ABSTRACT
Most bacterial cells are enclosed in a single macromolecule of the cell wall polymer, peptidoglycan, which is required for shape determination and maintenance of viability, while peptidoglycan biosynthesis is an important antibiotic target. It is hypothesized that cellular enlargement requires regional expansion of the cell wall through coordinated insertion and hydrolysis of peptidoglycan. Here, a group of (apparent glucosaminidase) peptidoglycan hydrolases are identified that are together required for cell enlargement and correct cellular morphology of Staphylococcus aureus, demonstrating the overall importance of this enzyme activity. These are Atl, SagA, ScaH, and SagB. The major advance here is the explanation of the observed morphological defects in terms of the mechanical and biochemical properties of peptidoglycan. It was shown that cells lacking groups of these hydrolases have increased surface stiffness and, in the absence of SagB, substantially increased glycan chain length. This indicates that, beyond their established roles (for example in cell separation), some hydrolases enable cellular enlargement by making peptidoglycan easier to stretch, providing the first direct evidence demonstrating that cellular enlargement occurs via modulation of the mechanical properties of peptidoglycan.
IMPORTANCE
Understanding bacterial growth and division is a fundamental problem, and knowledge in this area underlies the treatment of many infectious diseases. Almost all bacteria are surrounded by a macromolecule of peptidoglycan that encloses the cell and maintains shape, and bacterial cells must increase the size of this molecule in order to enlarge themselves. This requires not only the insertion of new peptidoglycan monomers, a process targeted by antibiotics, including penicillin, but also breakage of existing bonds, a potentially hazardous activity for the cell. Using Staphylococcus aureus, we have identified a set of enzymes that are critical for cellular enlargement. We show that these enzymes are required for normal growth and define the mechanism through which cellular enlargement is accomplished, i.e., by breaking bonds in the peptidoglycan, which reduces the stiffness of the cell wall, enabling it to stretch and expand, a process that is likely to be fundamental to many bacteria.
INTRODUCTION
In almost all bacteria, the major stress-bearing component of the cell envelope is a single, polymeric macromolecule of peptidoglycan. In order for an individual cell to grow (enlarge), new monomeric precursors are added to the peptidoglycan sacculus. These are inserted by penicillin binding proteins (PBPs), guided by a complex machinery involving many components (1). However, enlargement of the sacculus cannot occur solely through the addition of new material. Existing bonds must be cut in order that the sacculus can permanently expand via accommodation of more material. This activity is executed by peptidoglycan hydrolases. These break specific amide and glycosidic bonds in the sacculus, prompting the idea that some of these enzymes are required for the enlargement of individual bacterial cells and, consequently, for bacterial population growth.
While there is likely to be great variation in cellular enlargement mechanisms across the array of bacterial species, there is a broadly held assumption that new peptidoglycan monomers are inserted such that they do not initially experience stress derived from turgor pressure. The idea is that stress is subsequently placed on this new material due to the breaking of bonds within older peptidoglycan, allowing the sacculus as a whole to expand. While completely reasonable and therefore widely accepted, this assumption is largely unsupported by experimental evidence. This concept underpins the two major models of cellular enlargement, one of which is for elongation of rod-shaped Gram-positive species and the other for Escherichia coli. In rod-shaped Gram-positive bacteria, “inside-to-outside growth” is proposed (2). New monomers are applied at a high surface density close to the inner surface of the cell wall (the only region accessible to the membrane-bound PBPs). As subsequent layers are added, the older material is enzymatically degraded, causing stress to be applied to the new material and the sacculus to expand. In E. coli, the “three for one” model states that three new peptidoglycan monomers are added in a loop around an existing monomer (3). When the bonds attaching the existing monomer to its neighbors are broken, stress is applied to the new loop, enabling expansion of the sacculus. While they differ in detail, both of these models invoke breaking of bonds within peptidoglycan and, thus, hydrolase activity.
The necessity of hydrolases in cellular enlargement models suggests that removing hydrolase activity should arrest this process, causing the cell to stop dividing and ultimately terminating the growth of the population, but surprisingly, there is only one example of a hydrolase-encoding gene that is individually essential for survival, pcsB in Streptococcus pneumoniae (4, 5). However, there are groups of hydrolases that are synthetically essential. In Bacillus subtilis, removal of YvcE (CwlO) and LytE terminates cell elongation and, thus, population growth (6), and in E. coli, loss of Spr, YdhO, and YebA leads to an increasingly ellipsoid cell shape and synthetic lethality (7). The existence of individually or collectively essential hydrolases in these diverse species strongly suggests a general phenomenon applicable to many other bacteria: that bacterial cellular enlargement and thus, ultimately, division and population growth depend on the ability of cells to hydrolyze peptidoglycan (8). However, the detailed mechanisms by which hydrolysis enables individual cells to enlarge remain unclear.
In many species of bacteria, including E. coli, B. subtilis, and S. pneumoniae, cellular enlargement is accomplished by two machineries (groups of proteins that work together to execute a cellular process), one for elongation and another for division (9). In these species, hydrolases can potentially be attributed separately to either of these machineries (10). Staphylococcus aureus presents a simplified system in which to study the role of hydrolases in cell enlargement, as it is roughly spherical and does not have a specific elongation machinery. Insertion of peptidoglycan apparently occurs only during septation (11, 12), while cell volume increases at a constant rate throughout the cell cycle (13), before the process is repeated on a plane orthogonal to the two previous divisions (14). The septal disc is initially protected from the level of turgor-induced stress experienced by the rest of the cell wall, as it is formed inside the cell. For this reason and the absence of a preexisting layer of cell wall at the beginning of septation, the inside-to-outside-growth model cannot be applied. Maturation of the septal disc is accompanied by alterations in peptidoglycan architecture and mechanical properties. The nanoscale surface architecture of the peptidoglycan changes from a ring like (15) to a knobbly (punctate) pattern (11). This is accompanied by changes in the stiffness of the cell surface; Atomic Force Microscopy (AFM) has shown that recently revealed septal cell wall is stiffer than mature cell wall (16).
S. aureus has numerous genes encoding peptidoglycan hydrolases (17–19). Here, we focus on N-acetylglucosaminidases (glucosaminidases), a class of cell wall hydrolases that target the bond between the N-acetylglucosamine and N-acetylmuramic acid residues in the polysaccharide (glycan) backbone of peptidoglycan. S. aureus has very short glycan strands compared to many other Gram-positive species, attributed to prolific glucosaminidase activity (20, 21). Previously, the only glucosaminidase characterized in S. aureus was the bifunctional glucosaminidase N-acetylmuramyl-l-alanine amidase, Atl, thought to be primarily involved in cell division (22, 23).
