Abstract
We report a novel strategy to enzymatically release affinity-selected cells, such as circulating tumor cells (CTCs), from surfaces with high efficiency (~90%) while maintaining cell viability (>85%). The strategy utilizes single-stranded DNAs that link a capture antibody to the surfaces of a CTC selection device. The DNA linkers contain a uracil residue that can be cleaved.
The ability to release affinity-selected cells, such as circulating tumor cells (CTCs), from surfaces containing selection antibodies (Abs) without perturbing the cells’morphology, viability, molecular content and phenotype has been a major challenge. In spite of the challenge, compelling applications would result from the ability to release the selected cells such as securing molecular information from the cells that can be used for basic discovery and/or molecular diagnostics.
The challenges associated with current release strategies include inefficient release or damage imposed on the cell by the release process. For example, cell release by shear-based methods1,2 are highly dependent on the number of attachment points between the cells’ antigens (Ags) and surface immobilized Abs. Excessively high shear rates are required to release cells with multiple attachment points, which can damage them, especially fragile CTCs.3 Grafting methods are incompatible with microfluidic devices,4 which have proven to be an attractive platform for CTC selection.
Thermally responsive materials5–7 have been used to release selected cells with an efficiency of ~59% and viability of 90%.5 Polymer brushes on nanostructured surfaces were capable of isolating cells at 37 °C and releasing 90% of the selected cells at 4 °C, but the fabrication of the cell selection device and its functionalization was a challenge.8 Functionalized alginate hydrogels were able to isolate and release cells with 90% viability, but the purity of the isolated population was low.9,10 Lectins were reported to isolate and release lymphocytes; however, the specificity resulted in low purity of the selected cells with ~50% release efficiency.11 DNA aptamers have also been used for cell isolation; cells could be released using DNase (68%).12 Proteolytic digestion of selection Abs has been reported, for example using trypsin.13–16 While efficient cell release was demonstrated, the damage of extracellular domains of membrane Ags is possible limiting the ability to immunostain the cells. Light-triggered cell release using photocleavable linkers attached to quartz surfaces, which achieved 85% release efficiency, has been reported.17 Light-based release methods, however, can induce DNA damage that can confound diagnostic information.18
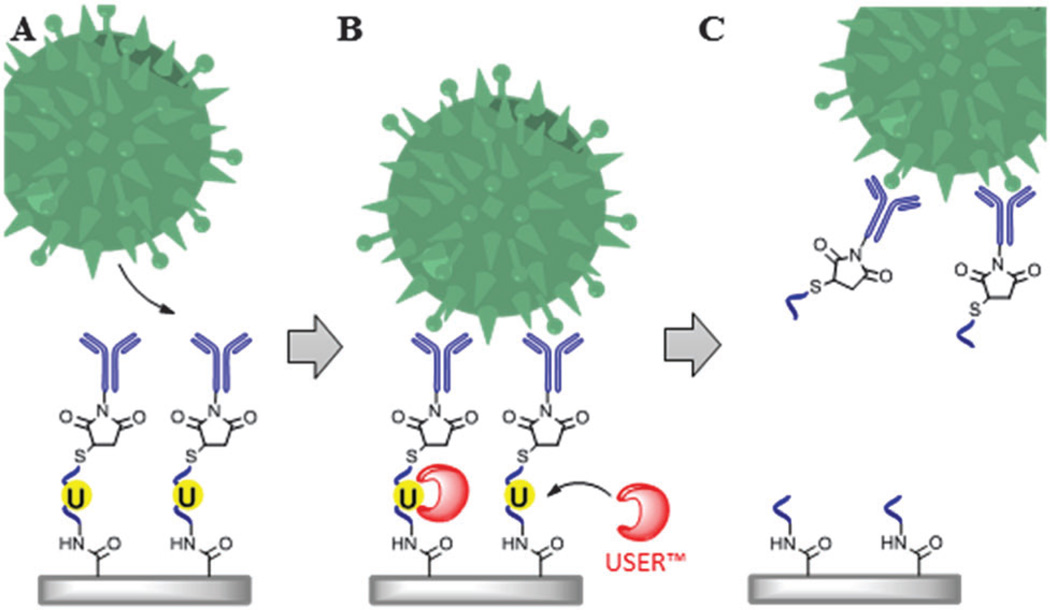
The work reported herein describes a unique assay (Scheme 1) for the positive selection of rare cells (see Fig. S1, ESI† for description of cell selection device) with their subsequent release for post-selection applications, such as the analysis of clinical CTCs, flow cytometry (FC) and fluorescence in situ hybridization (FISH). Heterobifunctional linkers (Table S1, ESI†) were used to immobilize monoclonal Abs (mAbs) to a UV/O3-activated fluidic surface presenting carboxylic acids (−COOH; Scheme S1 in ESI†). mAbs were reacted with a sulfo-NHS ester of succinimidyl trans-4 (maleimidylmethyl) cyclohexane-1-carboxylate (SMCC), yielding a maleimide-labeled mAb (SMCC-mAb). Once purified, the SMCC-mAb could be covalently attached to the reduced 