmHen1) (15–18). In Drosophila, 2′-O-methylation by DmHen1 prevents the addition and trimming of 3′ nucleotides in Argonaute2 (Ago2)-associated siRNAs (19).
Liver-specific miR-122 is expressed at high levels in hepatocytes and some hepatoma cells, and plays an important role in hepatic function by regulating cholesterol and fatty-acid metabolism (20,21). MiR-122 is also a host factor required for hepatitis C virus replication (22). We reported previously that miR-122 is selectively stabilized by 3′-mono-adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2 (also referred to as PAPD4), and that the steady-state level of miR-122 is decreased markedly in the livers of GLD-2-null mice, indicating that one or more 3′ to 5′ exonucleases are involved in the metabolism of this miRNA (23). A genome-wide analysis of a human monocytic cell line revealed that GLD-2 is a primary adenylating enzyme for a wide variety of miRNAs (24). In primary fibroblasts, GLD-2 adenylates and stabilizes specific miRNAs directly, including miR-122, which regulates the expression of the mRNA encoding cytoplasmic polyadenylation element binding protein (CPEB) (25). CPEB recruits factors to the 3′ UTR of the p53 mRNA to enhance polyadenylation mediated by GLD-4 and promote its translation; therefore, p53-controlled cellular senescence is regulated by miR-122 (26,27).
The precise mechanism by which miR-122 turnover is controlled by 3′-adenylation remains elusive. Identification of factors involved in miRNA decay is critical to understanding the association between miRNA metabolism and various biological processes. Several factors responsible for the degradation of mature miRNAs were identified in model organisms, including small RNA degrading nucleases in plants (28) and XRN-1/XRN-2 in Caenorhabditis elegans (29,30). In humans, polynucleotide phosphorylase (PNPase or PNPT1) degrades miR-221 (31), and the exosome subunit RRP41 is required for the degradation of miR-382 (32). In addition, Wispy, a putative homolog of GLD-2 in Drosophila, adenylates maternal miRNAs. However, unlike GLD-2-mediated stabilization of miR-122, Wispy-mediated adenylation down-regulates maternal miRNAs, implying the involvement of some exonucleases (33).
In this study, we report that poly(A)-specific ribonuclease (PARN) is involved in the destabilization of miR-122. We also demonstrate that an RNA-binding protein, CUG-binding protein 1 (CUGBP1), specifies a subset of miRNAs, including miR-122, for decay.
MATERIALS AND METHODS
Preparation of the small RNA fraction and pyrosequencing
Total RNA isolated from Huh7 cells was separated by PAGE in 15% gels containing 7 M urea, and then stained with SYBR Gold (Invitrogen). The gel region corresponding to 14–40 nt was excised. RNA was eluted from the gel pieces by incubation in elution buffer (500 mM NaCl, 0.1% SDS and 1 mM EDTA) for 2 h at 37°C, and then precipitated with ethanol. Construction of a cDNA library from the small RNAs was performed as described previously (23). High-throughput pyrosequencing of the cDNA library was performed on a GS20 system (Roche).
Isolation of miRNAs and LC/MS analysis
Human miR-122 variants were isolated from total RNA by reciprocal circulating chromatography, as described previously (23,34). The isolated miRNAs were analyzed using a nano ESI/MS system (LTQ Orbitrap XL; Thermo Fisher Scientific) coupled with a capillary LC system (DiNa, KYA Technologies), as described previously (23,35).
Isolation of miRNA-binding proteins
The sequences of the synthetic miRNA probes fused to 5′-aminated oligodeoxythymidines (Hokkaido System Science) were as follows: 5′-ttttttttttuggagugugacaaugguguuug-3′ for miR-122 and 5′-ttttttttttuagcagcacguaaauauuggcg-3′ for miR-16. The probes were immobilized on N-hydroxysuccinimiide (NHS)-activated Sepharose 4 Fast Flow (GE Healthcare) by coupling the 5′-terminal amino group of the probe to the activated carboxyl group of the resin. The miRNA probe (91 μg) was immobilized onto 150 μl of NHS-activated Sepharose. Huh7 cells (1 × 107) were washed twice with ice-cold phosphate-buffered saline (PBS) and harvested by scraping into PBS containing protease inhibitors (Complete Mini Protease Inhibitor Cocktail Tablet; Roche) and RNase inhibitor (SUPERase-IN; Ambion). After removal of the PBS, the cells were resuspended in 1 ml of buffer L [50 mM Tris-HCl (pH 8.0), 150 mM KCl, 1 mM DTT, 0.5% Triton X-100, 10% glycerol, 1 μl of SUPERase-IN (Ambion) and Complete Mini Protease Inhibitor Cocktail Tablet (Roche)] and lysed by passing the suspension through 25-gauge needles attached to 1 ml syringes a total of 10 times. The lysates were centrifuged twice at 20,000 g for 20 min to remove the cell debris. The total cell extracts were then mixed with Sepharose resin and rotated for 2 h at 4°C. The supernatants were collected as precleared lysate, mixed with the resin-immobilized miRNA probe, and rotated for 12 h at 4°C. The resin was then washed four times with ice-cold buffer I [20 mM Tris-HCl (pH 7.5), 100 mM KCl, and 2 mM EDTA], and the associated proteins were resolved by SDS-PAGE. The bands of interest were excised, digested with trypsin, and identified by peptide mass finger printing using LC/MS/MS and MASCOT (36).
Immunoprecipitation of CUGBP1 and real-time RT-PCR
The Huh7 cells (1 × 107) were washed twice with ice-cold PBS and harvested by scraping into PBS containing protease inhibitors (Complete Mini Protease Inhibitor Cocktail Tablet; Roche) and RNase inhibitor (SUPERase-IN; Ambion). After removal of the PBS, the cells were resuspended in 1 ml of lysis buffer [10 mM Tris-HCl (pH 8.0), 100 mM KCl, 2.5 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, 1 μl of SUPERase-IN (Ambion), and Complete Mini Protease Inhibitor Cocktail Tablet (Roche)] and lysed by passing the suspension through 25-gauge needles attached to 1 ml syringes a total of 10 times. The lysates were centrifuged twice at 20,000 g for 20 min to remove the cell debris. The total cell extracts were then mixed with protein G Sepharose 4 Fast Flow (GE Healthcare) and rotated for 1 h at 4°C, and the supernatants were harvested as precleared lysate. The anti-CUGBP1 antibody (RN002MW; MBL) or normal mouse IgG (Upstate) (5 μg) was mixed with 20 μl (50% slurry) of protein G Sepharose 4 Fast Flow and rotated for 1 h at 4°C. The precleared lysate was mixed with the beads, and immunoprecipitation was performed at 4°C for 3 h. The beads were washed four times with lysis buffer, and the immunoprecipitates were treated with TriPure (Roche) to isolate the bound RNAs. The isolated RNAs were then treated with RQ1 DNase (Promega), and specifically bound mRNAs were analyzed by real-time RT-PCR, as described in Supplementary information. The following gene-specific primer sets were used: 5′-taccacgacgactcatacacag-3′ and 5′-caggctcggtttcaggag-3′ for jun B, and 5′-ctggcaccacaccttctac-3′ and 5′-ggcatacccctcgtagatg-3′ for ACTB. Specifically bound miRNAs were analyzed by real-time RT-PCR using TaqMan miRNA assays (Applied Biosystems), according to the manufacturer's instructions.
