Abstract
Ty1 Gag comprises the capsid of virus-like particles and provides nucleic acid chaperone (NAC) functions during retrotransposition in budding yeast. A subgenomic Ty1 mRNA encodes a truncated Gag protein (p22) that is cleaved by Ty1 protease to form p18. p22/p18 strongly inhibits transposition and can be considered an element-encoded restriction factor. Here, we show that only p22 and its short derivatives restrict Ty1 mobility whereas other regions of GAG inhibit mobility weakly if at all. Mutational analyses suggest that p22/p18 is synthesized from either of two closely spaced AUG codons. Interestingly, AUG1p18 and AUG2p18 proteins display different properties, even though both contain a region crucial for RNA binding and NAC activity. AUG1p18 shows highly reduced NAC activity but specific binding to Ty1 RNA, whereas AUG2p18 shows the converse behavior. p22/p18 affects RNA encapsidation and a mutant derivative defective for RNA binding inhibits the RNA chaperone activity of the C-terminal region (CTR) of Gag-p45. Moreover, affinity pulldowns show that p18 and the CTR interact. These results support the idea that one aspect of Ty1 restriction involves inhibition of Gag-p45 NAC functions by p22/p18-Gag interactions.
INTRODUCTION
The Saccharomyces Ty1 element belongs to a widely disseminated group of retrotransposons with pronounced structural and functional similarities to retroviruses, except Ty1 transposition is not infectious (1,2). Ty1 contains GAG and POL genes bracketed by long-terminal repeats (LTRs). The element is transcribed from LTR to LTR, forming a genome-length terminally redundant RNA that serves as templates for reverse transcription and protein synthesis (3). Dimeric Ty1 RNA is packaged into virus-like particles (VLPs) (4) comprised of the capsid protein Gag and Gag-Pol, a large precursor synthesized by a programmed +1 frameshift event that occurs at overlapping leucine codons present in GAG and POL (5). POL encodes protease (PR), reverse transcriptase (RT) and integrase (IN), which are required for protein maturation (6), replication and integration, respectively. The process of Ty1 retrotransposition is completed by integration of a linear cDNA near genes transcribed by RNA polymerase III (7).
A universal feature of retrotransposon and retrovirus propagation is a series of nucleoprotein interactions mediated by Gag or its mature products. However, Ty1 Gag lacks homology with the subdomains present in Gag from well studied avian, murine and human retroviruses (8). In particular, Ty1 Gag does not contain the zinc-finger motif present in all retroviral nucleocapsid (NC) proteins with the exception of Foamy virus (9,10) and the Drosophila Gypsy retroelement (11). The nucleic acid chaperone (NAC) function of Gag and mature NC are required for RNA dimerization, packaging, annealing of the tRNA primer to retroviral RNA, strand-transfer during reverse transcription, retroviral recombination and cDNA integration (10). NAC proteins function in an adenosine triphosphate (ATP) independent manner. They bind nucleic acids (NA) with broad specificity, destabilize kinetically trapped, misfolded structures and facilitate their folding to the thermodynamically most favored form. Once the most stable NA structure is reached, their binding is no longer required (12). NAC proteins generally do not share common sequence motifs or specific 3D structures. Thus, biochemical assays are required to identify proteins with NAC activity. Since they may act in the sequence independent manner, oligonucleotides derived from HIV-1 genome are usually utilized in chaperone assays. In vitro studies demonstrated that key features of retroviral NAC activities are NA aggregation, destabilization and rapid binding kinetics (13–15).
The primary Ty1 Gag translation product is a 440 amino acid protein (p49), which undergoes C-terminal cleavage by PR to form the mature capsid (p45) and a small peptide (p4) that has not been detected in VLP preparations or cell extracts (1). Gag variants with altered mobility are also present. These Gag-related products migrate between p49 and p45 and may result from post-translational modifications or additional cleavage events (16), but have not been functionally defined. Mutational analyses have revealed Gag residues that contribute to VLP assembly and morphology and immunological probing suggests that the Gag N-terminal region (NTR) is located on the surface of VLPs, while the C-terminus is internal (17,18). Alignments of Gag proteins from the Pseudovirus superfamily reveal three conserved blocks of amino acids, termed A, B and C, as well as several invariant amino acids (19), which may be important for protein–protein interactions required for VLP assembly and are located in the C-terminal half of Gag. A nucleocapsid functionality may also reside in the N-terminus of PR that is distinct from proteolytic cleavage (20). Cristofari et al. used theoretical charge profiles to identify a short C-terminal segment of Gag-p45 containing three stretches of basic amino acids (21). A 103 residue synthetic peptide (TYA1-D) from Asn299-His401 derived from this region possesses NAC activity in vitro using model substrates. Peptide TYA1-D promotes annealing of initiator tRNAiMet to the Ty1 primer-binding site (PBS).
Recent work has shown that a truncated form of Gag (p22) or a form processed by PR (p18) inhibits Ty1 transposition and mediates copy number control (CNC) by preventing assembly of functional VLPs (22). Therefore, the self-encoded inhibitor from Ty1 is conceptually similar to the Gag-like restriction factors derived from mammalian retroviruses (23,24). An internally initiated Ty1 transcript (Ty1i) encodes p22, however, two closely spaced AUG codons AUG1 and AUG2 may be utilized to initiate p22 synthesis. Co-expression of Ty1 and p22 from AUG1 results in association of p22 with VLPs and Gag p49/p45, the appearance of morphologically aberrant VLPs, and a dramatic inhibition of Ty1 movement. p22 lacks the conserved A–C domains but contains the TYA1-D peptide sequence, and therefore, may possess an RNA chaperone activity involved in CNC.
In this work, we have taken a multifaceted approach to further understand the functional organization of GAG with emphasis on the NAC domain and how p22/p18 inhibits Ty1 transposition. We show that p22 synthesis initiates at either of two closely spaced AUG codons both in vivo and in vitro, translation of Ty1i RNA is cap-dependent, and only the p22 region is capable of strongly inhibiting transposition. Information from these studies was used to produce recombinant Gag-derived proteins for RNA interaction analyses in vitro. We demonstrate that the C-terminal disordered region (Asn355-His401) is critical for RNA binding and NAC activity of Ty1 Gag derivatives. Biochemical assays reveal that p18 proteins are less effective chaperones than the C-terminal region (CTR) of Gag-p45. Moreover, the chaperone activity of p18 proteins synthesized from AUG1 (AUG1p18) and AUG2 (AUG2p18) differs significantly. Despite those differences, the p18 proteins and CTR bind within the same regions of Ty1 RNA. Finally, we provide evidence suggesting that the Ty1 restriction factor inhibits NAC activity of the CTR, physically interacts with the CTR and decreases Ty1 RNA dimerization and packaging into VLPs.
MATERIALS AND METHODS
Genetic techniques, media and plasmid construction
Standard yeast media and microbiological procedures were used (25). Ty1 nucleotide coordinates correspond to the retrotransposon Ty1-H3 sequence (Genbank M18706.1). For expression studies, segments of Ty1-H3 GAG were amplified by polymerase chain reaction (PCR) using Phusion DNA Polymerase and conditions recommended by the manufacturer (Thermo Fisher Scientific). The primers used for constructing expression plasmids are listed in Supplementary Table S1. For pGPOLΔ related plasmids, the XhoI/BglII region (Ty1 238–1702) was replaced with AUG1 and/or AUG2 containing fragments created by overlap PCR using the A-outGAL1F and D-PR1R primer sets. EcoRI/XhoI GAG segments were subcloned into pYES2 (Life Technologies, Thermo Fisher), a multicopy URA3-based expression vector containing the GAL1 promoter. For His-tagged protein purification in BL21 Escherichia coli, NdeI/XhoI GAG segments were inserted into pET15TEV (Novagen EMD). For pulldown analysis, XbaI/HindIII CTR segment was subcloned into pEG(KT) (pGST/URA3, 2 μ) (26). pBDG1608 (pAUG1p18/TRP1, 2 μ) and pBDG1612 (psAUG1/TRP1, 2 μ) were made by converting URA3 on pAUG1p18 and psAUG1 to TRP1 by homologous recombination. PCR products were verified by DNA sequencing.
Ty1his3-AI mobility
Ty1 retrotransposition was monitored using the selectable indicator gene his3-AI (27). Ty1HIS3 insertions usually occur by retrotransposition following splicing of the artificial intron. Since His+ cells can also result from recombination of Ty1HIS3 cDNA with genomic Ty1 elements or solo LTRs (28,29), the term Ty1 ‘mobility’ is used to describe both types of insertion. pGPOLΔ/URA3, 2 μ or related plasmids were transformed into a Ty1-less Saccharomyces paradoxus strain [DG2196; (MATa his3-Δ200hisG ura3 trp1 Ty1his3-AI(96)] repopulated with 1 Ty1his3-AI element (30). pGTy1his3-AI/TRP1,Cen and pYES2 containing various segments of GAG were transformed into the Ty1-less S. paradoxus strain [DG3582; (MATα, his3-Δ200hisG, ura3, trp1) (22)]. The frequency of Ty1his3-AI mobility was determined as described previously (22). Ty1 mobilities were averaged from 4 single colonies (8 cultures) for strains expressing segments of GAG and pGTy1his3-AI and from 34 colonies (68 cultures) for the control strain that contained an empty pYES2 vector and pGTy1his3-AI.
Northern analysis
DG3582 containing pGTy1his3AI and a pYES2-GAG plasmid were inoculated in 1 ml of SC-Ura-Trp + 2% Raffinose and grown overnight at 30°C. Four-hundred microliters of the overnight culture was inoculated into 10 ml SC-Ura-Trp + 2% Galactose and grown at 22°C for 2 days. Total RNA was extracted using MasterPure yeast RNA purification kit (Epicenter Biotechnologies). Eight micrograms samples of RNA were separated by electrophoresis on a 1.5% agarose-formaldehyde gel at 120V for 2.5 h and blotted onto Hybond-N+ (GE Healthcare). To detect Ty1 and ACT1 mRNAs, strand-specific riboprobes were labeled with [α-32P] UTP (3000 Ci/mmol) (Perkin Elmer) by transcription in vitro using MAXIscript (Ambion), according to the manufacturer's instructions. Filter hybridization was performed at 65°C overnight and detected using a Storm 840 phosphoimager (GE Healthcare).
Immunoblotting
Total protein was extracted by trichloroacetic acid (TCA) as described previously with minor modifications (20). Two-milliliter cultures induced with galactose for 2 days were resuspended in 120 μl 17% TCA and vortexed with the same amount of glass beads (Sigma) for 5 min. After adding 1 ml 5% TCA, 1 ml of the supernatant was centrifuged at 10 000 rpm for 10 min at 4°C. The pellet was resuspended in 150 μl of Laemmli buffer (BioRad) containing 0.5 M Tris–HCl, pH 8.0. Two microliter protein samples were boiled for 8 min, separated by electrophoresis on a 15% sodium dodecyl sulphate (SDS)-Polyacrylamide gel at 120V for 1.5 h and transferred to Immun-Blot PVDF membrane by Trans-blot SD Semi-Dry Transfer Cell system (BioRad). The membranes were blocked in TBST buffer (50 mM Tris–HCl, 150 mM NaCl, 0.1% Tween-20, pH 7.5) with 5% (w/v) powdered milk (LabScientific) and incubated with a rabbit polyclonal antiserum against AUG2p18 (Gag M259-H401) (22) or a mouse monoclonal antiserum against Gag E25-D34 (Sigma). Immune complexes were detected by ECL (Amersham) and HyBlot ES film (Denville).
Purification of Gag derivatives
Synthesis of recombinant proteins was induced in E. coli BL21 cells carrying pET15TEV-GAG derivatives by adding 0.15 mM IPTG in 800 ml LB + 100 μg/ml ampicillin (Sigma) at 16°C for 24 h. The collected cells were resuspended in 50 ml lysis buffer (1 M NaCl, 50 mM phosphate buffer, pH 7.8) and lysed by sonication. The His-tagged Gag derivatives were purified by metal-chelate column chromatography using Talon affinity resin (Clontech) and eluted with lysis buffer containing 300 mM imidazole (Sigma). The eluted products were dialyzed against storage buffer (10% glycerol, 1 M NaCl and 25 mM Tris–HCl, pH 8.0) for 12 h at 4°C. Proteins were stored at −75°C. A synthetic peptide corresponding to nucleotide residues 1038–1068 of GAG (MKILSKSIEK) was purchased from ThermoFisher Scientific.
