Abstract
The mer operon confers bacterial resistance to inorganic mercury (Hg2+) and organomercurials by encoding proteins involved in sensing, transport and detoxification of these cytotoxic agents. Expression of the mer operon is under tight control by the dual-function transcriptional regulator MerR. The metal-free, apo MerR binds to the mer operator/promoter region as a repressor to block transcription initiation, but is converted into an activator upon Hg2+-binding. To understand how MerR interacts with Hg2+ and how Hg2+-binding modulates MerR function, we report here the crystal structures of apo and Hg2+-bound MerR from Bacillus megaterium, corresponding respectively to the repressor and activator conformation of MerR. To our knowledge, the apo-MerR structure represents the first visualization of a MerR family member in its intact and inducer-free form. And the Hg2+-MerR structure offers the first view of a triligated Hg2+-thiolate center in a metalloprotein, confirming that MerR binds Hg2+ via trigonal planar coordination geometry. Structural comparison revealed the conformational transition of MerR is coupled to the assembly/disassembly of a buried Hg2+ binding site, thereby providing a structural basis for the Hg2+-mediated functional switching of MerR. The pronounced Hg2+-induced repositioning of the MerR DNA-binding domains suggests a plausible mechanism for the transcriptional regulation of the mer operon.
INTRODUCTION
Bacteria thriving in heavy metal-containing environments rely on the activities of metal-specific resistance systems for survival. While different heavy metals exhibit distinct physical-chemical properties, the majority of these detoxification systems operate on a simple mechanism, consisting of a plasma membrane-embedded efflux transporter for removing its target metal ion from the cytoplasm, and a metal-responsive transcriptional regulator for controlling transporter expression (1,2). In this regard, the bacterial mercury resistance operon (mer operon) appears exceptionally delicate and sophisticated by harboring genes involved in the sensing, capture, transport and transformation of inorganic mercury (Hg2+) and organomercurials (3). Being located on a transposable element, the mer operon is widely distributed among Gram-positive and Gram-negative bacteria, and its expression effectively protects the hosts against high-dose mercury exposure (4–6). Key components encoded by the Gram-negative mer operon include MerC, MerP and MerT for recognition and uptake of Hg2+, one (or more) mercurial lyase MerB that cleaves carbon-mercury bond in organomercurials to yield Hg2+, a cytosolic mercuric reductase MerA that catalyzes an NADPH-dependent reduction of Hg2+ into the volatile and less toxic elemental Hg0 (3,7) and a transcriptional regulatory protein MerR that coordinates the expression of the operon (8–10).
With a capacity to undergo an Hg2+-dependent functional switching from a transcriptional repressor to an activator, MerR allows the expression of the mer operon to be tightly regulated (11–14). Unlike typical bacterial promoters whose −10 and −35 elements are spaced by 17±1 base pairs to favor a productive interaction with RNA polymerase (15,16), the mer promoter contains a longer (19∼20 base pairs) spacer (17,18), which reduces the basal transcription activity by misaligning the two polymerase-binding elements. Binding of the metal-free, repressor form of MerR to the operator sequence located between the −10 and −35 elements further represses transcription by interfering with open complex formation at the mer promoter. Expression of the mer operon can be stimulated upon the conversion of MerR into a transcriptional activator by Hg2+ (10,14,17,19). Various biochemical and biophysical studies indicate that Hg2+-binding likely triggers a structural change in MerR, in turn affecting the conformation of the bound DNA to facilitate transcription initiation by restoring the alignment between the two polymerase binding elements (14,20–22). Although MerR was first identified over three decades ago, only low-resolution solution structures derived from small-angle X-ray scattering data are currently available (20); its three-dimensional structures have not yet been resolved by either X-ray crystallography or nuclear magnetic resonance (NMR). Therefore, several outstanding questions, such as the structure and residue composition of the Hg2+ binding site, the nature and mechanism of Hg2+-induced conformational change of MerR and the structural basis of Hg2+-dependent transcriptional regulation by MerR, have remained to be addressed in greater detail.
The uniqueness of MerR is further underscored by being a dual-function transcriptional regulator and the founding member of the MerR family of transcription factors (17,23–24). Members of the MerR family are best known for their high sensitivity and selectivity toward environmental stimuli, ranging from heavy metals, xenobiotics, to reactive oxygen species (17). Earlier structural studies of some MerR family members revealed that the DNA and inducer-binding domains are located at the N- and C-terminus, respectively, and the two domains are connected by a long α-helix which also acts as the dimerization helix (25–31). Antiparallel packing of the two dimerization helices produces a functional homodimer for interacting with the pseudo-palindromic operator sequences. Moreover, dimerization allows the DNA-binding domain of one subunit to pair up with the inducer-binding domain of the opposing subunit, resulting in the formation of a composite inducer binding site composed of residues from both subunits. The crystal structures of BmrR and SoxR in complexes with the respective inducer and operator DNA illustrate significantly bending and twisting of the bound DNA, which reorients and shortens the spacing between the −10 and −35 regions to promote transcription initiation (26,29). Despite these progresses, how MerR family members undergo conformational switching upon inducer binding has remained speculative due to the lack of any three-dimensional structures of the apo forms.
In this study, we determined the crystal structures of MerR in both the metal-free as well as the Hg2+-bound forms, which correspond to the repressor and activator state of MerR, respectively. Structural comparison revealed how Hg2+ drives the formation of the metal binding site, and how Hg2+ regulates the conformational switching of MerR. Modeling of the MerR-DNA interactions suggests a structural basis for the Hg2+-regulated expression of the mer operon.
MATERIALS AND METHODS
Protein expression and purification
The construction of the expression plasmid for producing tag-free, full-length MerR from the Gram-positive bacterium Bacillus megaterium MB1 was described previously (32). For expression, the plasmid-harboring Escherichia coli BL21(DE3) was cultured in LB medium at 37°C to OD600 = 0.8, IPTG was added to a final concentration of 1.0 mM, and protein expression was induced at 20°C for 16 h. Bacteria were harvested by centrifugation and stored at −80°C until further use. For purification, bacteria were resuspended in buffer A (50 mM Tris (pH 7.5), 5% glycerol, 10 mM DTT) and the cells were disrupted by sonication. The crude cell lysate was clarified by centrifugation at 27 216 x g for an hour (repeat three times) at 4°C and applied to a pre-equilibrated heparin column. The column was washed to baseline, and the protein was eluted in a linear gradient over 10 column volumes with buffer B (buffer A containing 1 M ammonium sulfate). The eluted fractions were pooled and applied to a phenyl column pre-equilibrated with buffer C (buffer A containing 0.5 M ammonium sulfate). Buffer A was used for elution. Eluted fractions containing MerR were pooled and further purified on a size-exclusion column (Hi-Load Superdex 200) in gel filtration buffer (50 mM Tris (pH 7.5), 250 mM sodium chloride,1 mM PMSF, 5% glycerol, 10 mM DTT). The dimer-form protein was collected and concentrated to 5.0 mg/ml for crystallization. Expression of selenomethionyl MerR was achieved at 20°C after inducing for 16 h with 1.0 mM IPTG in E. coli strain B834(DE3) in M9 medium supplemented with 40 μg/ml seleno-L-methionine.
