Abstract
The reduced expression levels and functional impairment of global miRNAs are related to various human diseases, including cancers. However, relatively little is known about how global miRNA function may be upregulated. Here, we report that global miRNA function can be enhanced by Rho-associated, coiled-coil-containing protein kinase (ROCK) inhibitors. The regulation of miRNA function by ROCK inhibitors is mediated, at least in part, by poly(A)-binding protein-interacting protein 2 (PAIP2), which enhances poly(A)-shortening of miRNA-targeted mRNAs and leads to global upregulation of miRNA function. In the presence of a ROCK inhibitor, PAIP2 expression is enhanced by the transcription factor hepatocyte nuclear factor 4 alpha (HNF4A) through increased ROCK1 nuclear localization and enhanced ROCK1 association with HNF4A. Our data reveal an unexpected role of ROCK1 as a cofactor of HNF4A in enhancing PAIP2 transcription. ROCK inhibitors may be useful for the various pathologies associated with the impairment of global miRNA function.
INTRODUCTION
MicroRNAs (miRNAs) are endogenous ∼22-nucleotide RNAs that mediate important gene-regulatory events by base-pairing with mRNAs, directing their repression (1) and leading to decreased translational efficiency and decreased mRNA levels. Destabilization of target mRNAs is a major means by which protein expression of the targeted mRNAs is reduced by miRNAs (2).
While the precise molecular mechanisms of mammalian miRNA-mediated mRNA decay have not been fully uncovered, recognition of mRNAs by miRNA-induced silencing complexes results in rapid deadenylation of target mRNAs (3–5). Among the miRNA-induced silencing complexes, Argonaute (Ago) and TNRC6 (also known as GW182) are key proteins that induce deadenylation catalyzed by CAF1-CCR4-NOT deadenylase complexes, followed by Dcp1–Dcp2-complex-directed decapping. TNRC6 interacts with poly(A)-binding protein (PABP), which is required for the deadenylation and decay of miRNA targets (5–9). PABP antagonizes miRNA silencing, partly due to the antagonism of target mRNA deadenylation, without affecting bulk protein synthesis (10). Consistently, elevated levels of PABP-interacting protein 2 (PAIP2), which negatively regulates PABP function (11,12), enhance miRNA efficacy (10). While the exact role of PABP in miRNA-directed deadenylation remains controversial (13), studies indicated that poly(A) shortening is important in miRNA-directed mRNA decay (14–16).
Although specific miRNAs can function as tumor suppressors or oncogenes, a general reduction in miRNA expression and impaired miRNA processing are commonly observed in human and experimental cancers (17–19). Mutations in Dicer, a ribonuclease (RNase) III enzyme required for the production of mature miRNAs in the cytoplasm, were clinically identified in tumors (20–22). A heterozygous Dicer1 null mutation in mice leads to oncogenesis via reduced expression of miRNAs (23), consistent with the notion that globally reduced expression or function of miRNAs may be related to tumorigenesis. Thus, enhancers of miRNA function may be useful for the treatment of pathological conditions caused by reduced function of miRNAs.
We screened a comprehensive drug library and identified a Rho-associated, coiled-coil-containing protein kinase (ROCK) inhibitor as an enhancer of overall miRNA function. Enhancement of miRNA function by a ROCK inhibitor was found to be mediated by shortening the poly(A) length of targeted mRNAs by miRNAs. We showed that use of a ROCK inhibitor enhances the interaction between ROCK1 and HNF4A, a transcription factor involved in PAIP2 transcription. Our results reveal a new way of enhancing miRNA function, which may help prevent the pathological conditions caused by reduced miRNA function.
MATERIALS AND METHODS
Cells and reagents
Details of the cells and reagents used are provided in the Supplementary Materials and Methods.
Primers
Primers used in this study are listed in Supplementary Table S1.
Plasmids, viral transduction, transfection, luciferase assays and drug screening
Plasmid construction, lentiviral transduction and drug screening are described in the Supplementary Materials and Methods. Transfection and dual luciferase assays were performed as described previously (24).
RNA isolation, qRT-PCR and northern blotting
RNA isolation and qRT-PCR analysis were performed as described previously (24). All values were normalized to the mRNA level of the housekeeping gene, GAPDH, the expression of which was unaffected by ROCK inhibition in the cDNA microarray analyses. Relative expression was calculated according to the ΔΔCT method: ΔΔCT = ΔCTsample - ΔCTGAPDH. Construction of Tet-regulated β-globin plasmids with three let-7 recognition sites (let-7wt) or mutated let-7 recognition sites (let-7mut) in the 3′-UTR was described previously (4). miRNAs bound to Ago2 protein were purified by immunoprecipitation using an Ago2-microRNA Isolation Kit (Wako, Osaka, Japan). The northern blotting procedure is described in the Supplementary Materials and Methods. Northern blot analyses of miRNAs were performed as described previously (24).
cDNA oligoarrays and miRNA microarrays
cDNA microarray analysis to determine transcriptional changes after ROCK inhibitor treatment was performed using cDNA oligo chips (Toray Industries, Tokyo, Japan). Data were deposited in a public database (GEO; accession number GSE32024). miRNA microarray analysis was performed using miRNA oligo chips (Toray Industries). The data were deposited in a public database (GEO; accession number GSE33876).
Subcellular fractionation and western blot analysis
Western blotting was performed as described previously (24). Protocols for subcellular fractionation and the antibodies used are described in the Supplementary Materials and Methods.
mRNA degradation assay
mRNA degradation was determined as described previously (25). Briefly, cells were pretreated with 10 μM Y27632 in phosphate-buffered saline (PBS) or PBS only (negative control), which was added to cells for 6 h, then treated with Actinomycin D for 0, 3, 6 or 12 h to block transcription. mRNA degradation was determined by qRT-PCR.
EMSA, ChIP and FACS
Nuclear extracts were prepared as described previously (26). Electrophoretic mobility shift assay (EMSA), ChIP and FACS methods are described in detail in the Supplementary Materials and Methods.
Transcription factor search
The search for possible transcription factors was conducted using the MATCH (Matrix Search for Transcriptional Factor Binding Sites) database (http://www.gene-regulation.com) (27).
Immunoprecipitation, in vitro binding assay and immunocytochemistry
Detailed protocols for immunoprecipitation, binding assay and immunocytochemistry are provided in the Supplementary Materials and Methods.
Statistical analysis
Significant differences between groups were determined by Welch's t-test. P-values <0.05 were considered to indicate statistical significance.
RESULTS
ROCK inhibition enhances miRNA function
We used cell-based reporter systems for in vitro monitoring of miRNA function to identify compounds that enhance miRNA-mediated gene silencing. Two tandem miR-122 binding sites were inserted into the 3′ UTR of the CMV promoter-driven firefly luciferase gene. This reporter construct was transiently co-transfected with miR-122-precursor-overexpressing plasmids into 293T cells. Luciferase values were monitored to determine whether a given compound enhanced or inhibited miRNA-mediated gene suppression. After three rounds of initial screening using a chemical library containing 1280 compounds, we selected 30 compounds that reduced the relative luciferase values by at least 75% or to less than one-quarter of the original value as potential enhancers of miRNA function (Supplementary Table S2). Further analysis revealed that most of these compounds caused cell death or inhibited cell growth, with the exceptions of Y27632, a ROCK inhibitor, and MNS, a Src and Syk kinase inhibitor (Supplementary Table S2). Similar screening was performed using another independent chemical library containing 322 compounds; we again selected potential enhancers of miRNA function and identified four ROCK inhibitors (Supplementary Table S3). Although H1152, a ROCK inhibitor, had slightly negative effects on cell viability, no significant effects of Y27632 were observed (Supplementary Table S3). To confirm these results in other cell lines, additional screening was performed using transiently transfected human colon cancer Caco2 cells, with reporter constructs containing let-7-binding sites and a let-7-precursor-expressing plasmid. ROCK inhibitors exhibited reproducible enhancement of miRNA function (Supplementary Table S4). Finally, we examined the selected compounds in HCT116 cells, another colon cancer cell line, with the let-7 reporter and precursor-expressing constructs, and confirmed the effects of ROCK inhibition on miRNA function (Supplementary Table S4). The ROCK inhibitor Y27632, which consistently enhanced miRNA function without affecting cell viability, was used to confirm enhanced miRNA-mediated gene expression by testing its effects on inhibition mediated by various miRNAs (Figure 1A). The response was miRNA-dependent since it was not observed in Dicer-knockout Mouse Embryonic Fibroblasts (MEFs) (Figure 1B and Supplementary Table S5). The effect of the ROCK inhibitor was found in all cell lines tested regardless of whether or not the reporter mRNA was targeted by overexpressed miRNAs or endogenous miRNAs (Figure 1C and D, and Supplementary Table S5). We also analyzed the effect of ROCK inhibition on levels of the endogenous protein CAT-1, whose expression is regulated mainly by endogenous miR-122 (28). ROCK inhibition reduced the expression of CAT-1 in Huh7 cells, likely by enhancing miRNA-122 function (Figure 1E). To confirm that inhibition of ROCK was responsible for the enhancement of miRNA function we assessed another clinically used ROCK inhibitor, fasudil, and found that it also enhanced miRNA-mediated repression of gene expression without appreciable effects on cell viability (Supplementary Figure S1).
