Abstract
Polyamines are ubiquitous cations that are involved in regulating fundamental cellular processes such as cell growth and proliferation; hence, their intracellular concentration is tightly regulated. Antizyme and antizyme inhibitor have a central role in maintaining cellular polyamine levels. Antizyme is unique in that it is expressed via a novel programmed ribosomal frameshifting mechanism. Conventional computational tools are unable to predict a programmed frameshift, resulting in misannotation of antizyme transcripts and proteins on transcript and genomic sequences. Correct annotation of a programmed frameshifting event requires manual evaluation. Our goal was to provide an accurately curated and annotated Reference Sequence (RefSeq) data set of antizyme transcript and protein records across a broad taxonomic scope that would serve as standards for accurate representation of these gene products. As antizyme and antizyme inhibitor proteins are functionally connected, we also curated antizyme inhibitor genes to more fully represent the elegant biology of polyamine regulation. Manual review of genes for three members of the antizyme family and two members of the antizyme inhibitor family in 91 vertebrate organisms resulted in a total of 461 curated RefSeq records.
INTRODUCTION
The Reference Sequence (RefSeq) database at the National Center for Biotechnology Information (NCBI) is a collection of annotated genomic, transcript and protein sequence records for genomes across a wide taxonomic spectrum (1). The RefSeq transcript and protein records for higher eukaryotes, including vertebrates, are generated by automatic and manual processing of sequence data submitted to the International Nucleotide Sequence Database Consortium (INSDC, (2)), which includes GenBank at NCBI, the European Nucleotide Archive (ENA) and the DNA Databank of Japan (DDBJ). The subset of RefSeqs subject to curation, referred to as ‘known’, have an accession prefix starting with ‘N’ and can be distinguished from the ‘model’ RefSeqs generated by the NCBI's eukaryotic genome annotation pipeline (http://www.ncbi.nlm.nih.gov/books/NBK169439/), which have accession prefix starting with ‘X’. The known RefSeqs may be subjected to different levels of manual curation, as guided by our in-house QA analyses (3), review requests from public users and collaborators, and targeted curation of gene and protein families. RefSeqs with a ‘validated’ or ‘reviewed’ status are considered curated. Particular emphasis is given to curation of records from human, mouse, and other vertebrates due to their importance for biomedical research. A reviewed record has undergone our highest level of manual curation, which includes: nomenclature and literature review, providing a brief RefSeq summary of a gene, adding biologically relevant attributes (4) and feature annotations, in addition to full sequence review that may involve creating additional RefSeq records for supported transcript variants. The RefSeq data set is pivotal for additional resources and processes at NCBI, including Gene, Genomes, HomoloGene and Map Viewer, and the known RefSeq subset is an important reagent for NCBI's evidence-based eukaryotic genome annotation pipeline. This data set is also considered a gold standard by many in the scientific community and is extensively used for basic and biomedical research, as well as for large scale genome annotation and analyses.
Polyamines are ubiquitous, organic cations that are involved in many diverse processes, including cell growth and proliferation; and high polyamine levels are associated with transformation and tumorigenesis (5). Because of the wide-ranging effects of polyamines, their intracellular concentration is tightly regulated. The two major players involved in maintaining polyamine homeostasis within the cell are ornithine decarboxylase antizyme (OAZ, antizyme for short) and antizyme inhibitor (AZIN) (Figure 1). High polyamine levels induce +1 ribosomal frameshifting (detailed below), which results in increased antizyme production. Antizyme in turn decreases polyamine levels via three mechanisms: inhibiting ornithine decarboxylase (ODC), the key enzyme catalyzing the first and rate-limiting step in polyamine biosynthesis, by disrupting the active ODC homodimers; targeting ODC monomers for ubiquitin-independent degradation by the 26S proteasome; and inhibiting polyamine transporter-mediated polyamine uptake into cells. In contrast, antizyme inhibitor functions as a positive regulator of polyamine levels by binding to antizyme and suppressing its ODC-inhibitory effects. Antizyme inhibitor expression in turn is repressed by polyamines via a novel mechanism involving an upstream open reading frame (uORF, (6)). Thus, both antizyme and antizyme inhibitor are part of opposing autoregulatory circuits designed to maintain optimal levels of polyamine within a cell.
Figure 1.
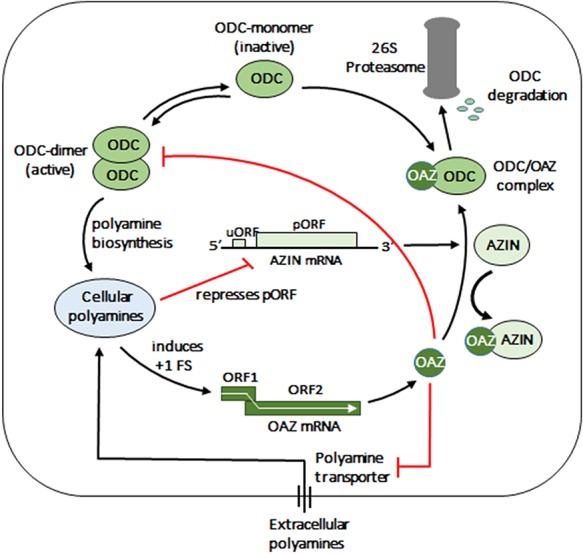
Schematic showing autoregulation of intracellular polyamine levels by antizyme and antizyme inhibitor. Key: ODC, ornithine decarboxylase; OAZ, antizyme; AZIN, antizyme inhibitor; FS, frameshift; uORF, upstream ORF; pORF, primary ORF; red lines indicate inhibition/repression.
The antizyme gene in rat was the first cellular gene in eukaryotes shown to employ +1 programmed ribosomal frameshifting for recoding its mRNA for protein expression (7). This mechanism for expression of antizyme has since been found to be conserved from yeast to mammals (8). All vertebrate antizymes are encoded by two partially overlapping open reading frames (ORFs) (Figure 2): a short ORF1 with a translation initiation codon and an in-frame stop codon at the frameshift site; and a longer ORF2, which lacks a start codon, that is in +1 frame with respect to ORF1, and encodes a larger portion of the antizyme protein. Therefore, the synthesis of full-length, functional antizyme requires translation initiation at the ORF1 start codon, followed by +1 ribosomal frameshift just before the ORF1 stop codon, and translation termination at the ORF2 stop codon. The +1 ribosomal frameshifting is stimulated by polyamines via a mechanism that is not completely understood; however, signals embedded within the mRNA (which include the ORF1 stop codon, an element 5′ and a pseudoknot 3′ of the frameshift site) are essential for polyamine sensing and stimulating the level of +1 frameshifting (7,9,10).