In vitro translation
All DNA templates for in vitro translation studies were amplified from pBDG433 using following forward primers F-AUG1AUG2, F-GCG1AUG2, F-AUG1GCG2 and reverse primer R-p22 (Supplementary Table S2). Forward primers for GCG1AUG2 and AUG1GCG2 introduced point mutations (AUG to GCG) at AUG1 or AUG2, respectively. The transcription reaction using MEGAscript T7 Transcription Kit was performed as recommended by the manufacturer (Invitrogen). The RNA transcripts were purified using Direct-zol RNA MiniPrep Kit (Zymo Research). The integrity of each transcript was confirmed by agarose gel electrophoresis. Capped RNA transcripts were synthesized using ARCA Cap Analog (Invitrogen) or non-physiological ApppG cap (Ambion). In vitro translation experiments were carried out in a wheat germ extract (WGE; Promega) following the manufacturer's protocol. Briefly, the reaction mixture containing 12.5 μl of WGE, 80 μM amino acid mixture minus methionine, 1.25 μl of [35S]-methionine (1000 Ci/mmol) (Hartmann Analytic), 79 mM potassium acetate, 20 units of RNasin Ribonuclease Inhibitor (Promega), 1 pmol of capped (ARCA or ApppG) or uncapped RNA in the final volume of 25 μl was incubated for 60 min at 25°C. For experiments with the cap analog as an inhibitor of cap-dependent translation, WGE was pre-incubated for 15 min at 25°C with 500 μM cap analog and equimolar amounts of magnesium acetate. Translation products were resolved on SDS-polyacrylamide gels followed by radioisotope imaging with FLA 5100 image analyzer (FujiFilm).
DNA and RNA substrates
TAR(−) DNA [5′-GGG TTC CTT GCT AGC CAG AGA GCT CCC GGG CTC GAC CTG GTC TAA CAA GAG AGA CC-3′] and TAR(+) DNA [5′- GGT CTC TCT TGT TAG ACC AGG TCG AGC CCG GGA GCT CTC TGG CTA GCA AGG AAC CC-3′] corresponding to the trans-acting responsive (TAR) element sequence of HIV-1MAL were purchased from Genomed. The templates for transcription of HIV-1 TAR RNA and tRNAiMet were PCR-generated and RNAs were synthesized using T7-MEGAshortscript (Ambion). Transcripts were purified by denaturing gel electrophoresis (8 M urea) in 1× Tris-Borate-EDTA (TBE) buffer, eluted from the gel matrix and concentrated by ethanol precipitation. Purified RNAs were dissolved in sterile water and stored at −20°C. TAR(−) DNA was 32P-labeled at the 5′-end using [γ-32P] ATP (3000 Ci/mmol) and T4 polynucleotide kinase (Fermentas) following manufacturer's protocol and subsequently purified on NucAway Spin Columns (Life Technologies). The in vitro synthesized, unmodified yeast tRNAiMet was 3′-end labeled with [α-32P] pCp using T4 RNA ligase (Fermentas) and purified on G50 columns (GE Healthcare). Templates for in vitro transcription of Ty1 mini RNA (560 nt) (31,32) were obtained by PCR amplification of the fragment corresponding to the RNA nt +1–560 from the pBDG433 using a forward primer F-miniRNA (Supplementary Table S2) containing an SP6 promoter sequence followed by 5′ Ty1 RNA sequence and a reverse primer R-miniRNA. RNA was synthesized using SP6-MEGAscript (Ambion) and purified using Direct-zol RNA MiniPrep Kit (Zymo Research). Purified RNAs were stored at −20°C until use.
Sedimentation assays
A total of 10 nM 32P-labeled Ty1 mini RNA was incubated with different amounts of Ty1 Gag derived proteins (0–1 μM) in a buffer containing 50 mM Tris–HCl pH 7.5, 20 mM NaCl and 0.2 mM MgCl2 at 24°C for 10 min. Subsequently, the mixtures (10 μl) were centrifuged for 20 min at 12 000 g. Supernatants (2 μl) were collected and subjected to scintillation counting. For the assay using heterologous substrates, 10 nM 32P-labeled TAR1(−) was combined with 40 nM complementary unlabeled TAR1(+). Ty1 Gag derived proteins (0–1 μM) were added and sedimentation was performed as described above.
Filter binding assays
All reactions were carried out in binding buffer containing 50 mM Tris–HCl pH 7.5, 40 mM KCl, 2 mM MgCl2, 0.01% Triton X-100 and different concentrations of NaCl (10, 50, 100, 150, 200, 250, 500 mM). 32P-labeled Ty1 mini RNA (0.4 nM) was incubated for 5 min at 95°C without magnesium ions and Triton X-100 and slowly cooled to 37°C. MgCl2 and Triton X-100 were added to the final concentration of 2 mM and 0.01% respectively, followed by incubation for 10 min at 37°C. The Ty1 protein solutions were prepared by diluting an appropriate amount of protein stock into binding buffer. Further dilutions were obtained by sequential two-fold dilutions. The binding reaction was initiated by mixing the same volumes of RNA and Ty1 protein solution in a microplate. Final concentration of RNA in the reaction was 0.2 nM. The reactions were incubated for 15 min at 24°C and then 30 μl of each binding reaction was filtered and washed with 400 μl of binding buffer. A 96-well filter block (Minifold, Whatman) was used to determine the dissociation constant for Ty1 proteins binding to Ty1 mini RNA. Membranes used were top nitrocellulose (Protran, Whatman) and bottom charged nylon (Hybond N+, GE Healthcare). Both membranes were soaked in binding buffer prior to use. Binding reactions were performed in rows of 8 in a 96-well microplate. After filtration the membranes were dried and exposed to a phosphoimager screen. Data were fitted to the Hill equation using Origin (OriginLab) software.
Hydroxyl radical footprinting
RNA samples (5 pmol) were refolded by heating at 95°C for 1 min followed by incubation for 25 min at 37°C in a buffer containing 40 mM Tris–HCl, pH 8.0, 130 mM KCl, 0.5 mM EDTA and 5 mM MgCl2. Folded Ty1 mini RNA samples were diluted 20× with 20 mM Tris–HCl, pH 8.0. Subsequently, 50 or 100 pmols of CTR, AUG2p18 or AUG1p18 (6 μl in the protein buffer containing 50 mM Tris–HCl, pH 8.0, 1 M NaCl, 10 mM β-mercaptoethanol, 2.5 mM DTT) were added to a 70 μl reaction. As a control for non-specific cleavage, buffer was added instead of protein. RNA/protein complexes were formed by incubating 20 min at 0°C. To initiate footprinting reactions, 1.5 μl of 2.5 mM (NH4)Fe(SO4)2, 50 mM sodium ascorbate, 1.5% H2O2 and 2.75 mM EDTA were applied on the wall of the tube followed by centrifugation (33). Reactions were incubated for 15 s at 24°C and quenched by addition of thiourea and EDTA to final concentrations of 20 mM and 40 mM, respectively. RNA were purified using Direct-zol RNA MiniPrep Kit (Zymo Research).
Detection of cleavage products and data processing
A total of 10 pmols of fluorescently labeled primer PR3 (Supplementary Table S2) was added to the 2 pmols of RNA [Cy5 (+reagent) and Cy5.5 (−reagent)] and 12 μl of primer-template solutions were incubated at 95°C for 3 min, 37°C for 10 min and 55°C for 2 min and reverse transcribed as described previously (34). Sequencing ladders were prepared using primers labeled with WellRed D2 (ddA) or LicorIR-800 (ddT) and a Thermo Sequenase Cycle Sequencing kit (Affymetrix) according to the manufacturer's protocol. Samples and sequencing ladders were purified using ZR DNA Sequencing Clean-up Kit (Zymo Research) and analyzed on a GenomeLab GeXP Analysis System (Beckman-Coulter). Electropherograms were processed as described (35). At least four repetitions were obtained for each read.
TAR annealing assays
Substrates were refolded by heating at 95°C for 2 min and 37°C for 10 min following addition of buffer A (20 mM Tris–HCl, pH 7.2, 30 mM NaCl, 0.1 mM MgCl2, 10 μM ZnCl2 and 5 mM DTT). To the 32P-labeled antisense TAR(−) DNA oligonucleotide (1 nM), complementary unlabeled RNA or sense DNA (6 nM) was added. Reactions were incubated in buffer A with increasing Ty1 protein concentrations (0–0.6 μM) at 24°C for 5 min. The reactions (10 μl) were chilled on ice. Subsequently, 5 μl of Stop Solution (20% glycerol, 20 mM EDTA pH 8.0, 0.2% SDS, 0.25% bromophenol blue and 0.4 mg/ml yeast tRNA) was added to denature proteins and induce their release from the oligonucleotides. Samples were analyzed by native polyacrylamide gel electrophoresis (8%) in 0.5× TBE at 4°C (DNApointer, Biovectis).
tRNAiMet/Ty1 RNA annealing assays
The 32P-labeled tRNAiMet and Ty1 mini RNA (560 nt) were separately refolded in 50 mM Tris–HCl (pH 7.5) by heating at 95°C for 1 min and slow cooling to 60°C, followed by addition of MgCl2 to 2 mM, placement on ice and subsequent incubation at 37°C for 10 min. Refolded RNAs were placed on ice and tRNAiMet (5 nM) was combined with Ty1 mini RNA (10 nM) in a solution containing 50 mM Tris–HCl, pH 7.5, 20 mM NaCl and 1 mM MgCl2. Initially, RNAs were incubated at 37°C for 10 min. Upon addition of protein, the annealing reaction proceeded at 37°C for another 10 min. Reactions were quenched by incubation with 1% (w/v) SDS at room temperature for 3 min. The samples were phenol/chloroform-extracted twice, mixed with 2 μl of 50% glycerol and separated on an agarose gel in 1× TBE-SDS buffer at 22°C.
GST-pulldown
Glutathione S-transferase (GST) pulldown analysis was performed as described previously (22). Briefly, pGST [pEG(KT)], pGST-CTR (pBDG1496) and pAUG1p18 (pBDG1608; pAUG1p18/TRP1) or psAUG1 [(deleted C-DR p18) pBDG1612; psAUG1/TRP1] were introduced into DG3582 by transformation. Cells were grown in SC-Ura-Trp + 2% Galactose for 2 days at 22°C after pre-culturing in SC-Ura-Trp + 2% Raffinose overnight at 30°C. To determine if the CTR interacts with p18, GST-complexes were analyzed by immunoblotting using mouse monoclonal antibody GST/B-14 (Santa Cruz Biotech) or rabbit polyclonal p18 antibody (22).
Ty1 RNA dimerization assays
The 32P-labeled Ty1 RNA (1 pmol) and tRNAiMet (2 pmols) were refolded separately in 50 mM Tris–HCl (pH 7.5) by heating at 95°C for 1 min and slow cooling to 60°C, followed by addition of MgCl2 to 2 mM, placement on ice and subsequent incubation at 37°C for 10 min. RNAs were combined in the solution 25 mM Tris–HCl, pH 7.5, 30 mM NaCl, 0.25 mM MgCl2. Upon addition of proteins, at the indicated concentrations, reactions were incubated at 37°C for 30 min. Reactions were quenched by incubation with 1% (w/v) SDS at 24°C for 3 min. The samples were phenol/chloroform-extracted twice, mixed with 2 μl of 50% glycerol and separated by agarose gel electrophoresis in 0.5× TB buffer at 24°C.