Protein crystallization
For crystallization of apo-MerR, 1 μl of purified native or selenomethionine-labeled MerR (5 mg/ml in gel filtration buffer) was mixed with an equal amount of reservoir solution (50 mM sodium citrate (pH 6.2), 0.1 M ammonium phosphate) and equilibrated against 200–500 μl of reservoir solution at 4°C. Native apo-MerR crystals of suitable size (∼0.15 × 0.05 × 0.03 mm) can be obtained within two weeks. For crystallization of Hg2+-MerR, 7.8 μl of HgCl2 stock (100 mM) was added into 1.0 ml of purified MerR (5 mg/ml in gel filtration buffer). After 10 min incubation on ice, 1 μl of native or selenomethionine-labeled Hg2+-MerR sample was mixed with an equal amount of reservoir solution (0.2 M ammonium citrate dibasic (pH 5.1), 20% PEG3350) and equilibrated against 200 μl of reservoir solution at 4°C. Native Hg2+-MerR crystals reached full size (∼0.05 × 0.05 × 0.01 mm) in about two weeks. In both cases, crystals of selenomethionine-labeled MerR used for data collection were obtained using the microseeding technique by adding seeds prepared from the respective native crystals to the crystallization drops. Crystals were harvested by transferring into mother liquor containing either 20% 1,6-hexanediol (for apo-MerR) or 25% ethylene glycol (for Hg2+-MerR) for about 10 s before looping and flash-freezing in liquid nitrogen for data collection.
Structure determination
For apo-MerR, a 2-wavelength multiwavelength anomalous dispersion (MAD) data set and a higher resolution native data set were collected at SPring-8 beamline SP12B2. For Hg2+-MerR, a 2-wavelength MAD data set and a single-wavelength anomalous dispersion (SAD) data set with optimized Hg anomalous signal were collected at NSRRC beamlines 13B1 and 15A1, respectively (Table 1). All diffraction data sets were processed using HKL2000 (33). For the MAD data sets, raw images from each wavelength were indexed according to the same crystal orientation matrix, but integrated and scaled independently. Initial MAD phases of the apo-MerR data were determined by SOLVE/RESOLVE at 2.75 Å and extended to 2.61 Å by applying non-crystallographic symmetry (NCS) averaging and solvent flattening implemented in RESOLVE (34–36). A starting model for apo-MerR was obtained with RESOLVE, which then underwent rounds of manual model rebuilding, water assignment, and refinement with Coot (37) and Phenix (38). The phase problem of the Hg2+-MerR structure was solved by a combination of Se-MAD and Hg-SAD at 2.56 Å using Phenix. The resulting electron density maps were of excellent quality and the bound Hg2+ ions were readily added into the initial model produced by Phenix. Coot and Phenix were then used for model rebuilding, water assignment and refinement. For both structures, refinement was conducted by applying torsion NCS and individual B-factor restraints in combination with Translation–Libration–Screw (TLS) refinement, with the parameters being selected based on TLSMD analysis (39). The following residues are omitted from the final refined structures due to disorder. For apo-MerR: 1–3, 34–39, 131–132 in chain A, 1–3, 12, 14, 31–39, 131–132 in chain B, 1–14, 25–43, 132 in chain C, 1–3, 11–14, 31–41, 132 in chain D, 1–5, 34–39, 132 in chain E, 1–4, 32–39, 132 in chain F, 1–7, 32–39, 132 in chain G and 1–7, 13–15, 31–43 in chain H. For Hg2+-MerR: 1–2, 32–34, 41–42, 132 in chain A and 1–2, 33–34, 41–42, 132 in chain B. The refinement statistics are summarized in Table 1. The figures were generated in Pymol (https://www.pymol.org/).
Table 1. Summary of crystallographic analysis for MerR.
| Protein | Se_MerR | apo-MerR | Se_Hg2+-MerR | Hg2+-MerR | ||
|---|---|---|---|---|---|---|
| Space group | P21 | P21 | P222 | P222 | ||
| Unit cell dimensions | ||||||
| a (Å) | 80.63 | 80.54 | 70.00 | 69.89 | ||
| b (Å) | 94.89 | 94.80 | 70.55 | 70.96 | ||
| c (Å) | 86.88 | 87.06 | 71.61 | 71.38 | ||
| Data collection | Se-inflection | Se-remote | Se-inflection | Se-remote | ||
| Wavelength (Å) | 0.9791 | 0.9537 | 1.0000 | 0.9792 | 0.9610 | 0.9224 |
| Resolution (Å) | 30–2.75 | 30–2.75 | 30–2.61 | 30–3.7 | 30–3.7 | 30–2.56 |
| No. of observed reflections | 140,766 | 151,989 | 137,710 | 37,272 | 37,100 | 137,888 |
| Unique reflections | 28,526 | 28,752 | 38,485 | 3.957 | 3.937 | 11,808 |
| Completeness (last shell)a (%) | 85.4(66.0) | 86.1(70.5) | 99.0(99.9) | 97.1(98.6) | 96.9(98.6) | 99.7(98.5) |
| Multiplicity | 5.3 | 5.3 | 3.6 | 9.4 | 9.4 | 11.7 |
| Rsym (last shell)a,b (%) | 8.3(40.5) | 8.7(37.5) | 8.3(40.5) | 21.8(48.9) | 23.2(50.1) | 13(52.1) |
| Refinement | ||||||
| Resolution range (Å) | 30–2.61 | 30–2.56 | ||||
| No. of reflection in working set (test set) | 36,559(1962) | 10630(1178) | ||||
| Rcryst (last shell)c | 22.5(28.5) | 22.5(22.3) | ||||
| Rfree (last shell)c | 25.7(30.7) | 28.7(34.5) | ||||
| Rmsd from ideal | ||||||
| Bond lengths (Å) | 0.003 | 0.011 | ||||
| Bond angle (deg.) | 0.67 | 1.67 | ||||
| Ramachandran outliers (%) | 0 | 0 | ||||
| Number of macromolecules | 7166 | 1943 | ||||
| Ligands | 0 | 2 | ||||
| H2O | 97 | 38 | ||||
| Average B-factor (protein/water/ligand) | 58.4(58.5/50.3/NA) | 39.5(39.6/36.9/37.7) | ||||
aStatistics for data from the resolution shell of 2.85–2.75 Å (Se_apo-MerR data), 2.68–2.61 Å (apo-MerR data), 3.99–3.7 Å (Se_Hg2+-MerR data) and 2.68–2.56 Å (Hg2+-MerR data).
bRsym = (Σ|Ihkl - <I>|)/(ΣIhkl), where the average intensity <I>is taken overall symmetry equivalent measurements and Ihkl is the measured intensity for any given refection.
cRcryst = (Σ∥Fo| - k|Fc∥)/(Σ|Fo|). Rfree = Rcryst for a randomly selected subset (5%) of the data that were not used for minimization of the crystallographic residual.
Structural modeling of the MerR-DNA and Hg2+-MerR-DNA complexes
The structural models of the mer operator DNA in complexes with either apo-MerR or Hg2+-MerR were constructed using Discovery Studio 4.1 (http://accelrys.com/). Initial models of the protein-DNA binary complexes were constructed by superimposing apo-MerR onto the TipAN DNA taken from the TipAN-tipA promoter DNA complex (PDBid: 2VZ4) and Hg2+-MerR onto the SoxR-soxS promoter DNA complex (26), followed by manual adjustments of the DNA backbone conformations to optimize the shape complementarity and contacting surface between DNA and the recognition helices of MerR. Energy minimization using CHARMm with harmonic restraints was then carried out to improve the stereochemical quality of the models. The flanking, non-operator DNA segments which extend toward the −35 and −10 elements were modeled to adopt a standard B-DNA conformation using the ‘Marcomolecules module’ implemented in Discovery Studio.