Figure 2.
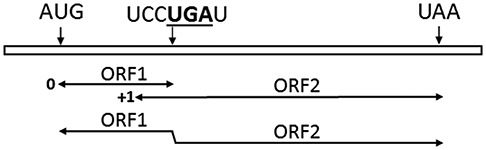
Schematic representing +1 ribosomal frameshifting in antizymes. The antizyme mRNA is shown as an open black bar. The overlapping 0 and +1 translational reading frames, corresponding to ORF1 and ORF2, respectively, and the +1 frameshifted ORF combining ORF1 and ORF2 are indicated. The AUG initiation codon of ORF1, as well as the stop codons of ORF1 (in bold and underlined as part of the frameshift site, UCCUGAU) and ORF2 (UAA) are shown. The figure is drawn approximately to scale.
Multiple antizyme paralogs exist in vertebrates, with mammals having three well-characterized paralogs: antizyme 1 (OAZ1), 2 (OAZ2) and 3 (OAZ3). OAZ1, the first member discovered in rat, is the prototype of the antizyme family. The OAZ1-3 genes are structurally similar and all three antizymes inhibit ODC activity and polyamine uptake. However, there are differences (summarized in Table 1); the most notable is that while OAZ1 and OAZ2 are ubiquitously expressed in somatic tissues, OAZ3 is expressed in a tissue and cell-specific manner, predominantly in the haploid germ cells in testis (11,12). The restricted expression pattern, together with studies showing infertility in transgenic male mice overexpressing ODC (13) and in homozygous Oaz3 gene-disrupted male mice (14), suggest a distinct role for OAZ3 in regulating ODC during spermatogenesis.
Table 1. Comparison of members of the mammalian antizyme gene family.
| Gene | Frameshift site in mRNA | Recoding signalsa | Expression | ODC degradationb |
|---|---|---|---|---|
| OAZ1 | UCCUGAU | yes | ubiquitous | in vivo and in vitro |
| OAZ2 | UCCUGAU | yes | ubiquitous | in vivo only |
| OAZ3 | UCCUGAG | no | testis-specific | no |
Antizyme inhibitors are homologs of ODC that have lost the ability to decarboxylate ornithine, but retain the ability to bind to antizymes. Antizyme inhibitors, in fact, have a higher affinity for antizymes than the antizymes have for ODC; hence, they prevent binding of antizymes to ODC. Several paralogs of antizyme inhibitor have been reported (15): the two best characterized in vertebrates are antizyme inhibitor 1 (AZIN1) and 2 (AZIN2). The AZIN1-2 genes are structurally similar and both antizyme inhibitors bind to and inhibit all three members of the antizyme family (16,17). AZIN1 is ubiquitously expressed and localized in the cytoplasm and nucleus, while AZIN2 is expressed predominantly in the brain and testis and localized in the endoplasmic reticulum-golgi intermediate compartment, suggesting distinct roles for the two antizyme inhibitors.
Canonical decoding of the genetic code requires translating ribosomes to linearly convert nucleotide (nt) triplets (codons) into amino acid (aa) sequence. The +1 ribosomal frameshifting needed for antizyme biosynthesis complicates automatic detection and annotation of its full-length coding sequence (CDS) on transcripts and genomic sequences by standard sequence analysis tools, as they lack the ability to predict sites of programmed ribosomal frameshifting. While manual curation and annotation is a time-consuming process, it is essential for accurate representation of antizyme encoding genes. Our goal, therefore, was to provide an accurately curated and annotated set of antizyme transcript and protein records across a wide taxonomic scope that can serve as reagents for accurate annotation of antizyme gene products on vertebrate genomes by NCBI's eukaryotic genome annotation pipeline. Antizyme inhibitor genes are expressed via standard translation; however, given the complementary role of antizyme inhibitors in polyamine regulation, we also curated antizyme inhibitor genes to more fully represent the biology of polyamine regulation.
MATERIALS AND METHODS
Identification of antizyme and antizyme inhibitor genes in vertebrates
Several different approaches were used to identify antizyme and antizyme inhibitor homologs in vertebrates:
Targeted literature search with a gene symbol (e.g. oaz*) or protein name (e.g. antizyme*) as query. This yielded limited results, as published reports of antizyme genes are available for only a few well-studied organisms; however, it was useful for finding paralogs and novel homologs in zebrafish.
Review of annotation on provisional RefSeq records, which are created by automatic processing based on primary sequence data from INSDC. This allowed identification and curation of genes in organisms for which annotated primary data were available (such as human, mouse, rat, chicken). Curated RefSeq records from these organisms facilitated identification and curation of homologs in other organisms based on similarity.
BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) search of publicly available databases at NCBI with related sequences. Several different BLAST programs, such as BLASTN and TBLASTN, were used with default parameters to search a suite of nucleotide databases (e.g. Nucleotide collection (nr/nt), Expressed sequence tags (EST), Transcriptome Shotgun Assembly (TSA) and NCBI Genomes (chromosome)) to find homologs. Blast of the organism-specific genomes (using discontiguous megablast for more dissimilar sequences) was most helpful because it enabled the genomic context of a BLAST search to be displayed in Map Viewer (http://www.ncbi.nlm.nih.gov/mapview/), which allowed determination of synteny and assessment of problems (if any) in the genomic region.
Review of orthologous models predicted by the NCBI's eukaryotic genome annotation pipeline. The models are predicted based on alignments of known RefSeq transcripts and proteins, INSDC transcripts and UniProtKB/Swiss-Prot proteins, and RNA-seq data; and orthologs are identified based on protein homology and local synteny. This approach proved to be most useful for identifying and curating orthologs in organisms that have little or no species-specific transcript support, which is the case for the majority of less-well-studied genomes (http://www.ncbi.nlm.nih.gov/genome/).
Analysis, assembly and annotation of RefSeq
NCBI's Genome Workbench application (Gbench; http://www.ncbi.nlm.nih.gov/tools/gbench/), customized for RefSeq curation was used to analyze, create and edit RefSeq records. The in-house version of Gbench is integrated with a custom tool named RADAR (RefSeq analysis display and recommendation) that displays stored transcript alignments of publically available sequences (such as transcripts, ESTs, known and model RefSeqs) to the genomic region of interest, and is used by the curators to analyze the alignments and create or update RefSeq records using transcript and/or genomic sequences. New and updated RefSeqs generated in the RADAR tool are uploaded to internal databases, where they are subjected to a suite of QA tests (3) and to further curation of the metadata associated with each reviewed gene (e.g. nomenclature, RefSeq summary text, attributes, publications, etc.). Additional feature annotation on the sequence record is added using Sequin (http://www.ncbi.nlm.nih.gov/Sequin/).