Figure 1.
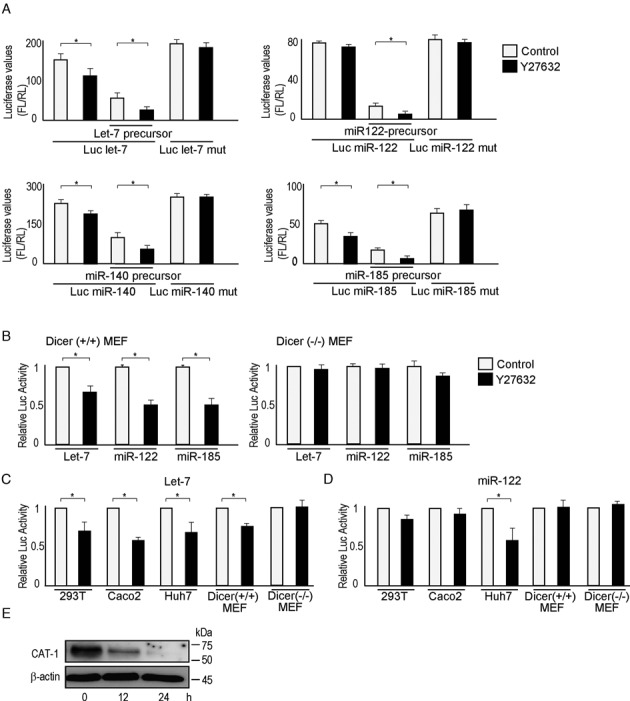
ROCK inhibition enhances miRNA function. (A) ROCK inhibitor Y27632 enhanced miRNA-mediated gene expression-suppressive effects. A reporter containing let-7-responsive elements was transfected together with a let-7-precursor overexpression plasmid into 293T cells. A reporter containing mutated let-7-responsive elements, which principally do not respond to let-7, was also included as a control. The values shown represent firefly luciferase values divided by those of renilla luciferase to normalize for transfection efficiency. Data represent the means ± S.D. of three experiments. *P< 0.05. Similar trends were observed when using miR-122, miR-140–5p or miR-185. (B) The effects of Y27632 were miRNA-dependent. The enhancement of miRNA function by Y27632 was observed in Dicer (+/+) MEFs, but not in Dicer (–/–) MEFs. Relative luciferase activity was calculated by adjusting the value (firefly luciferase value divided by renilla luciferase value) from the control sample as ‘1’. Data represent the means ± S.D. of three experiments. *P< 0.05. White bars, control; black bars, Y27632 treatment. (C, D) The effects of Y27632 on endogenous cellular miRNA function. Cells were transfected with reporter constructs to determine the effects of Y27632 on endogenous miRNA function. A let-7 reporter (C) and miR-122 reporter (D) were used. Because miR-122 was expressed only in Huh7 cells, its effects were evident only in this cell line (D). Relative luciferase values were calculated as described in (B). Data represent the means ± S.D. of three experiments. *P< 0.05. White bars, control; black bars, Y27632 treatment. (E) Protein expression of CAT-1, a miR-122 target gene, was suppressed by Y27632 treatment in Huh7 cells. Huh7 cells were treated with Y27632 for the indicated times. CAT-1 protein expression was determined by western blot analysis. Representative images from three independent experiments are shown.
ROCK inhibition enhances the deadenylation of miRNA-targeted mRNAs
To determine the mechanisms by which ROCK inhibition enhances miRNA function, we examined the expression of the miRNA pathway-related molecules Drosha, DGCR8, Dicer, Ago2, Gemin3 (DDX20), Gemin4 and KSRP in the presence or absence of Y27632; no significant differences in protein expression were evident (Figure 2A). Next, we examined the expression and maturation of miRNAs and found no ROCK-inhibitor-induced changes in the expression levels of mature endogenous let-7b, miRNA-122, miRNA-140–5p or miRNA-185 by quantitative RT-PCR (Supplementary Figure S2A). Northern blotting of overexpressed miR-122 confirmed these findings (Figure 2B). Comprehensive miRNA microarray analysis showed that ROCK inhibition did not significantly alter the expression levels of mature miRNAs (Supplementary Figure S2B).
Figure 2.
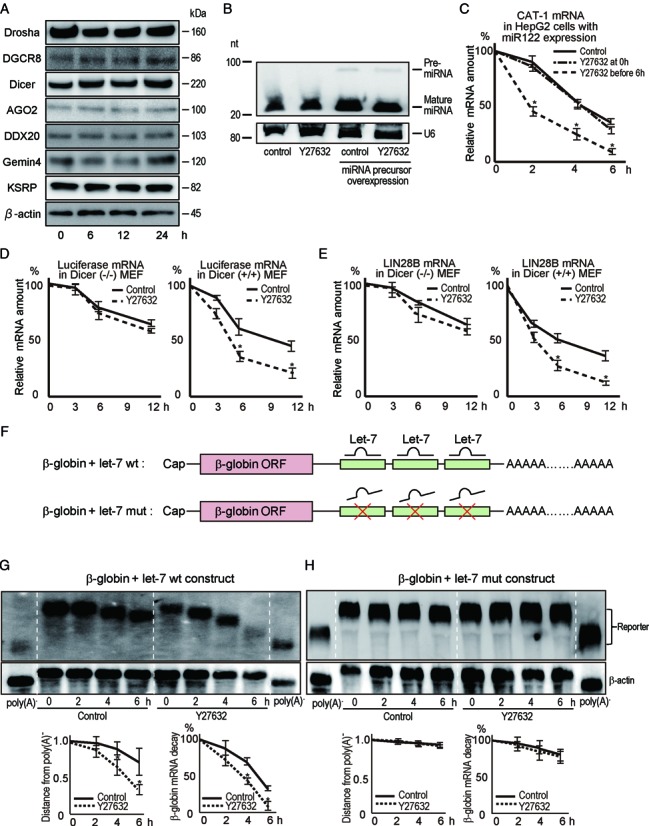
ROCK inhibition enhances deadenylation of miRNA-targeted mRNAs. (A) Huh7 cells were treated with Y27632. Western blotting results shown are representative of two independent experiments. Similar results were obtained using Caco2 cells. (B) Levels of mature miR122 and pre-miR-122 were measured by northern blotting and normalized to U6 snRNA levels. Total RNA from lysates of non-transfected Huh7 cells or Huh cells transfected with a miR-122 precursor-expressing plasmid were used. nt, nucleotides. The results shown are representative of three independent experiments. (C) HepG2 cells were transfected with a miR-122 precursor-expressing plasmid. Y27632 was applied either simultaneously or 6 h before treatment with ActD. Levels of CAT-1 mRNA after ActD treatment were determined at the indicated time points by qRT-PCR. Data represent the means ± S.D. of three experiments. *P< 0.05. (D) The depression of miRNA target mRNA expression by Y27632 was miRNA-dependent. Levels of luciferase mRNA expressed from a luciferase reporter construct with miR-122-responsive elements in its 3′-UTR were determined after transfection with a miR-122 precursor-expressing plasmid into Dicer (–/–) (left panel) and Dicer (+/+) (right panel) MEFs as described in (C). Data represent the means ± S.D. of three experiments. *P< 0.05. (E) The effects of Y27632 were detected using natural target sites and endogenous miRNA. A luciferase reporter construct carrying a region of the 3′-UTR of Lin28B mRNA targeted by let-7 was transfected; luciferase mRNA levels were determined as described in (D). Data represent the means ± S.D. of three experiments. *P < 0.05. (F) Constructs used to determine the kinetics of deadenylation. β-globin ORFs with three wild-type let-7 target sites (let-7 wt) or mutated let-7 target sites (let-7 mut) were used. The transcription of these constructs in HeLa-Tet-off cells was regulated by doxycycline. (G, H) Northern blots showing deadenylation and decay of let-7 wt (G) and let-7 mut (H) β-globin constructs after transcription was stopped in cells that were pretreated (or not) with Y27632. Cells were treated with Y27632 for 6 h and then transcription from the constructs was stopped using doxycycline. β-actin mRNA levels were used for normalization. Representative blotting images are shown. Plots below the blotting images show the poly(A) shortening by indicating the distances from the position of poly(A)− and the β-globin reporter mRNA levels after normalization. Poly(A)− RNA was prepared in vitro by treating the RNA sample with oligo(dT) and RNase H. Data represent the means ± S.D. of five independent experiments. *P < 0.05.