Overexpression of CUGBP1 and PARN, and immunoblotting
The cDNAs encoding human PARN and C-terminally Flag-tagged human CUGBP1 were cloned into the pDEST12.2 vector (Invitrogen). Huh7 cells cultured in 10-cm dishes were transfected with 6 μg of each vector using FuGENE reagent (Roche). As a control, a mock-transfection was performed without plasmid. At 72 h post-transfection, the cells were treated with ISOGEN reagent (Wako) to isolate total RNA. The steady-state level of each mature miRNA was measured using TaqMan miRNA assays (Applied Biosystems), according to the manufacturer's instructions. The expression level of CUGBP1 was determined by immunoblotting using anti-CUGBP1 (RN002MW; MBL) and anti-Flag (F1804; Sigma) antibodies. The expression levels of PARN and ACTB (control) were determined using anti-PARN (3899; Cell Signaling Technology) and anti-ACTB (SAB1403520; Sigma) antibodies, respectively. Ago2 was detected by immunoblotting using an anti-Ago2 antibody (4G8; Wako).
In vitro deadenylation
5′-32P-labeled RNA (3000 cpm/μl) was incubated with PARN in buffer comprising 20 mM Tris-HCl (pH 7.5), 50 mM KCl, 2.5 mM MgCl2, 50 μg/ml BSA, 1 U/μl SUPERase-IN (Ambion) and 250 nM non-labeled miR-21 (for Figure 5, Supplementary Figure S7). A portion of each sample was separated on a 20% denaturing acrylamide gel, and the radioactivity was visualized using a FLA7000 Image Analyzer (Fujifilm). The concentrations of His-PARN were 690 pM for Figures 1d and 6a, 40.8 pM for Supplementary Figure S3, and 10.2 pM for Figure 5a–d and Supplementary Figure S7a–c. The concentrations of CUGBP1-Flag were 9.6 nM for Figure 5a, 0–9.6 nM for Figure 5b and 4.8 nM for Figure 5c, d and Supplementary Figure S7.
Figure 5.

CUGBP1 enhances PARN-mediated degradation of miR-122 specifically. (A) In vitro deadenylation and degradation of miR-122 (27-nt, 3′-a5) in the presence (+) or absence (−) of recombinant PARN (10.2 pM) or recombinant CUGBP1 (9.6 nM). The reaction was stopped after 30 min of incubation. (B) In vitro deadenylation and degradation of miR-122 (27-nt, 3′-a5) in the presence (+) or absence (−) of recombinant PARN (10.2 pM) and increasing concentrations (0, 0.53, 1.6, 4.8 and 9.6 nM) of recombinant CUGBP1. The reaction was stopped after 30 min of incubation. The degradation ratio for each experiment was calculated from the band intensities of the 22–27 nt RNA. Data in the graph are represented as the mean ± SD (n = 3 independent experiments). (C, D) In vitro exonucleolytic degradation of miR-122 (27-nt, 3′-a5) (c) and miR-21 (d) in the presence (+) or absence (−) of recombinant PARN (10.2 pM) and recombinant CUGBP1 (4.8 nM). Aliquots of the reaction mixtures were collected and analyzed at the indicated time periods. The degradation ratio for each experiment was calculated from the band intensities of the 22–27 nt (c) or intact RNA (d). Data in the graphs are represented as the mean ± SD (n = 3 independent experiments).
Figure 1.
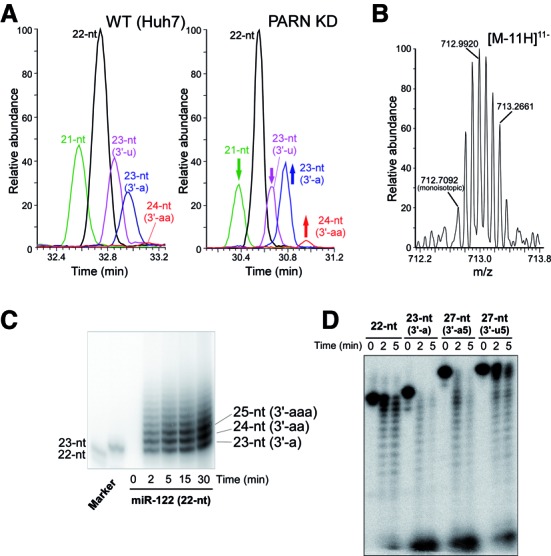
PARN is responsible for the 3′ deadenylation of miR-122. (A) Mass chromatograms of the 21-nt (MW 6847.84, green), 22-nt (MW 7192.88, black), 23-nt (3′-u) (MW 7498.91, magenta), 23-nt (3′-a) (MW 7521.94, blue) and 24-nt (3′-aa) (MW 7850.99, red) miR-122 variants isolated from the WT (Huh7, left panel) and PARN KD (right panel) cells. Each chromatogram was traced with [M-8H]8− and [M-9H]9− ions. The changes in intensity upon KD of PARN are indicated by arrows. (B) Mass spectrum of the [M-11H]11− ions of the 24-nt (3′-aa) variant isolated from PARN KD cells. Mass spectra in the retention time, 30.94–31.00 min, of the chromatogram shown in (a) (right panel) were extracted and integrated. The monoisotopic ion is indicated. (C) In vitro 3′-adenylation of miR-122 by recombinant GLD-2. Synthetic miR-122 (22-nt variant) was incubated with 5×Flag-GLD-2 (immunoprecipitated from HEK293T cells) in the presence of [α-32P] ATP for the indicated times. Synthetic 5′-32P-labeled 22-nt and 23-nt (3′-a) variants of miR-122 were used as markers. (D) In vitro exonucleolytic degradation of miR-122 variants by recombinant PARN. 5′-32P-labeled synthetic 22-nt, 23-nt (3′-a), 27-nt (3′-a5) and 27-nt (3′-u5) variants of miR-122 were incubated with recombinant PARN (690 pM) at 37°C for the indicated times. The reaction products were resolved by PAGE under denaturing conditions.
Figure 6.
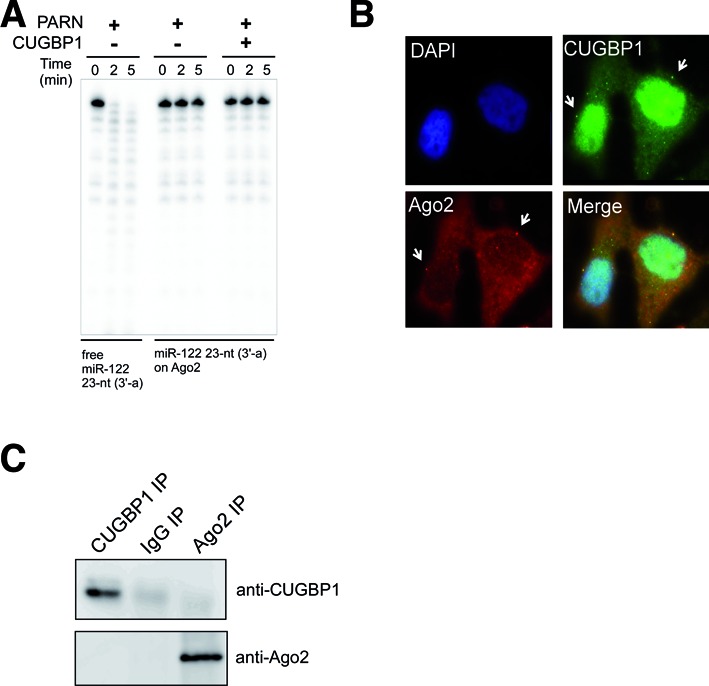
Ago2 protects miRNAs from degradation by PARN. (A) Free miR-122 or miR-122 loaded onto 5×Flag-Ago2 was incubated with recombinant PARN (690 pM) in the absence or presence of CUGBP1 (10.4 nM). Aliquots of the reaction mixture were collected and analyzed at the indicated time period. (B) Analysis of the subcellular localizations of endogenous CUGBP1 (green) and Ago2 (red) in Huh7 cells by immunofluorescence staining. Nuclei were stained with DAPI (blue). All images were superimposed to generate the merged panel. (C) Immunoprecipitation and immunoblot analyses showing the lack of interaction between CUGBP1 and Ago2 in Huh7 cells. As a control, immunoprecipitation was performed using non-specific IgG.