RESULTS
Challenges to curation and annotation of antizyme genes
Use of primary sequences from INSDC for RefSeq creation was often problematic, as shown for bovine OAZ1 gene (Figure 3A and B). AY911315.1 is annotated with the ORF1 CDS encoding a truncated protein of 68 aa, which renders it a candidate for nonsense-mediated mRNA decay (NMD, (18)). This misannotation was propagated to the provisional RefSeq transcript (NM_001025323.1) that was based on AY911315.1, and had not yet undergone manual review. Subsequent manual curation led to the suppression of the erroneous record and creation of NM_001127243.2 with an accurately annotated CDS encoding the full-length antizyme 1 protein of 227 aa (NP_001120715.1). Also illustrated here are challenges to accurate antizyme representation by automated genome annotation pipelines. XM_001790610.1, a model transcript predicted by the NCBI's genome annotation pipeline and structurally supported by transcript and RNA-seq data, was computationally modified to remove the ‘T’ of the ORF1 stop codon at the frameshift site. This ‘correction’ enables translation of a full-length model protein (XP_001790662.1) of the expected length, but does not accurately represent the antizyme transcript and biology. The Ensembl pipeline produced a transcript (ENSBTAT00000024656) that lacks 4 nt around the frameshift site, which allows connection of the 0 and +1 frames without a frameshift and translation of a read-through protein of 226 aa (ENSBTAP00000024656). These examples emphasize that standard computational annotations fail to correctly represent antizyme transcript and CDS. To ensure accurate annotation of the CDS feature of the antizyme genes on the RefSeq records, we manually add the split CDS range and a flag indicating ribosomal slippage using feature annotation details approved by INSDC (Figure 3C). This is the most accurate way of representing the antizyme CDS.
Figure 3.
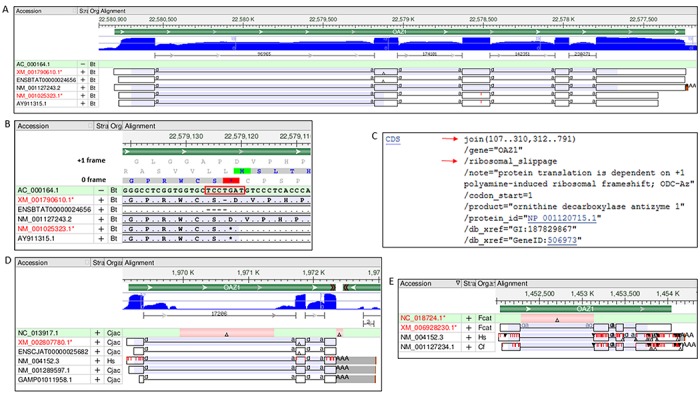
Manual curation and annotation of antizyme gene products. (A) Gbench display showing alignment of a select set of transcripts to the bovine genome at the location of OAZ1 gene. The annotated CDS is highlighted in purple, and the site of deletion in XM_001790610.1 and ENSBTAT00000024656 is indicated by a triangle. The histogram in blue and the bracketed regions beneath show exon coverage and interpreted intron features, respectively, based on RNA-seq data. (B) Expansion of the region around the triangle to the sequence level shows the frameshift site (TCCTGAT, boxed in red), which includes the ORF1 stop codon, TGA (highlighted in red in the 0 frame of the 3-frame translation track). (C) GenBank format display of manually added join CDS and ribosomal_slippage feature annotation (red arrows) on the public RefSeq record NM_001127243.2. (D) Gbench display showing alignment of a select set of transcripts to the marmoset genome at the location of OAZ1 gene. The site and extent of the two deletions in the genomic sequence (NC_013917.1); one in intron 1 and another downstream of exon 3, are shown by pink highlighted regions with a triangle. The unaligned region at the 3′ ends of indicated transcripts is shown in gray. (E) Gbench display showing alignment of a select set of transcripts to the cat genome at the location of OAZ1 gene. The site and extent of a deletion in the genomic sequence (NC_018724.1) between exons 1 and 3 is shown by the pink highlighted region with a triangle.
The orthologous model RefSeqs were a useful starting source for curated records for many organisms. However, model prediction in part depends on the quality of the genome. For organisms with high quality genome assemblies, full-length models of the type shown for bovine OAZ1 gene in Figure 3 were predicted, which allowed creation of curated RefSeqs using genomic sequence components (in the absence of species-specific transcripts) and manually setting the CDS feature (e.g. gibbon OAZ1, GeneID: 100580887). It was also possible to provide curated RefSeqs for organisms with lesser quality genomes if species-specific transcripts were available to fill-in for sequence missing in the genomic region of interest. For example, the marmoset genome (assembly accession GCF_000004665.1) has two gaps: a ∼460 basepair (bp) gap in the first intron and a ∼100 bp gap downstream of exon 3 of the OAZ1 gene (Figure 3D); the latter prevented alignment of the 3′ ends of orthologous transcripts (e.g. human NM_004152.3) and prediction of this region in models. Thus, the NCBI's (XM_002807780.1) and Ensembl's (ENSCJAT00000025682) models encode C-terminally truncated proteins of 155 aa and 153 aa, respectively. The availability of marmoset TSA sequences (e.g. GAMP01011958.1) containing the 3′ exonic sequence made it possible to create NM_001289597.1 encoding a full-length protein of 227 aa (NP_001276526.1). The OAZ1 gene is currently incompletely annotated on the marmoset genome; however, the availability of the RefSeq transcript and protein records will enable accurate annotation of the OAZ1 gene in the future when an improved genome assembly becomes available. There were instances when no species-specific transcript sequence was available to compensate for the problematic genomic region; hence, it was not possible to create full-length RefSeqs, as illustrated for the cat OAZ1 gene (Figure 3E). A gap of ∼850 bp in the cat genome (assembly accession GCF_000181335.1) between exons 1 and 3 of the OAZ1 gene prevented alignment of the exon 2 transcribed region (containing the frameshift site) of orthologous transcripts from human (NM_004152.3) and dog (NM_001127234.1). The predicted model (XM_006928230.1) encodes a truncated protein of 179 aa containing the expected N- and C- termini, but missing an internal exon-2 encoded 49 aa protein segment. In the absence of any cat-specific transcript sequence containing the missing region, it was not possible to create a RefSeq for the cat OAZ1 gene (GeneID: 101080766). In a few extreme cases, for example, the OAZ1 gene in horse, it was not even possible to define an OAZ1 locus because of a large gap in the syntenic region on chromosome 7 (assembly accession GCF_000002305.2); hence horse lacks both a GeneID and a RefSeq for OAZ1 gene.