RESULTS AND DISCUSSION
Structure determination of apo and Hg2+-bound MerR
To understand how MerR exhibits a high specificity toward Hg2+ and how Hg2+-binding converts MerR from a transcriptional repressor to an activator, we determined the crystal structures of MerR in the absence or presence of Hg2+, which correspond respectively to the repressed and activated state of MerR. Due to the expected structural similarity among the metal-binding subset of the MerR family members, it was anticipated that molecular replacement might be a feasible method for structure determination. However, in this study both the metal-free MerR (apo-MerR) and Hg2+-bound MerR (Hg2+-MerR) structures were solved by the MAD method using selenomethionine-labeled MerR crystals to acquire unbiased structural information regarding the Hg2+ binding sites and Hg2+-induced protein structural rearrangement. Initial MAD phases for apo-MerR and Hg2+-MerR were obtained to 2.75 and 3.7 Å resolution and subsequently extended to 2.61 and 2.56 Å using higher resolution native data sets. The apo-MerR crystallized with eight protomers per asymmetric unit arranged into four non-crystallographic symmetry related dimers, providing multiple crystallographically independent views of apo-MerR in monomeric as well as dimeric forms (Supplementary Figure S1). For the Hg2+-MerR crystal, the asymmetric unit consists of one dimer with both metal binding sites fully occupied by Hg2+ (Supplementary Figure S2). Data collection and structural determination statistics are summarized in Table 1.
Overall structure of apo-MerR and Hg2+-bound MerR
The three-dimensional structures of the metal-responsive MerR family members ZntR and CueR had been characterized in the presence of their cognate metal ions (28). In contrast, no corresponding apo structures are currently available in the PDB. Even for the drug-binding and redox-sensing subset of MerR family members (25–27,29–30), the apo structures were determined with the C-terminal inducer-binding motif being either absent or truncated (PDBids: 3GPV, 3HH0 and 2VZ4). To our knowledge, the apo-MerR structure reported here provides the first visualization of a MerR family of transcriptional regulator in its intact repressor form. Together with the Hg2+-MerR structure, which represents the transcriptionally active state of MerR, makes it suitable to delineate the molecular basis of Hg2+-induced structural and functional transition of MerR in detail.
Similar to the previously determined structures of CueR, ZntR and SoxR (26,28), the secondary structure connectivity ‘H1-H2-β1-β2-H3-H4-H5-(H6)-H7’ is largely preserved in MerR, except that H2 and H3 is connected by a hairpin-like loop structure. The MerR N-terminal fragment (H1 through H4) adopts a DNA-binding winged helix-turn-helix fold in which H2 functions as the recognition helix (19,21), followed by a long dimerization helix (H5) and a C-terminal metal binding motif (H6–H7) (Figure 1) (17,19,40). Two MerR subunits associate to form a stable homodimer via an antiparallel coiled-coil packing between the two H5 helices (Figure 2). Dimerization is essential for MerR to exert its regulatory function by defining the spacing and orientation between the two DNA-binding domains, which allows MerR to interact with the pseudo-palindromic mer operator sequence as well as modulate the operator DNA conformation upon binding. Moreover, dimerization leads to the assembly of two composite and structurally equivalent Hg2+ binding sites; each site is consisted of three strictly conserved Hg2+-ligating cysteine residues (Supplementary Table S1), with Cys79 originating from one subunit and Cys114 and Cys123 from the other subunit. The juxtaposition between the DNA-binding domain and the Hg2+ binding site (Supplementary Figure S3A) appears well suited for mediating metal-induced repositioning of the DNA-binding domain. Indeed, structural comparison of apo- and Hg2+-MerR revealed extensive Hg2+-induced rearrangement of the metal and DNA-binding modules (Supplementary Figure S3B). A detailed description of this metal-dependent conformational change and the possible causes and functional significance of the structural transition will be provided in the following sections.
Figure 1.
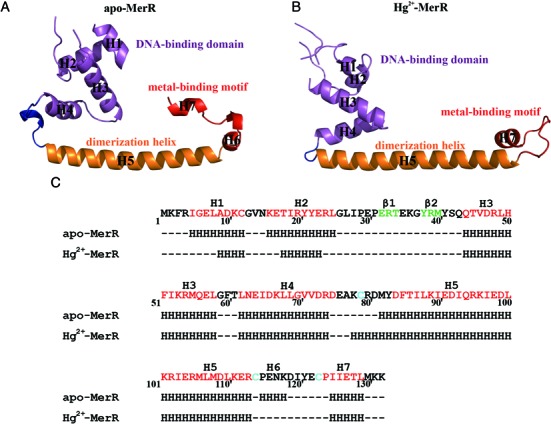
Secondary and tertiary structures of apo and Hg2+-bound MerR. Ribbon representations of the MerR protomer in apo (A) and Hg2+-bound (B) states. For both structures, the DNA-binding domain, dimerization helix and metal-binding motif are in purple, orange and red, respectively. The H4–H5 loop is highlighted in blue. The apo and Hg2+-bound MerR structures were first aligned by superimposing on the backbone atoms of the dimerization helices (H5) before being displayed in separate panels. (C) Secondary structures of MerR in the apo and Hg2+-bound state. The Hg2+-ligating cysteine residues are marked cyan in the MerR sequence. The α-helices and β-strands suggested by the PredictProtein server (https://www.predictprotein.org/) are colored red and green in the sequence for comparison.
Figure 2.
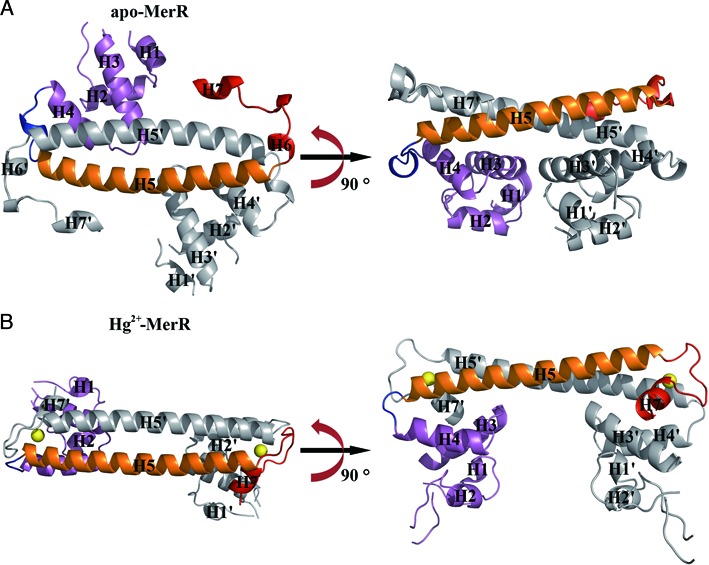
Overall structures of apo and Hg2+-bound MerR homodimer. Ribbon representations of the MerR homodimer in apo (A) and Hg2+-bound (B) state. Two orthogonal views of each structure related by a 90° rotation about the coiled-coil dimerization region are provided. For both structures, the DNA-binding domain, dimerization helix and metal-binding motif of one subunit are in purple, orange and red, respectively, the second subunit is in gray. The H4–H5 loop is highlighted in blue. The two metal ions (Hg2+) in Hg2+-MerR are shown as yellow spheres. Labels belonging to the second protein subunit are flagged by a prime. The apo and Hg2+-bound MerR structures were first aligned by superimposing on the backbone atoms of the dimerization helices (H5 and H5’) before being displayed in separate panels.