Electrophoretic mobility shift assays
5′-32P-labeled miRNAs (40 nM) were mixed with different concentrations of CUGBP1-Flag (1.25–1280 nM) in a mixture containing 20 mM HEPES-KOH (pH 7.6), 100 mM KCl, 2 mM MgCl2, 8% glycerol and RNasin ribonuclease inhibitor (Promega). The mixture was incubated at 30°C for 30 min. After adding a 25% volume of loading dye [50 mM Tris-HCl (pH 8.0) and 5 mM Mg(OAc)2], each sample was analyzed by 5% native PAGE at 4°C and visualized using an FLA7000 Image Analyzer (Fujifilm).
Other methods are described in the Supplementary Information.
RESULTS
PARN is responsible for the 3′ deadenylation of miR-122
GLD-2 was originally characterized as a cytoplasmic poly(A) polymerase that regulates the length of the poly(A) tail of mRNAs containing a CPE in their 3′ UTR (37–39). CPEB binds to CPEs in target mRNAs and recruits GLD-2, as well as other factors involved in regulating the length of mRNA poly(A) tails. As a component of the CPEB complex, PARN is also involved in shortening of the poly(A) tails (40). This knowledge prompted us to examine whether PARN is responsible for deadenylation and destabilization of miR-122. First, PARN was knocked down in human Huh7 hepatoma cells using a short hairpin RNA (shRNA). The steady-state level of the PARN mRNA in knockdown (KD) cells was 12% of that in untreated wild-type (WT) cells. In addition, the level of endogenous PARN was undetectable by western blotting in the KD cells (Supplementary Figure S1a). The small RNA fractions containing miRNAs were then isolated from WT and PARN KD cells, and analyzed by pyrosequencing. As shown in Table 1, 3′-oligoadenylated miR-122 variants containing two to five adenosines were detected in the PARN KD extract, while no oligoadenylated variants were detected in the WT extract, as reported previously (23). The populations of miR-122 variants were then isolated from the PARN KD and WT cells using reciprocal circulating chromatography (34), and analyzed by liquid chromatography/nano electrospray ionization mass spectrometry (miRNA-MS) (23,35). Using this technique, five miR-122 variants were detected in the WT cells (Figure 1a and Table 1), namely, the 21-nt, 22-nt, 23-nt (3′-u), 23-nt (3′-a) and 24-nt (3′-aa) variants. KD of PARN altered the profile of the miR-122 variants (Figure 1a); specifically, when normalized to the level of the 22-nt variant, the relative level of the 23-nt (3′-a) variant increased, whereas those of the 21-nt and 23-nt (3′-u) variants decreased. In addition, the relative level of the 3′-di-adenylated 24-nt (3′-aa) variant was markedly higher in the PARN KD cells than in the WT cells (Figure 1a); however, the oligoadenylated variants longer than 25-nt observed by pyrosequencing (Table 1 and Supplementary Table S1) were not detected in either cell type, most likely due to the limited detection sensitivity of miRNA-MS. A series of isotopes of the 11-charged negative ion of the 24-nt (3′-aa) variant were clearly observed in the PARN KD cells (Figure 1b). The relative steady-state levels of the 24-nt variant calculated from the area of the mass chromatogram peaks (Figure 1a) were estimated to be 0.6% in WT cells and 2.1% in PARN KD cells. These data indicate that PARN is responsible for the 3′ deadenylation of miR-122.
Table 1. Read numbers of miR-122 variants in WT and PARN KD cells.
| miR-122 variants | Sequences | Reads | |
|---|---|---|---|
| WT | PARN KD | ||
| 21-nt | UGGAGUGUGACAAUGGUGUUU | 95 | 180 |
| 22-nt | UGGAGUGUGACAAUGGUGUUUG | 131 | 463 |
| 23-nt (3'-u) | UGGAGUGUGACAAUGGUGUUUGu | 125 | 185 |
| 23-nt (3'-a) | UGGAGUGUGACAAUGGUGUUUGa | 51 | 93 |
| 24-nt (3'-aa) | UGGAGUGUGACAAUGGUGUUUGaa | 0 | 10 |
| 25-nt (3'-aaa) | UGGAGUGUGACAAUGGUGUUUGaaa | 0 | 2 |
| 26-nt (3'-aaaa) | UGGAGUGUGACAAUGGUGUUUGaaaa | 0 | 15 |
| 27-nt (3'-aaaaa) | UGGAGUGUGACAAUGGUGUUUGaaaaa | 0 | 3 |
| other variants | 28 | 60 | |
| total miR-122 | 430 (2.60%) | 1011 (3.18%) | |
| total sequence reads | 16 517 | 31 793 | |
Complete data for the miR-122 variants are shown in Supplementary Table S1. Bold values represent read numbers of adenylated miR-122.
We reported previously that immunoprecipitated GLD-2 mono-adenylates the 3′-terminus of miR-122 in vitro (23); however, the activity of the immunoprecipitated GLD-2 was quite low, likely due to an insufficient yield. Therefore, we examined the in vitro adenylation of miR-122 by recombinant 5×Flag-GLD-2 overexpressed in HEK293T cells (Supplementary Figure S2a). The isolated recombinant GLD-2 protein was capable of oligoadenylating the 3′-terminus of miR-122 in vitro (Figure 1c), suggesting that the oligoadenylation of this miRNA in PARN KD cells is catalyzed by GLD-2. It is also possible that the oligoadenylation of miR-122 by GLD-2 requires a specific adaptor protein. Next, we performed exonucleolytic degradation of miR-122 in vitro using recombinant PARN (Supplementary Figure S2b). Although recombinant PARN degraded both the 22-nt and 23-nt (3′-a) variants of miR-122, the adenylated variant was degraded faster than the non-adenylated variant (Figure 1d). Furthermore, 3′-oligoadenylated miR-122 (27-nt) was degraded much faster than the 3′-oligouridylated (27-nt) variant (Figure 1d). By contrast, 3′-oligoadenylated miR-122 (27-nt) was not degraded when incubated with a catalytically inactive mutant of PARN (D28A) (Supplementary Figures S2b and S3). This result rules out possible contamination of the preparation with Escherichia coli nucleases. In addition, these results are consistent with the fact that 3′-oligoadenylated RNA is the preferred substrate for PARN, although 3′ tails containing other nt can be degraded by PARN to some extent (41).