Competitive tRNAiMet/Ty1 RNA annealing
The 32P-labeled tRNAiMet and Ty1 mini RNA were prepared as described for tRNAiMet/Ty1 RNA annealing assays. Proteins were combined in the buffer containing 0.5 M NaCl and 25 mM Tris–HCl, pH 8.0 and incubated together for 5 min at 24°C. After addition of the proteins to the RNAs annealing reaction proceeded at 37°C for another 10 min. Reactions were quenched by incubation with 1% SDS at 24°C for 3 min. The samples were phenol/chloroform-extracted twice, mixed with 2 μl of 50% glycerol and separated on an agarose gel in 1× TBE-SDS buffer at 24°C. All gels and membranes were autoradiographed and quantitatively analyzed by phosphoimaging using FLA-5100 phosphoimager with MultiGaugeV 3.0 software (FujiFilm). In all cases, at least three independent experiments were performed and the data presented are representative of the whole.
VLP nuclease protection
DG3582 containing pGTy1his3-AI alone and pAUG1p22 or pAUG1p18 plasmids were grown as described above, except galactose cultures were incubated at 20°C. Nuclease protection was performed as described previously (36,37). Briefly, cells from 10 ml galactose-induced culture were disrupted with acid-washed glass beads and vortexing in lysis buffer (10 mM Tris–HCl pH 7.5, 100 mM KCl, 10 mM EDTA) containing 60 units/ml of RNasin Plus RNase Inhibitor (Promega) and protease inhibitor cocktail (0.125 mg/ml Aprotinin, Leupeptin, Pepstatin and 1.6 mg/ml PMSF) at 4°C. Whole cell extract, obtained after centrifuging the lysate at 500 g for 5 min was divided into two aliquots. One aliquot was treated with 75 units of benzonase (EMD Millipore) in reaction buffer (10 mM Tris–HCl pH 7.5, 10 mM MgCl2, 50 mM NaCl) while the other was not. Both aliquots were incubated at 22°C for 11 min. Reactions were terminated by adding 5 μl of 0.5 M EDTA. Total RNA extraction and northern analysis were performed as described above. Nuclease protection assays were repeated three times and representative results are shown. Whole cell extracts used for nuclease protection was also immunoblotted to detect Gag proteins as described previously (22).
RESULTS
p22 synthesis initiates at either of two closely spaced start codons
We discovered the Ty1 restriction factor p22 using plasmid pGPOLΔ that is repressed for Ty1 mRNA transcription but allows synthesis of Ty1i RNA (22). pGPOLΔ is a multicopy plasmid containing GAG fused to the regulatable GAL1 promoter and is deleted for most of POL (Figure 1A). Ty1-less cells repopulated with a single Ty1 element tagged with the retrotransposition indicator gene his3-AI (38) and harboring an empty vector, pGPOLΔ or a series of mutations affecting translation of p22 from either AUG1 (nt 1038) or AUG2 (nt 1068) were grown in medium containing the repressing carbon source glucose and analyzed for Ty1his3-AI mobility (Figure 1B) and production of p22 (Figure 1C). AUG1 and AUG2 were mutated to GCG-alanine individually or together, and a +1 frameshift mutation was introduced between AUG1 and AUG2 (pAUG1fs-PΔ; +C1040), which introduced a stop codon upstream of AUG2. As expected, Ty1his3-AI mobility decreased about 70-fold (Table 1) when cells containing pGPOLΔ were compared with an empty vector control or when both AUG's were converted to GCG-alanine (22,30). However, pGCG1AUG2-PΔ, pAUG1GCG2-PΔ and pAUG1fs-PΔ inhibited Ty1his3-AI mobility between 26- and 91-fold. Immunoblotting was performed with total protein extracts from the strains with antisera against p18 (anti-p18), the processed form of p22 (22), which also reacts with full-length Gag p49/p45) or histidyl tRNA synthetase (anti-Hts1), which was used as a loading control (Figure 1C). p22 was detected in all strains except those containing an empty vector or pGCG1GCG2-PΔ. Although we could not detect p18 due to the low level of Ty1 PR expressed from a chromosomal Ty1 element (38), p22 and p18 are both potent inhibitors of Ty1 transposition (22). Full length Gag from the chromosomal Ty1his3-AI element was also detected in the strains. Together, our results show that both AUG codons expressed from Ty1i mRNA are used to produce the p22 restriction factor.
Figure 1.
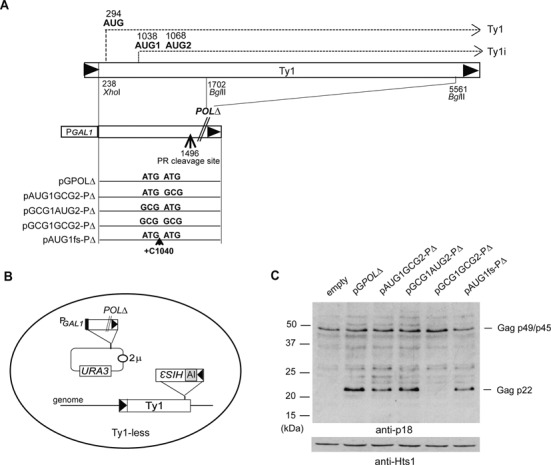
Mutational analysis of Ty1i RNA initiation codons AUG1 and AUG2. (A) Map of the Ty1 and pGTy1 mutant derivatives used to assess translation of Ty1i RNA from two closely spaced initiation codons AUG1 and AUG2. pGPOLΔ lacks most of POL (22), but contains GAG, including sequences required for transcribing Ty1i RNA and cleavage of p22 to p18 by Ty1 protease (PR). (B) pGPOLΔ (URA3, 2μ) plasmids were introduced into a Ty1-less Saccharomyces paradoxus strain containing a single chromosomal Ty1his3-AI element. Cells propagated on glucose are repressed for transcription of Ty1 mRNA from GAL1 promoter, but allow synthesis of Ty1i RNA and p22/p18. Ty1his3-AI mobility analyses using this assay are shown in Table 1. (C) Total cell protein isolated after growth in SC-Ura medium for 2 days at 22°C was immunoblotted with p18 antiserum (anti-p18). Histidyl tRNA synthetase (anti-Hts1) served as a loading control.
Table 1. Effect of endogenous p22/p18 expression on Ty1his3-AI mobility.
| Plasmida | Ty1his3-AI mobilityb x 10−6 (SD) | Fold decrease |
|---|---|---|
| pGAL1/2μ | 100 (14) | 1 |
| pGPOLΔ | 1.4 (0.4) | 71 |
| pAUG1GCG2-PΔ | 1.8 (0.5) | 56 |
| pGCG1AUG2-PΔ | 3.8 (0.8) | 26 |
| pGCG1GCG2-PΔ | 90 (7) | 1 |
| pAUG1fs-PΔ | 1.1 (0.2) | 91 |
aRefer to Figure 1A for additional information.
bRefer to ‘Materials and Methods’ section for additional information.
Segments of GAG capable of inhibiting Ty1his3-AI mobility in trans
To determine if additional Gag regions inhibited Ty1his3-AI mobility, phylogenetic comparisons between Gag proteins from different Pseudoviridae (19), predicted protein-folding patterns using XtalPred (http://ffas.burnham.org/XtalPred-cgi/xtal.pl) (39) and location of the p22/p18 restriction factor (22) and NAC region (21) were used to choose segments suitable for producing recombinant proteins (Figure 2). Phylogenetic comparisons show that homology blocks A, B and C are located in the C-terminal half of Gag with a highly conserved tryptophan (W) at position 184 (Figure 2A) in block A (19). However, we were unable to assign Ty1 amino acid coordinates to the homology blocks due to divergence of Ty1 Gag sequence when compared with other Pseudoviridae Gag proteins. The protein folding predictions suggest that Gag consists of a disordered NTR and an α-helical CTR followed by another disordered segment (C-DR), which is present in the NAC region. When mature Gag-p45, the NTR, the CTR or a shortened (s) CTR lacking C-DR residues from the NAC region were expressed from the GAL1 promoter on a multicopy expression plasmid (pYES2) along with pGTy1his3-AI (Figure 2B), Ty1 mobility decreased modestly if at all when compared with an empty pYES2 vector control (Table 2). These results suggest that the inhibitory effect conferred from expression of GAG or GAG segments is specific for the p22 region and is not a general property associated with the capsid gene.
Figure 2.
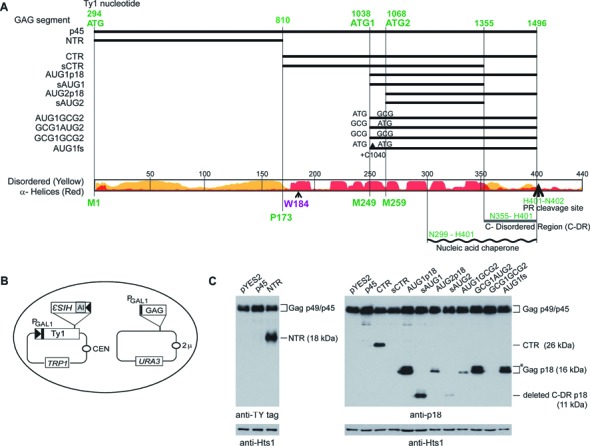
Functional organization of GAG and coexpression of subgenomic segments with pGTy1his3-AI. (A) At the top is the mature Gag (p45) coding sequence with selected ATG codons highlighted (green) and below are segments expressed ectopically from the pYES2 GAL1 promoter. At the bottom, XtalPred (http://ffas.burnham.org/XtalPred-cgi/xtal.pl) was used to predict Gag disordered (yellow) and α-helical (red) regions. Also shown is an invariant tryptophan residue (W184) found in Pseudoviridae Gag proteins (19), the position of the nucleic acid chaperone region (21), a C-terminal disordered region (C-DR), and the Ty1 protease (PR) cleavage site (H401-N402). An ATG codon was added adjacent to P173 for expression of the CTR and sCTR. (B) Two plasmids, pGTy1his3-AI (TRP1, CEN) and GAG segment under pYES2 (URA3, 2 μ) were induced from the GAL1 promoter in a Ty1-less Saccharomyces paradoxus strain to determine whether different Gag proteins inhibited Ty1his3-AI mobility (Table 2). (C) Total cell protein from induced cultures was immunoblotted with p18 antiserum (anti-p18) or TY-tag (anti-TY-tag). Histidyl tRNA synthetase (anti-Hts1) served as a loading control.
Table 2. Ty1 mobility resulting from co-expression of GAG segments and Ty1his3-AI.
| GAG segmenta | Ty1his3-AI mobilityb x 10−3 (SD) | Fold decrease |
|---|---|---|
| pYES2 | 32 (12) | 1 |
| p49 | 28 (17) | 1.1 |
| p45 | 34 (17) | 0.8 |
| pNTR | 24 (14) | 1.7 |
| pCTR | 4.7 (2.4) | 6.7 |
| psCTR | 5.8 (3.0) | 6.8 |
| pAUG1p22 | 0.00077 (0.00047) | 42 000 |
| pAUG1p18 | 0.00082 (0.00050) | 39 000 |
| psAUG1 | 0.0037 (0.0025) | 8600 |
| pAUG2p22 | 0.34 (0.26) | 94 |
| pAUG2p18 | 1.2 (0.65) | 27 |
| psAUG2 | 1.5 (0.76) | 21 |
| pAUG1GCG2 | 1.6 (0.79) | 43 |
| pGCG1AUG2 | 0.0063 (0.0035) | 5100 |
| pGCG1GCG2 | 15 (7.7) | 2 |
| pAUG1fs | 0.0030 (0.0020) | 11 000 |
aRefer to Figure 2A for additional information.
bThe pYES2 control strain was an average of 17 trials and the Ty1 GAG expression strains were an average of two trials.
Refer to ‘Materials and Methods’ section for additional information.