Curation of antizyme (OAZ) gene family
OAZ1 is found in all vertebrate species. While the OAZ1 orthologs are not conserved throughout the length of the protein, there are regions of high conservation (Figure 4); such as the 20 aa of ORF1 at the N-terminus, the sequence surrounding the frameshift site, and the ODC-binding domain in the C-terminal half (not shown), which is important for antizyme function (10). OAZ1 is unique amongst the antizyme family members in that its mRNA contains two potential in-frame AUG translation initiation codons, the positioning of which is very similar in OAZ1 orthologs (aa 1 and 34 in mammals). Studies in rat showed that alternative use of the two translation initiation sites resulted in N-terminally distinct protein isoforms with different subcellular localization (7,19). The N-terminus of the longer isoform resulting from the use of the first AUG contains a mitochondrial localization signal, which targets this isoform to the mitochondria. The shorter isoform derived from the use of the second AUG lacks the mitochondrial localization signal, and is localized in the cytoplasm. Based on the published evidence, separate RefSeq transcript records were created to represent the two isoforms: NM_139081.2 (encoding the long isoform of 227 aa; NP_620781.1) and NM_001297557.1 (encoding the short isoform of 194 aa; NP_001284486.1). Attributes (Figure 5A) and miscellaneous feature (misc_feature) annotations (Figure 5B) were also manually added to the RefSeq records to highlight the biologically relevant features of the OAZ1 gene.
Figure 4.
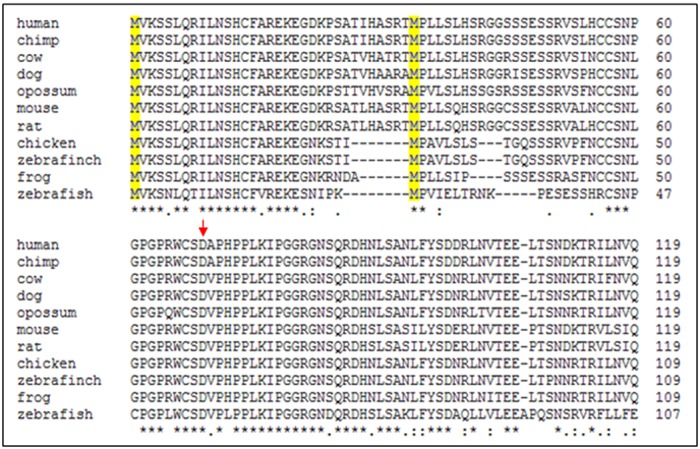
Comparison of amino acid sequence of the N-terminal half of antizyme 1 from phylogenetically distant organisms. The sequences used are as follows: human, NP_004143.1; chimp, NP_001120840.1; cow, NP_001120715.1; dog, NP_001120706.1; opossum, NP_001278356.1; mouse, NP_032779.2; rat, NP_620781.1; chicken, NP_990247.1; zebra finch, NP_001166235.1; frog, NP_001080045.1; zebrafish, NP_919240.2. The positions of the two in-frame translation initiation sites (M) are highlighted in yellow, and the location of the frameshift site is indicated by the red arrow. The multiple protein sequence alignment was performed using Clustal Omega, version 1.2.1 (http://www.ebi.ac.uk/Tools/msa/clustalo/) using default settings. An ‘*’ indicates identical residues, and ‘:’ and ‘.’ indicate conserved and weakly conserved amino acid substitutions, respectively.
Figure 5.
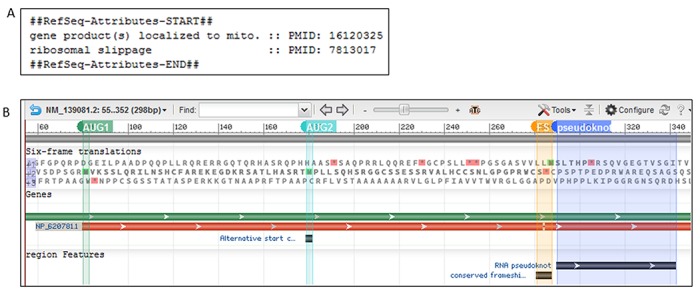
Reporting biological attributes and feature annotations on RefSeq record. (A) A portion of the COMMENT section of the rat Oaz1 RefSeq record (NM_139081.2) is displayed showing two attributes with supporting publications, relevant to the biology of Oaz1 gene. (B) NM_139081.2 is shown using the Graphics display setting on the nucleotide record. The display was configured to show 3-frame translation on the sense strand, and to add four markers highlighting the annotated translation initiation codon (AUG1, nt 80–82), the second in-frame AUG codon located 33 aa downstream (AUG2, nt 179–181), the conserved frameshift site (FS, nt 281–287) and the RNA pseudoknot (pseudoknot, nt 290–342).
The OAZ2 mRNA (e.g. NM_002537.3) has only one AUG translation initiation codon and encodes a relatively shorter protein (189 aa, lacking a mitochondrial localization signal; NP_002528.1) compared to antizyme 1. The OAZ2 ORF1 is shorter, accounting for the shorter protein length, which is similar in size to the shorter isoform of OAZ1 (NP_001284486.1, 194 aa). Antizyme 2 (NP_002528.1) is only 56% identical and 70% similar to antizyme 1 (NP_004143.1) in human. It is expressed at lower levels than OAZ1 (20), but is more conserved, as seen by the higher 99% identity between human and mouse (NP_035082.1) OAZ2 orthologs, compared to 84% identity between human and mouse (NP_032779.2) OAZ1 orthologs.
OAZ3 is more divergent from the other two paralogs. Antizyme 3 (NP_057262.2) is only 36% and 38% identical, and 53% and 56% similar to antizyme 1 and antizyme 2, respectively, in human. Another distinguishing feature is that OAZ3 mRNA contains a highly conserved non-AUG (CUG) codon, in-frame and upstream of an AUG codon (8), use of which would extend the N-terminus of the human antizyme 3 by 48 aa. Evidence for the use of CUG as the start codon in rat OAZ3 mRNA (DQ431008.1) was shown by N-terminal protein sequencing (21), which is corroborated by mass spectrometry data in ‘The Human Proteome Map’ database that identifies the N-terminus of human antizyme 3 (NP_057262.2) as MPCKRCRPSVYSLSYIK (22). Non-AUG start codons are not identified by conventional tools; hence, primary transcript sequences for human (AF175296.1) and mouse (AF175297.1, AB016275.1) OAZ3 genes deposited in INSDC were annotated with AUG as the start codon, and these annotations were propagated to un-reviewed versions of human (NM_016178.1) and mouse (NM_016901.1) OAZ3 RefSeq transcript records. Based on the described evidence, the RefSeq transcript for OAZ3 gene was annotated with CUG as the start codon, and the location of the downstream AUG indicated as a misc_feature (e.g. NM_016178.2).