3′-disulfide group (sulfhydryl) of a single-stranded oligonucleotide (ssDNA) linker that was immobilized to the activated surface using EDC–NHS coupling of the ssDNA linker’s 5′-amino group to the surface-confined COOHs. Incorporating uracil (dU) into the ssDNA linker enabled enzymatic nicking by the USER™ (Uracil-Specific Excision Reagent) system and thus, cell release (Scheme 1). USER™ consists of a mixture of uracil DNA glycosylase (UDG) and DNA glycosylase-lyase endonuclease VIII. UDG catalyzes the excision of dU forming an abasic site. Endonuclease VIII breaks the phosphodiester bond of the abasic site, cleaving the ssDNA linker and releasing the selected cell from the capture surface. The advantage of ssDNA linkers lies in their stability, low cost, and ease of covalent and ordered attachment to a variety of surfaces with a high load.19–23 Moreover, USER™ is active at physiological temperatures and in a variety of buffers, such as PBS, both of which can maintain cell viability.24,25
Scheme 1.
Cell selection and release assay. (A) mAbs immobilized to surfaces using oligonucleotide linkers containing a uracil residue are used for the positive selection of target cells. (B) Incubation of the selected cells and ssDNA linker with the USER™ enzyme system. (C) Removal of the uracil residue results in release of the selected cells.
The efficiency and specificity of USER™ cleavage was tested by nicking a 34 nt ssDNA substrate in a microtube at 37 °C and monitoring the appearance of a 26 nt product electrophoretically (Fig. S2, ESI†). Electropherograms indicated that after a 5 min reaction in PBS (pH 7.4), most of the 34 nt substrate was converted into the 26 nt product as a result of enzymatic nicking. Reaction times of 15 min completely nicked the substrate (Fig. S2, ESI†), demonstrating that USER™ specifically cleaved the oligonucleotide at only the dU residue.
To evaluate the ability to select cells and then release the selected cells using ssDNA linkers containing a dU residue, a microfluidic device used for cell selection fabricated in cyclic olefin copolymer (COC) consisting of an array of sinusoidal channels (25 µm wide, 150 µm deep and 3 cm in length; see Fig. S1, ESI†) was used as the model system. Using this microfluidic device and direct attachment of the selection mAbs to the activated COC surface (i.e., no ssDNA linker used), CTC recoveries of 98% have been noted.13,15 COC was used as the material of choice because of its low propensity for showing non-specific adsorption artifacts and its high loads of COOH groups following UV/O3 activation.26
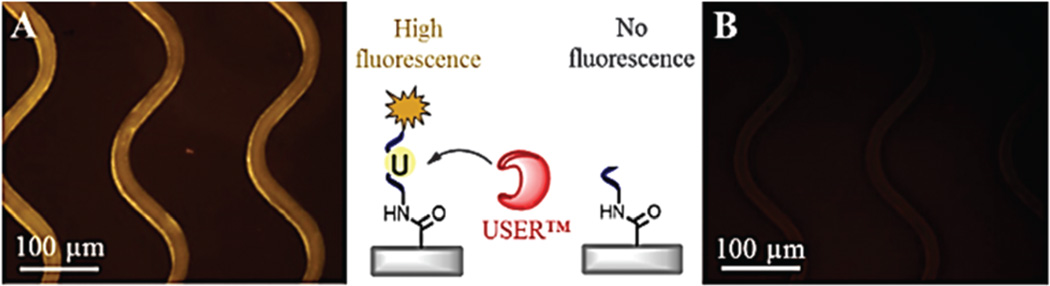
To initially evaluate the ability to cleave a ssDNA linker when immobilized to the surface of the COC microchannels, a dye-labeled oligonucleotide containing an internal dU residue was immobilized to the microchannel surfaces, visualized (Fig. 1A), and nicked by infusing a solution of USER™ (2 U per 10 µL) into the device. Successful oligonucleotide nicking was confirmed by microscopy, which indicated a loss of fluorescence within the microchannels (Fig. 1B) and the presence of fluorescence in the effluent. Fluorescence measurements indicated 5.2 ± 0.4 pmol cm−2 of the oligonucleotide was cleaved from the surfaces (1.9 × 1013 linkers per device). Based on the size of a 25 nt ssDNA with a random sequence,27,28 the theoretical oligonucleotide surface density was estimated to be 13.2 pmol cm−2, which is close to the measured load of the ssDNA linker. It