We determined the effect of ROCK inhibition on the loading of miRNAs onto RNA-induced silencing complexes (RISCs). We isolated Ago2-containing RISCs through immunoprecipitation from ROCK inhibitor-treated or non-treated 293T, Huh7 and Caco2 cells and quantified miRNAs associated with Ago2 (Supplementary Figure S2C and D). qRT-PCR revealed that when similar amounts of Ago2 protein were immunoprecipitated, the amount of miRNA associated with Ago2-containing RISCs was not altered by Y27632 (Supplementary Figure S2D), suggesting that ROCK inhibition does not affect the loading of miRNAs onto RISCs.
Finally, we examined whether ROCK inhibition affected the stability of miRNA-targeted mRNAs. mRNA expression of the miR-122-targeted gene CAT-1 was reduced by ∼50% after treatment with ROCK inhibitor for 12 h (Supplementary Figure S2E), correlating with the reduction in CAT-1 protein level (Figure 1E). To exclude the possibility that ROCK inhibitor treatment affects CAT-1 promoter activity, we measured the CAT-1 mRNA decay rate in the presence or absence of Y27632. Actinomycin D (ActD) was used to stop all cellular transcription after preincubation with Y27632 for 6 h. CAT-1 mRNA decay was enhanced by Y27632 in miR-122-overexpressing HepG2 cells (Figure 2C). In contrast, Y27632 had no effect on the stability of CAT-1 mRNA in non-miR-122–overexpressing HepG2 cells, suggesting that the effect of Y27632 was miR-122-dependent, consistent with reported data that HepG2 cells express only trace levels of miR-122 (29) (Supplementary Figure S2F). The enhanced effect was also Dicer-dependent because miR-122-targeted luciferase reporter mRNA decay was unaffected by Y27632 in Dicer-knockout MEFs (Figure 2D), and, more importantly, the reporter carrying mutations in miR-122-target sequences was unaffected by Y27632 (Supplementary Figure S2G). An endogenous let-7-targeted LIN28B reporter mRNA was also analyzed in wild-type and Dicer-/- MEFs (Figure 2E). Y27632 increased the decay of LIN28B mRNA in a Dicer-dependent manner. Thus, our data suggest that ROCK inhibition promotes miRNA-mediated degradation of mRNA targets.
Deadenylation of target mRNAs plays an initial role in miRNA-mediated mRNA decay (3,4,15,30). To monitor miRNA-mediated deadenylation and mRNA decay, we used a construct with a tetracycline (Tet)-promoter-driven β-globin open reading frame (ORF) containing three let-7 recognition sites in its 3′-UTR (4) (Figure 2F). After transfection of this construct into HeLa-Tet-off cells, addition of Tet derivative doxycycline (Dox) to the cells resulted in rapid and complete cessation of transcription from this construct, but did not affect the cell viability or expression of other genes (Supplementary Figure S2H and I). Northern blotting showed that Y27632 increased deadenylation and mRNA decay in cells treated with Dox (Figure 2G). These effects were let-7-dependent because mutation of let-7 recognition sites in the globin 3′-UTR (Figure 2F) abrogated ROCK-inhibitor-mediated enhancement of deadenylation (Figure 2H). Similar tendencies were identified in the changes in poly(A) length of HMGA2 mRNA, an endogenous let-7 target gene (Supplementary Figure S2J), by a ligation-mediated poly(A) test assay. These results suggest that Y27632 selectively enhances deadenylation of miRNA-targeted mRNAs, but not non-targeted reporter mRNAs.
Increased PAIP2 expression is involved in ROCK inhibitor-mediated mRNA deadenylation
To date, there is no information on the relationship between ROCK inhibition and miRNA function. We noticed that prolonged pretreatment with Y27632 was required to detect increased decay of miRNA-targeted mRNA when ActD was used to block global transcription (Figure 2C). Y27632 had no detectable effect when applied simultaneously with ActD. One possibility is that ROCK inhibition induces de novo protein synthesis and that newly synthesized proteins function to enhance miRNA-targeted mRNA degradation. To evaluate this, we compared the decay of a β-globin reporter mRNA driven by a Tet-off promoter after addition of Dox or ActD with or without Y27632. When transcription of a β-globin reporter mRNA containing wild-type let-7 binding sites was blocked by Dox, the ROCK inhibitor enhanced mRNA decay; however, when transcription was inhibited by ActD, the ROCK inhibitor had no detectable effect (Figure 3A). Thus ROCK inhibitors most likely require de novo protein synthesis to enhance miRNA function.
Figure 3.
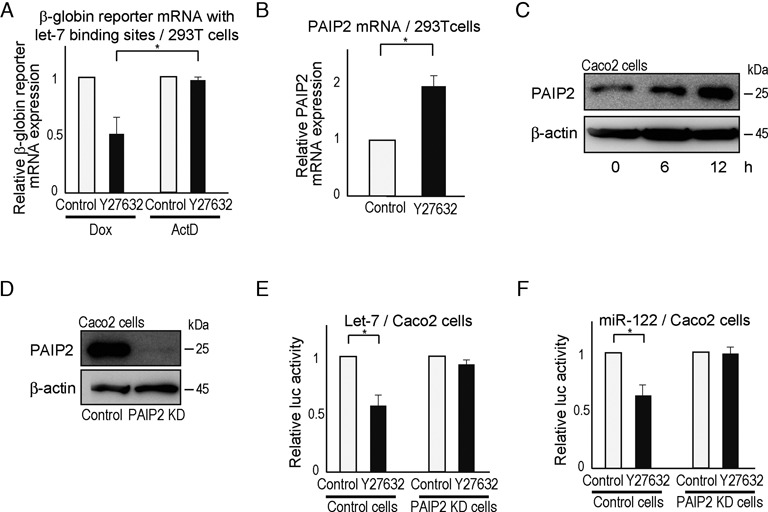
Enhancement of miRNA function by Y27632 is mediated by increased PAIP2 expression. (A) β-globin reporter mRNA levels were determined using a β-globin construct carrying let-7 wt binding sites. Let-7-mediated mRNA decay was enhanced by Y27632 when transcription from this construct was stopped by doxycycline, but not when ActD was applied. β-globin reporter mRNA levels in 293T cells were determined by qRT-PCR 8 h after adding Y27632 with Dox or ActD treatment. Data represent the means ± S.D. of three experiments. *P< 0.05. (B) PAIP2 mRNA levels in 293T cells were determined by qRT-PCR at 6 h after Y27632 treatment. Data represent the means ± S.D. of three experiments. *P< 0.05. Similar results were obtained using Caco2 cells. (C) PAIP2 protein levels in Caco2 cells treated with Y27632 were determined at the indicated time points by western blotting. (D) Confirmation of reduced PAIP2 protein levels in PAIP2-knockdown Caco2 cells. (E, F) Y27632 did not enhance miRNA function in PAIP2-knockdown Caco2 cells. Cells were transfected with a reporter construct and the corresponding miRNA precursor-expressing plasmid [let-7 in (E) and miR-122 in (F)] and the effect of Y27632 was determined. Data represent the means ± S.D. of three experiments. *P< 0.05.