MerR undergoes extensive secondary, tertiary and quaternary structural changes upon Hg2+ binding
Consistent with the notion that MerR undergoes Hg2+-induced functional switching from a transcription repressor to an activator (11–13,20–21,41), pronounced differences in secondary, tertiary and quaternary structures were recognized between apo-MerR and Hg2+-MerR (Figures 1 and 2). Specifically, compared to the apo structure, in Hg2+-MerR the N-terminus of H5 where Cys79 resides extends by one helical turn, the H6 which flanks Cys114 undergoes a helix-to-loop transition and residues adjacent to Cys123 form an alternative loop structure by adopting a new set of main-chain dihedral angles. The lengthening of H5 upon Hg2+-binding is further accompanied by the straightening of the helical axis, increasing the surface area of the dimer interface from 1794 to 2153 Å2 (calculated with the PISA server, http://www.ebi.ac.uk/pdbe/pisa/). Additional changes in the MerR tertiary and quaternary structure include a rigid body movement of the DNA-binding domain and significant relocation of the metal-binding motif with respect to the H5 coiled-coil dimerization domain (Figure 1 and Supplementary Figure S4). As a result, the spacing and orientation between the two DNA-binding domains are significantly altered (Supplementary Figure S5), and the two MerR conformers would interact differently with the mer operator DNA (Supplementary Figure S6).
Most unexpectedly, in apo-MerR the Hg2+-ligating cysteine residues are located apart from each other and not suitable for coordinating simultaneously to Hg2+ (Figure 3A). In contrast, these residues become juxtaposed in Hg2+-MerR with the three side-chain thiolate groups being triangularly arranged around the bound Hg2+ (Figure 3B). Since the formation of metal binding site is strictly Hg2+-dependent, it can be inferred that the conformational transition of MerR is coupled to the assembly of the Hg2+ binding site, and that the presence or absence of Hg2+ would specify the structural state of MerR.
Figure 3.
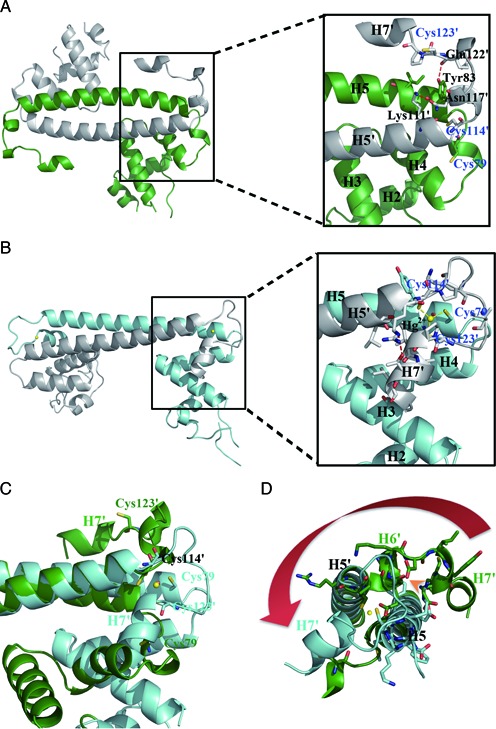
Detailed views of the Hg2+-induced movement of the metal-binding motif and assembly of the Hg2+ binding site. (A) Ribbon representation of apo-MerR homodimer (left) with an enlarged view (right) showing residues involved in the anchoring of the metal-binding motif as well as the spatial arrangement of the three Hg2+-ligating cysteine residues. (B) Ribbon representation of Hg2+-MerR homodimer (left) with an enlarged view (right) showing the coordination of Hg2+ (yellow sphere) by three cysteine residues and the involvement of H7 in Hg2+-binding. (C) Superimposition of apo-MerR (green) and Hg2+-MerR (cyan) shows that H7 and the three Hg2+-ligating cysteine residues undergo large repositioning upon Hg2+-binding. (D) The Hg2+-induced lengthening of H5 would result in steric clashes with the H5–H6 loop and H6 seen in apo-MerR structure (indicated by the orange arrowhead), causing conformational change in the metal-binding motif (highlighted by the red arrow). Labels belonging to the second protein subunit are flagged by a prime.
Superposition of the two structures revealed a plausible molecular mechanism by which the tertiary and quaternary conformational changes of MerR can be triggered upon Hg2+ binding (Figure 3C). Specifically, among the three Hg2+-ligating cysteine residues, Cys79 and Cys114’ (the ‘prime’ symbol specifies a residue from the other subunit) are more closely spaced (∼9.2 Å) and most likely responsible for mediating the initial engagement with Hg2+ (Figure 3A). Compared to Cys114’, which is structurally more constrained by being attached to the coiled-coil dimerization module, Cys79 is located in the more flexible H4–H5 loop in apo-MerR and may thus be more suitable to relocate, such as moving toward Cys114’ (Figure 3C). The resultant presentation of two thiolate groups in proximity is expected to facilitate the capture of incoming Hg2+ via the formation of a transient bidentate binding site, likely with a linear coordination geometry observed in the MerP-Hg2+ complex (42,43). The H4–H5 loop may then undergo a metal-induced loop-to-helix transition to adopt the conformation seen in Hg2+-MerR, extending the N-terminus of H5 by an extra helical turn. In the context of the apo-MerR dimer, the elongated H5 would clash with the N-terminal portion (H6 and H5–H6 loop) of the metal-binding motif from the opposing MerR subunit and cause its detachment from the coiled-coil module, thus allowing Cys123’ to approach the bound Hg2+ to complete the assembly of the composite and tridentate binding site (Figure 3D).
Because Cys123 is located at the N-terminus of H7, the coordination of this residue to Hg2+ would bring H7 toward the bound metal ion (Figure 3C and D). Being surrounded by three negatively charged thiolate groups, the tricoordinated Hg2+-thiolate center is expected to carry one net negative charge (28), which appears ideal for mediating the positioning of H7 via a charge-helix dipole interaction (Figure 3B). Such an interaction between the positive helix dipole and negatively charged metal-sulfur center has been suggested as a secondary metal-binding component (44). Importantly, the presence of H7 at this new position cannot be accommodated in the apo-MerR structure due to steric clashes with H3 and H4, which explains why the DNA-binding domain must undergo an Hg2+-induced rigid body movement in Hg2+-MerR (Figure 4A and B). The interactions observed between H3 and H7 in Hg2+-MerR indicate that H7, besides its role in driving the Hg2+-induced conformational change, also assists the anchoring of the DNA-binding domain (Figure 4C). Taken together, the structural rearrangement of the metal-binding motif not only enables the assembly of Hg2+ binding site but also triggers the functional switching of MerR by reorienting the DNA-binding domain. The identification of two distinct conformational states of MerR also allows the functional consequences of known MerR mutants (45–47) to be explained in structural terms (Supplementary Table S1).
Figure 4.
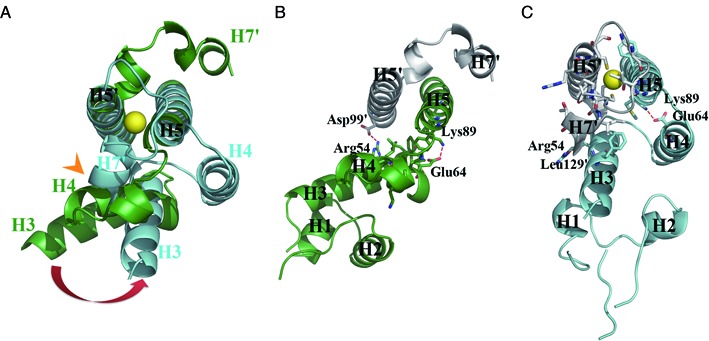
The DNA-binding domains of MerR homodimer undergo Hg2+-induced repositioning. (A) Superimposition of apo-MerR (green) and Hg2+-MerR (cyan) shows the Hg2+-thiolate-tethered H7 in Hg2+-MerR would clash with H3 and H4 seen in apo-MerR (indicated by the orange arrowhead), which is expected to trigger the Hg2+-induced relocation of the DNA-binding domain. The DNA-binding domain is anchored by distinct set of stabilization interactions in apo-MerR (B) and Hg2+-MerR (C). Yellow sphere represents the bound Hg2+.