is important to note that during immobilization, ssDNA linkers can cross-link to the surface with multiple points of attachment due to the presence of primary amines on some of the DNA bases. To mitigate this, ssDNA linkers were designed (see Table S1, ESI†) containing poly dT sequences at the 3′ and 5′ ends to eliminate crosslinking. Secondary structures, such as homo-dimers and hairpins were unlikely based on the sequences of the linkers, therefore ensuring accessibility of the linker’s 3′-sulfhydryl with the maleimide-labeled Ab.
Fig. 1.
Fluorescence images of sinusoidal microchannels (A) before and (B) after 5 min enzymatic cleavage of a 3′-Cy3 modified 25 nt oligonucleotide containing a dU residue. The microchannel surfaces were activated using UV/O3 exposure and functionalized with 20 mg mL−1 EDC and 2 mg mL−1 NHS in 0.1 M MES (pH 4.8). After 20 min of the EDC–NHS reaction, a 40 µM solution containing the 3′-Cy3 oligonucleotide was infused into the device. Before imaging, the device was rinsed with 0.1% SDS and nuclease-free water.
We used three cancer cell lines to evaluate the efficiency of cell isolation and release, SKBR3 and Hs578T (both adherent), which express EpCAM and FAPα (fibroblast activation protein alpha, FAPα), respectively, and acute myeloid leukemia KG-1 cells (non-adherent) expressing CD34. mAbs directed against these Ags were used for cell isolation. Cell recoveries were determined using a “self-referencing method” (Fig. S3, ESI†). The self-referencing method uses multiple cell selection devices connected in series to deplete the input cells with the recovery determined from the number of cells collected by the first device with respect to the total number of cells collected in the series. We found that the self-referencing method yielded similar calculated recovery values to seeding experiments, but the relative standard deviation in the measurements were significantly lower (35% versus 6%, see ESI†). Recoveries using the cleavable ssDNA linkers were compared to a previously reported direct attachment approach,15 where mAbs were covalently attached to the UV/O3 activated microchannel surfaces that bear COOH groups using EDC–NHS coupling chemistry (Table S2, ESI†).
The ssDNA linkers demonstrated similar recoveries for the three cell lines investigated when compared to the direct attachment protocol. Recoveries were normalized with respect to the anti-EpCAM mAb recovery of the SKBR3 cells isolated via the 40dX linkers. Statistically similar results were observed for cell recovery via direct attachment when compared to attachment using the ssDNA linkers for SKBR3 cells, 96 ± 12% (n = 4). FAPα Hs578T cells were recovered with slightly higher efficiency when mAbs were directly attached to the surface, 90 ± 9% (n = 8), compared to the 34dX or 40dX linkers, 74 ± 7% (n = 3) and 80 ± 6% (n = 5), respectively. Between the ssDNA linkers tested, the data did not indicate a strong dependence of recovery on linker length, sequence, or the nature of the chemical group between the 5′-amino group and the ssDNA linker (C6 for 34dX and C12 for 40dX). The recovery of CD34 KG-1 cells did not statistically differ between direct attachment, 81 ± 6% (n = 7), and attachment using 40dX, 40dT, or 20dT linkers, 76 ± 5% (n = 5), 74 ± 7% (n = 9), and 77 ± 5% (n = 5), respectively. We concluded that the linkers used in this study were able to generate accessible mAbs on the microchannel surfaces, irrespective of the linker length and sequence.
