SUMMARY
Malaria caused by Plasmodium falciparum is a major cause of global infant mortality, and there is currently no licensed vaccine that provides protection against infection or disease. Several P. falciparum vaccine targets have undergone early testing, but many more candidates remain with little data to support their development. Plasmodium falciparum Merozoite Surface Protein 6 (PfMSP6) is a candidate of particular interest because it is a member of the PfMSP3 multi-gene family, raising the possibility that vaccine-induced immune responses could cross-react across multiple family members. However, few immunoepidemiological studies of PfMSP6 have been carried out to measure domain-specific anti-PfMSP6 responses. This study investigated anti-PfMSP6 responses in P. falciparum infected individuals from the Peruvian Amazon, using two different PfMSP6 N-terminal allele antigens and a single C-terminal domain antigen, and compared the responses with both PfMSP6 genotyping data and anti-PfMSP3 response data that had been previously generated for the same samples. Anti-PfMSP6 responses were detected despite the low transmission setting, but were less frequent and of considerably lower intensity than anti-PfMSP3 responses. There was a positive correlation between anti-PfMSP3 and PfMSP6 responses, suggesting that the possibility that PfMSP3 family antigens could induce cross-reactive responses requires further detailed investigation.
Keywords: malaria, vaccine, Plasmodium falciparum, merozoite, PfMSP6, immunity, cross-reactivity
INTRODUCTION
With the growth in genomic sequence information for Plasmodium parasites that infect humans (1–3) there has been a concomitant increase in the number of potential Plasmodium vaccine candidates. For many of these antigens there is little data to support their candidacy other than theoretical considerations based on their predicted function or expression profile. Given the complexity of the Plasmodium life-cycle, even careful and exacting analysis can generate a significant number of reasonable candidates (4), with an urgent need for data to support or eliminate them from further development before large amounts of money are invested in them. Measuring immune responses to candidate antigens in naturally infected individuals is one commonly used and powerful tool to compare and contrast potential vaccine candidates. However, to be most useful, such studies need to address issues of genetic diversity whenever possible. Diversity is one of the most significant challenges facing P. falciparum vaccine development (5), particularly for blood stage antigens, which are exposed to the adaptive immune system and therefore can be under strong selection pressure (6). Comparing responses to more than one allele variant is becoming an increasingly critical part of immunoepidemiological studies if they are to assess the allele-specificity of immune responses and provide clearer guidance for vaccine development.
P. falciparum Merozoite Surface Protein 6 (PfMSP6) is a potential blood stage vaccine antigen that has been examined in relatively few studies. PfMSP6 is expressed on the surface of the merozoite, the sole extra-erythrocytic stage of the P. falciparum blood cycle, and forms a complex with the major surface GPI anchored protein, PfMSP1 (7, 8). PfMSP6 consists of two major domains, an N-terminal domain that is predicted to form coiled-coils and a glutamic-acid rich C-terminal domain, with the two domains separated by a PfSUB1 proteolytic cleavage site (9). PfMSP6 is a dimorphic antigen, with the two allele classes, 3D7-like and K1-like, being named for the strain in which they were first identified (10). Differences between the alleles are largely restricted to a series of indels in the N-terminal domain, but also include single nucleotide polymorphisms (SNPs) in both the N-terminal and C-terminal domains. Studies of PfMSP6 diversity have shown that many variants within each allele class exist at a global level (11), and PfMSP6 allele frequencies can vary significantly over time even under low transmission conditions (12).
PfMSP6 is encoded by one of a family of related genes arrayed along P. falciparum chromosome 10, raising the possibility that vaccines against one family member could raise responses that cross-react with others (13). Within the PfMSP3 gene family PfMSP6 is most closely related to PfMSP3, and analysis of PfMSP3 as a vaccine candidate has been extensive (14–18), including several human trials (19–22). By contrast study of PfMSP6 in this context has been much more limited. It is the C-terminal domain that is of most interest for vaccine development as it is the most conserved fragment across the PfMSP3 gene family, and is therefore a focus of development for antigens capable of inducing responses that cross-react across multiple family members (13, 23, 24). One previous PfMSP6-based study using sera from 30 patients from Cote D’Ivoire focused primarily on the C-terminal domain (25), while a second using serum samples from Vietnam compared responses to the N- and C-terminal domains (26). However, no immunoepidemiological study of PfMSP6 to date has compared responses between different PfMSP6 allele variants, and studies of cross-reactivity between PfMSP3 family members have been limited to relatively small numbers of serum samples.
In this study the prevalence, strength and isotype specificity of anti-PfMSP6 responses were measured in 342 samples from the ongoing MIGIA (Malaria Immunology and Genetics in the Amazon) cohort study near Iquitos, Peru (27). P. falciparum transmission in this study site is low, with an infection rate of less than one infection per person per year. Importantly, these P. falciparum infections are relatively genetically simple with few cases of multi-allele infections, unlike high transmission environments where infections are more genetically complex and overlapping. The serum samples used for this study were from individuals whose P. falciparum infections had previously been genotyped for their infecting PfMSP6 allele (12), and using different PfMSP6 allele antigens, which allowed for direct comparison of responses to both the infecting and non-infecting PfMSP6 allele. The same samples had also been used to measure anti-PfMSP3 responses in a previous study (28), which allowed for direct comparison of anti-PfMSP6 and anti-PfMSP3 immune responses These comparisons revealed both notable similarities and differences between anti-PfMSP3 and anti-PfMSP6 responses, and suggest priorities for future immunoepidemiological studies.
MATERIALS AND METHODS
Study Site, Subjects and Sample Collection
A detailed description of the MIGIA cohort study has been previously described (27). Briefly, the study involves the residents of the Zungarococha community, a cluster of 4 villages located south of Iquitos in the Peruvian Amazon. The village residents have homogenous housing construction, income levels, and access to healthcare, provided by the MIGIA cohort physicians at a community health post. Travel outside the community is rare, with the most frequent travel being to the city of Iquitos, where malaria transmission is nonexistent. The Zungarococha community was chosen as the focus of the MIGIA cohort because of the presence of continuing stable hypo-endemic transmission of both P. vivax and P. falciparum, with frequency of infection rates for P. falciparum being < 0.5 infections/person/year.