Structure of the Hg2+ binding site and structural basis of metal selectivity
While sequence analysis and mutagenesis studies of MerR have long established that each Hg2+ binding site is composed of three strictly conserved cysteine residues (28,31,48–50), it has remained unresolved whether the three metal-ligating residues are arranged in trigonal planar, trigonal pyramid or T-shaped coordination geometry. NMR analysis of MerR in complex with a stable magnetic mercury isotope (199Hg) indicated that, compared to the pyramidal coordination geometry, the trigonal planar model appears more consistent with the observed chemical shift values (50). By determining the crystal structure of Hg2+-MerR, we provide unequivocal evidence that MerR binds Hg2+ via trigonal planar coordination geometry and offer the first visualization of tricoordinated Hg2+ in a metalloprotein in two crystallographically independent views (Figure 5A).
Figure 5.
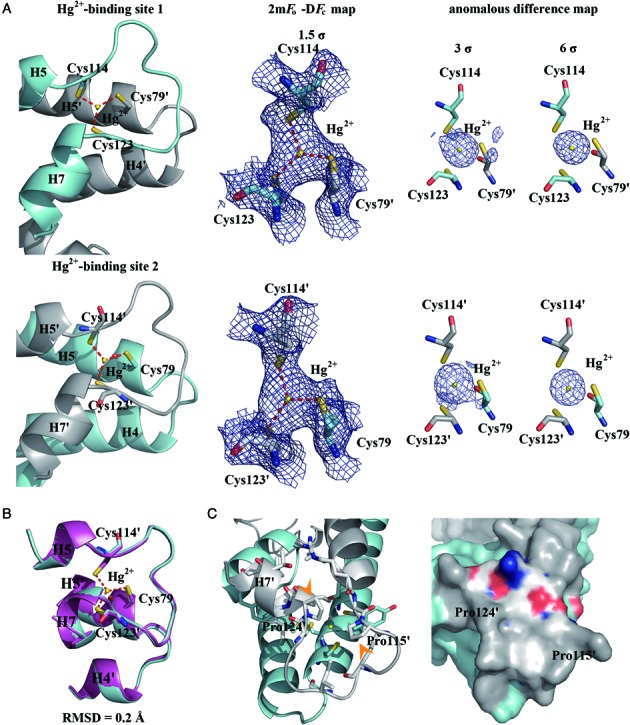
MerR binds Hg2+ in a trigonal planar coordination geometry. (A) Structures and electron density map of the two crystallographically independent Hg2+ binding sites in the MerR homodimer; each Hg2+ (yellow sphere) is coordinated by three cysteine residues to form a trigonal-planar Hg2+-thiolate complex. The averaged Hg2+-S bond length (red dashed lines) is ∼2.4 Å. The 2mFo-DFc map (middle, purple meshes) and the anomalous difference maps (right, purple spheres) are contoured at 1.5 and 3/6 σ, respectively, above mean level. (B) The two Hg2+ binding sites are structurally similar and can be superimposed with an RMSD of 0.2 Å over all equivalent atom pairs. (C) Being surrounded by the pyrrolidine rings of Pro115 and Pro124 (indicated by the orange arrowheads), H3, H4, H7 and H5-H7 loop, the Hg2+-thiolate center is fully buried inside MerR and thus shielded from the solvent. Except for the N-terminus of H7, all other second shell Hg2+-contacting atoms are predominantly hydrophobic.
The structures of the two Hg2+ binding sites are essentially identical; superimposition of equivalent residues constituting or flanking the binding site (residues 69∼85 and 109∼128) produced an RMSD of ∼0.2 Å (Figure 5B). The 3-fold symmetry specified by the trigonal planar coordination geometry applies to the positions of the three Hg2+-chelating thiolate groups and the S-Hg2+ bond lengths (all refined to ∼2.4 Å), but not the thiolate-bearing Cα atoms and beyond owing to the unique set of main-chain and side-chain dihedral angles adopted by each metal-ligating cysteine (Supplementary Table S2). Unlike MerR family members ZntR and SoxR in which the metal ions are located in partially exposed environments (26,28), in MerR the bound Hg2+ is fully buried and secluded from solvent by having the pyrrolidine rings (of Pro115’ and Pro124’) and helices (H4 and H5) blocking the axial faces and the N-terminus of H7 covering the side (Figure 5C). Similar arrangement of proline residues and helices around the bound metal ion is reminiscent of the Cu+ binding site of CueR (28). Since the N-terminus of H7 is the only polar element around the negatively charged trithiolate-Hg2+ center, the charge-helix dipole interaction observed in MerR and CueR may be further strengthened by the surrounding non-polar groups, thus fixing H7’ in position to drive the relocation of the DNA-binding domain.
Earlier studies measuring the MerR-dependent mer promoter activity showed that nanomolar level of Hg2+ is sufficient to activate transcription, whereas much higher concentrations of other metal ions (such as Cd2+, Zn2+, Au+, Au3+, Cu+, Cu2+) are required to produce even marginal activation (51). Theoretical calculations suggested that the high sensitivity and selectivity of MerR toward Hg2+ may be explained in part by the valance state, ionic radius and charge-accepting ability of the metal ion, as well as the net charge, charge-donating ability, dipole moment, polarizability and number of the metal-ligating atoms (44). Besides these physical-chemical considerations, the involvement of protein milieu has also been implicated, with the number of metal-ligating residues, environment of the metal binding site and length of the metal-binding motif being the potential determinants of binding specificity (28,52–53). Therefore, the preference of MerR in forming a buried and tricoordinated metal-binding center may favor the binding of Hg2+, which differs from the formation of bidentate Cu+ in CueR and binuclear metal clusters in ZntR and SoxR (26,28). Moreover, structural comparison between Hg2+-MerR and Cu+-bound CueR explains the length variation in the metal-binding loop (Supplementary Figure S7A), as measured by the spacing between the corresponding metal-ligating residues (Cys114 and Cys123 for MerR; Cys112 and Cys120 for CueR). The 7-residue spacer of CueR appears ideal to enclose the bithiolate-Cu+ center but cannot accommodate the bulkier trithiolate-Hg2+ center. In contrast, the 8-residue spacer of MerR favors the presence of tridentate Hg2+ by allowing the loop to bulge around the extra Hg2+-ligating residue (Cys79), such that potential steric clashes between Cys79 and Pro115’ can be avoided. A hydrogen bond formed between the ϵ-amino group of a MerR-specific and highly conserved lysine residue (Lys118’) and the main-chain carbonyl of Pro115’ may indirectly contribute to Hg2+-binding by stabilizing the bulge conformation (Supplementary Figure S7B–D). Although the metal binding site of MerR can sterically admit Cu+, the binding affinity may be significantly reduced due to the loss of tight packing around the metal center, which would disrupt the hydrophobic environment of the binding pocket and consequently destabilize the charge-dipole interaction with H7. The corresponding loops are even longer in ZntR and SoxR, hence extra metal-ligating residues (Cys and His) can be introduced to form the binuclear metal centers. Our work thus implicates the structural basis for the correlation between the length of metal-binding loop and the metal ion selectivity.
Functional implications for the MerR from Gram-negative bacteria and the prospective transcriptional regulator MerR2 of TnMERI1
The Bacillus MerR structures reported here suggest how the relocation of its DNA-binding domain is coupled to the formation of the Hg2+ binding site, and how the DNA-binding domain can be differentially oriented about the coiled-coil dimerization interface (Figure 2). Multiple sequence alignment of MerRs from Gram-positive and Gram-negative bacteria shows that residues involve in anchoring the DNA-binding domain, as seen in the apo and Hg2+-MerR structures, are largely conserved (Supplementary Figure S8). Thus, it appears that, regardless of their origin, the DNA-binding domain of all MerRs can adopt two spatially distinct positions. In addition, homology modeling of MerRs from Gram-negative bacteria indicated a buried Hg2+ binding site can be assembled in all MerRs (Supplementary Figure S9), thus H7 can be effectively tethered to the Hg2+-thiolate center to drive the movement of the DNA-binding domain. Taken together, we conclude that the molecular mechanism underlying the Hg2+-dependent conformational and functional switching of the Bacillus MerR is applicable to Gram-negative MerRs.