The efficiency of cell release following affinity selection for the three cell lines tested are summarized in Table S3 (ESI†) and illustrated in Fig. S4 (ESI†). We also evaluated two formats for the cell release process within the cell selection device: (i) continuous-flow (CF) of USER™ into the cell selection device; and (ii) incubation for a fixed time period after flooding the cell selection device with USER™. To determine the viability following release, unstained cells were released and stained with the LIVE/DEAD™ assay.
We determined that the incubation format provided more efficient release of selected cells using the microfluidic device compared to the CF reaction. In the CF reaction, 60 min was required to release ~84% of the selected cells, while the same release efficiency was achieved within 15 min using the incubation format; the release efficiency was 89 ± 3% (n = 5) for SKBR3 cells and 86 ± 4% for the Hs578T cells when the selection surface was modified with mAb and the 40dX linker. Additional advantages of the incubation format is low enzyme consumption and maintaining the enrichment factor because the cells were eluted into a smaller volume; ~10 µL. Enzymatic release of KG-1 cells with USER™ was carried out on live and fixed cells with mAb attached via 20dT and 40dT ssDNA linkers (Table S3, ESI†). Within 30 min, >80% release efficiency was observed with no significant increases in the release efficiency observed for longer incubation times (Fig. S4, ESI†).
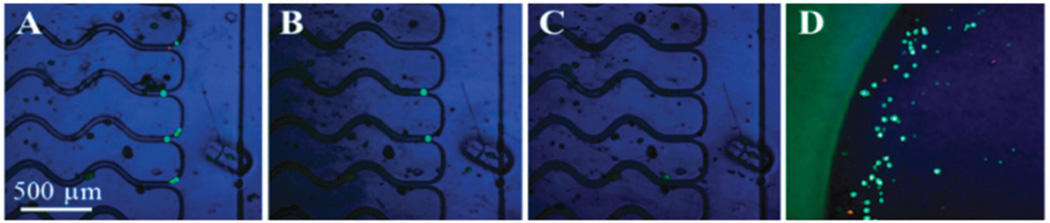
Fig. 2 presents images illustrating the release of viable KG-1 cells. High release efficiency was achieved for both viable and fixed cells. Proteolytic (i.e., trypsin) release was not possible for fixed cells following isolation for direct mAb attachment to the device surface due to Ag–Ab crosslinking. Interestingly, the rate of release was faster for the Hs578T and SKBR3 cells compared to the KG-1 cells; longer incubation times (30 min) were required for KG-1 cells to achieve the same release efficiency (see Table S3 and Fig. S4B, ESI†). We believe this result may reflect differences in the number of attachment points by the mAb–Ag associations. The density of CD34 antigens on KG-1 cells is significantly higher (89× isotype, see Table S4, ESI†) compared to FAPα on Hs578T cells (6× isotype) and EpCAM on SKBR3 cells (15× isotype; data not shown). Thus, the rate of release depends on the number of mAbs–Ag associations (i.e., the adhesion force).
Fig. 2.
Release of KG-1 cells isolated using the 40dT linker. Cells stained with (A) Live/Dead™ kit were released after (B) 15 min and (C) 30 min USER™ incubation. (D) Cells (eluted at 10 µL min−1) were enumerated. Live cells generate esterase-dependent calcein, fluorescing green (ex/em 488/515 nm). Dead cells were susceptible to a cell-impermeable, DNA-intercalating dye fluorescing red (ex/em 570 nm/605 nm).