PARN destabilizes miR-122 in human cells
To examine the effect of PARN-mediated deadenylation on the stability of miRNAs in Huh7 cells, the relative changes in miRNA levels following KD of GLD-2, PARN or both were determined using quantitative real-time RT-PCR analyses. Single KDs of GLD-2 and PARN reduced the steady-state levels of their mRNAs to 6.7% and 12% of the controls, respectively. When they were knocked down simultaneously, the GLD-2 and PARN mRNA levels were reduced to 9.8% and 11% of the controls, respectively. Although endogenous GLD-2 was not detected in Huh7 cells using the available antibodies, efficient KD of PARN was confirmed by immunoblotting (Supplementary Figure S1b). As expected, KD of PARN increased the miR-122 level significantly (Figure 2a), whereas the level of this miRNA was reduced following KD of GLD-2, as described previously (23). KD of five other major human deadenylases did not increase the steady-state level of miR-122 (Supplementary Figure S4). We also confirmed that the steady-state levels of various mRNAs that are targeted by miR-122 were down-regulated in PARN KD cells (Figure 2b and Supplementary Figure S1a). These decreases are consistent with increase of miR-122 in PARN KD cells. The increased miR-122 level in PARN KD cells was confirmed by a pyrosequencing analysis, which showed that the read fraction of miR-122 increased from 2.60% of total reads in the WT cells to 3.18% in the PARN KD cells (Table 1). The substantial change in the miR-122 level was not observed when GLD-2 and PARN were silenced simultaneously (Figure 2a), indicating that a delicate balance between these two factors determines the steady-state level of miR-122.
Figure 2.
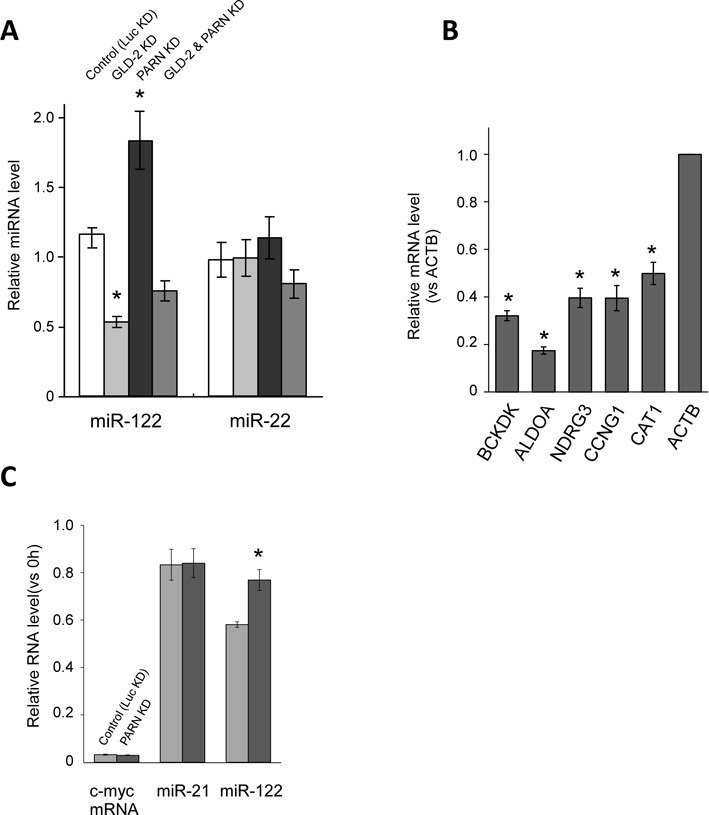
PARN is responsible for the degradation of miR-122. (A) The effects of shRNAs targeting GLD-2 and PARN on the steady-state levels of miR-122 and miR-22. KD of firefly luciferase (Luc KD) using a specific shRNA was used as a control. The values represent the ratios of the levels of each miRNA in the KD cells to those in the WT cells, as determined by real-time RT-PCR analyses, and the data were normalized to the ratio for miR-21. Data are presented as the mean ± SD (n = 3). *P < 0.001 versus control by Student's t-tests. (B) The steady-state levels of five mRNAs (BCKDK, ALDOA, NDRG3, Cyclin G1 and CAT1) targeted by miR-122 in PARN KD cells relative to those in WT cells, as determined by real-time RT-PCR analyses. The relative expression level of each mRNA was normalized to that of the ACTB mRNA. Data are presented as the mean ± SD (n = 3). *P < 0.01 versus WT cells by Student's t-tests. (C) Real-time RT-PCR analyses of the relative stabilities of miR-122, miR-21 and the c-myc RNA in Huh7 cells that were transfected with a siRNA targeting PARN or firefly luciferase (Luc KD), and treated with 5 μg/ml α-amanitin to inhibit transcription. The cells were harvested 20 h after the transcriptional inhibition. The relative stabilities of miR-122, miR-21 and the c-myc mRNA were normalized to the ratio for 7SK RNA. Data are presented as the mean ± SD (n = 3). *P < 0.01 versus control by a Student's t-test.
To determine whether PARN destabilizes miR-122 directly in human cells, PARN KD Huh7 cells were treated with α-amanitin, a specific inhibitor of RNA polymerase II. The c-myc mRNA, which has a short half-life, was degraded rapidly after transcription was inhibited (Figure 2c). By contrast, miRNA-122 and miR-21 showed relatively higher stability in Huh7 cells, which is consistent with the previous finding that miR-122 has a long half-life (42). Upon depletion of PARN, miR-122 showed even higher stability (Figure 2c), whereas the stabilities of miR-21 and the c-myc mRNA were not affected. This result suggests that PARN is directly involved in the destabilization of miR-122 in human cells.
To identify other miRNAs that are destabilized by PARN-mediated deadenylation, we analyzed the pyrosequencing data in more detail. The effects of PARN KD on the read numbers of individual miRNAs are shown in Supplementary Table S2. In addition to miR-122, some other miRNAs were also up-regulated following KD of PARN. Among these miRNAs, only miR-93 and miR-652–3p showed markedly increases in untemplated 3′-mono- and di-adenylation (Supplementary Table S1). For miR-93, only one read of the mono-adenylated 24-nt variant was detected in the WT cells, while up to 70 reads of the mono- or di-adenylated variants were detected in the PARN KD cells (Supplementary Table S1). The percentage of miR-93 reads in the PARN KD cells (2.58%) was much higher than that in the WT cells (0.49%) (Supplementary Table S2). Similarly, 3′-mono- or di-adenylated variants of miR-652–3p were detected only when PARN was silenced (Supplementary Table S1), and the percentage of miR-652–3p reads in the PARN KD cells (0.39%) was higher than that in the WT cells (0.15%) (Supplementary Table S2). The read percentage of miR-27b in the PARN KD cells (2.74%) was also much higher than that in the WT cells (1.14%) (Supplementary Table S2); however, the ratio of the 3′ adenylated variants in the total miR-27b reads was decreased following PARN KD (Supplementary Table S1), indicating that the steady-state level of miR-27b is not controlled by PARN-mediated deadenylation. No obvious changes in read percentages or 3′-adenylation were identified for other miRNAs following PARN KD (Supplementary Tables S1 and S2). Although the changes in miRNA levels detected in these analyses were limited due to the partial KD of PARN, these data indicate the involvement of PARN in deadenylation and destabilization of a subset of miRNA species, including miR-122, miR-93 and miR-652–3p. In addition to 3′-adenylated variants, the levels of some variants with 3′ additions of untemplated nucleotides, including monouridylated or oligouridylated variants, were increased slightly in PARN KD cells (Supplementary Table S1), indicating that PARN might have the ability to trim the 3′-termini of some miRNAs.