Cases of P. falciparum were detected by both active and passive means. Passive case detection involved symptomatic individuals presenting at a local health post located in Zungarococha village, where they were tested for malaria parasites by Geimsa-stained microscopy and underwent a comprehensive medical evaluation. PCR verification of the microscopy result was subsequently performed using species-specific primers. Active case detection involved random sampling of villagers over the course of the malaria transmission season. All confirmed cases of Plasmodium infection were treated with appropriate anti-malarial medication. Blood samples from confirmed P. falciparum infections were separated into a serum and packed RBC fraction by centrifugation and each was cataloged and stored at −80°C until needed.
Antigen Construction and Purification
Domain-specific PfMSP6 recombinant proteins were amplified from HB3 (HB3 N-terminal domain, C-terminal domain) or K1 (K1 N-terminal domain) genomic DNA. The forward and reverse primers used were 5’-TAGCGGATCCAATAACTTTATCAGAAATGAACTT-3’ and 5’-GCATAAGCTTTTAGTTTGCTTGTACAACTTG-3’, 5’-TAGCGGATCCAATAACTTTATCAGAAATGAACTT-3’ and 5’-TAGCAAGCTTTTAGTTTTCTTCTGCACCGTGTGT-3’, and 5’-TAGCGGATCCTCTGAAACAAATAAAAATCC-3’ and 5’-TAGCAAGCTTTTAATTATTACTAAATAGATG-3’ for the HB3 and K1 N-terminal domains, and HB3 C-terminal domain, respectively. Antigens were expressed as hexa-histidine tagged fusion proteins from pET15b (Novagen) or pRSETA (Invitrogen) vectors in the E. coli BL21(DE3)pLysS “Rosetta” strain (Novagen), and purified by high-temperature incubation (65°C for 25 minutes), followed by affinity and anion exchange chromatography, as previously described (29).
ELISA Assays
50ng of purified PfMSP6 antigen was coated onto each well and a 1:100 dilution of patient sera was used for each assay. Bound antibodies were detected by the addition of HRP-conjugated anti-IgG (Chemicon) at a dilution of 1:5,000, or HRP-conjugated IgG-isotype-specific (Southern Biotech) and IgM-specific (Fisher Scientific) secondary antibodies at a dilution of 1:1000. ChromoPure human IgG (Jackson ImmunoResearch Laboratories) was used to standardize antibody responses. All plates were read at 450nm using a Uniread 800 ELISA plate reader (GeneMate, Kaysville, UT). Serially-diluted positive pools were run concurrently with each set of ELISAs to ensure all OD450 measurements remained within the linear range and to facilitate ELISA normalization.
Statistical Analysis
Comparisons between proportions of ELISA antigens and allele infections were performed using the two-group chi-square test or Fisher’s exact test when the assumptions on the chi-square were not tenable. Comparisons between median IgG levels for ELISA antigens, separately for allele infections (Table 1) was performed using the Kruskal-Wallis test since IgG levels and antibody responses were determined to not be normally distributed. When a statistically significant overall result was obtained, the Dunn multiple comparisons