To determine if AUG1 or AUG2 were also responsible for p22/p18 synthesis in this context, wild-type and mutant versions of the p18 coding region (Figure 2A) were fused to the GAL1 promoter of pYES2, which also includes the GAL1 5′ untranslated region (UTR) and the CYC1 transcriptional terminator and analyzed for Ty1 mobility (Table 2). As expected from previous results (22) (Figure 1 and Table 1), including AUG1 and AUG2 in the expression plasmid (pAUG1p18) resulted in a 39 000-fold decrease in Ty1his3-AI mobility. However, expression from AUG2 (pAUG2p18) in the absence of AUG1 and the 30 intervening nucleotides decreased transposition only 27-fold, suggesting that either protein content or expression accounts for loss of over 1400-fold potency when compared with pAUG1p18. To further understand the sequence requirements responsible for loss of inhibition, GCG-alanine was substituted for either or both AUG codons (pAUG1GCG2, pGCG1AUG2 and pGCG1GCG2). Expressing pAUG1GCG2 inhibited Ty1his3-AI mobility 43-fold, while pGCG1AUG2 restored inhibition to over 5000-fold (Table 2), raising the possibility that AUG2 is used preferentially to initiate synthesis of p18 when fused to the pYES2 GAL1 promoter segment and the intervening nucleotides between AUG1 and AUG2 increase translation initiation. This view was reinforced by analyzing a AUG1p18 derivative containing a frameshift mutation (+C1040) (Figure 2A) that was inserted between AUG1 and AUG2 (pAUG1fs), as described above (Figure 1). Expressing pAUG1fs inhibited mobility 11 000-fold, which is comparable to that obtained with pAUG1p18 (Table 2). To determine the minimal p18 coding region inhibited Ty1his3-AI mobility, the C-DR was deleted from AUG1p18 (psAUG1) and AUG2 (psAUG2) (Figure 2). Even though the coding sequence was reduced to 96 codons (11 kDa) and lacked half the NAC region, robust inhibition was detected as long as the AUG1-AUG2 interval (psAUG1) was present. As expected, replacing both candidate start codons with GCG (pGCG1GCG2) abolished inhibition.
Immunoblot (Figure 2C and Supplementary Figure S1) and northern (Supplementary Figure S2) analyses were performed using cells induced for expression to detect the relative amounts of Gag proteins and transcripts from the pYES2-GAG derivatives and pGTy1his3-AI. Yeast Hts1 (histidyl tRNA synthetase) served as an internal standard for comparing different levels of Gag. Gag proteins were detected with either the TY-tag (40) or p18 (22) antisera in cells expressing the full-length Gag or Gag-derivatives, with the exception of the sCTR and the GCG1GCG2 mutant. Both the CTR and sCTR inhibited Ty1 mobility close to seven-fold (Table 2), however, the sCTR was not detected by immunoblotting whereas the CTR was (Figure 2C). These results raise the possibility that the sCTR is inhibitory but present in a low amount, perhaps due to the low level of mRNA produced from psCTR and instability of the sCTR protein. Mature Gag-p45 produced from pYES2 (p45) and pGTy1his3-AI are of the same size and therefore indistinguishable, but a shorter exposure of the filter revealed less Gag-p45 in the strain expressing just pGTy1his3-AI when compared with the strain expressing Gag-p45 from both p45 and pGTy1his3-AI (Supplementary Figure S1), suggesting both plasmids express Gag. A strain expressing only p45 also displayed full-length Gag (data not shown). The level of p18 and derivatives containing mutations in AUG1 (pGCG1AUG2), AUG2 (pAUG1GCG2) or the +C1040 frameshift (pAUG1fs), as well as the C-terminal truncations sAUG1 (psAUG1) and sAUG2 (psAUG2) correlated with the level of inhibition (Figure 2C, Supplementary Figure S1 and Table 2). For example, a low level of protein and inhibition of Ty1 mobility resulted from expression of pAUG2p18 and pAUG1GCG2 when compared with pAUG1p18, pGCG1AUG2 and pAUG1fs. p18 appeared in two forms (denoted by the asterisk, Figure 2C and Supplementary Figure S1) in several derivatives (pAUG1p18, pGCG1AUG2 and pAUG1fs): a major reactive protein and a minor slower migrating variant (see Supplementary Figure S1 for a shorter exposure of the filter). The composition of the minor variant will require further study, however, the variant probably does not derive from initiation at AUG1 since it is present in the pGCG1AUG2 and pAUG1fs mutants.
To determine the relative amount of Ty1 RNA from the pYES-GAG expression plasmids, total RNA isolated from galactose induced cells was subjected to northern analysis using a 32P-labeled riboprobe derived from Ty1 GAG (Supplementary Figure S2). The RNA levels varied over a three-fold range for most of the strains (pCTR, pAUG1p18, pAUG2p18, pAUG1GCG2, pGCG1AUG2, pGCG1GCG2 and pAUG1fs) when compared with the Ty1his3-AI transcript. The psCTR, psAUG1 and psAUG2 produced low levels of mRNA, which likely contributes to the reduced levels of p18 and inhibition of Ty1 mobility.
The level of inhibition and protein production observed from expression of different Gag regions (Figures 1C and 2C, Supplementary Figure S1, and Tables 1 and 2) guided our choice of recombinant proteins to analyze biochemically for RNA interactions in vitro and VLP assembly in vivo. Therefore, we purified the CTR, sCTR, AUG1p18, AUG2p18 and sAUG2 as N-terminal fusions to a 6x-His affinity tag in E. coli by metal chelate chromatography (Supplementary Figure S3). The recombinant proteins remained soluble in 1 M NaCl and were used to characterize RNA binding properties and NAC activity. The Ty1 CTR was used for NAC analysis, since we were unable to purify soluble full-length Ty1 Gag p45 at high concentration. This problem has only recently been overcome for producing soluble full-length HIV Gag-p55 (41).
Translation of p22 is cap-dependent in vitro
Translational activity of two closely spaced AUG1 and AUG2 in Ty1i RNA observed in vivo raised the possibility that alternative initiation events could be involved in p22 translation (Figure 1). To gain further insight on how AUG1 and AUG2 codons are utilized, we analyzed the requirement of the 5′ cap structure for translational activity in vitro (Figure 3). Model Ty1 RNA templates were synthesized that contain AUG1 and AUG2. Those templates start at the reported Ty1i initiation site (Ty1 nt 1000) (22) and end at the natural stop codon for Gag. Uncapped AUG1AUG2 RNA was poorly translated in WGE (Figure 3 Lane 2). However, the translation level of p22 increased almost 20-fold when a 5′ cap was present and approached 80% of the translation activity of a leaderless, capped Renilla Luciferase control RNA (Figure 3, Lane 1 and 3). This observation suggests that p22 translation occurs via a cap-dependent process. We corroborated this result by translation of AUG1AUG2 RNA capped with a non-physiological ApppG cap or in the presence of 500 μM m7GpppG cap analog in the translation reaction. ApppG cap reduces binding of the eIF4E initiation factor to the 5′ end of RNA while preincubation of WGE with m7GpppG results in eIF4E entrapment, which leads to inhibition of cap-dependent translation process (42,43). In both cases, translation was downregulated to a level comparable with the uncapped RNA, further supporting cap-dependence of p22 synthesis (Figure 3, Lane 4 and 5). To investigate whether both AUGs share similar cap-dependent translation activity, we mutated AUG1 (RNA GCG1AUG2) or AUG2 (RNA AUG1GCG2) by introducing GCG-alanine. Both mutant RNAs were also translated in a cap-dependent manner (Figure 3, Lanes 6–13). The level of p22 synthesis from both RNAs was similar. However, in both cases the combined translation level (RNA GCG1AUG2 + RNA AUG1GCG2) for capped transcripts reached only 30% of the initial value calculated for the capped AUG1AUG2 RNA. This result might be caused by perturbations in RNA structure of the region containing AUG1 and AUG2 codons.
Figure 3.
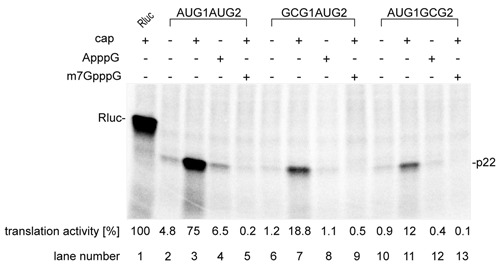
In vitro translation of Ty1 RNAs in wheat germ extract (WGE). In vitro transcribed RNA AUG1AUG2, GCG1AUG2 and AUG1GCG2 were translated in WGE in the presence of 35S-methionine followed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis electrophoresis and autoradiography. Quantitation of the translation products normalized to the level of Renilla Luciferase is shown below the gel (SD ≤ 0.09).
Aggregation of RNA by Gag-derived proteins
One of the principal components of NAC activity of retroviral nucleocapsid proteins is the ability to aggregate NA, leading to an increase in their local concentration (14,44,45). A sedimentation assay (21,46,47) was used to directly examine NA aggregation properties of Ty1 Gag-derived proteins. 32P-labeled Ty1 mini RNA (nt 1–560), which contains the cis-acting sequences required to initiate reverse transcription in vivo (31), was incubated at 24°C with increasing amounts of Ty1 proteins. We used a relatively low temperature for these reactions to approach the optimal temperature for Ty1 transposition (48). Aggregates were pelleted by centrifugation, whereas unbound RNA and protein remained in the supernatant. The CTR and AUG2p18 proteins aggregated Ty1 mini RNA with similar efficiency (Figure 4A). In both cases, ∼80% of RNA aggregated at 1:9 protein to nucleotide molar ratio. AUG1p18 showed slightly lower aggregation efficiency, however, the sCTR was unable to direct aggregation. To confirm that the NA aggregation properties of Ty1 Gag derived proteins are, as observed for NACs, sequence non-specific (44,45), sedimentation assays were also performed with NA derived from the HIV-1 TAR region (Figure 4B). Similar aggregation efficiency was observed for the CTR and AUG2p18 proteins, where ∼80% of RNA aggregated at 1:9 protein to nucleotide molar ratio. AUG1p18 showed lower aggregation efficiency and sCTR did not direct aggregation. As mentioned above (Figure 2A), the sCTR lacks almost half (48/103) of the NAC region defined previously (21). Deleting two basic amino acid clusters at C-terminus of the sCTR likely explains the differences seen between the sCTR and CTR, AUG1p18 and AUG2p18. For retroviral nucleocapsid proteins, an electrostatic model of protein-induced NA aggregation has been proposed (44,45). The N-terminal basic regions of HIV-1 and HIV-2 nucleocapsid proteins (NCp7 and NCp8) are thought to be crucial for both NA aggregation and chaperone activity. Deletions or mutations of basic residues led to significant decrease of chaperone activity (47,49–51).
Figure 4.
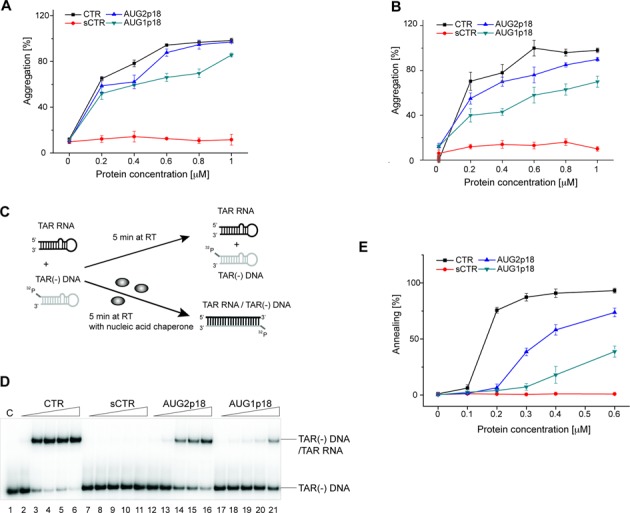
Nucleic acid aggregation and chaperone activities of Ty1 Gag-derived proteins. Percent of (A) Ty1 mini RNA (560-nt) and (B) TAR RNA and DNA aggregated by each protein at 24°C. Aggregation assays were performed as a function of protein concentration as indicated. Proteins concentrations correspond to 1:28; 1:14; 1:9; 1:7 and 1:5.6 protein to nt ratios, respectively. The graphs represent averaged data from three independent experiments. The error bars represent standard deviations. (C) Schematic of the DNA/RNA strand annealing reaction. The annealing assays were performed using TAR(−) DNA/TAR RNA substrates as a function of protein concentration (0; 0.1; 0.2; 0.3; 0.4; 0.6 μM). Protein concentrations correspond to 1:7.8; 1:4; 1:2.6; 1:2 and 1:1.3 protein to nt ratios. (D) A representative electrophoretic analysis of TAR(−) DNA/TAR RNA annealing. Lane 1 contains a control sample (denoted C) lacking protein and lanes 2–6, 7–11, 12–16 and 17–21 contain increasing amounts of CTR, sCTR, AUG2p18 and AUG1p18, respectively, as described above. (E) The graphs represent averaged data from three independent annealing experiments for each protein. The error bars represent standard deviations.