CUGBP1 recruits PARN to UG-rich miRNAs, including miR-122
To identify the factor(s) responsible for the miRNA specificity of PARN, we performed an affinity isolation of proteins that interact specifically with miR-122. In this experiment, Huh7 cell lysate was mixed with Sepharose beads conjugated to 5′-aminated miR-122 or miR-16, and the bound proteins were resolved by SDS-PAGE (Figure 3a). A band corresponding to 50 kDa that bound specifically to miR-122 was analyzed by peptide mass finger printing using LC/MS. The protein was identified as CUGBP1, a conserved multifunctional RNA-binding protein that specifically recognizes the GU-rich element (GRE) in target mRNAs (43,44). To confirm this finding in cells, endogenous CUGBP1 was immunoprecipitated using an anti-CUGBP1 antibody (Figure 3b), and bound RNAs were analyzed by real-time RT-PCR. As controls, the GRE-containing jun B mRNA (43) co-precipitated with CUGBP1, whereas the non-GRE-containing ACTB (β-actin) mRNA was not enriched in the precipitate (Figure 3c). A large amount of miR-122 co-precipitated with CUGBP1 and, compared with other miRNAs and small nucleolar RNAs, miR-93 and miR-652–3p were also enriched in the precipitate (Figure 3c). No miRNAs were precipitated by the control non-specific IgG (Figure 3c). Notably, miRNAs that had oligoadenylated 3′-ends in the PARN KD cells, such as miR-122, miR-93 and miR-652–3p, contain multiple UG-rich sequences, whereas non-adenylated miRNAs contain only limited UG regions (Supplementary Table S3). These data suggest that CUGBP1 recognizes mature miRNAs bearing UG-rich sequences. CUGBP1 interacts physically with PARN to stimulate poly(A) shortening of specific mRNAs bearing UG-rich elements (45); therefore, we hypothesized that CUGBP1 specifically recruits PARN to UG-rich miRNAs, including miR-122, miR-93 and miR-652–3p, to promote their deadenylation and destabilization.
Figure 3.
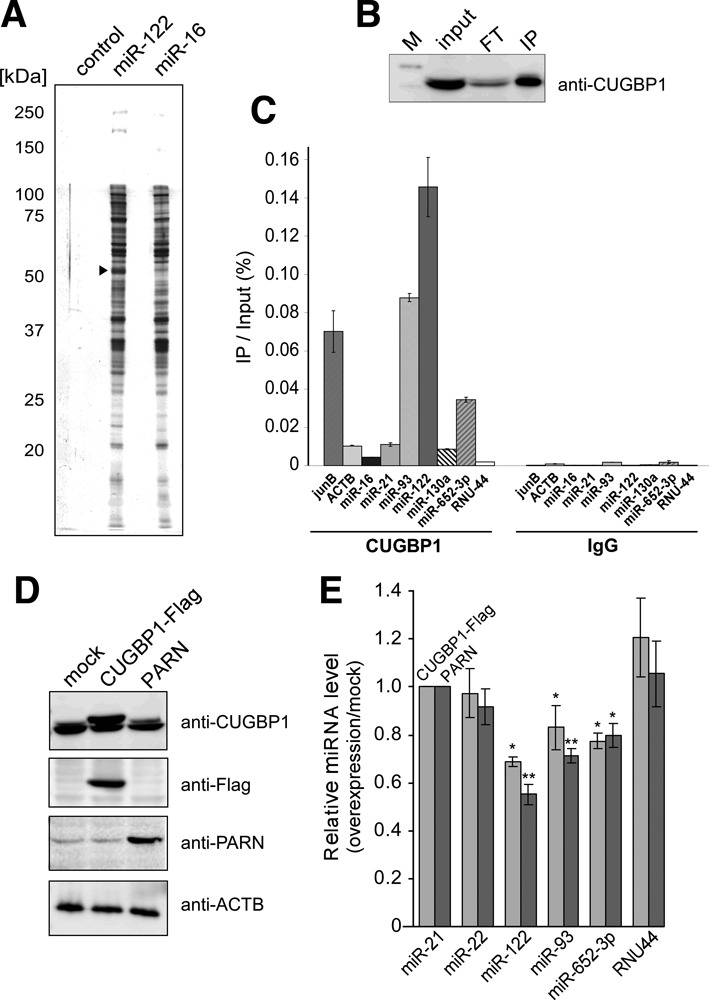
CUGBP1 interacts directly with miR-122. (A) Affinity isolation of miRNA-binding proteins. Proteins from Huh7 cell lysates that bound to control, miR-122- and miR-16-conjugated beads were resolved by SDS-PAGE. The arrowhead indicates CUGBP1 bound specifically to miR-122-conjugated beads. (B) Western blot analysis of immunoprecipitated endogenous CUGBP1 using an anti-CUGBP1 antibody. M, FT and IP indicate the marker, flow through and immunoprecipitate, respectively. (C) Real-time RT-PCR quantifications of mRNAs and miRNAs that co-immunoprecipitated with endogenous CUGBP1 or IgG as a control. The values represent the recovery rates of each mRNA or miRNA that co-immunoprecipitated with endogenous CUGBP1, normalized to the amount of input RNA in the cell lysate. Data are presented as the mean ± SD (n = 3). (D) Confirmation of overexpression of CUGBP1-Flag and PARN in Huh7 cells by immunoblotting using anti-CUGBP1, anti-FLAG and anti-PARN antibodies. Mock-transfected Huh7 cells were used as a negative control. The expression level of ACTB (β-actin) was detected as a loading control. (E) Quantitative real-time RT-PCR analyses of steady-state miRNA levels in Huh7 cells overexpressing CUGBP1 or PARN. The relative miRNA levels are expressed as the ratio of levels in the transfected cells to those in mock transfectants and are normalized to the ratio for miR-21. Data are presented as the mean ± SD (n = 3). *P < 0.05 and **P < 0.01 versus mock-transfected cells by Student's t-tests.
KD of CUGBP1 was not feasible, because siRNAs targeting the mRNA encoding this protein were cytotoxic to Huh7 cells, as reported previously for hepatocellular carcinoma (46) and lung cancer cells (47). Therefore, as an alternative approach, the effect of overexpression of CUGBP1 on miRNA stability was investigated. Huh7 cells were transiently transfected with a Flag-tagged CUGBP1 construct, and overexpression of the protein was confirmed by immunoblotting (Figure 3d). Cells overexpressing CUGBP1 had lower steady-state levels of miR-122, miR-93 and miR-652–3p than mock-transfected cells (Figure 3e), and a similar result was observed when PARN was overexpressed (Figure 3d and e). These results support the concept that CUGBP1 and PARN are involved in the down-regulation of UG-rich miRNAs.
To confirm that CUGBP1 recognizes UG-rich miRNAs specifically, we performed electrophoretic mobility shift assays (EMSAs) in which several 32P-labeled miRNAs were incubated with recombinant CUGBP1 protein (Supplementary Figure S2c). CUGBP1 retarded the motility of miR-122 and its 3′-adenylated species (3′-a and 3′-a5) in a dose-dependent manner, indicating a strong interaction between these species. Similarly, the EMSAs revealed that miR-93 and miR-652–3p also interacted with CUGBP1, albeit with lower efficiency than miR-122, whereas miR-16, miR-21, miR-130a and miR-20a did not (Figure 4a). These results indicate that CUGBP1 binds specifically to miR-122, miR-93 and miR-652–3p in vitro. Titration EMSAs revealed that the equilibrium dissociation constant (Kd) of the interaction between CUGBP1 and miR-122 was 59.3 ± 4.3 nM (Figure 4b). Similarly, the Kd values for miR-93 and miR-652–3p were 275 ± 64 nM and 139 ± 34 nM, respectively (Figure 4c and d). On the other hand, titration EMSAs of miR-20a and miR-130a confirmed that even high concentrations of CUGBP1 did not bind to miR-20a or miR-130a (Supplementary Figure S5a and b). To determine the element in miR-122 recognized by CUGBP1, we constructed four mutants of miR-122 containing different regions of miR-16 (1–4mt, 5–11mt, 12–16mt and 17–22mt), and one miR-122 mutant in which the 3′ region was replaced with oligocytidines (17–22C) (Figure 4e and Supplementary Table S3). The 5–11mt and 17–22C mutants lost the ability to bind to CUGBP1 (Figure 4e), while the other mutants showed weak but detectable interactions, indicating that CUGBP1 recognizes the UG-rich elements at positions 5–11 and 17–22 of miR-122, and that other regions of miR-122 also contribute to its stable interaction with CUGBP1. In addition, CUGBP1 did not bind to pre-miR-122 (Supplementary Figure S6), which is consistent with the fact that CUGBP1 binds to single-stranded RNAs (44).