procedure was used to determine which specific pairs of means were significantly different. Spearman correlation analyses were performed to examine the relationships between IgG levels of the ELISA antigens, separately for allele infections (Fig. 3, Fig. 4). All statistical tests were two-sided and were performed using a 5% significance level (i.e. alpha = 0.05). JMP software (version 9.0.1; SAS Institute, Inc., Cary, NC) was used to perform all statistical analyses.
Table 1. Naturally induced anti-PfMSP6 antibodies predominantly target the N-terminal domain.
Responses against PfMSP6 antigens in HB3-infected (n=309, Table 1A) and K1-infected (n=33, Table 1B) individuals. IgG antibody level [µg/ml] were calculated as described in Materials and Methods, and responses scored as positive (% Positive) if they exceeded the mean plus three standard deviations of the antigen-specific responses in a pool of control serum from non-P. falciparum infected individuals. Minimum (Min), Lower Quartile (LQ), Median, Upper Quartile (UQ) and Maximum (Max) values were generated for all responders.
| A | Total IgG Antibody Levels (ug/ml) |
||||||
|---|---|---|---|---|---|---|---|
| Antigen | Cut-off (ug/ml) |
% Positive | Min | LQ | Median | UQ | Max |
| HB3 N-Term | 0.492 | 40.1 | 0.191 | 0.271 | 0.400 | 0.718 | 319.786 |
| K1 N-Term | 0.370 | 37.2 | 0.188 | 0.237 | 0.319 | 0.459 | 48.307 |
| C-Term | 0.550 | 23.0 | 0.203 | 0.249 | 0.285 | 0.468 | 15.403 |
| B | Total IgG Antibody Levels (ug/ml) |
||||||
|---|---|---|---|---|---|---|---|
| Antigen | Cut-off (ug/ml) |
% Positive | Min | LQ | Median | UQ | Max |
| HB3 N-Term | 0.492 | 48.5 | 0.199 | 0.307 | 0.442 | 1.440 | 60.317 |
| K1 N-Term | 0.370 | 45.5 | 0.206 | 0.241 | 0.341 | 0.627 | 10.659 |
| C-Term | 0.550 | 24.2 | 0.203 | 0.233 | 0.281 | 0.400 | 5.001 |
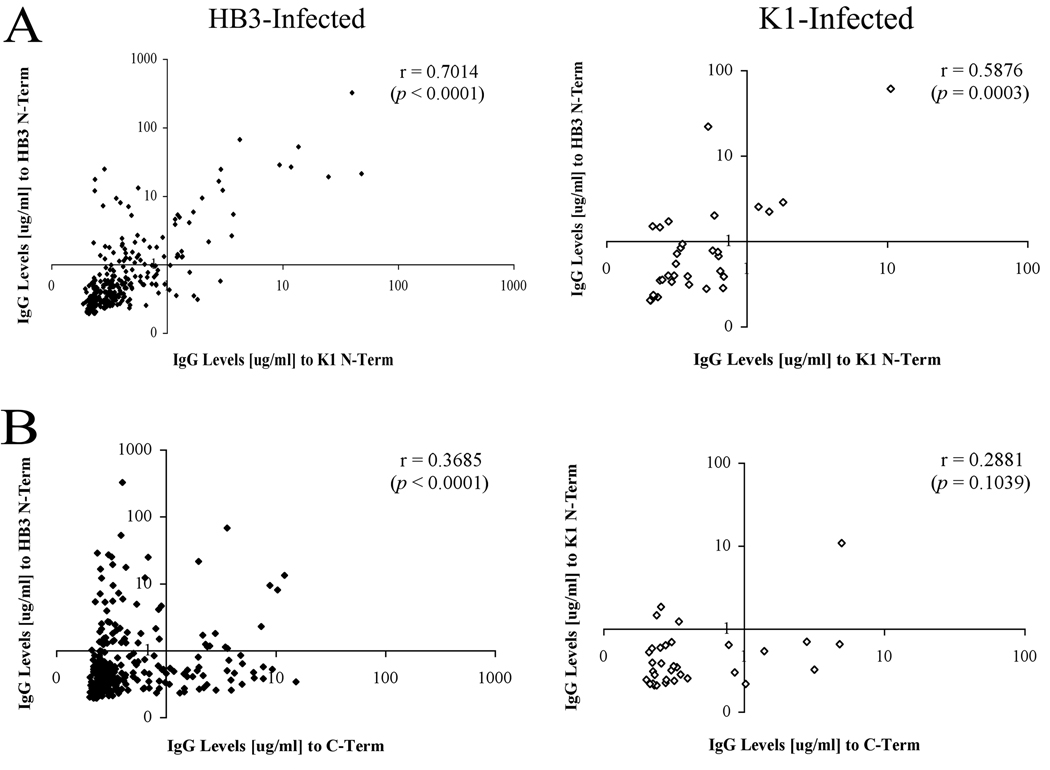
Figure 3. Comparison of PfMSP6 N-terminal domain responses across alleles.
Pair-wise correlation between responses to different PfMSP6 subdomains, with individuals separated on the basis of their infecting PfMSP6 allele. (A) Correlation of responses to HB3 and K1 N-terminal domains (upper panel) and HB3 N-terminal and C-terminal domains (lower panel) in HB3-infected individuals. (B) Correlation of responses to HB3 and K1 N-terminal domains (upper panel) and K1 N-terminal and C-terminal domains (lower panel) in K1-infected individuals.
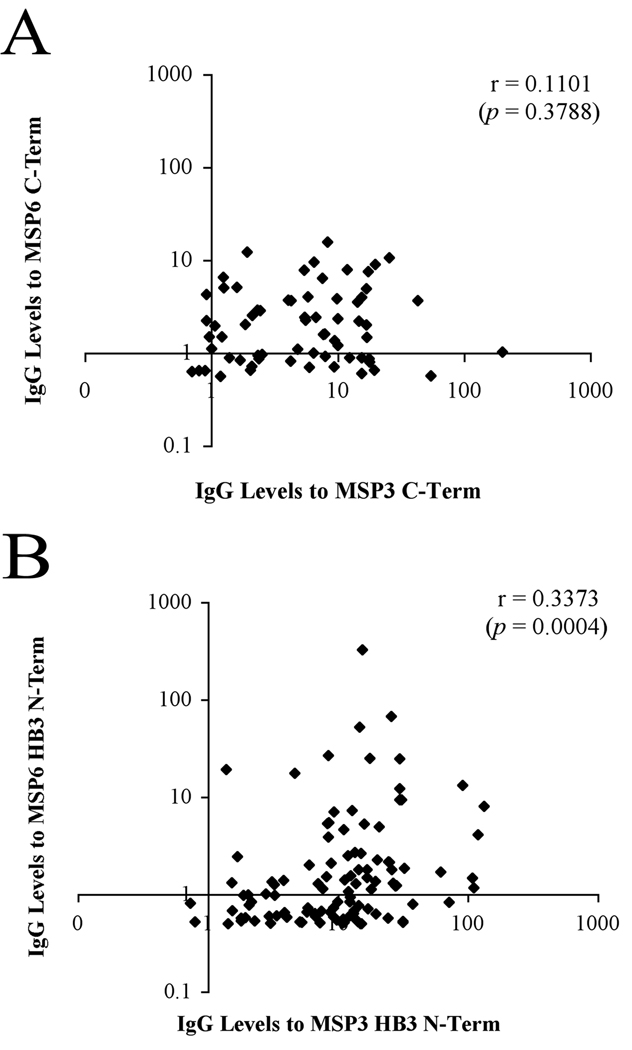
Figure 4. Comparison of anti-PfMSP6 and anti-PfMSP3 responses.
Pair-wise correlation between PfMSP6 and PfSMP3 responses. (A) Correlation of responses to PfMSP6 and PfMSP3 HB3 N-terminal domains. (B) Correlation of responses to PfMSP6 and PfMSP3 C-terminal domains.
Ethics committee approval
This study was approved by review boards of the University of Alabama at Birmingham, New York University, Universidad Peruana Cayetano Heredia, and the Peruvian Ministerio de Salud, Instituto Naccional de Salud. Written consent was obtained from all participants prior to study enrollment.