In this study, we report that a previously uncharacterized enzyme, SagB, is the major glucosaminidase for processing of glycan chains in S. aureus. We unravel the mechanisms connecting the biochemical activity of glucosaminidases to the mechanical properties of the cell wall, the cellular morphology and, ultimately, the capacity of the bacterial cells to enlarge and the population to grow. We propose that, as previously speculated, modulation of cell wall mechanical properties by hydrolases is a general mechanism for the enlargement of bacterial cells, with the timing and synchronicity of insertion and hydrolysis differing widely between species.
RESULTS
Shape changes and cell wall alteration throughout the cell cycle.
Cellular morphological dynamics were investigated using fluorescence microscopy of living cells stained with FM 1-43 to visualize the membrane. Of 100 cell cycle events scrutinized in detail, all cells exhibited a rapid splitting event within the 15-s interval between camera frames (Fig. 1a). This is consistent with very recently published data showing that this process occurs within milliseconds and that cell volume increase occurs at a constant rate throughout the cell cycle, leading to a prolate morphology (13). The splitting is preceded by the cell surface scission event previously observed during AFM imaging of cell division (24, 25).
FIG 1 .
Morphological dynamics during the cell cycle of S. aureus. (a) FM 1-43 labeling of living S. aureus SH1000 cells shows that the bacteria change shape rapidly immediately after division (see dashed boxes). (b) Images of S. aureus after labeling of the cell wall with Van-FL (~1.7-kDa) and WGA-AF350 (~38-kDa) fluorescent probe molecules. Arrowheads show cells where Van-FL is bound to regions from which WGA-AF350 has been excluded.
AFM imaging (11) and force measurements (16) indicate that septal peptidoglycan changes as it matures. To investigate this further, the accessibility of peptidoglycan to a large peptidoglycan-binding probe was assessed in S. aureus wild-type strain SH1000 cells (Fig. 1b). Wheat Germ Agglutinin-Alexa Fluor 350 conjugate (WGA-AF350; heterodimer of approximately 38 kDa) is a GlcNAc-binding lectin. Although the GlcNAc-MurNAc glycan motif is ubiquitous, in many instances, the WGA-AF350 complex labeled only part of the cell. Comparison with fluorescent vancomycin (Van-FL) labeling, which binds the pentapeptide that is prevalent in regions of newly inserted peptidoglycan and is thus a marker of nascent cell wall (26), revealed that WGA-AF350 preferentially bound the mature cell wall but was excluded from the septum. This suggests that the architecture of the nascent cell wall hinders access by the large WGA lectin, whereas in matured cells, the peptidoglycan is labeled homogenously. In contrast, the approximately 22-fold-smaller Van-FL (~1.5 kDa) could access the nascent peptidoglycan, even when daughter cells were not separated, further evidence of modification of the peptidoglycan network.
Glucosaminidases are critical for population growth in S. aureus.
Given that the short glycan chain length in S. aureus suggests a major role for glucosaminidases in overall peptidoglycan hydrolysis, we hypothesized that inactivation of all glucosaminidase activity would have an impact on population growth. Four enzymes with glucosaminidase activity (known and putative) were identified by BLAST searches against the known glucosaminidase domain of Atl (Fig. 2a; see also Fig. S1 in the supplemental material). Those identified are atl (23) and three additional glucosaminidase domain-encoding genes, for which the nomenclature sagA (SACOL2298) and sagB (SACOL1825), for S. aureus glucosaminidase A and B, respectively, and scaH (SACOL2666) is proposed. Gene inactivations were made in each of the four, and every combination of triple mutant constructed in S. aureus SH1000 (Table 1, Table 2, and Table 3). Despite repeated attempts, we were unable to obtain a strain in which all four putative glucosaminidase-encoding genes were inactivated, prompting the hypothesis that glucosaminidase activity is essential. To test this, a conditional quadruple glucosaminidase mutant was constructed by inserting an inducible sagB expression construct into the SH4611 (atl sagA scaH) triple mutant background. The resulting strain, SH4615 (Pspac-sagB atl sagA scaH), contains a truncated copy of sagB under the control of the native promoter and a full copy of sagB under the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter (see Fig. S2 in the supplemental material).
FIG 2 .
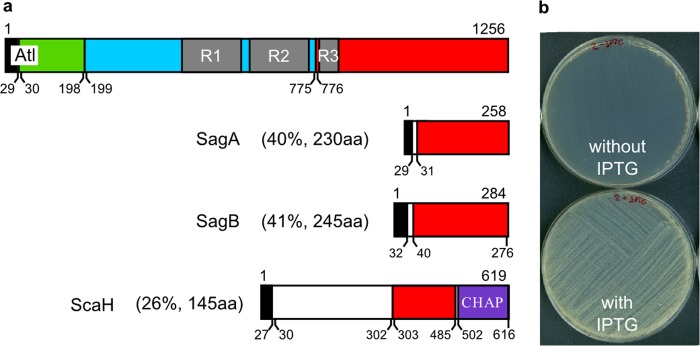
Role of glucosaminidases in population growth. (a) Physical map showing the domain structure of putative glucosaminidases of S. aureus. Percentage homology to Atl glucosaminidase domain and length of homologous amino acid sequence (aa) are indicated in brackets. Black, signal peptide; green, propeptide; grey, repeat domains R1, R2, and R3; blue, N-acetyl-muramyl-l-alanine amidase domain; red, N-acetyl-β-d-glucosaminidase domain; purple, cysteine, histidine-dependent amidohydrolase/peptidase (CHAP) domain. (b) Growth of SH4615 (Pspac-sagB atl sagA scaH) with or without IPTG.
TABLE 1 .