Figure 4.

CUGBP1 specifically recognizes GU-rich miRNAs. (A) EMSAs to determine the interactions of miR-122 variants and other miRNAs with CUGBP1. 5′-32P-labeled miRNAs (40 nM) were incubated with 0, 80 or 160 nM recombinant CUGBP1. (B–D) Titration EMSAs to determine the Kd values of the interactions of miR-122 (b), miR-93 (c) and miR-652–3p (d) with CUGBP1. 5′-32P-labeled miR-122 (40 nM) was incubated with the indicated concentrations of recombinant CUGBP1. The fraction of bound RNA was determined by measuring the radioactivity of the bound species and dividing by the total radioactivity of the lane. The Kd of the interaction was calculated by non-linear curve fitting using GraphPad Prism software. (E) EMSA to determine the interactions of miR-122 variants with CUGBP1. 5′-32P-labeled miR-122 variants (40 nM) were incubated with 0, 80 or 160 nM recombinant CUGBP1.
CUGBP1 enhances PARN-mediated degradation of miR-122
Next, we tested the hypothesis that CUGBP1 promotes PARN-mediated deadenylation and degradation of miR-122. As expected, the presence of CUGBP1 enhanced PARN-mediated deadenylation and degradation of the 27-nt, 3′-a5 variant of miR-122 in vitro (Figure 5a). Moreover, this effect was dose-dependent (Figure 5b), and PARN-mediated degradation of miR-122 (27-nt, 3′-a5) occurred much faster in the presence of CUGBP1 than in its absence (Figure 5c). We also performed the same experiment in the presence of a 44-nt spike RNA, and confirmed that the PARN-mediated degradation of miR-122 (27-nt, 3′-a5) was still stimulated by the addition of CUGBP1, whereas the stability of the spike RNA did not alter in the presence of CUGBP1 (Supplementary Figure S7a). CUGBP1 also enhanced PARN-mediated degradation of miR-122 (22-nt) without 3′-adenosine (Supplementary Figure S7b), but did not affect the stabilities of miR-21 or the 5–11mt miR-122 mutant, neither of which are recognized by CUGBP1 (Figure 5d and Supplementary Figure S7c). These results provide direct evidence that CUGBP1 binds specifically to miR-122 and promotes its PARN-mediated deadenylation and degradation.
Ago2 protects miRNAs from degradation by PARN
To determine whether PARN-mediated degradation of miRNAs occurs inside or outside miRISCs, the effects of PARN on miR-122 loaded onto immunoprecipitated 5×Flag-Ago2 (Supplementary Figure S2d) were examined in vitro. The isolated Ago2-miR-122 complex was subjected to PARN-mediated degradation in the presence or absence of CUGBP1 (Figure 6a). PARN-mediated degradation of miR-122 loaded onto Ago2 was markedly lower than PARN-mediated degradation of free miR-122, and was not affected by the addition of CUGBP1 (Figure 6a), suggesting that PARN-mediated miRNA degradation takes place outside miRISCs. The degradation of miRNAs outside RISCs by PARN is similar to the XRN-1/XRN-2-mediated degradation of miRNAs in C. elegans (29,30).
Cytoplasmic CUGBP1 localizes partially to P-bodies and interacts with Ago2 in epithelial cells (48). To determine its subcellular localization in Huh7 cells, endogenous CUGBP1 was detected by immunostaining, and Ago2 was used as a P-body marker (Figure 6b). This experiment revealed that a large proportion of CUGBP1 resides in the nucleus and that a part of cytoplasmic CUGBP1 co-localizes with Ago2 in cytoplasmic foci that appear to be P-bodies. To determine whether CUGBP1 and Ago2 interact directly in Huh7 cells, immunoprecipitation was performed using an anti-CUGBP1 antibody, and immunoblotting was performed using an anti-Ago2 antibody. This experiment did not detect an interaction between these proteins (Figure 6c), suggesting that cytoplasmic CUGBP1 resides close to Ago2 in P-bodies in hepatoma cells. This subcellular localization presumably facilitates the efficient recognition of UG-rich miRNAs after their release from Ago2, and the subsequent recruitment of PARN for degradation of the miRNAs.
DISCUSSION
A number of previous studies demonstrated that the stabilities of mature miRNAs are tightly regulated in some cellular contexts. For instance, rapid turnover of several miRNAs occurs in retinal hippocampal and cortical neurons, as well as differentiated neurons in vitro, and rapid decay and increased transcription of miRNAs are linked to neuronal activity (49). Members of the miR-16 family are constitutively unstable in mouse 3T3 cells (50), and active turnover of mature miRNAs by the 5′ to 3′ exonuclease XRN-2 was reported in C. elegans (30). Furthermore, the decapping scavenger enzyme DCS-1 forms a protein complex with XRN-1 to degrade miRNAs that are released from miRISCs (29). Unidentified factors in larval lysates promote the efficient release of miRNAs from miRISCs to expose them for degradation by XRN-1/XRN-2. Although Ago–miRNA complexes are thought to be extremely stable, a recent report suggested that guide RNAs in Ago2 can be dissociated easily upon binding to a highly complementary target RNA (51). After their release from miRISCs, sequence-specific recognition of miRNAs by RNA-binding proteins and ribonucleases is possible. In human cells, miR-328 interacts with and sequesters hnRNP E2 outside miRISCs, thereby releasing the translational repression of the CEPBA mRNA and promoting myeloid cell differentiation (52). Furthermore, miR-29 acts as a decoy that protects the tumor suppressor A20 mRNA from degradation by HuR (53). The results presented here demonstrate that CUGBP1 interacts with mature miR-122 and other UG-rich mature miRNAs outside miRISCs, and recruits PARN to induce their degradation. A recent study demonstrated that PARN is involved in the biogenesis of miR-451; specifically, the 3′ terminal region of Ago2-cleaved pre-miR-451 is trimmed by PARN (54). We here reveal for the first time that PARN degrades subsets of mature miRNAs in concert with CUGBP1.
CUGBP1 is a multifunctional RNA-binding protein that regulates the expression of multiple genes during post-transcriptional processes, including alternative splicing, deadenylation, mRNA decay and translation (44,55). This protein is associated with myotonic dystrophy type 1, which is caused by expanded CUG repeats in the mRNA encoding myotonic dystrophy protein kinase (56). CUGBP1 destabilizes a series of short-lived target mRNAs by binding to GREs in their 3′ UTRs (43), and interacts directly with PARN to stimulate the deadenylation of mRNAs (45). In this study, we identified UG-rich miRNAs as CUGBP1 targets for PARN-mediated destabilization. Based on a previous report that miRNA acts as a decoy (52), it is possible that miR-122 regulates the concentration of free CUGBP1 in hepatocytes and represses CUGBP1 functions, such as GRE-dependent mRNA decay or translational regulation. If CUGBP1 shuttles between the nucleus and cytoplasm, CUGBP1-mediated alternative splicing might also be affected. Because CUGBP1 is expressed at high levels in hepatocytes and has important roles in liver function (55), this observation on the metabolism of miR-122 might contribute in part to current understanding of the functions of CUGBP1 in the liver.