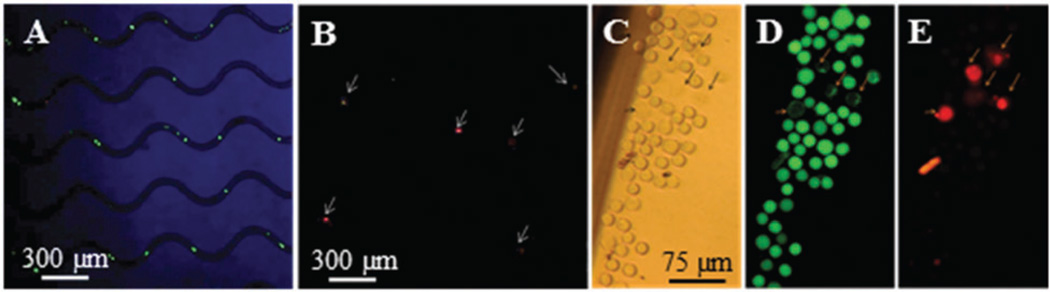
Cell viability was analyzed following isolation and also after release. Hs578T, SKBR3, and KG-1 cells freshly harvested from culture demonstrated a viability of 93 ± 3%. Following selection, 89 ± 5%, 92 ± 4%, and 92 ± 3% viability was found for Hs578T, SKBR3, and KG-1 cells, respectively, which was similar to the viability found from culture (Fig. 3A and B). Post-release viabilities of cell lines were also high, although slightly lower than post-isolation values (80 ± 3%, 82 ± 5% and 84 ± 4% viabilities for Hs578T and SKBR3 and KG-1 cells, respectively). The fluorescence images of released SKBR3 cells in a titer well are shown in Fig. 3C–E. In a control experiment, KG-1 cells were suspended in PBS (pH 7.4) within a fluidic device containing no mAb or subjected to release; the viability was 85 ± 7%, very similar to viabilities observed after enzymatic release. Therefore, the slight decrease in cell viability after release is caused by natural cell viability loss and not the release process. Clearly, the majority of cells were not damaged by the shear forces observed in the cell selection device26 and/or treatment with USER™.
Fig. 3.
Fluorescence images used to quantify cell viability (A, B) after isolation and (C–E) after release. Images C–E share the same scale bar.
The ability to culture selected and released cells was evaluated as well. Approximately 110 SKBR3 cells were collected into the well of a 24-well titer plate following selection and release. Media reagents were introduced and microscopic observations of cell growth were made (Fig. S5, ESI†). After 5 d, the number of cells doubled. The low seeding density caused a slower growth rate of these cells; at higher seeding densities, released SKBR3 cells divided more rapidly.
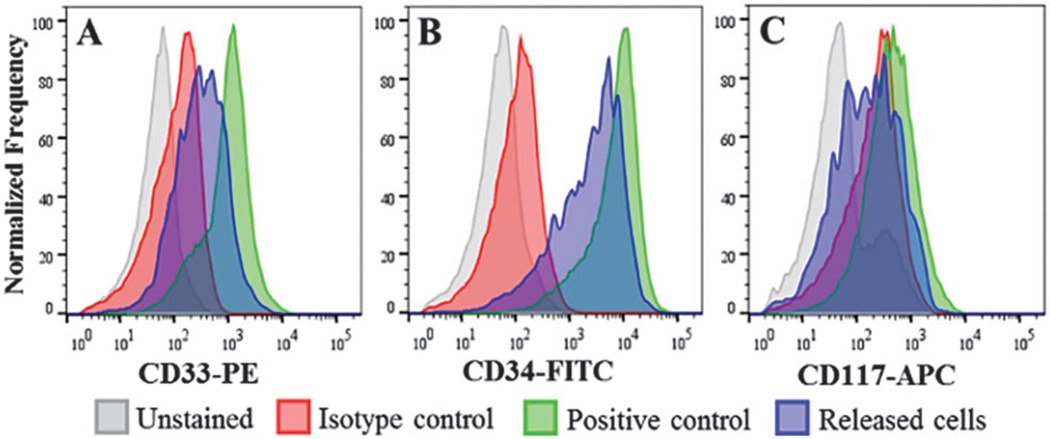
We performed flow cytometry of KG-1 cells to evaluate the ability to detect and quantify expression of immunostained cells following selection, on-chip staining and USER™ release. Histograms in Fig. 4 are shown for 3 antigens, CD33, CD34, and CD117. Cells selected and stained on-chip had fluorescence values 54 ± 7% compared to the KG-1 cells labeled in solution for all antigens indicating that approximately half of the cell’s surface Ags were inaccessible, possibly due to attachment and masking by the channel surface. Only for the weakly expressed CD117 was it difficult to discern these cells from an unstained population (see Table S4 and Fig. S6, ESI†). However, solid-phase staining provides advantages in terms of minimizing reagent consumption, easy washing of cells and eliminating cell loss or damage during washing steps (see ESI†).