RESULTS
Production of recombinant PfMSP6 antigens
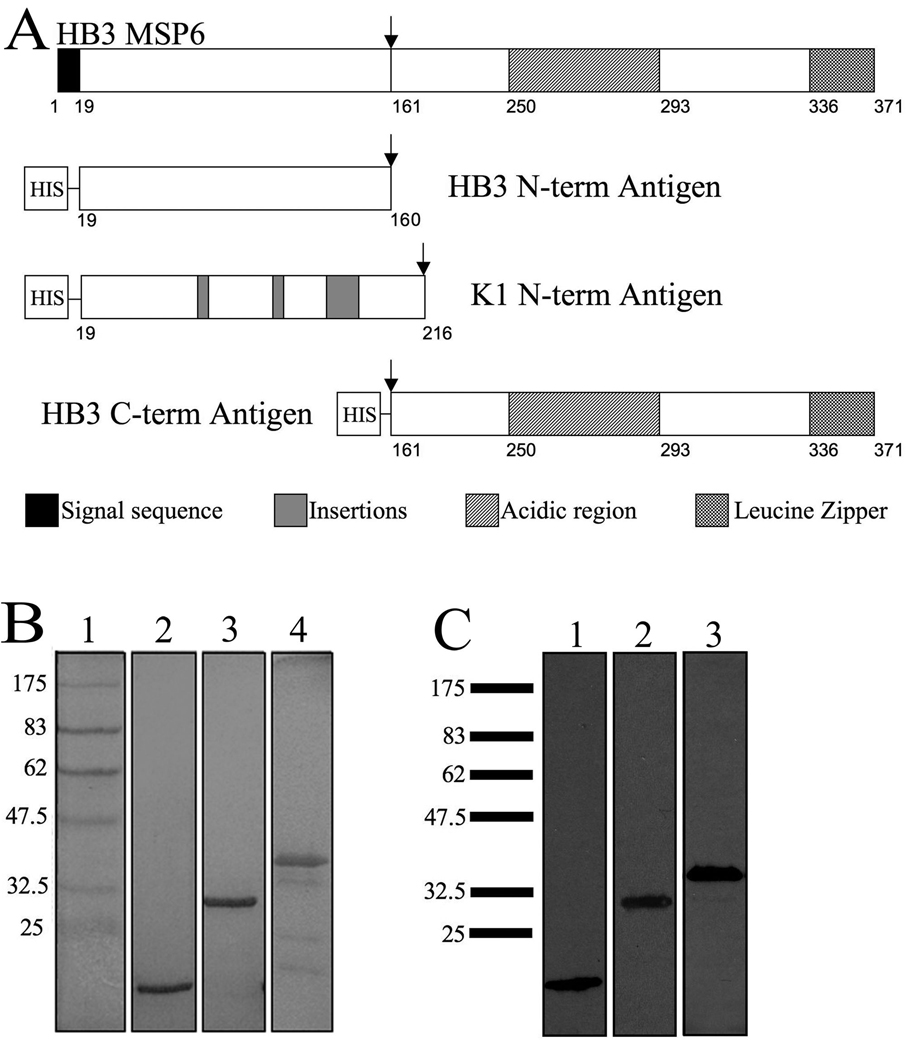
A previous genotyping study established that both PfMSP6 allele classes, 3D7-like and K1-like, were in circulation at the MIGIA study site, with 3D7-like alleles comprising 91.5% of 506 infections collected between 2003 and 2006 (12). The sequences of all circulating PfMSP6 alleles during 2003–2006 transmission seasons, as determined by direct sequencing, were identical to the previously published HB3 and K1 sequences, respectively (12). Based on this data, three recombinant antigens were generated to represent the PfMSP6 domains that P. falciparum infected individuals at the study site were exposed to: two N-terminal domain antigens, HB3 and K1, and one C-terminal domain (also referred to as MSP6–36 (7)) with the PfSUB1 cleavage site forming the in vivo boundary between the two domains (Fig. 1A). The HB3 C-terminal domain sequence was used, but there is only a single amino acid difference in the C-terminal domain between the HB3 and K1 sequences. Antigens were expressed in the E. coli BL21(DE3)pLysS “Rosetta” strain (Novagen), and purified using affinity and anion exchange chromatography, as previously described (29). Purification of these antigens yielded milligram quantities of >95% pure protein (Fig. 1B, 1C). While the N-terminal domain antigens appeared as a single protein band, the C-terminal band included several smaller bands (Fig. 1B), which were not recognized by an anti-his monoclonal antibody (Fig. 1C). Mass spectroscopy confirmed that all bands were fragments of PfMSP6, suggesting they were degradation products that lacked the N-terminal his tag (data not shown). Circular dichroism confirmed that the two N-terminal antigens had a high proportion of coiled-coil domains, just like PfMSP3 N-terminal antigens (29), suggesting that they were correctly folded. The presence of cleavage products in the C-terminal prep complicated such analysis, but the largest C-terminal antigen band runs at double its predicted molecular weight even under reducing conditions (Fig. 1C), arguing that it forms a homodimer, just like the PfMSP3 C-terminal domain, and again suggesting correct folding of the major C-terminal fragment.
Figure 1. Recombinant PfMSP6 antigens are highly pure.
(A) Schematic representation of HB3 PfMSP6. Black boxes indicate the signal sequence. Striped and checkered boxes indicate the acidic and leucine zipper regions, respectively. Insertions in the K1 sequence are indicated by the shaded boxes. The proteolytic cleavage site is indicated by the arrow. All constructs contained a hexahistidine tag at the N-terminal location to aid in purification. (B) Coomassie-blue stained SDS-PAGE gel showing the purified PfMSP6 antigens. Lane 1, marker; lane 2, HB3 N-term, lane 3, K1 N-term; lane 4, HB3 C-term. (C) Western blot of PfMSP6 antigens probed with mouse anti-His antibody. Lane 1, HB3 N-term; lane 2, K1 N-term; lane 3, C-term. The molecular mass markers (kDa) are listed to the left of each panel.
Anti-PfMSP6 immune responses primarily recognize the N-terminal domain
The three antigens were used to detect anti-PfMSP6 responses using time-of-infection sera samples from 342 P. falciparum infections collected between 2003 and 2006, all of which had previously been genotyped for the infecting PfMSP6 allele (12). The majority of samples came from different individuals; samples collected less than 60 days apart from a single individual were excluded to eliminate any recrudescent infections. PfMSP6 3D7-like alleles predominate at this study site, so samples were selected to reflect the observed allele frequencies; 309 from HB3 infections and 33 from K1 infections (which, given that K1 alleles are present in less than 10% of infections at the study site, included all K1 infections over a four year period that had sufficient sample volume for testing). Sera samples were tested against all three antigens, and ELISA results were normalized and converted into absolute concentration of anti-PfMSP6 IgG (see Materials and Methods). Individuals were scored as positive if their responses were greater than the calculated negative cutoff, defined as three standard deviations above the mean response of a negative control sera pool, which had been collected from Peruvian individuals not previously exposed to P. falciparum. Negative cutoff values against HB3 N-term, K1 N-term, and C-term antigens were 0.492, 0.370, and 0.550, respectively.