Strains used in this study
| Species | Strain | Relevant genotype or description | Source |
|---|---|---|---|
| E. coli | BL21 (DE3) | F− ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagen |
| TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80 lacZ ΔM15 ΔlacX74 recA1deoR araD139 Δ(ara-leu)7697 galK rpsL Strr endA1 nupG | Invitrogen | |
| SH3062 | BL21(DE3) pET24d+ sagB overexpression construct | ||
| SH3061 | E. coli BL21(DE3) pSRC002 | 36 | |
| SH2195 | E. coli BL21(DE3) pSRC003 | 36 | |
| S. aureus | SH1000 | Functional rsbU+ derivative of 8325-4 | 37 |
| SH1367 (atl) | SH1000 [atl::pAZ106 (ery lin)] | 23 | |
| SH4091 (atl::spc) | SA113 (atl::spc) | 38 | |
| SH4606 (sagA) | SH1000 (sagA::tet) | This study | |
| SH4607 (scaH) | SH1000 [scaH::tet(M)] | This study | |
| SH4608 (sagB) | SH1000 (sagB::kan) | This study | |
| SH4609 [sagB geh(sagB+)]a | SH1000 (sagB::kan geh::sagB) | This study | |
| SH4610 (atl sagA sagB) | SH1000 (atl::pAZ106 sagA::tet sagB::kan) | This study | |
| SH4611 (atl sagA scaH) | SH1000 [atl::spc sagA::tet scaH::tet(M)] | 39; this study | |
| SH4612 (sagA sagB scaH) | SH1000 [sagA::tet sagB::kan scaH::tet(M)] | This study | |
| SH4613 (atl sagB scaH) | SH1000 [atl::pAZ106 sagB::kan scaH::tet(M)] | This study | |
| SH4614 (Pspac-sagB) | SH1000 (Pspac-sagB) | This study | |
| SH4615 (Pspac-sagB atl sagA scaH) | SH1000 [Pspac-sagB atl::spc sagA::tet scaH::tet(M)] | This study | |
| SH4090 | RN4220 (Pspac-sagB Eryr) | This study | |
| RN4220 | Restriction-deficient derivative of 8325-4 | 40 | |
| RN6911 | agr::tet(M) derivative of RN6390 | 41 | |
| B. subtilis | 168 HR | Wild type | Howard Rogers |
Chromosomal sagB complementation construct abbreviated to geh(sagB+) in the main text.
TABLE 2 .
Plasmids used in this studya
| Plasmid | Relevant background/genotype/markers | Source |
|---|---|---|
| pAZ106 | Promoterless transcriptional lacZ fusion vector, used as a lacZ expression reporter plasmid; Ampr (E. coli), Eryr (S. aureus) | 42 |
| pDG1513 | Vector carrying tet cassette suitable for selection in Gram-positive bacteria; Minr Tetr | 43 |
| pET24d | His6 tag overexpression vector; Kanr | Novagen |
| pET24d-SagB | pET24d containing the sagB sequence, minus the signal sequence, upstream from the His6 tag | |
| pGL433b | Vector carrying kan cassette suitable for selection in Gram-positive bacteria; Kanr | J. Garcia-Lara and S. J. Foster, unpublished data |
| pInvSA | pUC19 containing a 2.4-kb fragment of the region spanning the sagB gene in which the sagB gene has been inactivated by a 600-bp deletion; Ampr | |
| pMUTIN4 | Insertion vector carrying IPTG-inducible Pspac promoter; Ampr (E. coli), Eryr (S. aureus) | 44 |
| pRW01 | pMUTIN4 insertion vector carrying Pspac promoter and 650-bp fragment of sagB; Ampr (E. coli), Eryr (S. aureus) | |
| pSA18Kan | pMUTIN4 containing EcoRI-BamHI-cut fragment of pInvSA and 1.5 kb Kanr cassette from pGL433 within disrupted sagB construct at KpnI restriction site; Ampr (E. coli), Eryr (S. aureus), Kanr | |
| pSA26Min2 | pSA26D with tet(M) cassette of S. aureus RN6911 inserted into KpnI site within the scaH gene; Ampr (E. coli), Eryr (S. aureus), tet(M) resistance gene | |
| pSA26D | pMUTIN4 containing 2.7-kb fragment of scaH and ~1 kb of flanking region; Ampr (E. coli), Eryr (S. aureus) | |
| pSRC002 | atl amidase domain overexpression construct | 36 |
| pSRC003 | atl glucosaminidase domain overexpression construct | 36 |
| pKASBAR | pUC18 vector containing attP and tet cassette (Ampr Tetr) | 45 |
| pKASBAR-sagB | pKASBAR containing a 1,357-bp fragment, including the sagB gene and promoter region, inserted between EcoRI and BamHI restriction sites; pUC19 E. coli cloning vector (Ampr) | NEB |
| pCR 2.1-TOPO | TOPO TA cloning vector; Ampr Kanr | Life Technologies |
Ampr, ampicillin resistance; Eryr, erythromycin resistance; Kanr, kanamycin resistance; Tetr, tetracycline resistance; Minr, minocycline resistance.
TABLE 3 .
Oligonucleotides used in this study
| Primer | Sequence (5′–3′)a | Restriction site |
|---|---|---|
| RW01_F | TTTTTTGAATTCAACAATGACCTAAGAGGTGTGGA | EcoRI |
| RW01_R | TTTTTTGGATCCCAACCATGCTTTTTAGC | BamHI |
| P1 | CGGGCTCTAGATAATCCACACAGCTGGCGTCTTAGC | XbaI |
| P2 | CGGCCGGTACCAGGATCTGTTTCGAATAATGATGTTGC | KpnI |
| FD2 | CGGCGGGTACCAACATCATTATTCGAAACAGATCCTAG | KpnI |
| RD2 | CGGGCAAGCTTTATTTACGTGCAAATGATATTAATC | HindIII |
| FKan | GGCGGGGTACCCAGCGAACCATTTGAGG | KpnI |
| RKan | GGGGCGGTACCAATTCCTCGTAGGCGCTCGG | KpnI |
| P6 | GGGCCGGATCCTTCAAGGTATAGTTTGAGCC | BamHI |
| P7 | TATTATGAGCTCTATCGTCGTATTCGGCTTAAG | SacI |
| MinFK3 | AACAAGGTACCAATATGCTCTTACGTGCT | KpnI |
| MinRK4 | AACAAGGTACCAGAAATATTGAAGCTAGT | KpnI |
| InvF1 | GCGCGGGGTACCAGAACATGAAGACTGAAGGAA | KpnI |
| InvR1 | GCGCGGGGTACCTTCAATCTTAATGTCGGAT | KpnI |
| SagA-F1 | CGACGGATCCTAACGGAACAATACCTACTC | BamHI |
| SagA-R1 | ATAACTGCGGCCGCGAGTGACATTCGCTGGGCAG | NotI |
| SagA-F2 | CCGGTACCTTCACGATGAGTAATACAGC | KpnI |
| SagA-R2 | ACATGAATTCAACCGCAGTACAGTGTTC | EcoRI |
| Tet-NotI | ATAACTGCGGCCGCGGATTTTATGACCGATGATGAAG | NotI |
| Tet-KpnI | CCGGTACCTGTTATAAAAAAAGGATCAAT | KpnI |
| rSagB_For | GCGCCCATGGTATCCGATCAGATATTTTTCAAACATGTT | NcoI |
| rSagB_Rev | GCGCCTCGAGCTTATTCAAATGTTTACTGTCATC | XhoI |
| 1825C_For | TTTTTTGAATTCGGTCAAATTGAAGGCACGAT | EcoRI |
| 1825C_Rev | TTTTTTGGATCCTTGCATTGGTGGGATTATCA | BamHI |
| Geh_For | GAGGTGCTGACAATGATGAAAA | |
| Geh_Rev | CCGATTAATTGAAAGAAGTCTGC |
Restriction sites included for cloning purposes are indicated in bold font.