Intriguingly, unlike the Tn21 and Tn501 mer operons of Gram-negative bacteria whose expression is exclusively regulated by MerR, the mer operon of Gram-positive bacteria encodes a second transcriptional regulatory protein termed MerR2 (10,18). Despite sharing ∼60% overall sequence similarity, the DNA- and metal-binding properties of MerR2 differ substantially from MerR. Sequence comparison and homology modeling analysis both reveal the replacement of the three Hg2+-ligating cysteines by residues with no metal-coordinating activities (Ile, Pro, Gly) in MerR2 (Supplementary Figure S10), consistent with its lack of affinity for Hg2+. Also we recognize significant divergence in residues involved in DNA-binding between MerR and MerR2, explaining their preference for binding to operator/promoter R1 and R2, respectively (32). The physiological role and the cognate ligand(s) of MerR2 remain to be explored.
Structural basis of MerR-mediated regulation of the mer operon
One of the most prominent structural differences between the apo-MerR and Hg2+-MerR structure is the repositioning of the two DNA-binding domains with respect to the coiled-coil region, which shortens the distance between the two symmetry-related DNA-recognition helices H2 and H2’ from 34 (apo) to 29 Å (Hg2+-bound) (Supplementary Figure S5). To understand how the expression of mer operon may be regulated by the conformational switching of MerR, modeling analysis was performed to investigate the functional consequences resulted from the binding of mer operator/promoter DNA by either apo-MerR or Hg2+-MerR. A solution NMR analysis of 199Hg2+-bound MerR revealed that the coordination environment was not affected by the addition of mer operator DNA (50), suggesting that DNA-binding does not significantly alter the overall structure of Hg2+-MerR. The lack of significant DNA-induced conformational changes implicates intrinsic structural rigidity of MerR family members, which may contribute to their activity in shaping DNA structure. Therefore, we constructed structural models of MerR-DNA complexes by keeping the structures of apo-MerR and Hg2+-MerR fixed while allowing adjustments of the DNA backbone conformations, such that shape complementarity and interacting surface between DNA and the recognition helices of MerR are optimized. In support of the validity of this approach, the Hg2+-MerR can be readily docked, without additional adjustment of the DNA conformation, onto a duplex taken from the binary complex formed by oxidized (activated) SoxR and DNA (26) with both recognition helices fitting snugly into the major groove to contact the operator sequences (Supplementary Figure S6).
For bacterial promoters, the typical spacing between the −10 and −35 elements is 17±1 base pairs, which favors productive binding by the RNA polymerase to initiate transcription (15,16,54,55) (Figure 6A). In contrast, optimal interaction between the mer promoter region and polymerase is perturbed by the longer (19∼20 base pairs) spacer, resulting in offsetting of the two polymerase-binding elements and much reduced transcription efficiency (14,19,41,56) (Figure 6B). The binding of apo-MerR to the mer operator sequence that is located within the spacer region apparently allows MerR to interact with and recruit RNA polymerase to the promoter to form a stable pre-initiation complex, suggesting that transcription of the mer operon can be promptly engaged once Hg2+ is present (57–60). Notably, our modeling analysis shows the bound apo-MerR may clash with the −10 element-binding region of the σ-factor (Figure 6C), which explains why only the −35, but not the −10 element, was protected in the presence of both apo-MerR and RNA polymerase (11). Therefore, the recruitment and preloading of RNA polymerase to the mer promoter by apo-MerR is mainly achieved via enhanced polymerase association with the −35 element. Binding of Hg2+ induces repositioning of the two DNA-binding domains of MerR, causing the bound DNA to be bent and twisted (Figure 6D) (14,19,22). The new DNA conformation restores the optimal spacing and alignment between the −10 and −35 elements to allow transcription initiation. In addition, Hg2+-MerR may enhance transcriptional efficiency by being properly positioned for mediating polymerase recruitment (57–60). Collectively, our modeling analysis suggests how MerR acts as an Hg2+-dependent switch to regulate the expression of mer operon.
Figure 6.
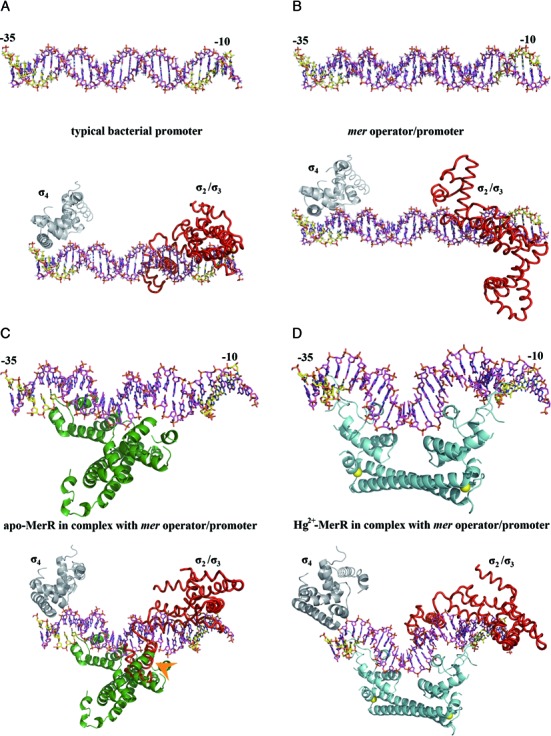
Structural basis of the Hg2+-mediated transcriptional regulation of the mer operon by MerR. (A) A typical bacterial promoter with the −10 and −35 elements (colored in yellow) being spaced by 17±1 base pairs, which allows productive association with the σ subunit of the RNA polymerase holoenzyme. The σ2/σ3 (red) and σ4 (gray) domains of the σ subunit are modeled onto the −10 and −35 elements, respectively, based on the crystal structures of the binary complexes formed by σ subunit and DNA.
It was proposed in earlier studies that the binding of one Hg2+ to MerR is sufficient to activate mer operon expression (49). However, the physiological significance of this claim is not obvious because no other MerR family members exhibit such a property, and all inducer-bound structures show the simultaneous presence of ligands in both binding sites (26,28–29). Given that the two Hg2+ binding sites are fully occupied in the Hg2+-MerR (Figure 5A), the validity and a structural explanation for the speculated anti-cooperativity between the two Hg2+-binding events remain unanswered. Nevertheless, it is conceivable that the conformational change resulted from the binding of one Hg2+ may produce a moderate transcription stimulation by partially restoring the alignment between the −10 and −35 elements.
CONCLUSION
This work reports the apo and Hg2+-bound conformation of the transcriptional regulatory protein MerR from B. megaterium, which correspond to the repressor and activator state of this dual-function transcription factor, respectively. The apo-MerR structure provides the first visualization of a MerR family member in its intact repressor form. And the Hg2+-MerR structure offers the first observation of tricoordinated Hg2+-thiolate center in protein and provides unequivocal evidence that MerR binds Hg2+ via trigonal planar coordination geometry. Structural comparison revealed that the conformational transition of MerR is tightly coupled to the assembly/disassembly of the metal binding site, and thus the structural state of MerR can be specified by the presence or absence of Hg2+. The Hg2+-induced relocation of the DNA-binding domains nicely explains how MerR may affect the conformation of mer operator-promoter DNA to regulate transcription. Nevertheless, due to the lack of DNA in our structures, we cannot rule out completely that the apo-MerR and Hg2+-MerR structures reported here may undergo additional conformational changes upon DNA binding. Structural characterizations of the mer operator DNA in complexes with either apo or Hg2+-bound MerR, or the ternary complexes formed by mer promoter, RNA polymerase, and either apo or Hg2+-bound MerR would serve as ultimate tests for our proposal.
ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited in the PDB with accession codes 4UA2 (apo-MerR) and 4UA1 (Hg2+-bound MerR).
Supplementary Material
Acknowledgments
Portions of this research were carried out at beamlines 15A1 and 13B1 of the National Synchrotron Radiation Research Center (Taiwan) and beamline SP12B2 of the SPring-8 (Japan). We thank Simon Silver, Le T. Phung, Ting-Yu Lee, Chun-Hua Hsu, Shiou-Ru Tzeng, Carmay Lim and the members of Chan lab for helpful discussion. We are grateful to the staffs of Technology Commons in College of Life Science and Center for Systems Biology, National Taiwan University.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology (NSC101–2911-I-002–303, 103–2113-M-002–010-MY3, 104–2911-I-002–302); National Research Program for Biopharmaceuticals (NSC101–2325-B-002–049); National Taiwan University (104R7614–3 and 104R7560–4); Ministry of Education, Taiwan ROC, under the ATU plan to NLC (National Chung Hsing University). Funding for open access charge: Research grants from the Ministry of Science and Technology, National Taiwan University, and Ministry of Education (Taiwan, ROC).
Conflict of interest statement. None declared.
REFERENCES
- 1.Silver S., Phung le T. A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. J. Ind. Microbiol. Biotechnol. 2005;32:587–605. doi: 10.1007/s10295-005-0019-6. [DOI] [PubMed] [Google Scholar]
- 2.Giedroc D.P., Arunkumar A.I. Metal sensor proteins: nature's metalloregulated allosteric switches. Dalton Trans. 2007;29:3107–3120. doi: 10.1039/b706769k. [DOI] [PubMed] [Google Scholar]
- 3.Barkay T., Miller S.M., Summers A.O. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 2003;27:355–384. doi: 10.1016/S0168-6445(03)00046-9. [DOI] [PubMed] [Google Scholar]
- 4.Huang C.C., Narita M., Yamagata T., Itoh Y., Endo G. Structure analysis of a class II transposon encoding the mercury resistance of the Gram-positive Bacterium bacillus megaterium MB1, a strain isolated from minamata bay, Japan. Gene. 1999;234:361–369. doi: 10.1016/s0378-1119(99)00184-5. [DOI] [PubMed] [Google Scholar]
- 5.Brown N.L., Misra T.K., Winnie J.N., Schmidt A., Seiff M., Silver S. The nucleotide sequence of the mercuric resistance operons of plasmid R100 and transposon Tn501: further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Mol. Gen. Genet. 1986;202:143–151. doi: 10.1007/BF00330531. [DOI] [PubMed] [Google Scholar]
- 6.Brown N.L., Ford S.J., Pridmore R.D., Fritzinger D.C. Nucleotide sequence of a gene from the Pseudomonas transposon Tn501 encoding mercuric reductase. Biochemistry. 1983;22:4089–4095. doi: 10.1021/bi00286a015. [DOI] [PubMed] [Google Scholar]
- 7.Fox B., Walsh C.T. Mercuric reductase. Purification and characterization of a transposon-encoded flavoprotein containing an oxidation-reduction-active disulfide. J. Biol. Chem. 1982;257:2498–2503. [PubMed] [Google Scholar]
- 8.Osborn A.M., Bruce K.D., Strike P., Ritchie D.A. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev. 1997;19:239–262. doi: 10.1111/j.1574-6976.1997.tb00300.x. [DOI] [PubMed] [Google Scholar]
- 9.Hobman J.L., Brown N.L. bacterial mercury-resistance genes. Met. Ions Biol. Syst. 1997;34:527–568. [PubMed] [Google Scholar]
- 10.Huang C.C., Narita M., Yamagata T., Phung le T., Endo G., Silver S. Characterization of two regulatory genes of the mercury resistance determinants from TnMERI1 by luciferase-based examination. Gene. 2002;301:13–20. doi: 10.1016/s0378-1119(02)01086-7. [DOI] [PubMed] [Google Scholar]
- 11.Frantz B., O’Halloran T.V. DNA distortion accompanies transcriptional activation by the metal-responsive gene-regulatory protein MerR. Biochemistry. 1990;29:4747–4751. doi: 10.1021/bi00472a001. [DOI] [PubMed] [Google Scholar]
- 12.O’Halloran T.V., Frantz B., Shin M.K., Ralston D.M., Wright J.G. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989;56:119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- 13.Lund P.A., Brown N.L. Regulation of transcription in Escherichia coli from the mer and merR promoters in the transposon Tn501. J. Mol. Biol. 1989;205:343–353. doi: 10.1016/0022-2836(89)90345-8. [DOI] [PubMed] [Google Scholar]
- 14.Ansari A.Z., Bradner J.E., O’Halloran T.V. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature. 1995;374:371–375. doi: 10.1038/374370a0. [DOI] [PubMed] [Google Scholar]
- 15.Moyle H., Waldburger C., Susskind M.M. Hierarchies of base pair preferences in the P22 ant promoter. J. Bacteriol. 1991;173:1944–1950. doi: 10.1128/jb.173.6.1944-1950.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harley C.B., Reynolds R.P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown N.L., Stoyanov J.V., Kidd S.P., Hobman J.L. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 2003;27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 18.Huang C.C., Narita M., Yamagata T., Endo G. Identification of three merB genes and characterization of a broad-spectrum mercury resistance module encoded by a class II transposon of Bacillus megaterium strain MB1. Gene. 1999;239:361–366. doi: 10.1016/s0378-1119(99)00388-1. [DOI] [PubMed] [Google Scholar]
- 19.Summers A.O. Untwist and shout: a heavy metal-responsive transcriptional regulator. J. Bacteriol. 1992;174:3097–3101. doi: 10.1128/jb.174.10.3097-3101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo H.B., Johs A., Parks J.M., Olliff L., Miller S.M., Summers A.O., Liang L., Smith J.C. Structure and conformational dynamics of the metalloregulator MerR upon binding of Hg(II) J. Mol. Biol. 2010;398:555–568. doi: 10.1016/j.jmb.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 21.Song L., Teng Q., Phillips R.S., Brewer J.M., Summers A.O. 19F-NMR reveals metal and operator-induced allostery in MerR. J. Mol. Biol. 2007;371:79–92. doi: 10.1016/j.jmb.2007.04.085. [DOI] [PubMed] [Google Scholar]
- 22.Ansari A.Z., Chael M.L., O’Halloran T.V. Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg-MerR. Nature. 1992;355:87–89. doi: 10.1038/355087a0. [DOI] [PubMed] [Google Scholar]
- 23.Hobman J.L. MerR family transcription activators: similar designs, different specificities. Mol. Microbiol. 2007;63:1275–1278. doi: 10.1111/j.1365-2958.2007.05608.x. [DOI] [PubMed] [Google Scholar]
- 24.Ni’Bhriain N.N., Silver S., Foster T.J. Tn5 insertion mutations in the mercuric ion resistance genes derived from plasmid R100. J. Bacteriol. 1983;155:690–703. doi: 10.1128/jb.155.2.690-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumaraswami M., Newberry K.J., Brennan R.G. Conformational plasticity of the coiled-coil domain of BmrR is required for bmr operator binding: the structure of unliganded BmrR. J. Mol. Biol. 2010;398:264–275. doi: 10.1016/j.jmb.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe S., Kita A., Kobayashi K., Miki K. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc. Natl. Acad. Sci. U.S.A. 2008;105:4121–4126. doi: 10.1073/pnas.0709188105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newberry K.J., Brennan R.G. The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus. J. Biol. Chem. 2004;279:20356–20362. doi: 10.1074/jbc.M400960200. [DOI] [PubMed] [Google Scholar]