Table 1 summarizes the naturally induced anti-PfMSP6 IgG responses at the MIGIA study site. Fewer than 50% of the samples contained positive responses against any PfMSP6 antigen. There were more positive responses against the N-terminal domain antigens than the C-terminal domain antigen. In HB3 infected individuals (Table 1A), this difference was highly statistically significant (p < 0.001, N=309) and K1-infected individuals (Table 1B) the increase trended toward significance (p = 0.071, N=33), but the power is clearly limited by the small number of K1-infected samples available. Within each infecting allele class there was no significant difference in the number of positive responses to either the infecting or non-infecting N-terminal allele (p=0.457 and p=0.805 for HB3- and K1-infected individuals, respectively), as might have been expected if there was allele-specific immunity. Responses against the non-infecting N-terminal allele were significantly higher than against the C-terminal domain (p < 0.001 for HB3-infected, p = 0.041 for K1-infected). The distribution of normalized IgG antibody levels [µg/ml] within each infecting allele class was analyzed to characterize the strength of anti-PfMSP6 responses. There was no statistically significant difference in the median responses detected when comparing the antigens.
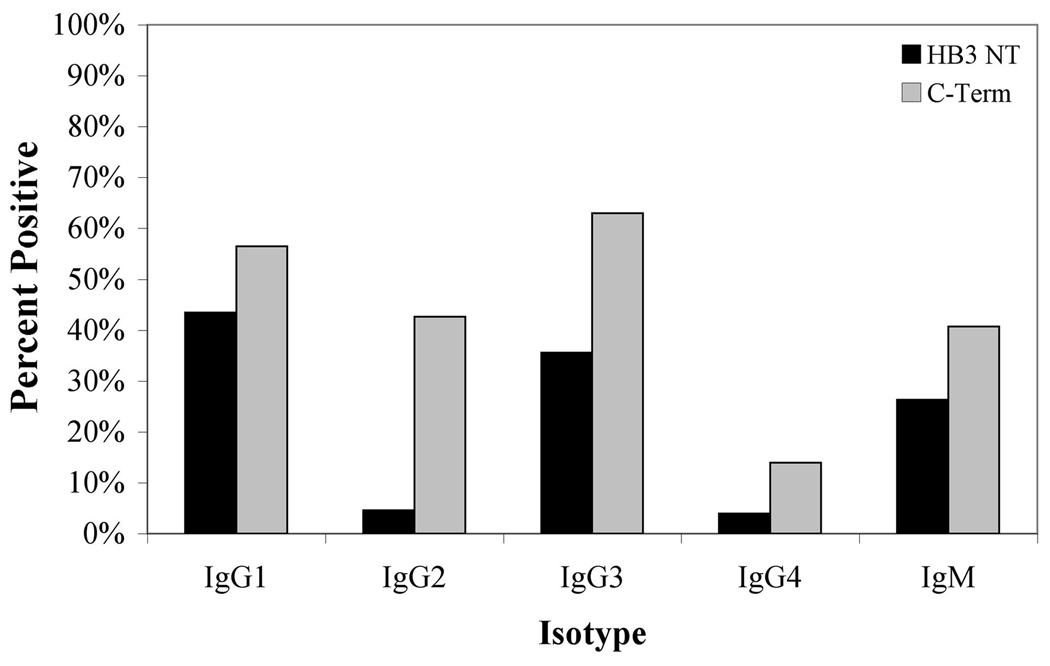
Anti-PfMSP6 antibodies are primarily IgG1, IgG3 and IgM subclasses
Because cytophilic IgG subclasses, IgG1 and IgG3, have been previously associated with protection against malaria (30–32), each antiserum sample that scored as positive by total IgG ELISA was tested for isotype class ELISA positivity using isotype-specific secondary antibodies. Because of sample volumes, isotype responses were established only for the antigens present in the infection, namely the infecting HB3 N-terminal domain and the more conserved C-terminal domain-individuals infected with K1 antigens were excluded due to inadequate sample size. Individuals were scored as positive for a given isotype if their responses were greater than the calculated negative cutoff for each isotype-specific secondary antibody, defined as three standard deviations above the mean response of a negative control sera pool. IgG1, IgG3 and IgM subclasses were the predominant subclasses detected against both PfMSP6 antigens (p < 0.0001), as was the case for responses against PfMSP3 in the same samples (28). 40% of samples (N=46) also had positive IgG2 responses against the C-terminus, significantly higher than the percent of IgG2 positive responses against the N-terminus (p<0.0001) (Fig. 2).
Figure 2. Isotype-specific responses against PfMSP6 antigens.
PfMSP6 percent positive isotype antibody responses (OD450 at 1:100 sera dilution) for IgG1–4 and IgM responses against HB3 N-terminal domain (n=152) and C-terminal domain (n=108) antigens. All HB3-infected samples that had positive total IgG responses were included. Results were considered positive if they exceeded the mean plus three standard deviations of the antigen-specific responses in a pool of control serum from non-P. falciparum infected individuals.
Responses to the PfMSP6 N-terminus correlate between allele classes
The overall prevalence of positive responses to the HB3 and K1 N-terminal domains was not significantly different in either HB3 or K1 infected individuals, suggesting that responses against the N-terminal domain may cross-react between allele classes. However, this population level analysis is an imperfect measure of cross-reactivity, as small populations of responses that are highly skewed to either antigen would go undetected. To more precisely test for evidence for cross-reactive responses, the strength of anti-N-terminal domain responses against the infecting and non-infecting allele class was compared for each individual infection (Fig 3). There was a clear positive correlation between the strength of response to both the infecting and non-infecting N-terminal domains in HB3-infected individuals (r = 0.7014, p < 0.0001, Fig 3A left panel) and K1-infected individuals (r = 0.5876, p = 0.0003, Fig 3A right panel). In support of this, of individuals who had a positive response against the HB3 N-terminal domain, 71% of those individuals had a positive response against the K1 N-terminal domain, while only 32% had a positive response to the C-terminal domain (data not shown). By contrast, there was no significant correlation comparing responses against the K1 N-terminal and C-terminal domains in K1-infected individuals (p = 0.1039, Fig 3B right panel). There was a significant correlation comparing responses against the HB3 N-terminal and C-terminal domains in HB3-infected individuals, but it was less strong than the association between N-terminal domain responses (r = 0.3685, p < 0.0001, Fig 3B left panel).