Fig. 4.
Flow cytometry analysis of KG-1 cells isolated, stained, and released from the cell selection device.
To demonstrate the utility of our assay for the selection and release of clinical CTCs, metastatic ovarian cancer (M-OVC) blood samples were processed using anti-EpCAM modified cell selection devices with the mAb attached to the surface via the ssDNA linkers. Isolated CTCs were stained with CD45, DAPI, and CK-Pan and enumerated (see Fig. S7, ESI†). Table S5 (ESI†) summarizes results secured from M-OVC blood samples. Similar cell counts from these samples were found using the ssDNA linker and direct attachment (Table S5, ESI†). The average viability of CTCs isolated from M-OVC blood samples was 93 ± 5% before release. Upon release, the viability was 87 ± 6% (Fig. S8, ESI†).
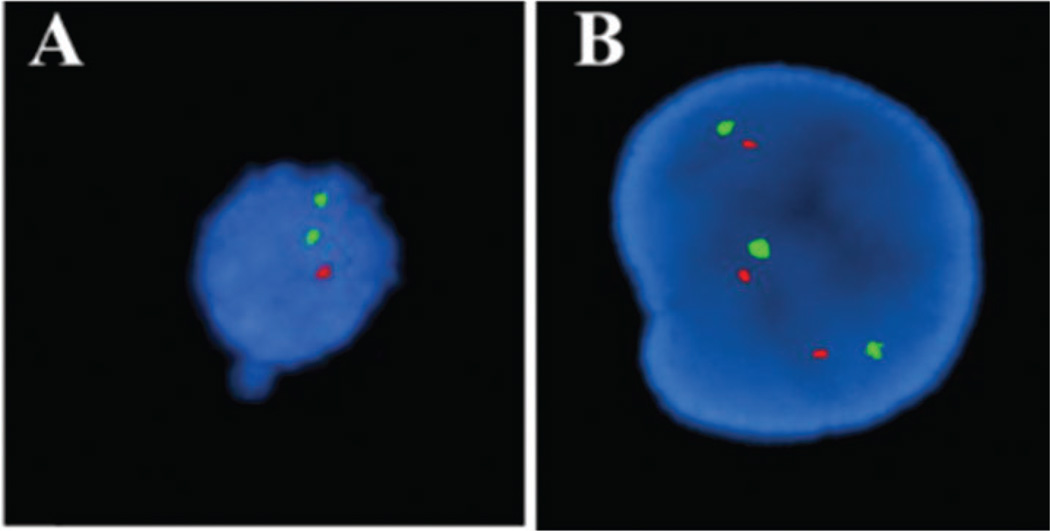
Circulating multiple myeloma cells (CMMCs) were isolated using anti-CD138 mAbs from multiple myeloma (MM) patients, released and subsequently tested by FISH for a possible deletion in chromosome 13, which is found in ~86% of MM patients. The presence of the deletion is typically associated with poor prognosis and the propensity of the disease to progress from an asymptomatic to symptomatic stage.29
Both mutated (Fig. 5A) and wild type (Fig. 5B) CMMCs were observed in patient’s blood. The deletion identified in chromosome 13 in the isolated CMMCs was also present in bone marrow from the same patient. Chromosomal analysis of CMMCs selected from whole blood and released intact allows for conventional cytogenetic analysis without the need for requiring the patient to undergo a bone marrow biopsy to provide more frequent testing.
Fig. 5.
FISH images with a 13q14 probe of patient CMMCs. Green signal indicates the presence of chromosome 13 and red signal indicates absence of the chromosome 13 deletion. The patient sample contained a mixture of CMMCs: (A) cell shows the presence of the deletion and (B) polyploidy cell without the deletion. Cells were isolated using anti-CD138 modified chip.