A putative fourth member of the antizyme family was originally isolated from a human brain library (AF293339.1) and tentatively named antizyme 4 (10), but it was not further characterized. In another study, a related EST (AI124747.1; 97% identical to AF293339.1) was used to show that the encoded protein interacted with both AZIN and ODC, and that binding to the latter inhibited its enzymatic activity, similar to other members of the antizyme family (16). Our blast analyses are consistent with a previous observation (8) that AF293339.1 (and AI124747.1, apparently originating from the same library) represents the Oryctolagus cuniculus (rabbit) OAZ1 gene, and was likely a contaminant in the human brain library; therefore, OAZ4 was not pursued further in our antizyme gene review.
To date, we have reviewed each of the three antizyme genes in 91 vertebrate organisms. This review included two antizyme 1 paralogs (oaz1a/azs and oaz1b/azl) in fish (23) and the novel, retina-specific antizyme (oaz2b/azr) in zebrafish (24). The GeneID and RefSeq identifiers and other related information for the genes reviewed in different organisms are provided in Supplemental Table S1, and general statistics of the data reviewed are summarized in Table 2. Supplemental Table S2 indicates the presence or absence (or undetermined status) for each of the reviewed genes in all 91 organisms, and also provides the genome assembly and annotation release information for each organism. This study revealed a few interesting details:
While the OAZ1 and OAZ2 genes were uniformly represented in all the major groups of organisms, the OAZ3 gene (outside of the placental mammals) was found only in the reptilian species (lizard, alligator and turtle) in the vertebrate group, but not in other lineages, such as birds, fish and amphibians, suggesting that the latter may have only two paralogs of antizyme.
Coelacanth is thought to be more closely related to lungfish and tetrapods than to the common ray-finned fishes (25). Consistent with this phylogenetic analysis, coelacanth (Latimeria chalumnae) was found to have only one paralog each of the antizyme 1 (OAZ1) and antizyme 2 (OAZ2) genes with greater similarity to land vertebrates, especially birds (78% and 74% identity, respectively), than to ray-finned fishes (64% and 55% identity, respectively) at the protein level. Coelacanth, like birds, also appears to lack the OAZ3 gene.
All the ray-finned fishes included in this report have two paralogs of antizyme 1 (oaz1a and oaz1b) and a single paralog of antizyme 2 (oaz2), except zebrafish, which has an additional novel antizyme gene, named azr, (24)) that shares highest similarity with fish and other vertebrate antizyme 2 (43–47% identity), and officially named oaz2b by the zebrafish nomenclature authority (ZFIN, (26)). The oaz2b gene was not identified in other fishes. As previously reported, the oaz2b/azr ortholog appears to be restricted to zebrafish and close related species (24). This could represent either a lineage specific duplication, or loss of the gene in other lineages shortly after the whole genome duplication event early in the evolution of teleost fish. Sequencing and analysis of more fish genomes will be needed to resolve these possibilities.
Table 2. Summary of curated RefSeqs for antizyme and antizyme inhibitor genes in the 91 vertebrate taxa reviewed.
| Gene | Number of genes with curated RefSeq | Genes without curated RefSeq | Number of curated RefSeq | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mammals | Primates | Rodents | Vertebrates | Total | Total | Totald | |||
| Birds | Fisha | Other | |||||||
| OAZ1 | 20 | 11 | 12 | 10 | 9 | 4 | 66 | 29 | 72 |
| OAZ2 | 30 | 11 | 12 | 7 | 6 | 5 | 71 | 21 | 83 |
| OAZ3 | 39 | 11 | 11 | 0 | 0 | 6 | 67 | 24b | 88 |
| AZIN1 | 39 | 12 | 11 | 12 | 8 | 9 | 91 | 4 | 142 |
| AZIN2 | 34 | 9 | 11 | 8 | 0 | 7 | 69 | 22c | 76 |
aThe fish count includes paralogs.
bThe total number without curated RefSeq for OAZ3 gene includes 19 taxa (12 birds, 2 amphibians and 5 fish) in which the OAZ3 gene could not be identified.
cThe total number without curated RefSeq for AZIN2 gene includes 5 fish species in which the AZIN2 gene could not be identified.
dThe total number of curated RefSeqs includes transcript variants
This review resulted in a total of 243 curated RefSeq records (including transcript variants) for 204 antizyme genes (including paralogs). It was not possible to provide curated RefSeqs for about 21% of the antizyme genes because of genome sequence or assembly problems and unavailability of species-specific transcripts.
Curation of antizyme inhibitor (AZIN) gene family
The AZIN1gene contains 13 exons and encodes a protein of 448 aa (NP_056962.2) in human. The expression of AZIN1 is regulated at multiple levels. A study in mouse showed that an uORF with a putative, rarely used non-AUG (AUU) start codon in the 5′ leader sequence of mouse Azin1 mRNA (NM_018745.5) mediates polyamine-induced translation repression of the downstream primary ORF (pORF) encoding antizyme inhibitor 1 (6). This uORF (named uORF-M) encodes a ∼50 aa peptide that is highly conserved in vertebrate AZIN1 orthologs (Figure 6A), especially at the N- and C- termini; the AUU codon and the C-terminal 10 aa were shown to be important for polyamine-induced pORF repression. We have recently started to report the presence of ‘regulatory uORF’ with supporting evidence, such as uORF-M, as a structured biological attribute (Figure 6B) and misc_feature annotation on RefSeq records (Figure 6C). AZIN1 mRNA in human and mouse was also shown to undergo post-transcriptional RNA editing, most predominantly in the liver tissue (27). The A>I RNA editing results in a serine-to-glycine (AGC>GGC) substitution at aa 367. The RNA-edited form induced greater cell proliferation, was abundantly found in tumor tissues, and was associated with pathogenesis of hepatocellular carcinoma. We also report RNA editing as an attribute (Figure 6B) and misc_feature annotation (not shown) on RefSeq records. Alternatively spliced transcript variants of AZIN1 gene with roles in disease and polyamine regulation have also been described. A transcript variant (named AZIN1 SV2; NM_001301668.1), alternatively spliced in the 3' coding region and encoding a shorter isoform with a distinct C-terminus (NP_001288597.1), was reported to play a role in reducing fibrogenicity of hepatic stellate cells (28). A novel transcript variant that is subject to NMD (named Azin1-X; represented as a non-coding variant, NR_125913.1) was described in mouse (29). This study showed that the ratio of Azin1-X to the full-length Azin1 mRNA was regulated by polyamines at the level of transcription and splice pattern selection, with lower polyamine levels stimulating Azin1 mRNA synthesis, and higher polyamine levels favoring Azin1-X synthesis.