Comparison of individual anti-PfMSP6 and PfMSP3 responses shows correlation between responses to PfMSP3 family members
A previous study using the same serum samples from the MIGIA cohort generated domain-specific antibody response data for PfMSP3, the closest homologue of PfMSP6 (28). Comparing anti-PfMSP3 and anti-PfMSP6 responses in the same individuals could be one indicator of antibody responses that cross-react against these two PfMSP3 family members, especially when comparing anti-C-terminal domain responses, which are more conserved between these two proteins (13), although this may also simply indicate the presence of two independent populations of antibodies in some samples. When responses against PfMSP6 and PfMSP3 antigens were compared, there was a statistically significant weak positive correlation between responses to the PfMSP6 and PfMSP3 HB3 N-terminal domains (r=0.3373, p=0.0004, Fig. 4A). Interestingly, no significant correlation was detected comparing PfMSP6 and PfMSP3 C-terminal domain antibody responses (r = 0.1101, p=0.3788, Fig. 4B).
DISCUSSION
PfMSP6 belongs to a cadre of potential P. falciparum blood stage vaccine antigens that have received little attention, but the disappointing results of recent trials with more high-priority targets (33, 34) argues that a wider net needs to be cast for potential candidates. This study is only the second large scale immunoepidemiological study investigating responses to PfMSP6, and comparing the data against previous PfMSP6 studies, as well as a study of anti-PfMSP3 responses carried out at the same study site, is instructive.
This study established that antibodies are raised against both PfMSP6 domains even in the relatively low exposure rates found at the MIGIA study site, but that fewer than 50% of the samples were positive for antibody responses against any PfMSP6 antigen. This is a broadly similar positivity rate to the only previous large immunoepidemiological study of anti-PfMSP6 responses, carried out on 174 samples from Vietnam, where a maximum of 50.6% of samples were positive for anti-PfMSP6 antibodies (26). However, the two studies differ in the proportion of positive responses to the N- and C-terminal domains, with more N-terminal positive responses in this Peruvian study, and more C-terminal positive responses in the Vietnam study (26). While strong caution always needs to be applied when comparing antibody responses between studies using different recombinant antigens, the antigens used in the Vietnam studies had the same domain boundaries for antigen construction as were used here, making the two studies more broadly comparable. It is therefore possible that anti-PfMSP6 responses in Peru may be more weighted to the N-terminal domain than those in Vietnam. It should also be noted that samples in this study were all taken from active P. falciparum infections, whereas the samples studied in Vietnam were taken from a cross-sectional survey. Anti-N-terminal domain responses may therefore be associated with active immune responses, but may be more short-lived than anti-C-terminal domains, and therefore less prevalent in cross-sectional studies.
One significant advantage of studying immune responses in the context of a longitudinal cohort study in a low transmission region, such as the MIGIA study, is that the low frequency and genetic simplicity of infections allows for direct comparison of the infecting genotype with the immune response, a comparison that is not possible in the complex infections present in high transmission environments. Comparing anti-PfMSP6 responses with the previously established PfMSP6 genotypes for each infection provided no evidence for allele-specific responses to the PfMSP6 N-terminal domains (12). At a population level, there was no significant difference in the number of positive responses to either the infecting or non-infecting N-terminal domain, and 71% of individuals who had a positive response against the HB3 N-terminus also had a positive response against K1 N-terminal domain. At an individual level, there was a clear positive correlation between the strength of response to both the infecting and non-infecting N-terminal domains in both HB3-infected individuals and K1-infected individuals. In theory, the presence of responses against both infecting and non-infecting N-terminal domain antigens could be explained by recent infections that were undetected in the cohort study, although the frequency of active sampling being carried out makes this extremely unlikely. Taking into account the predominance of 3D7-class infections (12) at the study site and the low frequency of infections overall, there is only a 0.84% probability that any given HB3-infected individual had an undetected asymptomatic K1 infection within the past 2 years. Undetected infections therefore clearly can not explain the data, and the most parsimonious explanation is that there is no evidence for allele-specificity in anti-PfMSP6 responses at this study site.
To establish the presence or absence of allele-specific anti-PfMSP6 responses unequivocally would require competition ELISA experiments, which were not possible in this study because of limiting serum volume. However it should be noted that the anti-PfMSP6 responses reported here are very different to anti-PfMSP3 responses reported previously for the same samples, where the evidence for allele-specific responses was very strong. Allele specificity in anti-PfMSP3 N-terminal responses is well-established (16, 17), just as it is for other merozoite surface antigens such as MSP2 (35). When responses to PfMSP3 antigens were tested in this same set of Peruvian samples, there was a very clear correlation between infecting genotype and immune responses to the N-terminus, with a significant increase in mean IgG responses to the infecting PfMSP3 allele compared to the non-infecting PfMSP3 allele (28). No such correlation was observed for anti-PfMSP6 responses in this study, suggesting that allele-specific responses may be less common against PfMSP6 than they are against PfMSP3. Competition ELISA experiments using PfMSP6 antigens to test this hypothesis should be performed as a matter of urgency.
Anti-PfMSP6 and anti-PfMSP3 responses in these Peruvian samples also differed in several other critical respects, being more frequent (90% of HB3-infected individuals has positive anti-PfMSP3 N-terminal responses, for example), and with a higher mean IgG level. There was a positive correlation between responses to the PfMSP3 and PfMSP6 HB3 N-terminal domains within the same individuals, but this is not necessarily any indication of cross-reactivity, and could simply indicate the presence of two independent populations of antibodies in some samples. No correlation was seen for anti-C-terminal PfMSP6 and PfMSP3 antibody responses, despite their homology, but cross-reactive responses have not been established for these domains even using polyclonal antibodies (13).