The lengths of the poly(A) tails of mRNAs containing CPEs in their 3′ UTR are regulated by the CPEB complex, and this regulation controls the initiation of translation required for various biological processes (38,39). In the CPEB complex, GLD-2 and PARN have opposing functions in regulating the polyadenylation of mRNAs (40). Similar to cytoplasmic mRNAs, we found that the 3′-terminus of miR-122 is oligoadenylated by GLD-2 and deadenylated by PARN, although the length of its poly(A) tail is much shorter than those of mRNAs. In miR-122, a GRE, rather than a CPE, is recognized by CUGBP1, which recruits PARN. The mechanism by which miR-122 is recognized by GLD-2 in the absence of a CPE remains to be elucidated. Because the results presented here show that RNA-binding proteins and enzymes that associate with the 3′ UTR are involved in miRNA metabolism, there may be proximity effects on sharing or competition for these factors by CPEs and miRNAs bound to their 3′ UTR.
Figure 7a and b show schematic diagrams depicting a proposed model for the regulation of miRNA levels by adenylation and deadenylation. In this model, the 3′-terminus of miR-122 is elongated by GLD-2-mediated oligoadenylation. The oligoadenylated 3′-terminus is concomitantly shortened to generate 22-nt and short variants by PARN-mediated deadenylation. This process is supported by CUGBP1, which recognizes the UG-rich elements at positions 5–11 and 17–22 of miR-122 (Figure 7a). Considering that CUGBP1 has three RNA-recognition motifs (RRMs) (57), the two UG-rich elements in miR-122 seem to be recognized by different RRMs, separately. The full-length sequence of miR-122 was required for stable interaction with CUGBP1 (Figure 4e), suggesting that the CUGBP1-miR-122 complex is destabilized once degradation is initiated. Although PARN preferentially degrades RNAs bearing a 3′-adenosine, it does have exonucleolytic activity towards other nucleotides (41); therefore, it is likely that the 22-nt variant of miR-122 lacking this 3′ nucleotide is also targeted by PARN. The results of the PARN-mediated degradation assay described in this study support the hypothesis that PARN has 3′ to 5′ exonuclease activity towards miRNAs; however, the involvement of other 3′ to 5′ exonucleases that work alongside PARN cannot be excluded. In fact, our data show that the reduced level of miR-122 upon GLD-2 KD was slightly restored when both GLD-2 and PARN were silenced simultaneously (Figure 2a), implying the contribution of other 3′ to 5′ exonucleases to regulate the miR-122 level. In addition, KD of some major deadenylases decreased the steady-state level of miR-122 (Supplementary Figure S4). Since these deadenylases are involved in global gene expression, the regulation of miRNA levels might be more complicated. Considering that Ago2 preferentially incorporates 21–23 nt miRNAs into miRISCs, miRNAs shorter than 21-nt may not be loaded into the complexes effectively and would therefore degrade rapidly (Figure 7b). This model explains why disruption of GLD-2 destabilized not only the 23-nt (3′-a) variant of miR-122, but all other variants also (23), and why inactivation of PARN resulted in the accumulation of all variants with altered expression profiles (Table 1, Supplementary Table S1, and Figure 1a). To support this model, we hypothesize that an active flux of mature miRNAs occurs between miRISCs and other components such as CUGBP1-PARN complexes. Mature miRNAs released from miRISCs are trapped by CUGBP1 for degradation or 3′-trimming; however, on some occasions, mature miRNAs may be reloaded into miRISCs. Recent reports showing that chemically modified single-stranded RNAs can form active RISCs with potent RNA interference activity suggest that direct loading of mature miRNAs into miRISCs is possible (58).
Figure 7.

Mechanistic insights into miRNA metabolism mediated by 3′ oligoadenylation and deadenylation. (A) Selective adenylation and deadenylation of miR-122 by GLD-2, PARN and CUGBP1. GLD-2 specifically stabilizes miR-122 by 3′-oligoadenylation. CUGBP1 recognizes the UG-rich region of miR-122 and recruits PARN onto the 3′-end of the miRNA. (B) Proposed model for the selective stabilization and degradation of miR-122 and other UG-rich miRNAs by 3′-oligoadenylation and deadenylation. In the cytoplasm, pre-miRNA is processed into the miRNA duplex by Dicer. After unwinding of the miRNA duplex, the 3′-terminus of the miRNA (22-nt) is elongated by GLD-2-mediated oligoadenylation and concomitantly shortened by PARN/CUGBP1-mediated deadenylation. PARN-mediated deadenylation takes place outside miRISCs. The balance between oligoadenylation and deadenylation determines the steady-state level of miR-122. In this model, the profile of miR-122 variants is determined by the fact that miRNAs of 21–23 nt in length are preferentially loaded onto Ago2 in the miRISC machinery.
In summary, a delicate balance between GLD-2-mediated oligoadenylation and PARN/CUGBP1-mediated deadenylation and degradation determines the variant profiles and steady-state levels of miR-122.