Figure 6.
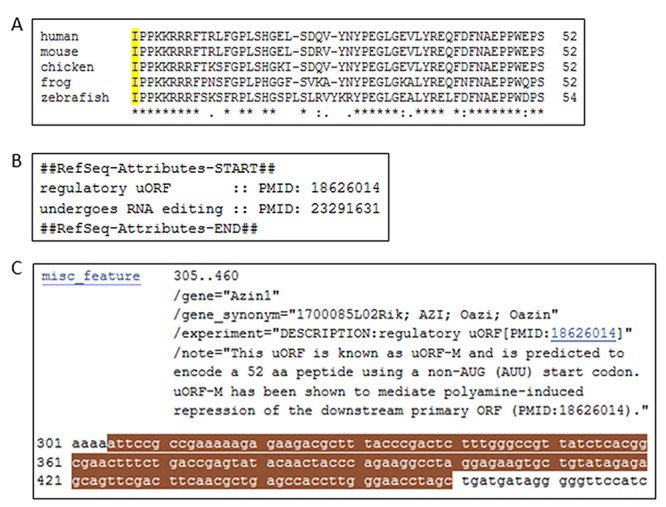
Regulatory uORF in antizyme inhibitor 1 mRNA. (A) Amino acid sequence comparison of the uORF-encoded peptide in the 5′ leader sequence of antizyme inhibitor 1 mRNA from five divergent species: human, nt 341–496 of NM_148174.3; mouse, nt 305–460 of NM_018745.5; chicken, nt 238–393 of NM_001008729.2; frog, nt 178–333 of NM_001094115.1; zebrafish, nt 94–255 of NM_001007159.2. The alignment was performed using Clustal Omega using default settings (see legend to Figure 4 for details). (B) A portion of the COMMENT section of the mouse Azin1 RefSeq record (NM_018745.5) is displayed, showing two attributes, including regulatory uORF, with supporting evidence. (C) A portion of the FEATURES section of NM_018745.5 is displayed showing annotation of regulatory uORF as a misc_feature with accompanying experimental evidence and comment. The sequence of the uORF (nt 305–460) encoding the 52 aa peptide is highlighted in brown.
AZIN2 was first identified in human brain and testis as an ODC paralog and named ODCp or ODC-like (30). A subsequent study established that ODCp was devoid of ODC activity; instead it functioned as an antizyme inhibitor, and proposed the name AZIN2 (31). The AZIN2 gene encodes a protein of 460 aa (NP_443724.1), which shares 45% identity and 67% similarity with antizyme inhibitor 1 in human. There has been confusion in literature and databases over the nomenclature of AZIN2 gene stemming from an earlier report that a human cDNA clone (identical to ODCp) had arginine decarboxylase (ADC) activity (32); however, this was subsequently proved to be incorrect (31,33). As a consequence, many ODCp-representing primary sequences deposited in the INSDC databases (e.g. BC010449.1, AK122697.1) were variously annotated with the ADC symbol, arginine decarboxylase description, and the related Enzyme Commission number (EC 4.1.1.19). These misannotations were automatically propagated to the provisional RefSeq records and to other orthologous records by the NCBI genome annotation pipeline. As part of this curation project, the ADC nomenclature was replaced by AZIN2 as the preferred nomenclature, and the E.C. number 4.1.1.19 (when found) was removed from curated RefSeq records. We coordinated with the official nomenclature authorities for human, mouse, rat, chicken and frog (34–37), as well as UniProt (38), to assure consistent data representation by all of NCBI's collaborating partners.
To date, we have reviewed each of the two antizyme inhibitor genes in the same set of 91 vertebrate organisms as used for the antizyme genes. This review included two antizyme inhibitor 1 paralogs, azin1a and azin1b (39), in fish. The curation details for AZIN1 and AZIN2 genes in different taxa are provided in Supplemental Tables S1 and S2, and general statistics of the reviewed data are summarized in Table 2. The AZIN1 and AZIN2 genes were uniformly represented in all major groups; however, the AZIN2 gene was not detected in any of the fishes reviewed (including coelacanth), suggesting that fish may lack this gene. This review resulted in a total of 218 curated RefSeq records (including transcript variants) for 160 antizyme inhibitor genes (including paralogs). It was not possible to provide curated RefSeqs for about 11.6% of the antizyme inhibitor genes (especially AZIN2) because of genome sequence or assembly problems and unavailability of species-specific transcripts.
DISCUSSION AND FUTURE PROSPECTS
Antizymes by their exceptional nature of protein expression offer challenges to accurate representation of their gene products. Conventional computational tools are unable to predict the +1 frameshifting event required for synthesis of antizymes; hence, are not suitable for automatic detection of full-length CDS in antizyme mRNAs. While tools such as OAF (ODC antizyme finder) have been developed (40), their use for genome annotation is not widespread. Our data set of manually curated and annotated full-length antizyme transcript and protein products can serve as valuable reagents for individual research projects, large scale computational analyses, and genome annotation. It greatly expands on the vertebrate antizyme data set tracked by other repositories of translational recoding events, such as Recode-2 (41) and FSDB (Frameshift Signal Database, (42)).
The data set described here is unique for its large taxonomic scope, expanded functional annotation, and representation of additional products resulting from alternative splicing and use of alternative translation initiation sites. The large taxonomic scope enabled comparison of antizyme orthologs across many divergent vertebrate species, which revealed that the OAZ3 and AZIN2 genes may be missing from some vertebrate lineages. The functional annotation provided on the RefSeq records in the form of attributes and/or misc_features highlight novel properties of a gene, its mRNA or protein, and allow visualization of the annotated features in the context of the whole sequence. The many tiered regulation of AZIN1 gene, captured here through annotation and transcript variants, illustrates the central role of this gene in maintaining optimal intracellular polyamine levels. Usage of non-AUG and downstream in-frame AUG start codons are largely underrepresented in databases because standard computational tools cannot distinguish between the dual functionality of a non-AUG codon (e.g. CUG, which normally codes for leucine), and usually assign just the first in-frame AUG as the start codon. Our representation of alternate translation initiation sites are based on published experimental evidence, as shown for OAZ1 and OAZ3 genes. The example of OAZ1 also demonstrates the importance of representing alternate protein isoforms as they can serve different functions.