Annealing of model substrates derived from HIV-1
NACs promote annealing of complementary or partially complementary NA strands. Annealing of HIV-1 TAR(−) DNA to a complementary TAR RNA or TAR(+) DNA is typically used as model assay to study the NAC activity (52–55) (Figure 4C). This in vitro model mimics the first strand transfer of reverse transcription. The TAR oligonucleotides can fold to form stable hairpin structures but spontaneous formation of the TAR(−) DNA/TAR RNA duplex in vitro is extremely slow at physiological conditions. However, addition of a NAC protein significantly accelerates the annealing rate (56,57). We tested the effect of CTR, sCTR, AUG2p18 and AUG1p18 proteins on annealing of HIV-1 TAR(−) DNA to TAR RNA (Figure 4D and E). The oligonucleotides were incubated at 24°C for 5 min with increasing protein/nucleotide molar ratios and products were separated on non-denaturing gels. The CTR protein effectively accelerated the formation of TAR(−) DNA/TAR RNA duplex and 75% of annealing was observed at a 1:4 CTR to nucleotide ratio. A slight increase in CTR concentration (1:3) resulted in a 90% annealing efficiency. As predicted from the aggregation assay, the sCTR failed to accelerate annealing of the TAR hairpins (Figure 4D and E). AUG2p18 displayed reduced annealing activity when compared with the CTR, despite the presence of the NAC region or if AUG2p18 was added at a higher molar ratio (1:1). Interestingly, the annealing activity of AUG1p18 was much lower than that of AUG2p18 and at the same molar ratio (1:1), even though AUG1p18 and AUG2p18 only differ by 10 residues at their N-terminus.
To compare our results with data obtained previously for Gag peptide TYA1-D (21), we determined the effect of the CTR and AUG2p18 in the annealing of TAR(−) DNA and TAR(+) DNA at 30°C for 10 min. Both proteins effectively facilitated annealing of this simplest substrate pair, however the difference in their activity was still evident (Supplementary Figure S4). More than 70% of TAR DNA strands were annealed at the CTR to nt molar ratio of 1:8, but only 20% at the same concentration of AUG2p18. Studies of the retroviral NCp7 protein demonstrated that 1 protein per 5–8 nt is required for optimal chaperone activity (15,52,58,59), which is comparable to our results with the CTR.
Distinct RNA binding properties of the p18 proteins
One of the key features of RNA chaperone proteins is their ability for non-specific interactions with NA that can be altered by competing cations (59,60). Since Ty1 proteins derived from Gag possess different chaperone activities, perhaps it is because they bind RNA differently. Therefore, we determined the average dissociation constants for RNA/protein complexes using a double-membrane filter-binding assay (Figure 5A–D) (61). The binding reactions contained 32P-labeled Ty1 mini RNA, the CTR (Figure 5A), AUG2p18 (Figure 5B) or AUG1p18 (Figure 5C) and NaCl concentrations ranging from 10–500 mM. The three Gag derivatives displayed similar binding affinities to Ty1 mini RNA (Kd ∼7 nM) in 10 and 50 mM NaCl (Figure 5D and Supplementary Table S3). However, at salt concentrations between 100–250 mM, a trend emerged where the CTR had the lowest Kd, AUG1p18 was intermediate and AUG2p18 was highest (∼46, ∼72, ∼650 nM at 250 mM NaCl, respectively). Furthermore, the binding of AUG2p18 was most sensitive to salt since its Kd varied more than 100-fold with increasing salt concentration. In contrast, the change in Kd for AUG1p18 with increasing salt resembled the CTR rather than AUG2p18. The Kd in increasing salt represents the binding conditions where electrostatic interactions are increasingly masked by salt and the higher contribution to binding comes from specific contacts such as hydrogen bonds or aromatic residue stacking (62). Together, these results suggest that AUG2p18 binding is relatively non-specific and focuses our attention on the role that the 10 additional residues (MKILSKSIEK) present in AUG1p18 play in RNA binding. However, the chemically synthesized peptide (p1; MKILSKSIEK) did not bind Ty1 RNA even at 10 μM concentration (Supplementary Figure S5), suggesting that multiple protein regions contribute to modulating the RNA binding by the NAC region. None of the proteins bound RNA at 500 mM NaCl. Moreover, neither the sCTR nor sAUG2 bound Ty1 mini RNA even in low salt (Supplementary Figure S5), demonstrating critical role of C-DR in RNA binding.
Figure 5.
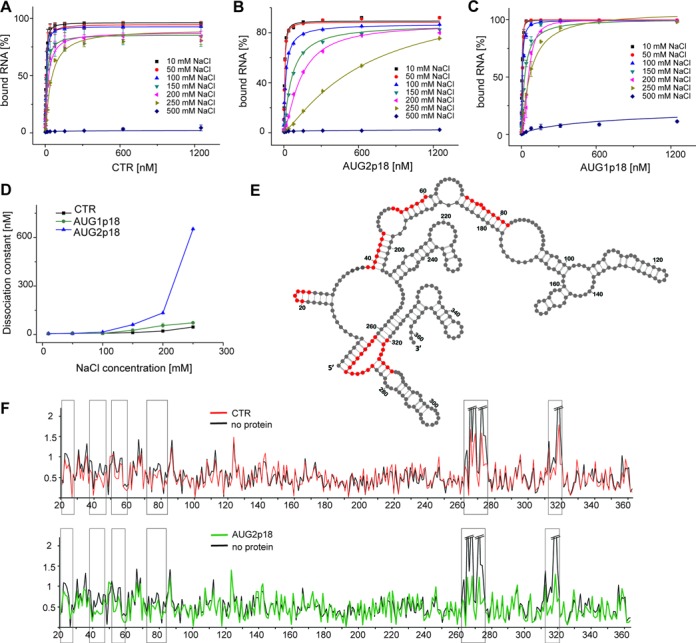
RNA binding properties of CTR, AUG2p18 and AUG1p18 proteins. Data plots of the filter-binding assay performed in different concentrations of NaCl (10–500 mM) for Ty1 mini RNA and (A) CTR, (B) AUG2p18 and (C) AUG1p18. The lines correspond to the best fit of the data. (D) Data plot of the dissociation constant measured for CTR, AUG2p18 and AUG1p18 as a function of NaCl concentration. The error bars represent standard deviations. (E) 2D structure model of +1–362 region (35) of Ty1 mini RNA with the positions protected from hydroxyl radical cleavage in the presence of the Ty1 Gag derived proteins marked (red). (F) Reactivity plots of protein free Ty1 mini RNA (black) in comparison with RNA probed in the presence of CTR (red) and p18 (green). Regions showing consistent increased reactivity over several nucleotides are boxed.
To further investigate differences in binding of Ty1 Gag derived proteins to the Ty1 mini RNA, binding sites were mapped using hydroxyl radical probing (Figure 5E and F). Overall reactivity profiles for RNA in the presence or absence of CTR, AUG1p18 or AUG2p18 were compared to reveal protected sequences. Several regions displayed decreased susceptibility to hydroxyl radical cleavage in the presence of CTR. The strongest effect was observed in the RNA strands forming a pseudoknot structure, which is critical for the Ty1 retrotransposition (32,35). However several other regions displayed smaller but reproducible reactivity differences spanning several consecutive nucleotides (Figure 5F, Supplementary Dataset 1). Those motifs correspond well with the proposed Ty1 Gag binding sites revealed by probing RNA in VLPs and in the protein-free state using SHAPE (35). Interestingly, we found reactivity differences within the same regions of Ty1 mini RNA in the presence of p18 proteins (Figure 5E and F, Supplementary Dataset 1). Therefore, our data suggest that all analyzed proteins bind within the same regions of Ty1 mini RNA in vitro.
p18 activity in the tRNAiMet annealing to Ty1 RNA is compromised
Ty1 uses a retroviral-like mechanism to initiate reverse transcription, where yeast tRNAiMet anneals with a multipartite PBS on Ty1 RNA (35) and acts as a primer (63). To further investigate NAC activity of Ty1 Gag derived proteins, we used an assay that mimics the annealing of tRNAiMet to Ty1 RNA during Ty1 retrotransposon replication. Unmodified tRNAiMet was 3′-end labeled with 32P and incubated with a Ty1 mini RNA in the presence of recombinant Ty1 Gag derived proteins. The annealed products were analyzed by agarose gel electrophoresis following removal of proteins from the reaction. The CTR enhanced annealing of primer tRNAiMet with the PBS of Ty1 RNA (Figure 6A and B). At a molar ratio of 1:4, about 50% of tRNAiMet annealed with Ty1 mini RNA. Further increasing the molar ratio of the CTR (1:2) raised the annealing efficiency to ∼80%. In contrast, at the same concentration only 40% annealing was observed for AUG2p18 and <15% for AUG1p18. Although both restriction proteins enhanced tRNAiMet annealing, AUG1p18 displayed significantly weaker NAC activity. Furthermore, even increasing the molar ratio of AUG2p18 or AUG1p18 to 1:1 did not increase tRNAiMet annealing to a level comparable with that observed for the CTR. As expected, the sCTR was unable to promote tRNAiMet annealing, supporting the key role of the C-terminal 48 residues in the NAC activity of Ty1 Gag-p45.
Figure 6.
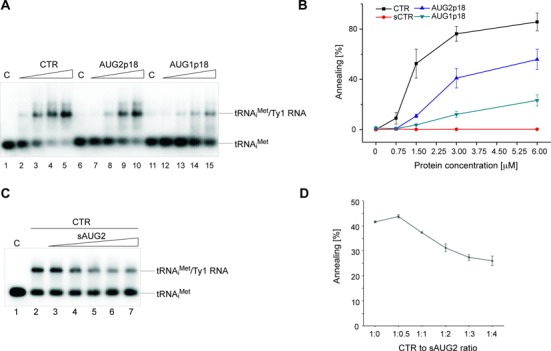
Protein-mediated annealing of tRNAiMet with Ty1 RNA. (A) A representative electrophoretic analysis in the presence of increasing concentrations (0; 0.75; 1.5; 3; 6 μM) of CTR, AUG2p18 or AUG1p18. Proteins concentrations correspond to 1:8; 1:4; 1:2 and 1:1 protein to nt ratios. Lanes 1, 6 and 11 (denoted C) represent control samples that lack protein and lanes 2–5, 7–10 and 12–15 contain increasing amounts of CTR, AUG2p18 and AUG1p18, respectively, as described above. (B) The graph representing the averaged percent of annealed tRNAiMet from three independent experiments for CTR, AUG2p18, AUG1p18 or sCTR protein. The error bars represent standard deviations. (C) A representative electrophoretic analysis performed in the presence of CTR (constant concentration of 1.5 μM) and increasing concentrations of sAUG2 (0.75, 1.5, 3, 6 μM). Lane 1 (denoted C) is a control sample lacking protein and lanes 2–7 contain increasing amounts of sAUG2 relative to CTR protein, as described above. (D) The graph representing the averaged percent of annealed tRNAiMet in the presence of both proteins. Respective molar protein ratios are indicated.
p18 proteins impact the CTR chaperone activity in vitro
Since forms of p18 inhibit Ty1his3-AI mobility in vivo (Tables 1 and 2), we determined whether recombinant AUG1p18, AUG2p18 or sAUG2 inhibited NAC activity of the CTR in vitro (Figure 6C, D and Supplementary Figure S6). To simplify the experiment, the CTR was preincubated with increasing amounts of sAUG2, which lacks chaperone activity and RNA binding (Supplementary Figure S5) but retains restriction activity (Table 2) and then the NAC activity of the mixture was assessed by annealing 32P-labeled tRNAiMet with Ty1 mini RNA (Figure 6C and D). At a 1:4 CTR to sAUG2 ratio, the efficiency of tRNAiMet annealing decreased from 40 to 25%. Since AUG1p18 and AUG2p18 possessed some NAC activity (Figure 4), we hypothesized that there may be an additive increase in tRNAiMet annealing if the CTR and p18 proteins did not interact and were present in non-saturating conditions with respect to RNA binding (refer to Figure 6A and B). For example, 1.5 μM CTR and 3 μM AUG2p18 each enhance annealing to about 40%, therefore, the mixture should result in 80% annealing if the proteins do not interact. However, no increase in annealing was detected when AUG1p18 or AUG2p18 were mixed with the CTR in several different molar ratios (Supplementary Figure S6). Therefore, when p18 and CTR proteins were both present, 7.5 μM protein concentration was required to observe the same annealing activity as was displayed by CTR alone at 1.5 μM. Taken together, our results support the view that p18 inhibits NAC activity of the CTR, and that inhibition is mediated by p18-CTR interactions.