- 28.Changela A., Chen K., Xue Y., Holschen J., Outten C.E., O’Halloran T.V., Mondragon A. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 29.Heldwein E.E., Brennan R.G. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature. 2001;409:378–382. doi: 10.1038/35053138. [DOI] [PubMed] [Google Scholar]
- 30.Godsey M.H., Baranova N.N., Neyfakh A.A., Brennan R.G. Crystal structure of MtaN, a global multidrug transporter gene activator. J. Biol. Chem. 2001;276:47178–47184. doi: 10.1074/jbc.M105819200. [DOI] [PubMed] [Google Scholar]
- 31.Zeng Q., Stalhandske C., Anderson M.C., Scott R.A., Summers A.O. The core metal-recognition domain of MerR. Biochemistry. 1998;37:15885–15895. doi: 10.1021/bi9817562. [DOI] [PubMed] [Google Scholar]
- 32.Chen C.Y., Hsieh J.L., Silver S., Endo G., Huang C.C. Interactions between two MerR regulators and three operator/promoter regions in the mercury resistance module of Bacillus megaterium. Biosci. Biotechnol. Biochem. 2008;72:2403–2410. doi: 10.1271/bbb.80294. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Terwilliger T.C. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terwilliger T.C. Maximum-likelihood density modification. Acta Crystallogr. D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terwilliger T.C., Berendzen J. Discrimination of solvent from protein regions in native Fouriers as a means of evaluating heavy-atom solutions in the MIR and MAD methods. Acta Crystallogr. D. 1999;55:501–505. doi: 10.1107/S0907444998012657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Painter J., Merritt E.A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 40.Shewchuk L.M., Verdine G.L., Walsh C.T. Transcriptional switching by the metalloregulatory MerR protein: initial characterization of DNA and mercury (II) binding activities. Biochemistry. 1989;28:2331–2339. doi: 10.1021/bi00431a052. [DOI] [PubMed] [Google Scholar]
- 41.Outten C.E., Outten F.W., O’Halloran T.V. DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J. Biol. Chem. 1999;274:37517–37524. doi: 10.1074/jbc.274.53.37517. [DOI] [PubMed] [Google Scholar]
- 42.Veglia G., Porcelli F., DeSilva T., Prantner A., Opella S.J. The structure of the metal-binding motif GMTCAAC is similar in an 18-residue linear peptide and the mercury binding protein MerP. J. Am. Chem. Soc. 2000;122:2389–2390. [Google Scholar]
- 43.Steele R.A., Opella S.J. Structures of the reduced and mercury-bound forms of MerP, the periplasmic protein from the bacterial mercury detoxification system. Biochemistry. 1997;36:6885–6895. doi: 10.1021/bi9631632. [DOI] [PubMed] [Google Scholar]
- 44.Dudev T., Lim C. Competition among metal ions for protein binding sites: determinants of metal ion selectivity in proteins. Chem. Rev. 2014;114:538–556. doi: 10.1021/cr4004665. [DOI] [PubMed] [Google Scholar]
- 45.Parkhill J., Lawley B., Hobman J.L., Brown N.L. Selection and characterization of mercury-independent activation mutants of the Tn501 transcriptional regulator, MerR. Microbiology. 1998;144:2855–2864. doi: 10.1099/00221287-144-10-2855. [DOI] [PubMed] [Google Scholar]
- 46.Shewchuk L.M., Helmann J.D., Ross W., Park S.J., Summers A.O., Walsh C.T. Transcriptional switching by the MerR protein: activation and repression mutants implicate distinct DNA and mercury(II) binding domains. Biochemistry. 1989;28:2340–2344. doi: 10.1021/bi00431a053. [DOI] [PubMed] [Google Scholar]
- 47.Ross W., Park S.J., Summers A.O. Genetic analysis of transcriptional activation and repression in the Tn21 mer operon. J. Bacteriol. 1989;171:4009–4018. doi: 10.1128/jb.171.7.4009-4018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wright J.G., Tsang H.T., Pennerhahn J.E., Ohalloran T.V. Coordination chemistry of the Hg-Merr metalloregulatory protein - evidence for a novel tridentate Hg-cysteine receptor-site. J. Am. Chem. Soc. 1990;112:2434–2435. [Google Scholar]
- 49.Helmann J.D., Ballard B.T., Walsh C.T. The MerR metalloregulatory protein binds mercuric ion as a tricoordinate, metal-bridged dimer. Science. 1990;247:946–948. doi: 10.1126/science.2305262. [DOI] [PubMed] [Google Scholar]
- 50.Utschig L.M., Bryson J.W., O’Halloran T.V. Mercury-199 NMR of the metal receptor site in MerR and its protein-DNA complex. Science. 1995;268:380–385. doi: 10.1126/science.7716541. [DOI] [PubMed] [Google Scholar]
- 51.Ralston D.M., O’Halloran T.V. Ultrasensitivity and heavy-metal selectivity of the allosterically modulated MerR transcription complex. Proc. Natl. Acad. Sci. U.S.A. 1990;87:3846–3850. doi: 10.1073/pnas.87.10.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reyes-Caballero H., Campanello G.C., Giedroc D.P. Metalloregulatory proteins: metal selectivity and allosteric switching. Biophys. Chem. 2011;156:103–114. doi: 10.1016/j.bpc.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen P.R., He C. Selective recognition of metal ions by metalloregulatory proteins. Curr. Opin. Chem. Biol. 2008;12:214–221. doi: 10.1016/j.cbpa.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 54.Murakami K.S., Masuda S., Campbell E.A., Muzzin O., Darst S.A. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 55.Campbell E.A., Muzzin O., Chlenov M., Sun J.L., Olson C.A., Weinman O., Trester-Zedlitz M.L., Darst S.A. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 56.Parkhill J., Brown N.L. Site-specific insertion and deletion mutants in the mer promoter-operator region of Tn501; the nineteen base-pair spacer is essential for normal induction of the promoter by MerR. Nucleic Acids Res. 1990;18:5157–5162. doi: 10.1093/nar/18.17.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kulkarni R.D., Summers A.O. MerR cross-links to the alpha, beta, and sigma 70 subunits of RNA polymerase in the preinitiation complex at the merTPCAD promoter. Biochemistry. 1999;38:3362–3368. doi: 10.1021/bi982814m. [DOI] [PubMed] [Google Scholar]
- 58.Livrelli V., Lee I.W., Summers A.O. In vivo DNA-protein interactions at the divergent mercury resistance (mer) promoters. I. Metalloregulatory protein MerR mutants. J. Biol. Chem. 1993;268:2623–2631. [PubMed] [Google Scholar]
- 59.Lee I.W., Livrelli V., Park S.J., Totis P.A., Summers A.O. In vivo DNA-protein interactions at the divergent mercury resistance (mer) promoters. II. Repressor/activator (MerR)-RNA polymerase interaction with merOP mutants. J. Biol. Chem. 1993;268:2632–2639. [PubMed] [Google Scholar]
- 60.Heltzel A., Lee I.W., Totis P.A., Summers A.O. Activator-dependent preinduction binding of sigma-70 RNA polymerase at the metal-regulated mer promoter. Biochemistry. 1990;29:9572–9584. doi: 10.1021/bi00493a011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