The importance of glucosaminidase activity was assessed by plating the conditional mutant on solid medium in the presence of 1 mM IPTG and then streaking single colonies onto solid medium with or without IPTG (Fig. 2b). In the absence of IPTG, little growth was observed. Thus, it is not the individual enzymes that are required, as they are functionally redundant, but more likely glucosaminidase activity itself.
Although inactivation of sagB alone did not affect growth on solid medium (data not shown), in liquid medium, inactivation of sagB alone led to a substantial increase in doubling time [for SH1000, 30 ± 1 min (mean ± standard error), and for SH4608 (sagB), 43 ± 2 min] and yield (see Fig. S3 in the supplemental material). Strain SH4615 (Pspac-sagB atl sagA scaH) without IPTG exhibited a longer doubling time (50 ± 4 min) than SH4608 (sagB), SH4611 (atl sagA scaH), and SH4615 (Pspac-sagB atl sagA scaH) with IPTG.
Cells lacking glucosaminidases have morphological defects.
Exponential-phase cells (optical density at 600 nm [OD600] of ~0.3) were labeled with Van-FL prior to fixation, to visualize the cell wall and plane of septation. In wild-type SH1000, the normal range of roughly spherical-to-prolate shapes with or without septa were identified (11). However, in SH4615 (Pspac-sagB atl sagA scaH) without IPTG, roughly hemispherical cells distinct from previously observed wild-type morphologies were observed (Fig. 3a and b). These hemispherical cells were not attached to their sisters, and in some cases, these were bisected by a nascent septum despite not yet having expanded into the mature morphology. This shows that normal cellular enlargement has failed to take place prior to the cell attempting to initiate another round of division. Hemispherical cells were also observed at lower prevalence in SH4615 (Pspac-sagB atl sagA scaH) with IPTG and in SH4611 (atl sagA scaH) (Fig. 3c), suggesting that expression from the Pspac promoter is insufficient to give native levels of SagB. In SH4608 (sagB), very few hemispherical cells were observed (see Fig. S4 in the supplemental material). To summarize, cells impaired in glucosaminidase activity are also impaired in their ability to increase in size after division and adopt the correct mature shape.
FIG 3 .
Morphological defects in S. aureus cells lacking glucosaminidases. (a) Images of fixed, Van-FL-labeled cells showing altered morphology. Arrowheads indicate hemispherical cells. Cells of this shape are not found in wild-type populations unless attached to a sister cell. (b) Examples of hemispherical cells. In some cells, septa are visible in bacteria that have not completed the shape change to the mature spherical morphology. This shows that correct shape change is not taking place within the duration of the cell cycle. (c) Quantification of the proportion of hemispherical cells in each sample. P values are the result of Fisher’s exact tests comparing the wild type with each mutant.
SagB modulates cell wall elasticity.
In order to investigate the relationship between the ability of cells to enlarge and assume correct morphology and cell wall mechanical properties, the stiffness of the cell wall was measured using AFM in several glucosaminidase mutants. This enabled exploration of the possibility that increased stiffness (i.e., more force must be applied to result in the same amount of stretching of the cell wall) is associated with impaired glucosaminidase activity (Fig. 4). With this approach, a force is applied by an AFM tip to a surface of interest and the resulting deflection of the cantilever and, thus, indentation of the cell surface is measured. By measuring the gradient of a tangent to this force-displacement curve in the region of low deformation, a relative measure of cell surface stiffness is obtained independently of overall cell deformation and turgor. More sophisticated contact mechanics models were not employed for reasons described previously (16). The measurements were taken from multiple points on the surface of the cell (Fig. 4a) in regions not specifically identified as recently having been part of the septal plate, i.e., regions lacking ring or spiral surface architecture.
FIG 4 .

Mechanical properties of the S. aureus cell wall. (a) AFM heights and effective spring constants (stiffness maps) of SH1000 and SH4608 (sagB) derived from force maps. In the height map, regions with a lighter color are higher than darker regions. In the stiffness map, regions with a lighter color are stiffer than darker regions. Scale bars, 200 nm; height scale, 500 nm; stiffness scale, 0.010 to 0.018 Nm−1. (b) Stiffness of the cell wall of wild-type and glucosaminidase mutant strains, derived from AFM force maps.
In strains lacking any combination of three glucosaminidase-encoding genes, the median cell wall stiffness was significantly increased (Wilcoxon rank sum test) compared with that of the wild-type strain SH1000 (Fig. 4b; also see Table S1 in the supplemental material). In all strains combining other mutations with sagB, the cell wall stiffness was similar to that of SH4608 (sagB) cells. SH4611 (atl sagA scaH) cells had cell walls that were stiffer than those of SH1000 cells but less stiff than the cell walls of any strain lacking sagB. Chromosomal complementation of the sagB deletion using the native promoter led to almost complete restoration of wild-type stiffness. Thus, even though SagB has the most profound role in cell wall stiffness determination, there is an important contribution from the other three enzymes.
SagB regulates glycan chain length.
The bulk properties of a polymer (such as peptidoglycan) are a consequence of its nanoscale structure. Glucosaminidases might therefore modify the mechanical properties of peptidoglycan, making it less stiff, through reduction of glycan strand length. In order to establish whether the observed stiffness changes could be ascribed to altered chain length, we investigated the individual contribution of each glucosaminidase to glycan chain length regulation, using size exclusion chromatography to analyze the chain length of purified N-acetyl[14C]glucosamine ([14C]GlcNAc)-labeled glycans (Fig. 5; see also Fig. S5 and S6 in the supplemental material).
FIG 5 .

Role of glucosaminidase activity in glycan chain length determination in S. aureus. Strains lacking sagB had substantially longer glycan strands than did SH1000. The presence or absence of other glucosaminidase-encoding genes (atl, sagA, and scaH) had minimal effect on strand length. Annotations show proportions of glycan strands within ranges of numbers of disaccharides (DS); grey traces show the proportions in SH1000 cells for comparison. To compensate for the fact that longer glycan strands incorporate more [14C]GlcNAc, radioactivity counts (cpm) were divided by the corresponding theoretical molecular weight (see Materials and Methods). The glycan chain abundance is plotted normalized relative to the maximal ratio between radioactivity counts and theoretical molecular weight (cpm/MW).