Manual curation was essential for correcting errors in the representation of antizyme transcript, CDS and protein. Errors resulted from misannotated primary transcripts or incorrectly predicted models—in the latter, errors reflected the difficulty of correctly annotating a frameshift site, as well as problems related to the quality of genome assemblies. Genome assembly problems, especially seen with draft assemblies, also affect prediction of models for genes that use standard translation (e.g. AZIN2); hence, it is important to realize the limitations of automated genome annotation overall. Manual curation also included cleanup of incorrect nomenclature, as exemplified for AZIN2 gene, to assure propagation of correct nomenclature to RefSeq transcript, protein and genomic records. Manually curated data therefore are an important complement to computational genome annotation of structural, as well as associated metadata, by the NCBI's genome annotation pipeline. High quality annotation of a genome is extremely valuable to aid its interpretation, as the value of a genome is only as good as its annotation!
Since the curation of the data set presented here, 74 more vertebrate genome annotations have been released by the NCBI genome annotation pipeline. Our future goal is to continue to offer manually curated RefSeq records for the antizyme and antizyme inhibitor family members in these and other vertebrate species as their genomes become available, as well as to make adjustments in NCBI's annotation pipeline to better represent this complex biology in an automated fashion. We also currently have 392 manually curated selenoprotein genes (another example of a recoded gene) from 28 vertebrate organisms represented in the RefSeq collection, and plan to provide curated RefSeqs for complete selenoproteomes of well-studied model organisms in the future. Another future goal is to look into representation in RefSeq of other recoding events, such as stop codon readthrough, evidence for which was recently found in several mammalian genes (43). These efforts exemplify the ongoing value of manual curation to provide high quality annotations of important gene sets to serve as reference sequences for future research.
AVAILABILITY
The RefSeq data presented here are publicly available from several resources at NCBI (http://www.ncbi.nlm.nih.gov). Information about the antizyme and antizyme inhibitor genes, transcripts and proteins can be accessed from the Gene, Nucleotide and Protein databases, respectively, by query, and from reciprocal links on the Gene and RefSeq records. These data are also included in the bi-monthly comprehensive FTP release from: ftp://ftp.ncbi.nlm.nih.gov/refseq/release/. Examples of web queries relevant to this data set, and useful for retrieving subsets of related records are:
Query the Gene database with a gene symbol, such as ‘oaz1[sym]’, to retrieve the oaz1 ortholog set. The orthologs are displayed in a tabular format, and individual Gene records in the set can be retrieved via the link on the Name/GeneID column.
Query the Nucleotide or Protein database to retrieve all vertebrate antizyme RefSeq records annotated with the attribute ‘ribosomal slippage’: vertebrates[orgn] refseq[filter] ribosomal slippage[prop] antizyme[title].
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for this work and the open access charge: Intramural Research Program of the NIH, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakamura Y., Cochrane G., Karsch-Mizrachi I. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2013;41:D21–D24. doi: 10.1093/nar/gks1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M., et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kahana C. Regulation of cellular polyamine levels and cellular proliferation by antizyme and antizyme inhibitor. Essays Biochem. 2009;46:47–61. doi: 10.1042/bse0460004. [DOI] [PubMed] [Google Scholar]
- 6.Ivanov I.P., Loughran G., Atkins J.F. uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10079–10084. doi: 10.1073/pnas.0801590105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsufuji S., Matsufuji T., Miyazaki Y., Murakami Y., Atkins J.F., Gesteland R.F., Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanov I.P., Atkins J.F. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 2007;35:1842–1858. doi: 10.1093/nar/gkm035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petros L.M., Howard M.T., Gesteland R.F., Atkins J.F. Polyamine sensing during antizyme mRNA programmed frameshifting. Bioch. Biophys. Res. Commun. 2005;338:1478–1489. doi: 10.1016/j.bbrc.2005.10.115. [DOI] [PubMed] [Google Scholar]
- 10.Ivanov I.P., Gesteland R.F., Atkins J.F. Antizyme expression: a subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Res. 2000;28:3185–3196. doi: 10.1093/nar/28.17.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tosaka Y., Tanaka H., Yano Y., Masai K., Nozaki M., Yomogida K., Otani S., Nojima H., Nishimune Y. Identification and characterization of testis specific ornithine decarboxylase antizyme (OAZ-t) gene: expression in haploid germ cells and polyamine-induced frameshifting. Genes Cells. 2000;5:265–276. doi: 10.1046/j.1365-2443.2000.00324.x. [DOI] [PubMed] [Google Scholar]
- 12.Ivanov I.P., Rohrwasser A., Terreros D.A., Gesteland R.F., Atkins J.F. Discovery of a spermatogenesis stage-specific ornithine decarboxylase antizyme: antizyme 3. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4808–4813. doi: 10.1073/pnas.070055897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hakovirta H., Keiski A., Toppari J., Halmekyto M., Alhonen L., Janne J., Parvinen M. Polyamines and regulation of spermatogenesis: selective stimulation of late spermatogonia in transgenic mice overexpressing the human ornithine decarboxylase gene. Mol. Endocrinol. 1993;7:1430–1436. doi: 10.1210/mend.7.11.8114757. [DOI] [PubMed] [Google Scholar]
- 14.Tokuhiro K., Isotani A., Yokota S., Yano Y., Oshio S., Hirose M., Wada M., Fujita K., Ogawa Y., Okabe M., et al. OAZ-t/OAZ3 is essential for rigid connection of sperm tails to heads in mouse. PLoS Genet. 2009;5:e1000712. doi: 10.1371/journal.pgen.1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanov I.P., Firth A.E., Atkins J.F. Recurrent emergence of catalytically inactive ornithine decarboxylase homologous forms that likely have regulatory function. J. Mol. Evol. 2010;70:289–302. doi: 10.1007/s00239-010-9331-5. [DOI] [PubMed] [Google Scholar]