GST-pulldowns support an interaction between the CTR and p18
To further explore the interaction between Gag and p18 (22), a fusion protein consisting of the CTR tagged on its N-terminus with glutathione S-transferase from Schistosoma japonicum (64) was expressed from the GAL1 promoter (pGST-CTR) in a Ty1-less strain that also expressed pAUG1p18 (Figure 7A) or psAUG1 (deleted C-DR p18; Figure 7B). Free glutathione S-transferase (pGST) was coexpressed with pAUG1p18 or psAUG1 as negative controls. Protein extracts were immunoblotted using antisera specific for GST, p18 or Hts1 prior to mixing with glutathione-coated resin (Input) or released from GST complexes bound to resin after several washes in lysis buffer (Pull-down). Unexpectedly, we observed a faster migrating p18 derivative (∼12.5 kDa, denoted *), whose appearance correlated with coexpression of free GST or GST-CTR (Supplementary Figure S7). The aberrant p18 protein was not investigated further, however, expression of AUG1p18 alone yielded the expected product, as did expression of GST-CTR and GST alone. GST-CTR formed a complex with p18 that was detected with anti-p18 antibodies, whereas the aberrant form did not. To further define the region of p18 that interacts with the CTR, pull down analysis was performed with cells expressing GST-CTR and psAUG1 (Figure 7B). Note that psAUG1 produces a protein lacking the C-DR (Figure 2) and RNA binding/NAC activity, but inhibits Ty1 mobility more than 8000-fold (Table 2). The pulldown analysis showed that p18 deleted for the C-DR also interacted with GST-CTR. Hts1 was used as a control for non-specific trapping of cellular proteins in the GST complexes and, as expected, was detected only in the input samples. Our results suggest there is an interaction between the CTR and p18 and the interaction does not require the NAC region of p18.
Figure 7.
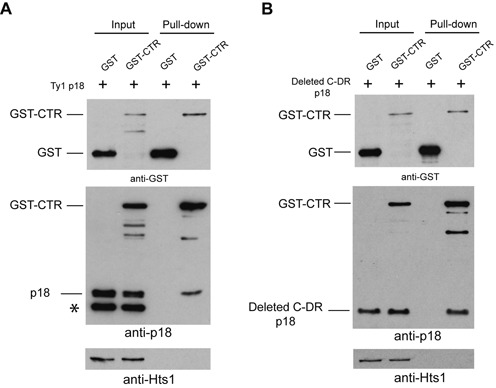
GST-CTR interacts with p18 or deleted CD-R p18. Protein extracts (Input) from a Ty1-less strain induced for expression of GST [pEG(KT); pGST], GST-CTR (pBDG1496; pGST-CTR) and (A) p18 (pAUG1p18) or (B) p18 lacking the C-DR (pBDG1612; psAUG1) were incubated with glutathione-coated resin. Bound proteins (Pull-down) were analyzed by immunoblotting with GST and p18 antisera to detect, GST, GST-CTR, p18 or deleted C-DR p18 after extensive washing with lysis buffer. A putative p18 degradation product of ∼12.5 kDa is noted in panel A (*; also refer to Supplementary Figure S7).
p22/p18 affects Ty1 RNA dimerization and packaging
Since the CTR interacts with p18, we determined if Ty1 RNA packaging might be affected by the Ty1 restriction factor. Ty1 RNA is dimeric in VLPs (4) and RNA dimerization is considered prerequisite for the RNA packaging (65), therefore, we performed in vitro dimerization assays to assess the influence of Gag derived proteins. Cristofari et al. demonstrated that TYA1-D peptide promotes Ty1 RNA dimerization when tRNAiMet is present in the reaction mixture (21). Therefore, we reconstituted the previously described in vitro dimerization system. 32P-labeled Ty1 mini RNA was combined with tRNA at 1:2 molar ratio and incubated with increasing concentrations of CTR, AUG1p18 or AUG2p18 (Figure 8A). At a molar ratio of 1:20 about 25% of Ty1 mini RNA dimer was observed with CTR but only about 10% with AUG2p18 and <5% with AUG1p18 (Figure 8A and B). Further increase in the molar ratio of the CTR to 1:7 raised the dimerization efficiency to ∼30%. In contrast, only 20% annealing was observed for AUG2p18 and about 10% for AUG1p18 at the same concentrations. Further increases in the protein concentration also did not promote dimer formation. A low level of dimeric RNA structures formed in the presence of p18 proteins and their lower chaperone activity support the notion that p18 proteins negatively influence RNA transactions required for VLP assembly.
Figure 8.
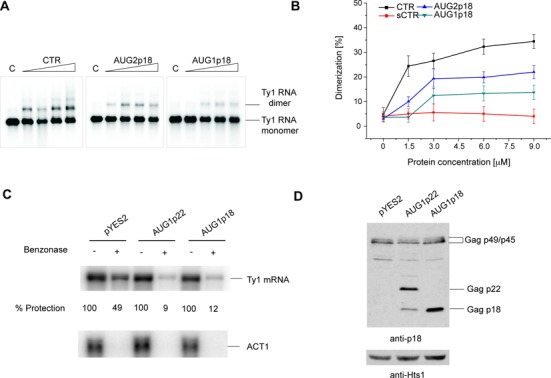
Impact of p22/p18 or p18 on RNA dimerization and packaging. (A) A representative electrophoretic analysis in the presence of increasing concentrations of CTR, AUG2p18 or AUG1p18. Proteins concentrations correspond to 1:20; 1:10; 1:7 and 1:5 protein to nt ratios. Lanes denoted C represent control samples that lack protein. (B) The graph representing the averaged percent of Ty1 mini RNA dimer from three independent experiments. The error bars represent standard deviations. (C) Nuclease protection of Ty1 mRNA in cells coexpressing pGTy1his3-AI alone and pAUG1p22 or pAUG1p18. Equal aliquots of whole-cell extracts from galactose-induced cells were incubated with (+) or without (−) the nuclease benzonase. RNA extracted from these samples was analyzed by Northern blotting using a 32P-labeled riboprobe specific for Ty1 mRNA. Protection from benzonase is expressed as a ratio of treated to untreated Ty1 mRNA. ACT1 was used as a control to confirm RNA degradation in the nuclease treated samples. (D) Total protein extracts used in (C) was immunoblotted with p18 antiserum (anti-p18). Histidyl tRNA synthetase (anti-Hts1) served as a loading control.
To examine Ty1 RNA packaging in vivo, cell extracts containing VLPs assembled in the absence or presence of p22/p18, which result from co-expressing pGTy1his3-AI and an empty vector, or pAUG1p22 and pAUG1p18, were treated with the nuclease benzonase. Total RNA (Figure 8C) and protein (Figure 8D) was subjected to northern and immunoblot analyses using Ty1 specific probes or antibodies. Hybridization signals were normalized to mock-treated samples treated in parallel. Benzonase activity was monitored by degradation of cellular ACT1 transcripts or control extracts spiked with Ty1 RNA synthesized in vitro (35). Equivalent levels of Gag-p49/p45 were present in the samples. p22 and its processed form p18 (22) were detected in extracts from cells expressing AUG1p22, as was p18 in cells expressing AUG1p18. The Hts1 protein served as a loading control. As expected, Ty1 RNA decreased about two-fold when extracts from cells lacking p22/p18 were treated with benzonase. However, co-expression of p22 or p18 with pGTy1his3-AI resulted in a 10-fold decrease in the level of protected Ty1 RNA. Our results suggest that p22/p18 prevents stable packaging of Ty1 RNA.
DISCUSSION
Here, we investigate the functional organization of Ty1 GAG, the NAC properties of several Gag derivatives and further define how p22/p18 interacts with Gag and inhibits retrotransposition. Although Ty1 Gag and p22 play opposing roles in the process of retrotransposition (22), the p22/p18 restriction factors are contained within Gag-p49/p45 and share a previously reported NAC domain (21). We demonstrate that p18 and its short derivatives sAUG1 and sAUG2 impact Ty1his3-AI mobility, and the level of inhibition correlates with the level of protein expression. In contrast, the NTR, CTR segments as well as mature Gag-p45 have a modest effect on Ty1 mobility despite a high level of protein production (Figure 2C and Table 2). Mutational analyses of translation initiation from either Ty1i RNA or when different GAG segments are fused to the GAL1-promoter and its 5′ UTR show that although translation initiates at either of two closely spaced AUG codons, the use of either AUG1 or AUG2 is context dependent. Together, these results support the idea that Ty1 GAG evolved a specific organization and expression strategy to produce products both essential and antagonistic for retrotransposon movement.
In vitro translation analyses were used to further understand how AUG1 and AUG2 codons are chosen for initiation. The most profound observation was almost a 20-fold stimulation of p22 synthesis when AUG1AUG2 RNA contained cap structure at the 5′ end (Figure 3), which suggests that the majority of the protein is synthesized in a cap-dependent manner. This finding was further supported by translation where cap-dependent formation of initiation complexes was compromised and p22 protein synthesis was inhibited to the level observed for uncapped RNA (Figure 3). Using transcripts with mutations of AUG1 (RNA GCG1AUG2) or AUG2 (RNA AUG1GCG2), we showed that both codons are translated in a cap-dependent manner. However, the combined levels of translation from the capped mutated transcripts reached only 30% of translation from capped AUG1AUG2 RNA (Figure 3).
Further studies are needed to explain how AUG selection occurs, however one of the possible reasons may be the contribution of the secondary and/or tertiary RNA structure present in the Ty1i 5′ UTR and in the 30 nt region spanning AUG1 and AUG2, and if the AUG mutations alter the structure. This idea is under investigation and is supported by the different levels of p22 detected in vivo from transcripts bearing a mutation in AUG1 or AUG2 and the presence of the Ty1i versus the GAL1 5′ UTRs (compare Figures 1 and 2). The Ty1i RNA 5′ UTR extends for about 38 nt upstream of AUG1 (22), but these sequences were replaced by the GAL1 5′ UTR to maximize expression from the well-studied GAL1 promoter (Figure 2). The ratio of p22 synthesis was about 1:20 for pAUG1GCG2 and pGCG1AUG2 respectively (Figure 2). However expression from constructs using the natural Ty1i 5′ UTR (pAUG1GCG2-PΔ and pGCG1AUG2-PΔ) resulted in equivalent amounts of p22, which was also visible in in vitro translation assay (Figures 1 and 3). These results suggest that proper folding of 5′ proximal region of Ty1i RNA may dictate initiation from AUG1 and AUG2.
The presence of two translationally active AUG codons in Ty1i RNA raises the possibility that an alternative mechanism of translation initiation is involved in p22 synthesis. Our results suggest that translation from AUG1 involves canonical cap-dependent scanning while leaky scanning is the most plausible mechanism for AUG2. Leaky scanning takes place when the first AUG codon is localized in a suboptimal context (66). Consequently, some of the initiation complexes skip the first AUG codon and translation starts from a downstream AUG present in an optimal context. In the case of Ty1i RNA, this rule is fulfilled since AUG2 is embedded in a better context for the yeast translational apparatus [5′-AAAAAAUGC-3′] than AUG1 [5′-GAUAUCAUGA-3′] (67). The leaky scanning mechanism is further supported by the expression of pAUG1fs-PΔ and pAUG1fs in vivo (Figures 1 and 2). These constructs contain frameshift mutation that introduces a stop codon upstream of AUG2. Hence, full-length p22 can be synthesized only from AUG2. The expression of p22 protein from pAUG1fs-PΔ and pAUG1fs indicate that a significant number of initiation complexes skip AUG1 and start translation from AUG2.