Microarray analysis was performed to identify genes whose expression in 293T cells changed 3 and 6 h after ROCK inhibitor treatment. Of the 24 267 genes examined, 369 were more than 2-fold upregulated and 371 were at least 2-fold downregulated 3 or 6 h after ROCK inhibition (GEO accession number GSE32024). Analysis of these 369 genes revealed that PABP-interacting protein 2 (PAIP2) was a candidate participant in ROCK-inhibitor-mediated enhancement of miRNA function (please see the Supplementary Text). As PAIP2 stimulates mRNA deadenylation by displacing PABP from poly(A) tails (11,12), its induction by Y27632 could be involved in Y27632-mediated enhancement of mRNA deadenylation. We first confirmed that ROCK inhibitor Y27632 treatment increased PAIP2 expression, PAIP2 mRNA and protein levels in 293T and Caco2 cells (Figure 3B and C, and Supplementary Figure S3A).
To assess the involvement of PAIP2 in miRNA function, we established stable PAIP2-knockdown 293T and Caco2 cells (Figure 3D and Supplementary Figure S3B). Enhancement of miRNA function by Y27632 was almost completely lost in PAIP2-knockdown cells, irrespective of the miRNA species examined (Figure 3E and F, and Supplementary Figure S3C and D), suggesting that PAIP2 is a key player in ROCK-inhibitor-enhanced miRNA function. Another PAIP2 knockdown construct showed similar results in Caco2 cells, excluding the possible off-target effects of knockdown constructs (Supplementary Figure S3E–G). Results from the PAIP2-knockdown experiments in Hela-Tet-off cells also demonstrated the same importance of PAIP2 in ROCK-inhibitor-mediated enhancement of deadenylation and reporter mRNA decay (Supplementary Figure S4A and B). We did not detect an interaction between PAIP2 and Ago2 (Supplementary Figure S4C), suggesting that PAIP2 does not function via interaction with RISC complex. While the increase in PAIP2 expression was modest (Figure 3B and C), PAIP2 overexpression at a level comparable to that observed in Y27632-treated Caco2 cells indeed enhanced miRNA function to a level similar to Y27632 treatment (Supplementary Figure S4D and Supplementary Table S5). These results are consistent with previous reports that PAIP2 affects miRNA function by enhancing deadenylation of miRNA-targeted mRNAs (10).
HNF4A participates in ROCK-inhibition-enhanced PAIP2 expression
To determine the mechanisms by which ROCK inhibition increases PAIP2 expression, we examined PAIP2 promoter activity by analyzing nucleotides –1754 to +100 of the PAIP2 promoter. Deletion construct analysis showed that the region from –1223 to –727 was responsible for Y27632-induced enhancement of PAIP2 promoter activity (Figure 4A and B). A computational search revealed that four transcriptional factors, hepatocyte nuclear factor 4A (HNF4A), cut homeobox 1 (CUX1), organic cation transporter 1 (Oct1) and aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator (AhR/Arnt), likely participate in the regulation of PAIP2 expression (Supplementary Figure S5A). An EMSA showed that a shift of only the HNF4A-binding sequence was enhanced by Y27632 (Figure 4C, and Supplementary Figure S5B and C). To determine whether DNA binding of the protein was increased by HNF4A, probes with artificial HNF4A-targeting sequences were used in a gel-shift assay, confirming that Y27632 enhanced the binding of HNF4A to DNA (Figure 4D). The binding was HNF4A-specific, because no shifted bands were observed using PAIP2 promoter probes with mutations at possible HNF4-binding sites (Supplementary Figure S5D). Additionally, binding of HNF4A to PAIP2 promoter regions was confirmed by chromatin immunoprecipitation (Figure 4E). HNF4A knockdown experiments further confirmed that HNF4A is a mediator of ROCK inhibition, because HNF4A knockdown cells did not exhibit enhanced miRNA function upon ROCK inhibition (Supplementary Figure S5E and Supplementary Table S5). Since levels of HNF4A were not affected by Y27632 treatment in the tested cells (Supplementary Figure S5F), we next examined the molecular mechanisms underlying the effect of ROCK inhibition on the transcriptional activity of HNF4A.
Figure 4.
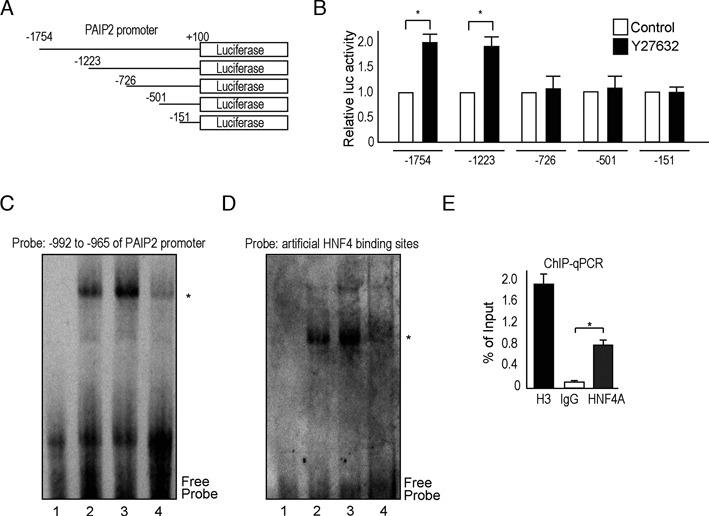
ROCK inhibition enhances PAIP2 promoter activity. (A) Deletion constructs carrying segments of the PAIP2 promoter conjugated to a firefly luciferase gene. (B) Y27632 activates the PAIP2 promoter at the –1223 to –727-bp region. (C, D) Y27632 increased protein binding to HNF4A binding sites, as determined by a gel-shift assay performed using Huh7 cell nuclear extracts and DNA probes with HNF4A binding sites. 1, no nuclear extract; 2, control nuclear extract; 3, nuclear extract from cells treated with Y27632 for 4 h; 4, as in (3), with an excess amount of cold probes (50-fold excess cold probes as competitors). DNA probes derived from –992 to –965 of the PAIP2 promoter containing two possible HNF4A binding sites were used in (C) and probes consisting of an artificial HNF4A binding site were used in (D). The results shown are representative of four independent experiments. (E) ChIP analysis results using Huh7 cells and anti-HNF4A, anti-H3 (positive control) or normal rabbit IgG (negative control) antibodies are shown. Primers designed against the HNF4A binding region in the PAIP2A promoter were used for subsequent RT-PCR. Data are presented as percentages of the input and represent means ± S.D. of three experiments. *P< 0.05.