- 16.Mangold U., Leberer E. Regulation of all members of the antizyme family by antizyme inhibitor. Biochem. J. 2005;385:21–28. doi: 10.1042/BJ20040547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Contreras A.J., Ramos-Molina B., Cremades A., Penafiel R. Antizyme inhibitor 2: molecular, cellular and physiological aspects. Amino Acids. 2010;38:603–611. doi: 10.1007/s00726-009-0419-4. [DOI] [PubMed] [Google Scholar]
- 18.Kervestin S., Jacobson A. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gandre S., Bercovich Z., Kahana C. Mitochondrial localization of antizyme is determined by context-dependent alternative utilization of two AUG initiation codons. Mitochondrion. 2003;2:245–256. doi: 10.1016/S1567-7249(02)00105-8. [DOI] [PubMed] [Google Scholar]
- 20.Ivanov I.P., Gesteland R.F., Atkins J.F. A second mammalian antizyme: conservation of programmed ribosomal frameshifting. Genomics. 1998;52:119–129. doi: 10.1006/geno.1998.5434. [DOI] [PubMed] [Google Scholar]
- 21.Fitzgerald C., Sikora C., Lawson V., Dong K., Cheng M., Oko R., van der Hoorn F.A. Mammalian transcription in support of hybrid mRNA and protein synthesis in testis and lung. J. Biol. Chem. 2006;281:38172–38180. doi: 10.1074/jbc.M606010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S., et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito T., Hascilowicz T., Ohkido I., Kikuchi Y., Okamoto H., Hayashi S., Murakami Y., Matsufuji S. Two zebrafish (Danio rerio) antizymes with different expression and activities. Biochem. J. 2000;345:99–106. doi: 10.1042/bj3450099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivanov I.P., Pittman A.J., Chien C.B., Gesteland R.F., Atkins J.F. Novel antizyme gene in Danio rerio expressed in brain and retina. Gene. 2007;387:87–92. doi: 10.1016/j.gene.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 25.Amemiya C.T., Alfoldi J., Lee A.P., Fan S., Philippe H., Maccallum I., Braasch I., Manousaki T., Schneider I., Rohner N., et al. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311–316. doi: 10.1038/nature12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradford Y., Conlin T., Dunn N., Fashena D., Frazer K., Howe D.G., Knight J., Mani P., Martin R., Moxon S.A., et al. ZFIN: enhancements and updates to the Zebrafish Model Organism Database. Nucleic Acids Res. 2011;39:D822–D829. doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L., Li Y., Lin C.H., Chan T.H., Chow R.K., Song Y., Liu M., Yuan Y.F., Fu L., Kong K.L., et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 2013;19:209–216. doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paris A.J., Snapir Z., Christopherson C.D., Kwok S.Y., Lee U.E., Ghiassi-Nejad Z., Kocabayoglu P., Sninsky J.J., Llovet J.M., Kahana C., et al. A polymorphism that delays fibrosis in hepatitis C promotes alternative splicing of AZIN1, reducing fibrogenesis. Hepatology. 2011;54:2198–2207. doi: 10.1002/hep.24608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murakami Y., Ohkido M., Takizawa H., Murai N., Matsufuji S. Multiple forms of mouse antizyme inhibitor 1 mRNA differentially regulated by polyamines. Amino Acids. 2014;46:575–583. doi: 10.1007/s00726-013-1598-6. [DOI] [PubMed] [Google Scholar]
- 30.Pitkanen L.T., Heiskala M., Andersson L.C. Expression of a novel human ornithine decarboxylase-like protein in the central nervous system and testes. Biochem. Biophys. Res. Commun. 2001;287:1051–1057. doi: 10.1006/bbrc.2001.5703. [DOI] [PubMed] [Google Scholar]
- 31.Kanerva K., Makitie L.T., Pelander A., Heiskala M., Andersson L.C. Human ornithine decarboxylase paralogue (ODCp) is an antizyme inhibitor but not an arginine decarboxylase. Biochem. J. 2008;409:187–192. doi: 10.1042/BJ20071004. [DOI] [PubMed] [Google Scholar]
- 32.Zhu M.Y., Iyo A., Piletz J.E., Regunathan S. Expression of human arginine decarboxylase, the biosynthetic enzyme for agmatine. Biochim. Biophys. Acta. 2004;1670:156–164. doi: 10.1016/j.bbagen.2003.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Contreras A.J., Lopez-Garcia C., Jimenez-Cervantes C., Cremades A., Penafiel R. Mouse ornithine decarboxylase-like gene encodes an antizyme inhibitor devoid of ornithine and arginine decarboxylating activity. J. Biol. Chem. 2006;281:30896–30906. doi: 10.1074/jbc.M602840200. [DOI] [PubMed] [Google Scholar]
- 34.Gray K.A., Yates B., Seal R.L., Wright M.W., Bruford E.A. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015;43:D1079–D1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eppig J.T., Blake J.A., Bult C.J., Kadin J.A., Richardson J.E. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015;43:D726–D736. doi: 10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimoyama M., De Pons J., Hayman G.T., Laulederkind S.J., Liu W., Nigam R., Petri V., Smith J.R., Tutaj M., Wang S.J., et al. The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 2015;43:D743–D750. doi: 10.1093/nar/gku1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burt D.W., Carre W., Fell M., Law A.S., Antin P.B., Maglott D.R., Weber J.A., Schmidt C.J., Burgess S.C., McCarthy F.M. The Chicken Gene Nomenclature Committee report. BMC Genomics. 2009;10(Suppl. 2):S5. doi: 10.1186/1471-2164-10-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.UniProt Consortium. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hascilowicz T., Murai N., Matsufuji S., Murakami Y. Regulation of ornithine decarboxylase by antizymes and antizyme inhibitor in zebrafish (Danio rerio) Biochim. Biophys. Acta. 2002;1578:21–28. doi: 10.1016/s0167-4781(02)00476-1. [DOI] [PubMed] [Google Scholar]
- 40.Bekaert M., Ivanov I.P., Atkins J.F., Baranov P.V. Ornithine decarboxylase antizyme finder (OAF): fast and reliable detection of antizymes with frameshifts in mRNAs. BMC Bioinformatics. 2008;9:178–188. doi: 10.1186/1471-2105-9-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bekaert M., Firth A.E., Zhang Y., Gladyshev V.N., Atkins J.F., Baranov P.V. Recode-2: new design, new search tools, and many more genes. Nucleic Acids Res. 2010;38:D69–D74. doi: 10.1093/nar/gkp788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moon S., Byun Y., Han K. FSDB: a frameshift signal database. Comput. Biol. Chem. 2007;31:298–302. doi: 10.1016/j.compbiolchem.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loughran G., Chou M.Y., Ivanov I.P., Jungreis I., Kellis M., Kiran A.M., Baranov P.V., Atkins J.F. Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 2014;42:8928–8938. doi: 10.1093/nar/gku608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