Expression of p18 is complicated by translation from two AUG codons, therefore, recombinant AUG1p18 and AUG2p18 were analyzed for NAC activity as well as RNA binding, along with the CTR and the shorter derivatives sCTR and sAUG2. The CTR displays comparable NAC activity to HIV-1 nucleocapsid protein (NCp7), a canonical chaperone (68,69) and extends previous analyses performed with Ty1 synthetic peptide TYA1-D from the CTR of Gag-p45 (21). Interestingly, p18 proteins contain less activity than the CTR even though all proteins contain sequences present in TYA1-D. In addition, p18 proteins and a derivative incapable of RNA binding (sAUG2) decrease the CTR chaperone activity. These results support the idea that one aspect of p18 restriction involves inhibition of Gag-p45 NAC functions by p18/Gag interactions. Our finding that the CTR and p18 interact in the yeast cells further reinforces this notion.
Gag-p45 is the major structural component of Ty1 VLPs (70) and also binds NA in vitro (21,71). The 103-amino acid, synthetic peptide TYA1-D is apparently necessary for RNA binding since a Ty1 Gag derivative lacking this part of the protein fails to bind RNA in vitro. Because TYA1-D displays NAC activity, it was proposed that Ty1 Gag helps regulate key steps during retrotransposition. In vitro evidence for its involvement in primer tRNAiMet annealing and reverse transcription (21,72), genome cyclization (73) and genomic RNA dimerization (21) was presented. Here, we compared the RNA binding properties and chaperone activity of the C-terminal part of Ty1 Gag-p45 (CTR–230 aa) and newly identified Ty1 proteins AUG2p18 and AUG1p18 that inhibit Ty1 retrotransposition by interfering with VLP assembly, maturation and function (22). Ty1 Gag contains three clusters of basic amino acids and present in TYA1-D that could mediate RNA binding and chaperone activity (21). Basic amino acids in the nucleocapsid-like region of the endogenous retroelement Gypsy, which like Ty1 lacks zinc-finger motifs, are required for RNA binding and chaperone activity (11). Our experiments with the sCTR and sAUG2 show that deleting clusters 2 and 3 abrogate RNA binding (Supplementary Figure S5) and NAC activity (Figures 4A, B, D, E, 6A and B), and strongly support the crucial role of the C-terminal 47 residues (Figure 2A; C-DR) in RNA/protein transactions. Interestingly, in the case of the retroviral-like NC from the Ty3 retrotransposon, substituting local sets of charged residues by alanines show that individual residues do not perform essential functions in retrotransposition but the clusters are necessary (74). The decreased chaperone activity of p18 proteins relative to the CTR suggests that N-terminal end of CTR is also important for activity. The N-terminus of the CTR coincides with C-terminus of the predicted disordered region of the NTR, whose specific functions are not understood. However, other than the C-DR, the rest of the CTR is not sufficient for RNA binding, since the sCTR lacks the C-DR (half of the NAC region) and fails to bind Ty1 RNA. We conclude that the N-terminal segment of the CTR may help regulate NAC activity, participate in VLP assembly most likely by protein–protein interactions and interact with p22/p18 (Figure 7) (Mitchell and Garfinkel, unpublished results).
The CTR, AUG2p18 and AUG1p18 display NAC chaperone activity in vitro, however, chaperone activity of the p18 proteins is lower than that of the CTR. The tRNAiMet annealing assays demonstrate larger differences in NAC activity between the CTR and p18 proteins (Figure 6) than the structurally simpler TAR substrates (Figure 4). Ty1 and Ty3 contain a bipartite PBS with extensive RNA architecture (35,73,75), potentially requiring more efficient chaperone functions. This might indicate that during Ty1 replication tRNAiMet annealing with PBS is one of the processes inhibited by p18. AUG1p18 and AUG2p18 exhibit different characteristics in RNA binding and chaperone activity. AUG1p18 shows tighter binding of Ty1 RNA but has low chaperone activity, whereas AUG2p18 shows the converse behavior. We do not understand the functional relevance of these reciprocal properties, however, mutational analyses suggest both AUG1p18 and AUG2p18 are produced in yeast and restrict Ty1 transposition (Figure 1). It will also be interesting to determine the function of the 10 amino acid extension on AUG1p18, since that is the only difference in primary sequence between the two proteins.
p18 proteins and the CTR bind Ty1 RNA with high affinity at a physiological salt concentration of 150 mM (Figure 5), suggesting the possibility that p18 and Gag compete for the same binding sites on Ty1 genomic RNA during the process of retrotransposition. Demonstrating that the CTR and p18 proteins bind Ty1 mini RNA in vitro within the same structural motifs further supports this observation and is also consistent with results obtained in vivo (35). p22/p18 may interact with unassembled Gag-p45 or VLP assembly intermediates, and therefore, may contain multiple protein–protein interaction domains capable of inhibiting the transactions required to form a functional particle (22). p18 proteins display lower activity in the Ty1 RNA dimerization (Figure 6), which is considered a prerequisite for encapsidation (65). Moreover, Ty1 genomic RNA in VLPs is much less protected from nuclease degradation when p22 or p18 are present during assembly (Figure 8). Importantly, the observations that p18 interacts with both Gag (22) and the CTR (Figure 7A), and sAUG2 inhibits CTR chaperone activity (Figure 6) but does not bind NA (Supplementary Figure S5) strengthens the notion that protein–protein interactions between N-terminal portion of p18 and Gag/CTR are major reasons for the transposition inhibition in vivo. In addition, the interaction between the CTR and the sAUG1 derivative of p18 lacking the C-DR (Figure 7B), which is responsible for RNA binding and NAC activity, suggests that an RNA bridge does not mediate their association. How p18 proteins inhibit the CTR or Gag-p45 NAC functions remains unclear, but it is tempting to speculate that p18 proteins antagonize the Gag–Gag transitions required to fold RNA into a biologically active structure, which is a feature of NACs (12). Our data also supports the idea that both strong and weaker defects mediated by p22/p18 collectively contribute to the robust inhibition of Ty1 movement and forms the basis of copy number control.
Supplementary Material
Acknowledgments
We thank Steven Hajduk and Zachary Wood for sharing equipment and reagents, and Ryszard W. Adamiak, Hyo Won Ahn, Jessica A. Mitchell, Zachary Wood for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Center Poland [2011/01/D/NZ1/03478, 2012/06/A/ST6/00384]; Foundation for Polish Science [HOMING PLUS/2012-6/12 to K.J.P.]; Ministry of Science and Higher Education Poland [0492/IP1/2013/72 to K.J.P.]; National Institutes of Health [GM095622 to D.J.G.]. Funding for open access charge: Ministry of Science and Higher Education Poland [0492/IP1/2013/72 to K.J.P.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Voytas D.F., Boeke J.D. Ty1 and Ty5 of Saccharomyces cerevisiae. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 614–630. [Google Scholar]
- 2.Curcio M.J., Lutz S., Lesage P. The Ty1 LTR-retrotransposon of budding yeast. Microbiol. Spectr. 2015;3:1–35. doi: 10.1128/microbiolspec.MDNA3-0053-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elder R.T., Loh E.Y., Davis R.W. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc. Natl. Acad. Sci. U.S.A. 1983;80:2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng Y.X., Moore S.P., Garfinkel D.J., Rein A. The genomic RNA in Ty1 virus-like particles is dimeric. J. Virol. 2000;74:10819–10821. doi: 10.1128/jvi.74.22.10819-10821.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belcourt M.F., Farabaugh P.J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Youngren S.D., Boeke J.D., Sanders N.J., Garfinkel D.J. Functional organization of the retrotransposon Ty from Saccharomyces cerevisiae: Ty protease is required for transposition. Mol. Cell. Biol. 1988;8:1421–1431. doi: 10.1128/mcb.8.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devine S.E., Boeke J.D. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- 8.Swanstrom R., Wills J.W. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. NY: Cold Spring Harbor; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 9.Mullers E. The foamy virus Gag proteins: what makes them different? Viruses. 2013;5:1023–1041. doi: 10.3390/v5041023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirambeau G., Lyonnais S., Gorelick R.J. Features, processing states, and heterologous protein interactions in the modulation of the retroviral nucleocapsid protein function. RNA Biol. 2010;7:724–734. doi: 10.4161/rna.7.6.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabus C., Ivanyi-Nagy R., Depollier J., Bucheton A., Pelisson A., Darlix J.L. Characterization of a nucleocapsid-like region and of two distinct primer tRNALys,2 binding sites in the endogenous retrovirus Gypsy. Nucleic Acids Res. 2006;34:5764–5777. doi: 10.1093/nar/gkl722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajkowitsch L., Chen D., Stampfl S., Semrad K., Waldsich C., Mayer O., Jantsch M.F., Konrat R., Blasi U., Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 13.Cruceanu M., Gorelick R.J., Musier-Forsyth K., Rouzina I., Williams M.C. Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein's nucleic acid chaperone function. J. Mol. Biol. 2006;363:867–877. doi: 10.1016/j.jmb.2006.08.070. [DOI] [PubMed] [Google Scholar]
- 14.Mirambeau G., Lyonnais S., Coulaud D., Hameau L., Lafosse S., Jeusset J., Justome A., Delain E., Gorelick R.J., Le Cam E. Transmission electron microscopy reveals an optimal HIV-1 nucleocapsid aggregation with single-stranded nucleic acids and the mature HIV-1 nucleocapsid protein. J. Mol. Biol. 2006;364:496–511. doi: 10.1016/j.jmb.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 15.Urbaneja M.A., Wu M., Casas-Finet J.R., Karpel R.L. HIV-1 nucleocapsid protein as a nucleic acid chaperone: spectroscopic study of its helix-destabilizing properties, structural binding specificity, and annealing activity. J. Mol. Biol. 2002;318:749–764. doi: 10.1016/S0022-2836(02)00043-8. [DOI] [PubMed] [Google Scholar]
- 16.Merkulov G.V., Swiderek K.M., Brachmann C.B., Boeke J.D. A critical proteolytic cleavage site near the C terminus of the yeast retrotransposon Ty1 Gag protein. J. Virol. 1996;70:5548–5556. doi: 10.1128/jvi.70.8.5548-5556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brookman J.L., Stott A.J., Cheeseman P.J., Adamson C.S., Holmes D., Cole J., Burns N.R. Analysis of TYA protein regions necessary for formation of the Ty1 virus-like particle structure. Virology. 1995;212:69–76. doi: 10.1006/viro.1995.1454. [DOI] [PubMed] [Google Scholar]
- 18.Martin-Rendon E., Marfany G., Wilson S., Ferguson D.J.P., Kingsman S.M., Kingsman A.J. Structural determinants within the subunit protein of Ty1 virus-like particles. Mol. Microbiol. 1996;22:667–679. doi: 10.1046/j.1365-2958.1996.d01-1716.x. [DOI] [PubMed] [Google Scholar]
- 19.Peterson-Burch B.D., Voytas D.F. Genes of the Pseudoviridae (Ty1/copia retrotransposons) Mol. Biol. Evol. 2002;19:1832–1845. doi: 10.1093/oxfordjournals.molbev.a004008. [DOI] [PubMed] [Google Scholar]
- 20.Lawler J.F., Merkulov G.V., Boeke J.D. A nucleocapsid functionality contained within the amino terminus of the Ty1 protease that is distinct and separable from proteolytic activity. J. Virol. 2002;76:346–354. doi: 10.1128/JVI.76.1.346-354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cristofari G., Ficheux D., Darlix J.L. The GAG-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J. Biol. Chem. 2000;275:19210–19217. doi: 10.1074/jbc.M001371200. [DOI] [PubMed] [Google Scholar]
- 22.Saha A., Mitchell J.A., Nishida Y., Hildreth J.E., Ariberre J.A., Gilbert W.A., Garfinkel D.J. A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control. J. Virol. 2015;89:3922–3938. doi: 10.1128/JVI.03060-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spencer T.E., Palmarini M. Endogenous retroviruses of sheep: a model system for understanding physiological adaptation to an evolving ruminant genome. J. Reprod. Dev. 2012;58:33–37. doi: 10.1262/jrd.2011-026. [DOI] [PubMed] [Google Scholar]