Consistent with previously published data (20, 21), wild-type S. aureus had predominantly short glycan chains (on average, 6 to 10 dissacharides), with approximately 30% of glycan chains exceeding 50 dissacharides in length (Fig. 5; see also Fig. S5 and S6 in the supplemental material). However, inactivation of sagB resulted in a substantial increase in the proportion of long glycan strands (52.5% had >50 disaccharides). Complementation of sagB restored the wild-type chain length distribution (32.9% had >50 disaccharides) (Fig. 5).
Of the strains lacking three glucosaminidases, cells of all strains carrying the sagB inactivation had an increased proportion of long glycan strands (48.4 to 59.3% had >50 disaccharides) (Fig. 5). In these cases, the proportion was even higher than for SH4608 (sagB). Strains with inactivations in both sagA and sagB have the highest proportion of long glycans. This suggests a modest additional impact on chain length from Atl, ScaH, and in particular, SagA. SH4611 (atl sagA scaH), in which SagB was the sole remaining glucosaminidase, had a glycan chain length distribution similar to that of SH1000 (32.5% had >50 disaccharides). Thus, SagB has a dominant enzymatic activity and is the major glucosaminidase responsible for the archetypical short glycan chain length of S. aureus. Furthermore, this activity is nonredundant, as the presence of functional atl, sagA, and scaH did not compensate for sagB inactivation. The glycan chain length distributions of SH1367 (atl), SH4606 (sagA), and SH4607 (scaH) were similar to that of SH1000 (see Fig. S5 in the supplemental material). Given the dominant role of SagB in glycan chain length reduction, its activity likely masks any more subtle combined, mutually redundant role of the other three enzymes.
The capability of SagB to hydrolyze peptidoglycan was confirmed in vitro (using B. subtilis peptidoglycan as a substrate) by zymogram assay (see Fig. S7a in the supplemental material). There was more complete hydrolysis when the assay was carried out at pH 5 than at pH 7.5. B. subtilis peptidoglycan is a useful and appropriate substrate, as it has previously been shown to have long glycan strands compared to those of S. aureus (21). B. subtilis purified glycan chains were digested with recombinant SagB or Atl (glucosaminidase domain) and analyzed by size exclusion chromatography (see Fig. S7b). Both SagB- and Atl (glucosaminidase domain)-digested material had a lower molecular weight than undigested glycan strands. Material digested by Atl (glucosaminidase domain) had an overall lower molecular weight than that digested by SagB. This indicates a partial digestion by SagB compared to the digestion by Atl (glucosaminidase domain), suggesting a preferential activity by SagB on longer glycan strands as substrates (i.e., an inability to hydrolyze shorter strands).
SagB has a minimal role in cell separation.
The effect of hydrolase inactivation on cell separation was investigated using flow cytometry and optical microscopy. Those strains lacking Atl exhibited higher levels of forward scatter than other strains (forward scatter tends to be higher for larger objects, i.e., larger clumps; see Fig. S8a in the supplemental material), a finding qualitatively confirmed by optical microscopy (see Fig. S8b), suggesting a less important role in this process for SagA, SagB, and ScaH. However, the highest level of forward scatter was observed for SH4611 (atl sagA scaH), demonstrating a combined effect.
DISCUSSION
The group of hydrolases we have studied here exhibit functional redundancy in terms of population growth, and only by inactivating sagA, atl, and scaH and depleting sagB expression do we see that it is critical for the bacteria to retain at least one of the products of these genes. We have also shown that it is only under conditions where glucosaminidase activity has been effectively removed that S. aureus cells are impaired in their ability to enlarge normally. It seems clear that this inability to enlarge at the cellular level explains the population growth defects.
In light of this redundancy, it is surprising to see that SagB is by far the dominant enzyme in terms of the effect on glycan chain length. S. aureus has short glycan chains relative to those of other Gram-positive bacteria for which size exclusion High-Performance Liquid Chromatography (HPLC) measurements have been made (20, 21, 27). Our data show that the processivity of enzymes that insert peptidoglycan into the sacculus by forming glycosidic bonds (PBP2, MGT, and SgtA [28]) is not responsible for the predominance of short glycan chains but, instead, that chains are subsequently processed by SagB. Inactivation of sagB alone is enough to increase cell surface stiffness, a phenomenon explicable simply in terms of the dependence of the bulk properties of a polymer on the number of cross-links between individual chains; this explanation is supported by the discovery that peptide cross-linking levels also affect stiffness (29). This establishes a clear relationship between glycan chain length and cell surface stiffness. However, inactivation of sagA, atl, and scaH together also led to an increase in stiffness, although not as great as that caused by inactivation of sagB. This is despite there being no apparent alteration from the wild-type chain length in SH4611 (atl sagA scaH). We interpret this as evidence that these hydrolases affect cell surface stiffness with minimal influence on the overall glycan chain length, most likely by breaking a small number of bonds, bonds in specific locations in the chains, or bonds in specific locations within the sacculus. Nevertheless, active sagB is unable to fully compensate for their absence. Ultimately, it seems that cellular enlargement depends on reduction of cell wall stiffness mediated by hydrolases, with a concomitant alteration to the peptidoglycan structure and architecture.
While there have been many proposals describing detailed mechanisms by which peptidoglycan is inserted into the cell wall during enlargement of bacterial cells, the overarching concept is that unstressed material is added before parts of the preexisting sacculus are hydrolyzed to enable expansion. We have shown a mechanistic basis for this in S. aureus, where hydrolysis of peptidoglycan modulates the mechanical properties of the cell wall, which enables irreversible expansion of the cell. A unifying model across the bacteria can be invoked in which dense, stiff regions of peptidoglycan are initially formed, becoming less dense and less stiff as they are hydrolyzed and, thus, enabling enlargement of the cell surface area; this model is independent of the detailed mechanism of monomer insertion and mode of hydrolysis. In E. coli, the insertion of new peptidoglycan is targeted to less dense, more porous regions of the cell wall (1, 30) via an established mechanism involving inner and outer membrane proteins (31, 32), making these regions more dense. Hydrolysis and expansion of these regions would allow for enlargement. In B. subtilis, the detailed mechanism of peptidoglycan insertion is less well understood (27, 33), but essential hydrolase activity is required for cell enlargement (6). All of the proposed growth modes are compatible with the general principle of reduction in peptidoglycan density and increase in elasticity through hydrolase activity to enable enlargement, as demonstrated here for S. aureus.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The S. aureus strains used in this study are listed in Table 1, plasmids are listed in Table 2, and primers in Table 3.
Growth conditions and media.