ROCK inhibition enhances the binding of HNF4 and ROCK1
Both phosphorylation and lysine acetylation of HNF4A affect its transcriptional activity (31–33). Therefore, we first examined changes in post-translational modifications of HNF4A after ROCK inhibition, but detected no changes in such modifications under our experimental conditions (Supplementary Figure S6A). Next, to determine the mechanism by which the transcriptional ability of HNF4A is enhanced by ROCK inhibition, we investigated the interaction between HNF4A and two ROCK paralogs by immunoprecipitation of overexpressing flag-tagged HNF4A. There was a clear interaction between HNF4A and cellular endogenous ROCK1 and not ROCK2 (Figure 5A). Moreover, the interaction between HNF4A and ROCK1 was significantly enhanced upon treatment with the ROCK inhibitor (Figure 5A). To confirm the binding between HNF4A and ROCK1, we used HCT116 cells transfected with a halo-tagged ROCK1-expressing plasmid and flag-tagged HNF4A-expressing plasmid. Endogenous ROCK1 and halo-tagged ROCK1 proteins were co-precipitated by immunoprecipitation of the flag-tagged HNF4A protein. Consistent with the data shown in Figure 5A, the interaction between HNF4A and ROCK1 was enhanced upon treatment with the ROCK inhibitor (Figure 5B).
Figure 5.
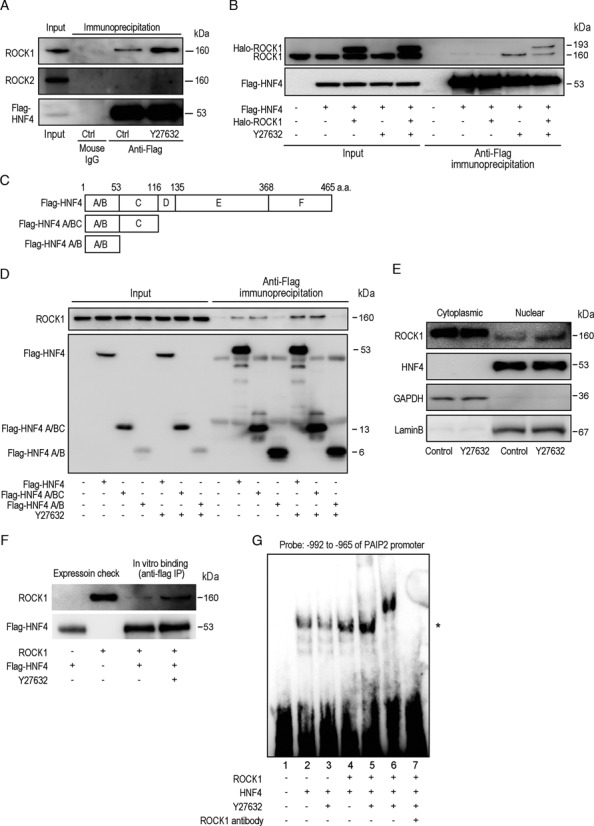
ROCK inhibition enhances the interaction between HNF4A and ROCK1. (A) ROCK1 binds HNF4A. HCT116 cells were transfected with flag-tagged HNF4A expressing plasmids. Cells were treated or non-treated with Y27632 for 6 h followed by immunoprecipitation using mouse anti-flag antibody. Mouse control IgG was used as a negative control. Precipitated lysates were subjected to western blotting. Five percent of the total cell lysate before immunoprecipitation was used as an internal control (‘input’). The results are representative of four independent experiments. (B) ROCK inhibition enhances the interaction between ROCK1 and HNF4A. HCT116 cells were transfected with flag-tagged HNF4A and Halo-tagged ROCK1-expressing plasmids as indicated. Cells were treated with Y27632 for 6 h before immunoprecipitation. Cell lysates were immunoprecipitated with an anti-flag antibody. Precipitated lysates were subjected to western blotting with anti-ROCK1 antibodies and anti-flag antibodies. Five percent of total cell lysate was used as ‘input.’ The results shown are representative of four independent experiments. (C) Deletion constructs of HNF4A. Plasmids expressing C terminal domain-deleted and flag-tagged HNF4A were constructed and named as indicated. (D) HCT116 cells were transfected with flag-tagged HNF4A expressing plasmids as indicated and subjected to immunoprecipitation using anti-flag antibodies. Y27632 was used for 6 h before immunoprecipitation in indicated cases. Endogenous ROCK1 was detected in the precipitated lysates. The results are representative of four independent experiments. (E) Cytoplasmic and nuclear fractions of Huh7 cells treated or non-treated with Y27632 for 6 h were blotted with anti-ROCK1 and anti-HNF4A antibodies. GAPDH (cytoplasmic marker) and Lamin B (nucleus marker) were blotted to confirm appropriate fractionation. The results are representative of four independent experiments. (F) Flag-tagged HNF4A and ROCK1 proteins were purified using 293T cell lysates transfected with flag-tagged HNF4A and Halo-tagged ROCK1 expressing plasmids by immunoprecipitation with anti-flag antibody and halo-tagged beads, followed by the elution using flag peptides and endoprotease, respectively. Equal amounts of protein were mixed in vitro and immunoprecipitated (IP) using anti-flag antibodies. Y27632 was added in the mixture for 6 h before the precipitation. Precipitated proteins, with 5% of purified proteins as an expression check, were subjected to western blotting using anti-ROCK1 and anti-flag antibodies. The results shown are representative of four independent experiments. (G) Y27632 increased the binding of HNF4A protein to the PAIP2 promoter. A gel-shift assay was performed by mixing DNA probes with purified HNF4A and ROCK1 proteins. 1, no protein; 2, HNF4A protein only; 3, HNF4A protein only with Y27632 treatment for 30 min; 4, HNF4A with ROCK1 protein; 5, as in (4), plus Y27632 treatment for 30 min; 6, as in (5), plus anti-ROCK1 antibody; 7, as in (4), with an excess amount of cold probes. * indicates the position of protein-bound DNA probe. n.s.; non-specific bands. DNA probes derived from –992 to –965 of the PAIP2 promoter were used as described in Figure 4. The results shown are representative of four independent experiments.
HNF4A is a member of the nuclear receptor family of transcriptional factors and has conserved domains: A/B (including activation function), C (DNA binding), D (hinge), E (ligand binding) and F (negative regulatory) (34,35). To identify the regions in HNF4A responsible for binding with ROCK1 protein, we constructed flag-tagged deletion constructs of HNF4A-protein-expressing plasmids (Figure 5C). Cellular ROCK1 protein was co-immunoprecipitated with the full-length HNF4A protein or the HNF4A protein containing the A/B and C regions, but not with HNF4A protein containing only the A/B regions (Figure 5D). This suggests that ROCK1 binds to the C region of HNF4A protein, which may enhance HNF4-DNA binding (please see below), similar to the interaction between CBP and HNF4A, which increases HNF4A DNA binding (33).
ROCK1 protein is localized mainly in the cytoplasm (36,37), whereas HNF4A is mainly in the nucleus (33); therefore, it is possible that these two molecules rarely interact naturally within cells. Thus, we re-examined the precise subcellular localization of these two molecules using confocal microscopy. HNF4A was located exclusively in the nucleus whereas ROCK1 was located mainly in the cytoplasm but detectable in the nucleus (Supplementary Figure S6B). Furthermore, upon treatment with the ROCK inhibitor, nuclear-localized ROCK1 increased, and co-localization of ROCK1 and HNF4A was detected in some cells (Supplementary Figure S6B). These localization changes were confirmed by western blotting using subcellular-fractioned lysates (Figure 5E). These results suggest that inhibition of ROCK enhances its translocation into the nucleus, which in turn could result in increased binding between HNF4A and ROCK1.
To further examine the effects of ROCK inhibition on the interaction between HNF4A and ROCK1, purified flag-tagged HNF4A and ROCK1 proteins were mixed in vitro with or without the ROCK inhibitor and immunoprecipitated using an anti-flag antibody. ROCK1 binding with HNF4A was enhanced in the presence of the ROCK inhibitor (Figure 5F), suggesting that a structural change within ROCK1, upon binding with its inhibitor, facilitates the ROCK1-HNF4A interaction. Furthermore, the amount of HNF4A protein bound to PAIP2 promoter DNA was increased slightly in the presence of ROCK1 protein, but increased significantly upon addition of the ROCK inhibitor, as determined by gel-shift assay using purified HNF4A and ROCK1 proteins (Figure 5G). The effects of ROCK inhibition were ROCK1-specific because there was no effect when only HNF4A protein was used (lanes 2 and 3 in Figure 5G). Collectively, our results suggest that HNF4A and ROCK1 interact in nuclei and that this interaction is enhanced by ROCK inhibition by both increasing the intra-nuclear levels of ROCK1 protein and enhancing the binding affinity of ROCK1 to HNF4A.