- 24.Sanz-Ramos M., Stoye J.P. Capsid-binding retrovirus restriction factors: discovery, restriction specificity and implications for the development of novel therapeutics. J. Gen. Virol. 2013;94:2587–2598. doi: 10.1099/vir.0.058180-0. [DOI] [PubMed] [Google Scholar]
- 25.Guthrie C., Fink G.R. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–863. [PubMed] [Google Scholar]
- 26.Pierce B.D., Wendland B. Sequence of the yeast protein expression plasmid pEG(KT) Yeast. 2009;26:349–353. doi: 10.1002/yea.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curcio M.J., Garfinkel D.J. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melamed C., Nevo Y., Kupiec M. Involvement of cDNA in homologous recombination between Ty elements in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:1613–1620. doi: 10.1128/mcb.12.4.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharon G., Burkett T.J., Garfinkel D.J. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol. Cell. Biol. 1994;14:6540–6551. doi: 10.1128/mcb.14.10.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garfinkel D.J., Nyswaner K., Wang J., Cho J.Y. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics. 2003;165:83–99. doi: 10.1093/genetics/165.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolton E.C., Coombes C., Eby Y., Cardell M., Boeke J.D. Identification and characterization of critical cis-acting sequences within the yeast Ty1 retrotransposon. RNA. 2005;11:308–322. doi: 10.1261/rna.7860605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Q., Purzycka K.J., Lusvarghi S., Li D., Legrice S.F., Boeke J.D. Retrotransposon Ty1 RNA contains a 5′-terminal long-range pseudoknot required for efficient reverse transcription. RNA. 2013;19:320–332. doi: 10.1261/rna.035535.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Somarowthu S., Legiewicz M., Keating K.S., Pyle A.M. Visualizing the ai5gamma group IIB intron. Nucleic Acids Res. 2014;42:1947–1958. doi: 10.1093/nar/gkt1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purzycka K.J., Pachulska-Wieczorek K., Adamiak R.W. The in vitro loose dimer structure and rearrangements of the HIV-2 leader RNA. Nucleic Acids Res. 2011;39:7234–7248. doi: 10.1093/nar/gkr385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purzycka K.J., Legiewicz M., Matsuda E., Eizentstat L.D., Lusvarghi S., Saha A., Le Grice S.F., Garfinkel D.J. Exploring Ty1 retrotransposon RNA structure within virus-like particles. Nucleic Acids Res. 2013;41:463–473. doi: 10.1093/nar/gks983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larsen L.S., Zhang M., Beliakova-Bethell N., Bilanchone V., Lamsa A., Nagashima K., Najdi R., Kosaka K., Kovacevic V., Cheng J., et al. Ty3 capsid mutations reveal early and late functions of the amino-terminal domain. J. Virol. 2007;81:6957–6972. doi: 10.1128/JVI.02207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dutko J.A., Kenny A.E., Gamache E.R., Curcio M.J. 5′ to 3′ mRNA decay factors colocalize with Ty1 gag and human APOBEC3G and promote Ty1 retrotransposition. J. Virol. 2010;84:5052–5066. doi: 10.1128/JVI.02477-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curcio M.J., Garfinkel D.J. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol. Cell. Biol. 1992;12:2813–2825. doi: 10.1128/mcb.12.6.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slabinski L., Jaroszewski L., Rychlewski L., Wilson I.A., Lesley S.A., Godzik A. XtalPred: a web server for prediction of protein crystallizability. Bioinformatics. 2007;23:3403–3405. doi: 10.1093/bioinformatics/btm477. [DOI] [PubMed] [Google Scholar]
- 40.Bastin P., Bagherzadeh Z., Matthews K.R., Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 1996;77:235–239. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- 41.McKinstry W.J., Hijnen M., Tanwar H.S., Sparrow L.G., Nagarajan S., Pham S.T., Mak J. Expression and purification of soluble recombinant full length HIV-1 Pr55Gag protein in Escherichia coli. Protein Expr. Purif. 2014;100:10–18. doi: 10.1016/j.pep.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 42.Blaszczyk L., Ciesiolka J. Secondary structure and the role in translation initiation of the 5′-terminal region of p53 mRNA. Biochemistry. 2011;50:7080–7092. doi: 10.1021/bi200659b. [DOI] [PubMed] [Google Scholar]
- 43.Martin F., Barends S., Jaeger S., Schaeffer L., Prongidi-Fix L., Eriani G. Cap-assisted internal initiation of translation of histone H4. Mol. Cell. 2011;41:197–209. doi: 10.1016/j.molcel.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 44.Le Cam E., Coulaud D., Delain E., Petitjean P., Roques B.P., Gerard D., Stoylova E., Vuilleumier C., Stoylov S.P., Mely Y. Properties and growth mechanism of the ordered aggregation of a model RNA by the HIV-1 nucleocapsid protein: an electron microscopy investigation. Biopolymers. 1998;45:217–229. doi: 10.1002/(SICI)1097-0282(199803)45:3<217::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 45.Stoylov S.P., Vuilleumier C., Stoylova E., De Rocquigny H., Roques B.P., Gerard D., Mely Y. Ordered aggregation of ribonucleic acids by the human immunodeficiency virus type 1 nucleocapsid protein. Biopolymers. 1997;41:301–312. doi: 10.1002/(SICI)1097-0282(199703)41:3<301::AID-BIP5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 46.Vo M.N., Barany G., Rouzina I., Musier-Forsyth K. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J. Mol. Biol. 2006;363:244–261. doi: 10.1016/j.jmb.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 47.Pachulska-Wieczorek K., Stefaniak A.K., Purzycka K.J. Similarities and differences in the nucleic acid chaperone activity of HIV-2 and HIV-1 nucleocapsid proteins in vitro. Retrovirology. 2014;11:54. doi: 10.1186/1742-4690-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paquin C., Williamson V. Temperature effects on the rate of Ty transposition. Science. 1984;226:53–55. doi: 10.1126/science.226.4670.53. [DOI] [PubMed] [Google Scholar]
- 49.Wu H., Mitra M., Naufer M.N., McCauley M.J., Gorelick R.J., Rouzina I., Musier-Forsyth K., Williams M.C. Differential contribution of basic residues to HIV-1 nucleocapsid protein's nucleic acid chaperone function and retroviral replication. Nucleic Acids Res. 2014;42:2525–2537. doi: 10.1093/nar/gkt1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levin J.G., Guo J., Rouzina I., Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 51.Levin J.G., Mitra M., Mascarenhas A., Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010;7:754–774. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.You J.C., McHenry C.S. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J. Biol. Chem. 1994;269:31491–31495. [PubMed] [Google Scholar]
- 53.Gabus C., Mazroui R., Tremblay S., Khandjian E.W., Darlix J.L. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Res. 2004;32:2129–2137. doi: 10.1093/nar/gkh535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ivanyi-Nagy R., Lavergne J.P., Gabus C., Ficheux D., Darlix J.L. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res. 2008;36:712–725. doi: 10.1093/nar/gkm1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuciak M., Gabus C., Ivanyi-Nagy R., Semrad K., Storchak R., Chaloin O., Muller S., Mely Y., Darlix J.L. The HIV-1 transcriptional activator Tat has potent nucleic acid chaperoning activities in vitro. Nucleic Acids Res. 2008;36:3389–3400. doi: 10.1093/nar/gkn177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Rocquigny H., Shvadchak V., Avilov S., Dong C.Z., Dietrich U., Darlix J.L., Mely Y. Targeting the viral nucleocapsid protein in anti-HIV-1 therapy. Mini Rev. Med. Chem. 2008;8:24–35. doi: 10.2174/138955708783331603. [DOI] [PubMed] [Google Scholar]
- 57.Beltz H., Clauss C., Piemont E., Ficheux D., Gorelick R.J., Roques B., Gabus C., Darlix J.L., de Rocquigny H., Mely Y. Structural determinants of HIV-1 nucleocapsid protein for cTAR DNA binding and destabilization, and correlation with inhibition of self-primed DNA synthesis. J. Mol. Biol. 2005;348:1113–1126. doi: 10.1016/j.jmb.2005.02.042. [DOI] [PubMed] [Google Scholar]
- 58.Stewart-Maynard K.M., Cruceanu M., Wang F., Vo M.N., Gorelick R.J., Williams M.C., Rouzina I., Musier-Forsyth K. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J. Virol. 2008;82:10129–10142. doi: 10.1128/JVI.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vo M.N., Barany G., Rouzina I., Musier-Forsyth K. Effect of Mg(2+) and Na(+) on the nucleic acid chaperone activity of HIV-1 nucleocapsid protein: implications for reverse transcription. J. Mol. Biol. 2009;386:773–788. doi: 10.1016/j.jmb.2008.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Webb J.A., Jones C.P., Parent L.J., Rouzina I., Musier-Forsyth K. Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: implications for viral genomic RNA packaging. RNA. 2013;19:1078–1088. doi: 10.1261/rna.038869.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fahlman R.P., Uhlenbeck O.C. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P- and A-sites. Biochemistry. 2004;43:7575–7583. doi: 10.1021/bi0495836. [DOI] [PubMed] [Google Scholar]
- 62.Record M.T. Jr, Lohman M.L., De Haseth P. Ion effects on ligand-nucleic acid interactions. J. Mol. Biol. 1976;107:145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- 63.Keeney J.B., Chapman K.B., Lauermann V., Voytas D.F., Aström S.U., von Pawel-Rammingen U., Bystrom A., Boeke J.D. Multiple molecular determinants for retrotransposition in a primer tRNA. Mol. Cell. Biol. 1995;15:217–226. doi: 10.1128/mcb.15.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitchell D.A., Marshall T.K., Deschenes R.J. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 65.Johnson S.F., Telesnitsky A. Retroviral RNA dimerization and packaging: the what, how, when, where, and why. PLoS Pathog. 2010;6:e1001007. doi: 10.1371/journal.ppat.1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hinnebusch A.G. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 2011;75:434–467. doi: 10.1128/MMBR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cavener D.R., Ray S.C. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muriaux D., Darlix J.L. Properties and functions of the nucleocapsid protein in virus assembly. RNA Biol. 2010;7:744–753. doi: 10.4161/rna.7.6.14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsuchihashi Z., Brown P.O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adams S.E., Mellor J., Gull K., Sim R.B., Tuite M.F., Kingsman S.M., Kingsman A.J. The functions and relationships of Ty-VLP proteins in yeast reflect those of mammalian retroviral proteins. Cell. 1987;49:111–119. doi: 10.1016/0092-8674(87)90761-6. [DOI] [PubMed] [Google Scholar]
- 71.Mellor J., Malim M., Gull K., Tuite M., McCready S., Dibbayawan T., Kingsman S., Kingsman A. Reverse transcriptase activity and Ty RNA are associated with virus-like particles in yeast. Nature. 1985;318:583–586. doi: 10.1038/318583a0. [DOI] [PubMed] [Google Scholar]
- 72.Pratico E.D., Silverman S.K. Ty1 reverse transcriptase does not read through the proposed 2′,5′-branched retrotransposition intermediate in vitro. RNA. 2007;13:1528–1536. doi: 10.1261/rna.629607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cristofari G., Bampi C., Wilhelm M., Wilhelm F.X., Darlix J.L. A 5′-3′ long-range interaction in Ty1 RNA controls its reverse transcription and retrotransposition. EMBO J. 2002;21:4368–4379. doi: 10.1093/emboj/cdf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Larsen L.S., Beliakova-Bethell N., Bilanchone V., Zhang M., Lamsa A., Dasilva R., Hatfield G.W., Nagashima K., Sandmeyer S. Ty3 nucleocapsid controls localization of particle assembly. J. Virol. 2008;82:2501–2514. doi: 10.1128/JVI.01814-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gabus C., Ficheux D., Rau M., Keith G., Sandmeyer S., Darlix J.L. The yeast Ty3 retrotransposon contains a 5′-3′ bipartite primer-binding site and encodes nucleocapsid protein NCp9 functionally homologous to HIV-1 NCp7. EMBO J. 1998;17:4873–4880. doi: 10.1093/emboj/17.16.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