All S. aureus strains were grown in brain heart infusion (BHI) broth at 37°C with aeration at 250 rpm unless otherwise stated. E. coli and B. subtilis strains were routinely grown in Luria-Bertani (LB) medium or Nutrient Broth, respectively, at 37°C with aeration at 250 rpm. For solid media, 1.5% (wt/vol) agar was added. Where required, selection for antibiotic resistance markers was carried out using the following concentrations of drugs: Ampr, ampicillin (100 µg/ml); Chlr, chloramphenicol (30 µg/ml); Eryr, erythromycin (5 µg/ml) with lincomycin (25 µg/ml); Kanr, kanamycin (50 µg/ml) with neomycin (50 µg/ml); Minr, minocycline (2 µg/ml); Spcr, spectinomycin (100 µg/ml); Tetr, tetracycline (5 µg/ml).
Genetic modification of bacteria.
Transformation by electroporation of E. coli or the restriction-deficient S. aureus RN4220 strain was performed according to published methods (34, 35). Phage transduction into the S. aureus SH1000 background using φ11 or φ85 was carried out as described previously (23). Details of construction of strains can be found in Text S1 in the supplemental material.
Overexpression and purification of recombinant enzymes.
An overnight culture was used to inoculate 1 liter of preheated LB containing appropriate antibiotics for maintenance of the overexpression plasmid to an OD600 of 0.05. At an OD600 of approximately 0.4, 1 mM IPTG was added, and the culture incubated for a further 4 h. Cells were harvested by centrifugation and stored as pellets at −80°C. Pellets were freeze-thawed three times in sodium phosphate buffer and sonicated on ice six times. Insoluble material was separated by centrifugation at 10,000 × g for 30 min. The supernatant was filter sterilized (0.45-µm filters) and purified using a 5-ml HiTrap column (Amersham) with a BioRad Econo gradient pump and fraction collector. The His-tagged proteins were eluted from the column using an isocratic gradient of 5-to-60% 0.5 M imidazole over 30 min. Eluted fractions were analyzed by SDS-PAGE. Fractions containing overexpressed protein were pooled, transferred to dialysis tubing, and dialyzed three times in phosphate-buffered saline (PBS) for 18 h in total. The identities of purified, overexpressed proteins were confirmed by N-terminal sequencing.
Analysis of autolysin activity by zymograms.
The lytic activity of the recombinant glucosaminidases was investigated by zymogram (23), using purified cell walls of vegetative B. subtilis as a substrate.
Purification of sacculi.
Bacterial cultures were grown to exponential phase (OD600 of ~0.5), and peptidoglycan purified as described previously (11). Briefly, cells were broken by mechanical shearing using a FastPrep homogenizer (S. aureus) or French press (B. subtilis). Sacculi were extracted by boiling in SDS (4% wt/vol), Pronase (2 mg/ml) treatment, and removal of accessory polymers by incubation in hydrofluoric acid (48% vol/vol) at 4°C for 48 h. Purified sacculi were washed extensively (at least six times) in water after SDS or hydrofluoric acid treatment. Long-term storage of sacculi was at −20°C. Radiolabeling with N-acetyl[14C]glucosamine ([14C]GlcNAc) was carried out as described previously (21). Briefly, exponential-phase (OD600 of 0.3) cultures were diluted in 50 ml of prewarmed LB containing 0.185 MBq [14C]GlcNAc (1.67 TBq/mmol; Hartmann Analytic) and 500 ml of nonradioactive medium to give a starting OD600 of 0.04. After three generations, the cells were harvested by centrifugation and sacculi purified.
Purification of glycan strands.
Glycan strands were purified as previously described (21). Radiolabeled peptidoglycan sacculi were digested by recombinant S. aureus Atl amidase domain (36). Typically, 1 mg of peptidoglycan was digested overnight with Atl at a concentration 5-fold greater than that required to solubilize more than 90% of the peptidoglycan. The enzyme was inactivated by boiling (3 min), and the supernatants were collected for further analysis.
Plating efficiency of conditional mutant.
A single colony of SH4615 (Pspac-sagB atl sagA scaH) was taken from an agar plate containing 1 mM IPTG and appropriate antibiotics using a sterile inoculation loop and was resuspended in 10 ml PBS. A cotton bud was then used to streak this suspension onto plates containing 1 mM IPTG or lacking IPTG.
Liquid growth of conditional mutant.
Colonies were taken from agar plates containing appropriate antibiotics [and 1 mM IPTG in the case of SH4615 (Pspac-sagB atl sagA scaH)] using a sterile inoculation loop. These were individually resuspended in 1 ml BHI. Subsequently, these suspensions were used to inoculate 50 ml BHI in 250-ml conical flasks to a calculated OD600 of 0.001, and the flasks were incubated at 37°C with agitation at 250 rpm, with optical density measurements taken periodically. Strains were sonicated prior to measurements of optical density and inoculations to reduce the potential effects of clumping.
Time-lapse microscopy.
Bacteria were grown overnight at 37°C with agitation at 250 rpm in BHI and then subcultured to an OD600 of ~0.05 and grown under the same conditions to an OD600 of ~0.3. Subsequently, 2 µl of this culture was pipetted onto an agarose pad containing 0.5 µg/ml FM 1-43 (Molecular Probes), allowed to partially dry, and then topped with a coverslip before being imaged using a Nikon Eclipse inverted epifluorescence microscope equipped with an incubator used to hold the experiment at 37°C.
Imaging cells labeled with fluorescent vancomycin or WGA.
BHI medium was used throughout. Strains were sonicated prior to measurements of optical density and inoculations to reduce the potential effects of clumping. Strains were grown overnight in the presence of appropriate antibiotics and, in the case of SH4615 (Pspac-sagB atl sagA scaH), 1 mM IPTG. Starter cultures [containing no antibiotics but with 10 µM IPTG in the case of SH4615 (Pspac-sagB atl sagA scaH)] were then inoculated. These were incubated at 37°C with agitation at 250 rpm until they reached an OD600 of ~1. A 10-ml sample was then washed once in prewarmed BHI and used to inoculate cultures from which microscopy samples would be taken. One millimolar IPTG was added to one culture of SH4615 (Pspac-sagB atl sagA scaH) to induce the expression of sagB, while another was left without IPTG. Samples for microscopy were taken from exponential-phase cultures and then labeled with Van-FL and imaged as previously described (11). Fluorescent WGA (Molecular Probes) labeling took place after Van-FL labeling but before fixing cells. Cells were resuspended in 250 µl distilled water containing 1 mM CaCl2 and 100 µg/ml fluorescent WGA and then washed three times by centrifugation.
Cell wall stiffness measurements.