What then are the implications for vaccine development? Again, it is important to point out that extreme caution needs to be employed when comparing the strength of antibody responses across different antigens. However, on the basis that the PfMSP6 and PfMSP3 studies were carried out using the same samples, secondary antibody reagents, expression methods and detection systems, this data can be interpreted to suggest that anti-PfMSP3 responses are more frequent but more allele-specific than anti-PfMSP6 responses are, at least at this study site. In both antigens responses against the N-terminal domain were more prevalent than reactions against the more conserved C-terminal domains, and there appears to be greater potential for generating responses that cross-react across multiple N-terminal variants than has been previously appreciated, particularly in the case of PfMSP6, where there was no evidence for allele-specific responses, at least based on the indirect measure of comparing the antigen-specificity of immune responses with the genotype of the infection being tested. This would argue that further careful testing of the PfMSP6 as a vaccine candidate is needed, and suggests that definitive experiments to establish whether responses against the N-terminal domain can be allele-transcending should be carried out as a matter of urgency.
Because the P. falciparum genome presents a large number of potential vaccine candidates, it is critical that go/no-go decisions based on solid evidence are applied to limit the number of candidates entering expensive later stage vaccine trials (4, 36, 37). As our appreciation of the depth of P. falciparum genetic diversity increases with the expansion of nextgen sequencing efforts, it is critical that immunoepidemiological studies become more complex to reflect the realities of in vivo infection. It is clearly not possible to establish priority between different vaccine candidates based solely on comparatively immunoepidemiological studies, but these data do suggest that responses against two related antigens, PfMSP3 and PfMSP6, are qualitatively different. Studies such as these, comparing multiple allelic variants and sub-domains of candidate antigens, will become increasingly critical if we are to move P. falciparum vaccine design forward in a rational, evidence-based manner.
Acknowledgements
The authors wish to thank Patrick Sutton and Eva Clark for help with sample processing and Mike Jablonsky for his help with the circular dichroism. We would like to thank all residents in the Zungarococha community who participate so willingly in the MIGIA cohort study. We thank all the staff of the MIGIA project, which is a strong collaboration with the Universidad Nacional de la Amazonia Peruana. We thank all for sample collection, clinic visits and management, and laboratory sample processing and care.
Financial Support
This work was supported by the National Institute of Health grants R21 AI072421 and R01 AI064849, and the UAB Sparkman Center for Global Health.
Footnotes
Disclosures:
The authors have no financial conflicts of interest.
References
- 1.Carlton JM, Adams JH, Silva JC, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardner MJ, Hall N, Fung E, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pain A, Bohme U, Berry AE, et al. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature. 2008;455:799–803. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coppel RL. Vaccinating with the genome: a Sisyphean task? Trends Parasitol. 2009;25:205–212. doi: 10.1016/j.pt.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Takala SL, Plowe CV. Genetic diversity and malaria vaccine design, testing and efficacy: preventing and overcoming 'vaccine resistant malaria'. Parasite Immunol. 2009;31:560–573. doi: 10.1111/j.1365-3024.2009.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ochola LI, Tetteh KK, Stewart LB, Riitho V, Marsh K, Conway DJ. Allele frequency-based and polymorphism-versus-divergence indices of balancing selection in a new filtered set of polymorphic genes in Plasmodium falciparum. Mol Biol Evol. 2010 doi: 10.1093/molbev/msq119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trucco C, Fernandez-Reyes D, Howell S, et al. The merozoite surface protein 6 gene codes for a 36 kDa protein associated with the Plasmodium falciparum merozoite surface protein-1 complex. Mol Biochem Parasitol. 2001;112:91–101. doi: 10.1016/s0166-6851(00)00350-9. [DOI] [PubMed] [Google Scholar]
- 8.Kauth CW, Woehlbier U, Kern M, et al. Interactions between merozoite surface proteins 1, 6, and 7 of the malaria parasite Plasmodium falciparum. J Biol Chem. 2006;281:31517–31527. doi: 10.1074/jbc.M604641200. [DOI] [PubMed] [Google Scholar]
- 9.Koussis K, Withers-Martinez C, Yeoh S, et al. A multifunctional serine protease primes the malaria parasite for red blood cell invasion. EMBO J. 2009;28:725–735. doi: 10.1038/emboj.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearce JA, Triglia T, Hodder AN, Jackson DC, Cowman AF, Anders RF. Plasmodium falciparum merozoite surface protein 6 is a dimorphic antigen. Infect Immun. 2004;72:2321–2328. doi: 10.1128/IAI.72.4.2321-2328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy SW, Weedall GD, da Silva RL, Polley SD, Ferreira MU. Sequence diversity and evolutionary dynamics of the dimorphic antigen merozoite surface protein-6 and other Msp genes of Plasmodium falciparum. Gene. 2009;443:12–21. doi: 10.1016/j.gene.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Neal AT, Jordan SJ, Oliveira AL, Hernandez JN, Branch OH, Rayner JC. Limited variation in vaccine candidate Plasmodium falciparum Merozoite Surface Protein-6 over multiple transmission seasons. Malar J. 2010;9:138. doi: 10.1186/1475-2875-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh S, Soe S, Weisman S, Barnwell JW, Perignon JL, Druilhe P. A conserved multi-gene family induces cross-reactive antibodies effective in defense against Plasmodium falciparum. PLoS One. 2009;4:e5410. doi: 10.1371/journal.pone.0005410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oeuvray C, Bouharoun-Tayoun H, Grass-Masse H, et al. A novel merozoite surface antigen of Plasmodium falciparum (MSP-3) identified by cellular-antibody cooperative mechanism antigenicity and biological activity of antibodies. Mem Inst Oswaldo Cruz. 1994;89(Suppl 2):77–80. doi: 10.1590/s0074-02761994000600018. [DOI] [PubMed] [Google Scholar]