Effects of ROCK inhibition are independent of its kinase activity
To further test whether the effects of ROCK1 on HNF4A by ROCK inhibition are dependent on its kinase activity, we tested a kinase-dead construct of ROCK1 (ROCK1-KA) (38), which showed a similar defect of kinase activity to that of wild-type ROCK1 treated with the ROCK inhibitor (Figure 6A and B). The binding of ROCK1 and HNF4A, as well as HNF4A and the PAIP2 promoter, was similar to that of wild-type ROCK1 when using the ROCK-KA construct (Figure 6C and D); ROCK inhibition still enhanced these effects (Figure 6C and D), suggesting that the observed effects are independent of kinase activity.
Figure 6.
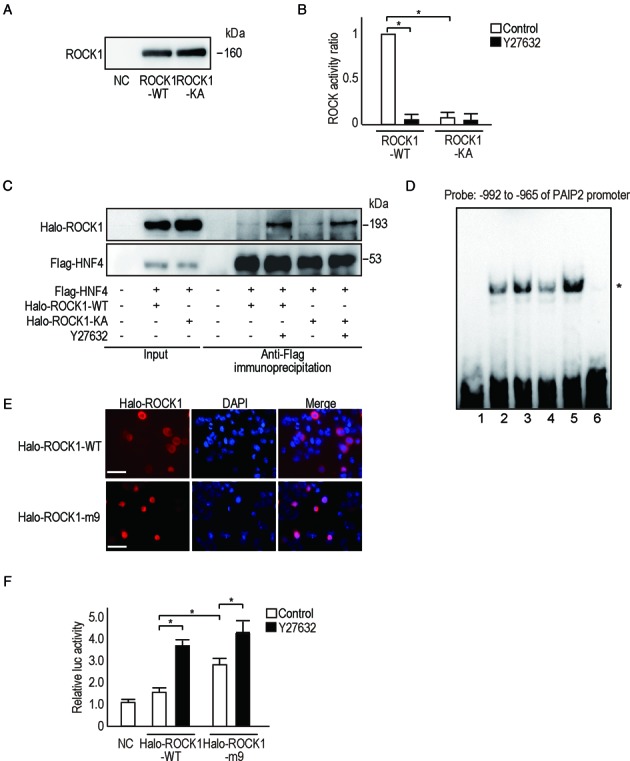
The effects of ROCK1 on the PAIP2 promoter are independent of its kinase activity. (A) ROCK1 proteins were purified from 293T cell lysates transfected with halo-tagged ROCK1 wild-type (ROCK1-WT) or ROCK1-KA-expressing plasmids. An equal amount of isolated protein was confirmed by western blotting. NC, negative control from non-transfected cell lysates. (B) ROCK activity of the isolated proteins (ROCK-WT and ROCK-KA) was determined in vitro. Y27632 was added, as indicated, at 10 μM for 6 h before measuring. Data represent the means ± S.D. of three experiments. *P < 0.05. (C) Binding between ROCK-KA and HNF4A was similar to that of ROCK1-WT and HNF4A. HCT116 cells were transfected with halo-tagged ROCK1-WT or ROCK-KA-expressing plasmids together with flag-tagged HNF4A-expressing plasmids as indicated. Y27632 was added for 6 h before immunoprecipitation with an anti-flag antibody. Precipitated lysates were subjected to western blotting using anti-Halo antibodies or anti-flag antibodies. Five percent of the total cell lysate was used as ‘input’ for comparison. The results shown are representative of four independent experiments. (D) A gel-shift assay using DNA probes derived from –992 to –965 of the PAIP2 promoter was performed to examine the binding between flag-tagged HNF4A to the DNA region in the presence of ROCK1-WT or ROCK-KA proteins. 1, no protein; 2, Flag-HNF4A with ROCK1-WT; 3, as in (2) plus treatment with Y27632 for 30 min; 4, Flag-HNF4A with ROCK1-KA; 5, as in (4) plus treatment with Y27632 for 30 min; 6, as in (2) with an excess amount of cold probes. * Indicates the position of the protein-bound DNA probe. n.s., non-specific bands. The results shown are representative of four independent experiments. (E) Halo-tagged ROCK1-m9 proteins translocate into the nucleus. HCT116 cells transfected with halo-tagged ROCK1-m9-expressing plasmids were immunostained using anti-halo antibodies. Nuclei were stained with DAPI. Images are representative of three independent experiments. Bar, 100 μm. (F) Halo-tagged ROCK1-m9 enhanced PAIP2 promoter activity more than halo-tagged ROCK1-WT. A luciferase assay was performed using HCT116 cell lysates transfected with halo-tagged ROCK1-WT or halo-tagged ROCK1-m9 with or without Y27632 to determine changes in PAIP2 promoter activity. Data represent the means ± S.D. of five experiments. *P < 0.05.
Next, to determine whether nuclear localization of ROCK1 is the sole cause of the effects on HNF4A, we constructed nuclear-localizing ROCK1 (ROCK1-m9) (Figure 6E) by fusing a nuclear localizing signal (m9) to its C-terminus (39). Although PAIP2 promoter activity was higher when expressing ROCK1-m9, it was further enhanced by ROCK inhibition (Figure 6F). These results confirm that ROCK inhibition enhances the effects of ROCK1 on HNF4A, in addition to changing its subcellular localization.
DISCUSSION
ROCK is a downstream kinase of RhoA (40) but here we show that ROCK inhibition enhances miRNA-mediated gene repression. We found that ROCK interacts with transcription factor HNF4A and ROCK inhibitors enhance this interaction. HNF4A acts as an effector of the ROCK inhibitor to enhance the expression of the deadenylation-related protein PAIP2, which promotes the degradation of miRNA-targeted mRNAs.
ROCK is known to mediate RhoA signaling in regulating various cellular functions, including smooth muscle contraction, cell adhesion and motility, cytokinesis and gene expression. Y27632 is widely used as a representative ROCK inhibitor. Y27632, developed as a pyridine derivative, is highly specific (41), and such specificity was confirmed in a subsequent study (42). In addition, structural studies have revealed that Y27632 binds to the inter-subdomain cleft where the substrate ATP is known to bind, and that Y27632-bound Rho kinase undergoes an induced-fit conformational change, providing the structural basis for its high selectivity (43). Results indicating that ROCK inhibition enhances miRNA-mediated gene repression found in this study were surprising. During the preparation of this manuscript, ROCK activity was reported to be important for the expression of a number of miRNA transcripts (44). However, in our microarray study, we detected no changes in mature miRNA expression levels due to ROCK inhibition. Rather, we found that ROCK inhibitors function by increasing HNF4A-dependent PAIP2 expression and enhanced miRNA-targeted mRNA deadenylation. Because PAIP2 displaces PABPs from poly(A) tails (11,45), it will be interesting to see if ROCK inhibitors may regulate the degradation of other decay pathway such as AU-rich element-mediated mRNA decay.
Consistent with our results, there is a report that PAIP2 knockdown and PABP overexpression diminished miRNA function, and the elevated levels of endogenous PAIP2 enhanced miRNA efficacy (10). However, whether PABP stabilizes mRNA adenylation is still controversial, and there are contradicting reports that PABP is required for CCR4-mediated deadenylation (46–48).