Cell wall stiffness measurements were carried out as described previously (16). Briefly, samples of cells of each strain to be studied were grown to exponential phase and then washed and immobilized on a microstructure (24). Bacteria were indented repeatedly under BHI medium, a curve from an equivalent indentation on an incompressible material subtracted, and the tangent to the resulting force versus indentation curves used to derive a relative effective spring constant to obtain a measure of stiffness.
HPLC separation of glycan strands.
Size exclusion chromatography of glycan strands was performed as described previously (21, 27). Approximately 20,000 cpm of each radiolabeled glycan strand fraction (corresponding to 50 to 100 µg peptidoglycan) was injected in a volume of 200 µl onto a TSKSW2000 (7.5 by 600 mm) size exclusion HPLC column (Tosoh) preequilibrated in 100-mM phosphate buffer (pH 6.0). Elution was carried out at a flow rate of 0.4 ml/min. Radiolabeled glycan strands were detected with a LabLogic (β-Ram model 4) radio flow detector using a 1:1 scintillation cocktail and a 100-µl solid cell. The gel filtration columns were calibrated as described previously (21), using dextran standards ranging from 1 kDa to 150 kDa (analytical standard grade for GPC; Sigma-Aldrich). Analysis of the glycan strand distribution was carried out as described previously (21).
Flow cytometry analysis
Bacteria were incubated overnight with agitation at 37°C. Ten milliliters of fresh BHI was inoculated with 100 µl of overnight culture (dilution of 1:100). Bacteria were incubated to an OD600 of 0.3 to 0.4 (early exponential phase) and then diluted 1:100 in PBS. The samples were analyzed by flow cytometry using an Attun autosampler and imaged using a Novex optical microscope.
SUPPLEMENTAL MATERIAL
Identification of four putative N-acetylglucosaminidases encoded by the genome of S. aureus COL. Download
Construction of SH4615 (Pspac-sagB atl sagA scaH). Download
Growth of glucosaminidase mutants in liquid medium. Download
Role of SagB in S. aureus morphology. Download
Impact of single glucosaminidase-encoding gene inactivation on glycan strand length in S. aureus as determined by gel filtration chromatography. Download
Impact of glucosaminidase inactivation on glycan strand length in S. aureus. Download
Enzymatic activities of Atl and SagB. Download
Analysis of cell clumps in glucosaminidase mutants. Download
Comparison of AFM stiffness measurements between strains.
Genetic modification of bacteria. Download
ACKNOWLEDGMENTS
This work was funded by Biotechnology and Biological Sciences Research Council (BBSRC grants BB/H011005/1 and BBL006162/1) and made use of facilities provided through the Medical Research Council (MRC)-funded SHIMA project (grant MR/K015753/1). S.M. was supported by a Marie Curie intra-European Fellowship (251336). The Royal Society funded equipment.
Footnotes
Citation Wheeler R, Turner RD, Bailey RG, Salamaga B, Mesnage S, Mohamad SAS, Hayhurst EJ, Horsburgh M, Hobbs JK, Foster SJ. 2015. Bacterial cell enlargement requires control of cell wall stiffness mediated by peptidoglycan hydrolases. mBio 6(4):e00660-15. doi:10.1128/mBio.00660-15.
REFERENCES
- 1.Typas A, Banzhaf M, Gross CA, Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koch AL, Doyle RJ. 1985. Inside-to-outside growth and turnover of the wall of gram-positive rods. J Theor Biol 117:137–157. doi: 10.1016/S0022-5193(85)80169-7. [DOI] [PubMed] [Google Scholar]
- 3.Höltje J-V. 1993. Three for one —a simple growth mechanism that guarantees a precise copy of the thin, rod-shaped murein sacculus of Escherichia coli, p 419–426. In de Pedro MA, Höltje J-V, Löffelhardt W (ed), Bacterial growth and lysis: metabolism and structure of the bacterial sacculus. Springer, New York, NY. [Google Scholar]
- 4.Bartual SG, Straume D, Stamsås GA, Muñoz IG, Alfonso C, Martínez-Ripoll M, Håvarstein LS, Hermoso JA. 2014. Structural basis of PcsB-mediated cell separation in Streptococcus pneumoniae. Nat Commun 5:3842. doi: 10.1038/ncomms4842. [DOI] [PubMed] [Google Scholar]
- 5.Sham L-T, Barendt SM, Kopecky KE, Winkler ME. 2011. Essential PcsB-putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proc Natl Acad Sci U S A 108:E1061–E1069. doi: 10.1073/pnas.1108323108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisicchia P, Noone D, Lioliou E, Howell A, Quigley S, Jensen T, Jarmer H, Devine KM. 2007. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol Microbiol 65:180–200. doi: 10.1111/j.1365-2958.2007.05782.x. [DOI] [PubMed] [Google Scholar]
- 7.Singh S, SaiSree, Amrutha R, Reddy M. 2012. Three redundant murein endopeptidases catalyze an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol Microbiol. 86:1036–1051. doi: 10.1111/mmi.12058. [DOI] [PubMed] [Google Scholar]
- 8.Vollmer W. 2012. Bacterial growth does require peptidoglycan hydrolases. Mol Microbiol 86:1031–1035. doi: 10.1111/mmi.12059. [DOI] [PubMed] [Google Scholar]
- 9.Pinho MG, Kjos M, Veening J-W. 2013. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol 11:601–614. doi: 10.1038/nrmicro3088. [DOI] [PubMed] [Google Scholar]
- 10.Uehara T, Bernhardt TG. 2011. More than just lysins: peptidoglycan hydrolases tailor the cell wall. Curr Opin Microbiol 14:698–703. doi: 10.1016/j.mib.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner R, Ratcliffe E, Wheeler R, Golestanian R, Hobbs J, Foster S. 2010. Peptidoglycan architecture can specify division planes in Staphylococcus aureus. Nat Commun 1:1–9. [DOI] [PubMed] [Google Scholar]
- 12.Pinho MG, Errington J. 2005. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol Microbiol 55:799–807. doi: 10.1111/j.1365-2958.2004.04420.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhou X, Halladin DK, Rojas ER, Koslover EF, Lee TK, Huang KC, Theriot JA. 2015. Mechanical crack propagation drives millisecond daughter cell separation in Staphylococcus aureus. Science 348:574–578. doi: 10.1126/science.aaa1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tzagoloff H, Novick R. 1977. Geometry of cell division in Staphylococcus aureus. J Bacteriol 129:343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Touhami A, Jericho MH, Beveridge TJ. 2004. Atomic force microscopy of cell growth and division in Staphylococcus aureus. J Bacteriol 186:3286–3295. doi: 10.1128/JB.186.11.3286-3295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey RG, Turner RD, Mullin N, Clarke N, Foster SJ, Hobbs JK. 2014. The interplay between cell wall mechanical properties and the cell cycle in Staphylococcus aureus. Biophys J 107:2538–2545. doi: 10.1016/j.bpj.2014.