- 15.Singh S, Soe S, Mejia JP, et al. Identification of a conserved region of Plasmodium falciparum MSP3 targeted by biologically active antibodies to improve vaccine design. J Infect Dis. 2004;190:1010–1018. doi: 10.1086/423208. [DOI] [PubMed] [Google Scholar]
- 16.Osier FH, Polley SD, Mwangi T, Lowe B, Conway DJ, Marsh K. Naturally acquired antibodies to polymorphic and conserved epitopes of Plasmodium falciparum merozoite surface protein 3. Parasite Immunol. 2007;29:387–394. doi: 10.1111/j.1365-3024.2007.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polley SD, Tetteh KK, Lloyd JM, et al. Plasmodium falciparum merozoite surface protein 3 is a target of allele-specific immunity and alleles are maintained by natural selection. J Infect Dis. 2007;195:279–287. doi: 10.1086/509806. [DOI] [PubMed] [Google Scholar]
- 18.Roussilhon C, Oeuvray C, Muller-Graf C, et al. Long-term clinical protection from falciparum malaria is strongly associated with IgG3 antibodies to merozoite surface protein 3. PLoS Med. 2007;4:e320. doi: 10.1371/journal.pmed.0040320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hisaeda H, Saul A, Reece JJ, et al. Merozoite surface protein 3 and protection against malaria in Aotus nancymai monkeys. J Infect Dis. 2002;185:657–664. doi: 10.1086/339187. [DOI] [PubMed] [Google Scholar]
- 20.Druilhe P, Spertini F, Soesoe D, et al. A malaria vaccine that elicits in humans antibodies able to kill Plasmodium falciparum. PLoS Med. 2005;2:e344. doi: 10.1371/journal.pmed.0020344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sirima SB, Nebie I, Ouedraogo A, et al. Safety and immunogenicity of the Plasmodium falciparum merozoite surface protein-3 long synthetic peptide (MSP3-LSP) malaria vaccine in healthy, semi-immune adult males in Burkina Faso, West Africa. Vaccine. 2007;25:2723–2732. doi: 10.1016/j.vaccine.2006.05.090. [DOI] [PubMed] [Google Scholar]
- 22.Lusingu JP, Gesase S, Msham S, et al. Satisfactory safety and immunogenicity of MSP3 malaria vaccine candidate in Tanzanian children aged 12–24 months. Malar J. 2009;8:163. doi: 10.1186/1475-2875-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daher LJ, Demanga CG, Prieur E, Perignon JL, Bouharoun-Tayoun H, Druilhe P. Toward the rational design of a malaria vaccine construct using the MSP3 family as an example: contribution of immunogenicity studies in models. Infect Immun. 2010;78:477–485. doi: 10.1128/IAI.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demanga CG, Daher LJ, Prieur E, et al. Toward the rational design of a malaria vaccine construct using the MSP3 family as an example: contribution of antigenicity studies in humans. Infect Immun. 2010;78:486–494. doi: 10.1128/IAI.01359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh S, Soe S, Roussilhon C, Corradin G, Druilhe P. Plasmodium falciparum merozoite surface protein 6 displays multiple targets for naturally occurring antibodies that mediate monocyte-dependent parasite killing. Infect Immun. 2005;73:1235–1238. doi: 10.1128/IAI.73.2.1235-1238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Crouch L, Richie TL, Nhan DH, Coppel RL. Naturally acquired antibody responses to the components of the Plasmodium falciparum merozoite surface protein 1 complex. Parasite Immunol. 2003;25:403–412. doi: 10.1111/j.1365-3024.2003.00647.x. [DOI] [PubMed] [Google Scholar]
- 27.Branch O, Casapia WM, Gamboa DV, et al. Clustered local transmission and asymptomatic Plasmodium falciparum and Plasmodium vivax malaria infections in a recently emerged, hypoendemic Peruvian Amazon community. Malar J. 2005;4:27. doi: 10.1186/1475-2875-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jordan SJ, Oliveira AL, Hernandez JN, et al. Malaria immunoepidemiology in low transmission: Correlation of infecting genotype and immune response to domains of Plasmodium falciparum Merozoite Surface Protein-3. Infect Immun. 2011 doi: 10.1128/IAI.01332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgess BR, Schuck P, Garboczi DN. Dissection of merozoite surface protein 3, a representative of a family of Plasmodium falciparum surface proteins, reveals an oligomeric and highly elongated molecule. J Biol Chem. 2005;280:37236–37245. doi: 10.1074/jbc.M506753200. [DOI] [PubMed] [Google Scholar]
- 30.Oeuvray C, Theisen M, Rogier C, Trape JF, Jepsen S, Druilhe P. Cytophilic immunoglobulin responses to Plasmodium falciparum glutamate-rich protein are correlated with protection against clinical malaria in Dielmo, Senegal. Infect Immun. 2000;68:2617–2620. doi: 10.1128/iai.68.5.2617-2620.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanisic DI, Richards JS, McCallum FJ, et al. Immunoglobulin G subclass-specific responses against Plasmodium falciparum merozoite antigens are associated with control of parasitemia and protection from symptomatic illness. Infect Immun. 2009;77:1165–1174. doi: 10.1128/IAI.01129-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor RR, Allen SJ, Greenwood BM, Riley EM. IgG3 antibodies to Plasmodium falciparum merozoite surface protein 2 (MSP2): increasing prevalence with age and association with clinical immunity to malaria. Am J Trop Med Hyg. 1998;58:406–413. doi: 10.4269/ajtmh.1998.58.406. [DOI] [PubMed] [Google Scholar]
- 33.Ogutu BR, Apollo OJ, McKinney D, et al. Blood stage malaria vaccine eliciting high antigen-specific antibody concentrations confers no protection to young children in Western Kenya. PLoS One. 2009;4:e4708. doi: 10.1371/journal.pone.0004708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sagara I, Dicko A, Ellis RD, et al. A randomized controlled phase 2 trial of the blood stage AMA1-C1/Alhydrogel malaria vaccine in children in Mali. Vaccine. 2009;27:3090–3098. doi: 10.1016/j.vaccine.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polley SD, Conway DJ, Cavanagh DR, et al. High levels of serum antibodies to merozoite surface protein 2 of Plasmodium falciparum are associated with reduced risk of clinical malaria in coastal Kenya. Vaccine. 2006;24:4233–4246. doi: 10.1016/j.vaccine.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 36.Greenwood B, Targett G. Do we still need a malaria vaccine? Parasite Immunol. 2009;31:582–586. doi: 10.1111/j.1365-3024.2009.01140.x. [DOI] [PubMed] [Google Scholar]
- 37.Holder AA. Malaria vaccines: where next? PLoS Pathog. 2009;5:e1000638. doi: 10.1371/journal.ppat.1000638. [DOI] [PMC free article] [PubMed] [Google Scholar]