In addition, the specificity of enhanced miRNA-mediated mRNA decay upon increased PAIP2 expression without affecting most mRNAs, regardless of miRNAs, remains a major question. Previous studies using PABP knockdown or PAIP2 expression also reported no significant reduction in bulk protein synthesis (10,45), suggesting that changes in PABP or PAIP2 expression levels may retain some specificity in gene expression regulation. While we can assume that miRNA-mediated mRNA decay may be more sensitive to increased PAIP2 expression levels than non-miRNA-targeted, nonspecific mRNA decay, the precise mechanisms of this specificity await further examination. Nontheless, although there is still some debate over the role of PABP in the deadenylation of miRNA-targeted mRNAs (9,10,13,14,49), our results, along with those from a previous report (4), suggest that inhibition of PABP activity upon increased PAIP2 expression augments miRNA silencing, due at least in part to enhanced deadenylation of target mRNA.
Transcription of PAIP2 was enhanced by increased binding of HNF4A, a transcription factor, to the PAIP2 promoter. Post-translational modifications of HNF4A affect its transcriptional activity (50,51) and ROCK1, as a kinase, may be involved in post-translational modification of HNF4A. Therefore, we examined these modifications, but found that they did not appear to play a major role. Rather, we found that ROCK1 binds to HNF4A, which enhances HNF4A binding to its target consensus DNA and PAIP2 promoter regions. HNF4A has a nuclear receptor domain organization composed of five domains (34,52). Our results indicate that ROCK1 binds to the DNA-binding domain (C domain), which may affect DNA-binding affinity (53). It has been reported that binding to this domain by some proteins such as protein arginine methyltransferase 1 (PRMT1) augments DNA-binding affinity (54). Our results suggest that the binding of ROCK1 to the C domain of HNF4A promotes binding to target DNA regions, similar to PRMT1. While the precise conformational changes of HNF4A due to ROCK1 docking remain to be elucidated, the importance of conformational changes induced by cofactors in the nuclear receptor family is currently under investigation (34).
HNF4A is a nucleoprotein and ROCK1 is normally present in the cytoplasm. However, in some circumstances, such as under TGFb stimulation, ROCK1 translocates into the nucleus (55,56). Translocation into the nucleus has been reported even under normal conditions, but more significantly upon application of a ROCK inhibitor, as in this study. Translocation was not observed in all cells, suggesting that it may depend on the cell cycle or other factors. Structural solutions of ROCK1 complexes with inhibitors, such as Y27632, fasudil and H-1152, should provide crucial insight into the understanding of ROCK1 inhibitor induced structural changes in ROCK1 (43,57). Moreover, since the binding of ROCK1 with HNF4A can be enhanced by a ROCK inhibitor in vitro, in addition to increased translocation of ROCK1 into the nucleus, ROCK inhibitors should also be able to enhance the binding affinity of ROCK1 for HNF4A. Both of these effectors could be mediated by ROCK inhibitor-induced conformational changes in ROCK1.
Our results suggest that ROCK inhibitors enhance miRNA function by promoting miRNA-mediated mRNA decay. These effects may be similar for any type of miRNA. As we and others have reported, a global reduction in miRNA levels is a general trait of human cancers and plays a causal role (17–19). Although failure of eyelid and ventral body wall closure were reported in ROCK1-knockout mice (58,59), these are related to embryonic developmental processes. The ROCK inhibitor fasudil is used clinically for the treatment of cerebral vasospasms; thus, its safety has already been demonstrated. The effects of the ROCK inhibitor-induced increase in global miRNA activity, in addition to the inhibitory effects on cell-autonomous promotion of tumor cell invasion and metastasis (57), may be useful for preventing carcinomas or other pathological conditions caused by a global reduction in, or insufficient activity of, miRNAs.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan (#25293076, #25860520 and #20390204 to M.O., T.Y., K.K.); National Institutes of Health (GM46454); Houston Endowment, Inc. (to A.S.); 973 program 2009CB522200 from the Ministry of Science and Technology of China and Natural Science Foundation of China (#91029304 to J.H.). Funding for open access charge: Japan Science and Technology Agency, Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu L., Fan J., Belasco J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl Acad. Sci. U.S.A. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C.Y., Zheng D., Xia Z., Shyu A.B. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun J.E., Huntzinger E., Fauser M., Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Chekulaeva M., Mathys H., Zipprich J.T., Attig J., Colic M., Parker R., Filipowicz W. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabian M.R., Cieplak M.K., Frank F., Morita M., Green J., Srikumar T., Nagar B., Yamamoto T., Raught B., Duchaine T.F., et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol. 2011;18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 9.Zekri L., Kuzuoğlu-Öztürk D., Izaurralde E. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. EMBO J. 2013;32:1052–1065. doi: 10.1038/emboj.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walters R.W., Bradrick S.S., Gromeier M. Poly(A)-binding protein modulates mRNA susceptibility to cap-dependent miRNA-mediated repression. RNA. 2010;16:239–250. doi: 10.1261/rna.1795410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khaleghpour K., Svitkin Y.V., Craig A.W., DeMaria C.T., Deo R.C., Burley S.K., Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 12.Karim M.M., Svitkin Y.V., Kahvejian A., De Crescenzo G., Costa-Mattioli M., Sonenberg N. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc. Natl Acad. Sci. U.S.A. 2006;103:9494–9499. doi: 10.1073/pnas.0603701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moretti F., Kaiser C., Zdanowicz-Specht A., Hentze M.W. PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat. Struct. Mol. Biol. 2012;19:603–608. doi: 10.1038/nsmb.2309. [DOI] [PubMed] [Google Scholar]
- 14.Huntzinger E., Kuzuoglu-Öztürk D., Braun J.E., Eulalio A., Wohlbold L., Izaurralde E. The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res. 2013;41:978–994. doi: 10.1093/nar/gks1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eulalio A., Huntzinger E., Nishihara T., Rehwinkel J., Fauser M., Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 17.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 18.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 19.Kumar M.S., Lu J., Mercer K.L., Golub T.R., Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 20.Hill D.A., Ivanovich J., Priest J.R., Gurnett C.A., Dehner L.P., Desruisseau D., Jarzembowski J.A., Wikenheiser-Brokamp K.A., Suarez B.K., Whelan A.J., et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rio Frio T., Bahubeshi A., Kanellopoulou C., Hamel N., Niedziela M., Sabbaghian N., Pouchet C., Gilbert L., O’Brien P.K., Serfas K., et al. DICER1 mutations in familial multinodular goiter with and without ovarian Sertoli-Leydig cell tumors. JAMA. 2011;305:68–77. doi: 10.1001/jama.2010.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heravi-Moussavi A., Anglesio M.S., Cheng S.W., Senz J., Yang W., Prentice L., Fejes A.P., Chow C., Tone A., Kalloger S.E., et al. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N. Engl. J. Med. 2012;366:234–242. doi: 10.1056/NEJMoa1102903. [DOI] [PubMed] [Google Scholar]
- 23.Kumar M.S., Pester R.E., Chen C.Y., Lane K., Chin C., Lu J., Kirsch D.G., Golub T.R., Jacks T. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kojima K., Takata A., Vadnais C., Otsuka M., Yoshikawa T., Akanuma M., Kondo Y., Kang Y., Kishikawa T., Kato N. MicroRNA122 is a key regulator of alpha-fetoprotein expression and influences the aggressiveness of hepatocellular carcinoma. Nat. Commun. 2011;2:338. doi: 10.1038/ncomms1345. [DOI] [PubMed] [Google Scholar]