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frankel MB, Hendrickx AP, Missiakas DM, Schneewind O. 2011. LytN, a murein hydrolase in the cross-wall compartment of Staphylococcus aureus, is involved in proper bacterial growth and envelope assembly. J Biol Chem 286:32593–32605. doi: 10.1074/jbc.M111.258863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frankel MB, Schneewind O. 2012. Determinants of murein hydrolase targeting to cross-wall of Staphylococcus aureus peptidoglycan. J Biol Chem 287:10460–10471. doi: 10.1074/jbc.M111.336404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stapleton MR, Horsburgh MJ, Hayhurst EJ, Wright L, Jonsson I-M, Tarkowski A, Kokai-Kun JF, Mond JJ, Foster SJ. 2007. Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J Bacteriol 189:7316–7325. doi: 10.1128/JB.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boneca IG, Huang Z-H, Gage DA, Tomasz A. 2000. Characterization of Staphylococcus aureus cell wall glycan strands, evidence for a new β-N-acetylglucosaminidase activity. J Biol Chem 275:9910–9918. doi: 10.1074/jbc.275.14.9910. [DOI] [PubMed] [Google Scholar]
- 21.Wheeler R, Mesnage S, Boneca IG, Hobbs JK, Foster SJ. 2011. Super-resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Mol Microbiol 82:1096–1109. doi: 10.1111/j.1365-2958.2011.07871.x. [DOI] [PubMed] [Google Scholar]
- 22.Oshida T, Sugai M, Komatsuzawa H, Hong YM, Suginaka H, Tomasz A. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-β-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc Natl Acad Sci U S A 92:285–289. doi: 10.1073/pnas.92.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foster SJ. 1995. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J Bacteriol 177:5723–5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kailas L, Ratcliffe EC, Hayhurst EJ, Walker MG, Foster SJ, Hobbs JK. 2009. Immobilizing live bacteria for AFM imaging of cellular processes. Ultramicroscopy 109:775–780. doi: 10.1016/j.ultramic.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 25.Turner RD, Thomson NH, Kirkham J, Devine D. 2010. Improvement of the pore trapping method to immobilize vital coccoid bacteria for high-resolution AFM: a study of Staphylococcus aureus. J Microsc 238:102–110. doi: 10.1111/j.1365-2818.2009.03333.x. [DOI] [PubMed] [Google Scholar]
- 26.Daniel RA, Errington J. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767–776. doi: 10.1016/S0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 27.Hayhurst EJ, Kailas L, Hobbs JK, Foster SJ. 2008. Cell wall peptidoglycan architecture in Bacillus subtilis. Proc Natl Acad Sci U S A 105:14603–14608. doi: 10.1073/pnas.0804138105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed P, Veiga H, Jorge AM, Terrak M, Pinho MG. 2011. Monofunctional transglycosylases are not essential for Staphylococcus aureus cell wall synthesis. J Bacteriol 193:2549–2556. doi: 10.1128/JB.01474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loskill P, Pereira PM, Jung P, Bischoff M, Herrmann M, Pinho MG, Jacobs K. 2014. Reduction of the peptidoglycan crosslinking causes a decrease in stiffness of the Staphylococcus aureus cell envelope. Biophys J 107:1082–1089. doi: 10.1016/j.bpj.2014.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner R, Hurd A, Cadby A, Hobbs J, Foster S. 2013. Cell wall elongation mode in gram-negative bacteria is determined by peptidoglycan architecture. Nat Commun 4:1496. doi: 10.1038/ncomms2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, Breukink E, den Blaauwen T, Gross CA, Vollmer W. 2010. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beeby M, Gumbart JC, Roux B, Jensen GJ. 2013. Architecture and assembly of the Gram-positive cell wall. Mol Microbiol 88:664–672. doi: 10.1111/mmi.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 35.Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett 73:133–138. [DOI] [PubMed] [Google Scholar]
- 36.Clarke SR, Brummell KJ, Horsburgh MJ, McDowell PW, Mohamad SA, Stapleton MR, Acevedo J, Read RC, Day NP, Peacock SJ, Mond JJ, Kokai-Kun JF, Foster SJ. 2006. Identification of in vivo-expressed antigens of Staphylococcus aureus and their use in vaccinations for protection against nasal carriage. J Infect Dis 193:1098–1108. doi: 10.1086/501471. [DOI] [PubMed] [Google Scholar]
- 37.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol 184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pasztor L, Ziebandt A-K, Nega M, Schlag M, Haase S, Franz-Wachtel M, Madlung J, Nordheim A, Heinrichs DE, Götz F. 2010. Staphylococcal major autolysin (Atl) is involved in excretion of cytoplasmic proteins. J Biol Chem 285:36794–36803. doi: 10.1074/jbc.M110.167312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohamad SAS. 2007. Ph.D. thesis University of Sheffield, Sheffield, South Yorkshire, United Kingdom. [Google Scholar]
- 40.Kreiswirth BN, Löfdahl S, Betley MJ, O’Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 41.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 12:3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kemp EH, Sammons RL, Moir A, Sun D, Setlow P. 1991. Analysis of transcriptional control of the gerD spore germination gene of Bacillus subtilis 168. J Bacteriol 173:4646–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guérout-Fleury AM, Shazand K, Frandsen N, Stragier P. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 44.Vagner V, Dervyn E, Ehrlich SD. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 45.Bottomley AL, Kabli AF, Hurd AF, Turner RD, Garcia-Lara J, Foster SJ. 2014. Staphylococcus aureus DivIB is a peptidoglycan-binding protein that is required for a morphological checkpoint in cell division. Mol Microbiol 94:1041–1064. doi: 10.1111/mmi.12813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of four putative N-acetylglucosaminidases encoded by the genome of S. aureus COL. Download
Construction of SH4615 (Pspac-sagB atl sagA scaH). Download
Growth of glucosaminidase mutants in liquid medium. Download
Role of SagB in S. aureus morphology. Download
Impact of single glucosaminidase-encoding gene inactivation on glycan strand length in S. aureus as determined by gel filtration chromatography. Download
Impact of glucosaminidase inactivation on glycan strand length in S. aureus. Download
Enzymatic activities of Atl and SagB. Download
Analysis of cell clumps in glucosaminidase mutants. Download
Comparison of AFM stiffness measurements between strains.
Genetic modification of bacteria. Download