Supplementary Material
Acknowledgments
We are grateful to the members of the Suzuki laboratory, especially Yuriko Sakaguchi, Kenjyo Miyauchi, Takeo Suzuki and Hiroki Ueda, for providing materials, technical assistance and fruitful discussions. Special thanks are also due to Dr Shin-ichi Hoshino (Nagoya City University, Japan) and Dr Andres Virtanen (Uppsala University, Sweden) for providing materials.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan (to T.S.); New Energy and Industrial Technology Development Organization (NEDO) (to T.S.). Funding for open access charge: Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 4.Kim Y.K., Heo I., Kim V.N. Modifications of small RNAs and their associated proteins. Cell. 2010;143:703–709. doi: 10.1016/j.cell.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 5.Thornton J.E., Chang H.M., Piskounova E., Gregory R.I. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA. 2012;18:1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang H.M., Triboulet R., Thornton J.E., Gregory R.I. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497:244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heo I., Joo C., Kim Y.K., Ha M., Yoon M.J., Cho J., Yeom K.H., Han J., Kim V.N. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Hagan J.P., Piskounova E., Gregory R.I. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones M.R., Quinton L.J., Blahna M.T., Neilson J.R., Fu S., Ivanov A.R., Wolf D.A., Mizgerd J.P. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat. Cell Biol. 2009;11:1157–1163. doi: 10.1038/ncb1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rau F., Freyermuth F., Fugier C., Villemin J.P., Fischer M.C., Jost B., Dembele D., Gourdon G., Nicole A., Duboc D., et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat. Struct. Mol. Biol. 2011;18:840–845. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- 11.Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren G., Chen X., Yu B. Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr. Biol. 2012;22:695–700. doi: 10.1016/j.cub.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y., Yu Y., Zhai J., Ramachandran V., Dinh T.T., Meyers B.C., Mo B., Chen X. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol. 2012;22:689–694. doi: 10.1016/j.cub.2012.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park W., Li J., Song R., Messing J., Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohara T., Sakaguchi Y., Suzuki T., Ueda H., Miyauchi K. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 16.Saito K., Sakaguchi Y., Suzuki T., Siomi H., Siomi M.C. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirino Y., Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 18.Horwich M.D., Li C., Matranga C., Vagin V., Farley G., Wang P., Zamore P.D. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 19.Ameres S.L., Horwich M.D., Hung J.H., Xu J., Ghildiyal M., Weng Z., Zamore P.D. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 21.Hu J., Xu Y., Hao J., Wang S., Li C., Meng S. MiR-122 in hepatic function and liver diseases. Protein Cell. 2012;3:364–371. doi: 10.1007/s13238-012-2036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 23.Katoh T., Sakaguchi Y., Miyauchi K., Suzuki T., Kashiwabara S., Baba T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burroughs A.M., Ando Y., de Hoon M.J., Tomaru Y., Nishibu T., Ukekawa R., Funakoshi T., Kurokawa T., Suzuki H., Hayashizaki Y., et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010;20:1398–1410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D’Ambrogio A., Gu W., Udagawa T., Mello C.C., Richter J.D. Specific miRNA stabilization by Gld2-catalyzed monoadenylation. Cell Rep. 2012;2:1537–1545. doi: 10.1016/j.celrep.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burns D.M., Richter J.D. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 2008;22:3449–3460. doi: 10.1101/gad.1697808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burns D.M., D’Ambrogio A., Nottrott S., Richter J.D. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature. 2011;473:105–108. doi: 10.1038/nature09908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramachandran V., Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosse G.D., Ruegger S., Ow M.C., Vasquez-Rifo A., Rondeau E.L., Ambros V.R., Grosshans H., Simard M.J. The decapping scavenger enzyme DCS-1 controls microRNA levels in Caenorhabditis elegans. Mol. Cell. 2013;50:281–287. doi: 10.1016/j.molcel.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chatterjee S., Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 31.Das S.K., Sokhi U.K., Bhutia S.K., Azab B., Su Z.Z., Sarkar D., Fisher P.B. Human polynucleotide phosphorylase selectively and preferentially degrades microRNA-221 in human melanoma cells. Proc. Natl Acad. Sci. U.S.A. 2010;107:11948–11953. doi: 10.1073/pnas.0914143107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bail S., Swerdel M., Liu H., Jiao X., Goff L.A., Hart R.P., Kiledjian M. Differential regulation of microRNA stability. RNA. 2010;16:1032–1039. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee M., Choi Y., Kim K., Jin H., Lim J., Nguyen T.A., Yang J., Jeong M., Giraldez A.J., Yang H., et al. Adenylation of Maternally Inherited MicroRNAs by Wispy. Mol. Cell. 2014;56:696–707. doi: 10.1016/j.molcel.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyauchi K., Ohara T., Suzuki T. Automated parallel isolation of multiple species of non-coding RNAs by the reciprocal circulating chromatography method. Nucleic Acids Res. 2007;35:e24. doi: 10.1093/nar/gkl1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki T., Ikeuchi Y., Noma A., Sakaguchi Y. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 2007;425:211–229. doi: 10.1016/S0076-6879(07)25009-8. [DOI] [PubMed] [Google Scholar]
- 36.Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 37.Wang L., Eckmann C.R., Kadyk L.C., Wickens M., Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- 38.Richter J.D. CPEB: a life in translation. Trends Biochem. Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 39.Barnard D.C., Ryan K., Manley J.L., Richter J.D. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 40.Kim J.H., Richter J.D. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 41.Henriksson N., Nilsson P., Wu M., Song H., Virtanen A. Recognition of adenosine residues by the active site of poly(A)-specific ribonuclease. J. Biol. Chem. 2010;285:163–170. doi: 10.1074/jbc.M109.043893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gatfield D., Le Martelot G., Vejnar C.E., Gerlach D., Schaad O., Fleury-Olela F., Ruskeepaa A.L., Oresic M., Esau C.C., Zdobnov E.M., et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vlasova I.A., Tahoe N.M., Fan D., Larsson O., Rattenbacher B., Sternjohn J.R., Vasdewani J., Karypis G., Reilly C.S., Bitterman P.B., et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dasgupta T., Ladd A.N. The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscip. Rev. RNA. 2012;3:104–121. doi: 10.1002/wrna.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moraes K.C., Wilusz C.J., Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y., Huang H., Yuan B., Luo T., Li J., Qin X. Suppression of CUGBP1 inhibits growth of hepatocellular carcinoma cells. Clin. Invest. Med. 2014;37:E10–18. doi: 10.25011/cim.v37i1.20864. [DOI] [PubMed] [Google Scholar]
- 47.Wu L.N., Xue Y.J., Zhang L.J., Ma X.M., Chen J.F. Si-RNA mediated knockdown of CELF1 gene suppressed the proliferation of human lung cancer cells. Cancer Cell Int. 2013;13:115. doi: 10.1186/1475-2867-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu T.X., Rao J.N., Zou T., Liu L., Xiao L., Ouyang M., Cao S., Gorospe M., Wang J.Y. Competitive binding of CUGBP1 and HuR to occludin mRNA controls its translation and modulates epithelial barrier function. Mol. Biol. Cell. 2013;24:85–99. doi: 10.1091/mbc.E12-07-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krol J., Busskamp V., Markiewicz I., Stadler M.B., Ribi S., Richter J., Duebel J., Bicker S., Fehling H.J., Schubeler D., et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 50.Rissland O.S., Hong S.J., Bartel D.P. MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol. Cell. 2011;43:993–1004. doi: 10.1016/j.molcel.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De N., Young L., Lau P.W., Meisner N.C., Morrissey D.V., MacRae I.J. Highly complementary target RNAs promote release of guide RNAs from human Argonaute2. Mol. Cell. 2013;50:344–355. doi: 10.1016/j.molcel.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eiring A.M., Harb J.G., Neviani P., Garton C., Oaks J.J., Spizzo R., Liu S., Schwind S., Santhanam R., Hickey C.J., et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balkhi M.Y., Iwenofu O.H., Bakkar N., Ladner K.J., Chandler D.S., Houghton P.J., London C.A., Kraybill W., Perrotti D., Croce C.M., et al. miR-29 acts as a decoy in sarcomas to protect the tumor suppressor A20 mRNA from degradation by HuR. Sci. Signal. 2013;6:ra63. doi: 10.1126/scisignal.2004177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoda M., Cifuentes D., Izumi N., Sakaguchi Y., Suzuki T., Giraldez A.J., Tomari Y. Poly(A)-specific ribonuclease mediates 3′-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013;5:715–726. doi: 10.1016/j.celrep.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones K., Timchenko L., Timchenko N.A. The role of CUGBP1 in age-dependent changes of liver functions. Ageing Res. Rev. 2012;11:442–449. doi: 10.1016/j.arr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Timchenko L.T., Miller J.W., Timchenko N.A., DeVore D.R., Datar K.V., Lin L., Roberts R., Caskey C.T., Swanson M.S. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edwards J.M., Long J., de Moor C.H., Emsley J., Searle M.S. Structural insights into the targeting of mRNA GU-rich elements by the three RRMs of CELF1. Nucleic Acids Res. 2013;41:7153–7166. doi: 10.1093/nar/gkt470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lima W.F., Prakash T.P., Murray H.M., Kinberger G.A., Li W., Chappell A.E., Li C.S., Murray S.F., Gaus H., Seth P.P., et al. Single-Stranded siRNAs Activate RNAi in Animals. Cell. 2012;150:883–894. doi: 10.1016/j.cell.2012.08.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