- 25.Mols J., van den Berg A., Otsuka M., Zheng M., Chen J., Han J. TNF-alpha stimulation inhibits siRNA-mediated RNA interference through a mechanism involving poly-(A) tail stabilization. Biochim. Biophys. Acta. 2008;1779:712–719. doi: 10.1016/j.bbagrm.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schreiber E., Matthias P., Müller M., Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matys V., Fricke E., Geffers R., Gössling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang J., Nicolas E., Marks D., Sander C., Lerro A., Buendia M.A., Xu C., Mason W.S., Moloshok T., Bort R., et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 29.Bhattacharyya S., Habermacher R., Martine U., Closs E., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 30.Djuranovic S., Nahvi A., Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z., Tavares-Sanchez O.L., Li Q., Fernando J., Rodriguez C.M., Studer E.J., Pandak W.M., Hylemon P.B., Gil G. Activation of bile acid biosynthesis by the p38 mitogen-activated protein kinase (MAPK): hepatocyte nuclear factor-4alpha phosphorylation by the p38 MAPK is required for cholesterol 7alpha-hydroxylase expression. J. Biol. Chem. 2007;282:24607–24614. doi: 10.1074/jbc.M611481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ktistaki E., Ktistakis N.T., Papadogeorgaki E., Talianidis I. Recruitment of hepatocyte nuclear factor 4 into specific intranuclear compartments depends on tyrosine phosphorylation that affects its DNA-binding and transactivation potential. Proc. Natl Acad. Sci. U.S.A. 1995;92:9876–9880. doi: 10.1073/pnas.92.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soutoglou E., Katrakili N., Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol. Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 34.Chandra V., Huang P., Potluri N., Wu D., Kim Y., Rastinejad F. Multidomain integration in the structure of the HNF-4α nuclear receptor complex. Nature. 2013;495:394–398. doi: 10.1038/nature11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daigo K., Kawamura T., Ohta Y., Ohashi R., Katayose S., Tanaka T., Aburatani H., Naito M., Kodama T., Ihara S., et al. Proteomic analysis of native hepatocyte nuclear factor-4α (HNF4α) isoforms, phosphorylation status, and interactive cofactors. J. Biol. Chem. 2011;286:674–686. doi: 10.1074/jbc.M110.154732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leung T., Manser E., Tan L., Lim L. A novel serine/threonine kinase binding the Ras-related RhoA GTPase which translocates the kinase to peripheral membranes. J. Biol. Chem. 1995;270:29051–29054. doi: 10.1074/jbc.270.49.29051. [DOI] [PubMed] [Google Scholar]
- 37.Matsui T., Amano M., Yamamoto T., Chihara K., Nakafuku M., Ito M., Nakano T., Okawa K., Iwamatsu A., Kaibuchi K. Rho-associated kinase, a novel serine/threonine kinase, as a putative target for small GTP binding protein Rho. EMBO J. 1996;15:2208–2216. [PMC free article] [PubMed] [Google Scholar]
- 38.Ishizaki T., Naito M., Fujisawa K., Maekawa M., Watanabe N., Saito Y., Narumiya S. p160ROCK, a Rho-associated coiled-coil forming protein kinase, works downstream of Rho and induces focal adhesions. FEBS Lett. 1997;404:118–124. doi: 10.1016/s0014-5793(97)00107-5. [DOI] [PubMed] [Google Scholar]
- 39.Siomi H., Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishizaki T., Maekawa M., Fujisawa K., Okawa K., Iwamatsu A., Fujita A., Watanabe N., Saito Y., Kakizuka A., Morii N., et al. The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO J. 1996;15:1885–1893. [PMC free article] [PubMed] [Google Scholar]
- 41.Uehata M., Ishizaki T., Satoh H., Ono T., Kawahara T., Morishita T., Tamakawa H., Yamagami K., Inui J., Maekawa M., et al. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- 42.Davies S.P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi H., Miwa Y., Kasa M., Kitano K., Amano M., Kaibuchi K., Hakoshima T. Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y-27632. J. Biochem. 2006;140:305–311. doi: 10.1093/jb/mvj172. [DOI] [PubMed] [Google Scholar]
- 44.Stiles J.M., Kurisetty V., Mitchell D.C., Bryan B.A. Rho kinase proteins regulate global miRNA expression in endothelial cells. Cancer Genomics Proteomics. 2013;10:251–263. [PubMed] [Google Scholar]
- 45.Yoshida M., Yoshida K., Kozlov G., Lim N.S., De Crescenzo G., Pang Z., Berlanga J.J., Kahvejian A., Gehring K., Wing S.S., et al. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J. 2006;25:1934–1944. doi: 10.1038/sj.emboj.7601079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simón E., Séraphin B. A specific role for the C-terminal region of the Poly(A)-binding protein in mRNA decay. Nucleic Acids Res. 2007;35:6017–6028. doi: 10.1093/nar/gkm452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao G., Chiang Y.C., Zhang C., Lee D.J., Laue T.M., Denis C.L. PAB1 self-association precludes its binding to poly(A), thereby accelerating CCR4 deadenylation in vivo. Mol. Cell. Biol. 2007;27:6243–6253. doi: 10.1128/MCB.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang C., Lee D.J., Chiang Y.C., Richardson R., Park S., Wang X., Laue T.M., Denis C.L. The RRM1 domain of the poly(A)-binding protein from Saccharomyces cerevisiae is critical to control of mRNA deadenylation. Mol. Genet. Genomics. 2013;288:401–412. doi: 10.1007/s00438-013-0759-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fabian M.R., Mathonnet G., Sundermeier T., Mathys H., Zipprich J.T., Svitkin Y.V., Rivas F., Jinek M., Wohlschlegel J., Doudna J.A., et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chellappa K., Jankova L., Schnabl J.M., Pan S., Brelivet Y., Fung C.L., Chan C., Dent O.F., Clarke S.J., Robertson G.R., et al. Src tyrosine kinase phosphorylation of nuclear receptor HNF4α correlates with isoform-specific loss of HNF4α in human colon cancer. Proc. Natl Acad. Sci. U.S.A. 2012;109:2302–2307. doi: 10.1073/pnas.1106799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iordanidou P., Aggelidou E., Demetriades C., Hadzopoulou-Cladaras M. Distinct amino acid residues may be involved in coactivator and ligand interactions in hepatocyte nuclear factor-4alpha. J. Biol. Chem. 2005;280:21810–21819. doi: 10.1074/jbc.M501221200. [DOI] [PubMed] [Google Scholar]
- 52.Petrescu A.D., Hertz R., Bar-Tana J., Schroeder F., Kier A.B. Role of regulatory F-domain in hepatocyte nuclear factor-4alpha ligand specificity. J. Biol. Chem. 2005;280:16714–16727. doi: 10.1074/jbc.M405906200. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto K.R. Steroid receptor regulated transcription of specific genes and gene networks. Annu. Rev. Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]
- 54.Barrero M.J., Malik S. Two functional modes of a nuclear receptor-recruited arginine methyltransferase in transcriptional activation. Mol. Cell. 2006;24:233–243. doi: 10.1016/j.molcel.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhowmick N.A., Ghiassi M., Aakre M., Brown K., Singh V., Moses H.L. TGF-beta-induced RhoA and p160ROCK activation is involved in the inhibition of Cdc25A with resultant cell-cycle arrest. Proc. Natl Acad. Sci. U.S.A. 2003;100:15548–15553. doi: 10.1073/pnas.2536483100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu J., Landerholm T.E., Wei J.S., Dong X.R., Wu S.P., Liu X., Nagata K., Inagaki M., Majesky M.W. Coronary smooth muscle differentiation from proepicardial cells requires rhoA-mediated actin reorganization and p160 rho-kinase activity. Dev. Biol. 2001;240:404–418. doi: 10.1006/dbio.2001.0403. [DOI] [PubMed] [Google Scholar]
- 57.Rath N., Olson M.F. Rho-associated kinases in tumorigenesis: re-considering ROCK inhibition for cancer therapy. EMBO Rep. 2012;13:900–908. doi: 10.1038/embor.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimizu Y., Thumkeo D., Keel J., Ishizaki T., Oshima H., Oshima M., Noda Y., Matsumura F., Taketo M.M., Narumiya S. ROCK-I regulates closure of the eyelids and ventral body wall by inducing assembly of actomyosin bundles. J. Cell Biol. 2005;168:941–953. doi: 10.1083/jcb.200411179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rikitake Y., Oyama N., Wang C.Y., Noma K., Satoh M., Kim H.H., Liao J.K. Decreased perivascular fibrosis but not cardiac hypertrophy in ROCK1+/- haploinsufficient mice. Circulation. 2005;112:2959–2965. doi: 10.1161/CIRCULATIONAHA.105.584623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