We have successfully demonstrated the use of ssDNA linkers engineered with a dU residue for attaching mAbs to surfaces for selecting clinically relevant cells with high recovery and when using the USER™ enzyme system, release the cells both efficiently (~90%) and rapidly (15–30 min) while maintaining their viability. Clinical CTCs and CMMCs were selected directly from patients’ peripheral blood then released to demonstrate assay integration into existing clinical processing pipelines while obviating the need for invasive biopsies. We have demonstrated that the selected cells can be fixed, stained, released, and subjected to conventional flow cytometry. The selected/released cells could also be subjected to FISH to determine chromosomal abnormalities using conventional cytogenetic assays. The USER™ release strategy preserved cell viability making them available for cultivation as well. An additional benefit of the ssDNA linkers and the USER™ enzyme system was the seamless integration with existing microfluidic devices used for rare cell selection and the general attachment chemistry, which can enable the application of these ssDNA linkers to a variety of solid-phase supports.
Supplementary Material
Acknowledgments
This work was supported through the NIH (IMAT; R21-CA173279), the University Cancer Research Fund (UNC), Lineberger Comprehensive Cancer Center (UNC), the Carlson Foundation, and the Triad Golfers Against Cancer.
Footnotes
Electronic supplementary information (ESI) available: Experimental details, Self-referencing method for cell recovery evaluation. Evaluation of affinity selected cell release, cell cultivation after release with USER™, KG-1 cell staining and flow cytometry analysis. See DOI: 10.1039/c4cc09765c
Notes and references
- 1.Cheung LSL, Zheng X, Stopa A, Baygents JC, Guzman R, Schroeder JA, Heimark RL, Zohar Y. Lab Chip. 2009;9:1721. doi: 10.1039/b822172c. [DOI] [PubMed] [Google Scholar]
- 2.Yu X, He R, Li S, Cai B, Zhao L, Liao L, Liu W, Zeng Q, Wang H, Guo S-S, Zhao X-Z. Small. 2013;9:3895. doi: 10.1002/smll.201300169. [DOI] [PubMed] [Google Scholar]
- 3.Parkinson D, Dracopoli N, Petty B, Compton C, Cristofanilli M, Deisseroth A, Hayes D, Kapke G, Kumar P, Lee J, Liu M, McCormack R, Mikulski S, Nagahara L, Pantel K, Pearson-White S, Punnoose E, Roadcap L, Schade A, Scher H, Sigman C, Kelloff G. J. Transl. Med. 2012;10:138. doi: 10.1186/1479-5876-10-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah AM, Yu M, Nakamura Z, Ciciliano J, Ulman M, Kotz K, Stott SL, Maheswaran S, Haber DA, Toner M. Anal. Chem. 2012;84:3682. doi: 10.1021/ac300190j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurkan UA, Anand T, Tas H, Elkan D, Akay A, Keles HO, Demirci U. Lab Chip. 2011;11:3979. doi: 10.1039/c1lc20487d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Z, Akiyama Y, Itoga K, Kobayashi J, Yamato M, Okano T. Biomaterials. 2012;33:7405. doi: 10.1016/j.biomaterials.2012.06.077. [DOI] [PubMed] [Google Scholar]
- 7.Liu H, Liu X, Meng J, Zhang P, Yang G, Su B, Sun K, Chen L, Han D, Wang S, Jiang L. Adv. Mater. 2013;25:922. doi: 10.1002/adma.201203826. [DOI] [PubMed] [Google Scholar]
- 8.Hou S, Zhao H, Zhao L, Shen Q, Wei KS, Suh DY, Nakao A, Garcia MA, Song M, Lee T, Xiong B, Luo S-C, Tseng H-R, Yu H-h. Adv. Mater. 2013;25:1547. doi: 10.1002/adma.201203185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatch A, Hansmann G, Murthy SK. Langmuir. 2011;27:4257. doi: 10.1021/la105016a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plouffe BD, Brown MA, Iyer RK, Radisic M, Murthy SK. Lab Chip. 2009;9:1507. doi: 10.1039/b823523f. [DOI] [PubMed] [Google Scholar]
- 11.Vickers DL, Hincapie M, Hancock W, Murthy S. Biomed. Microdevices. 2011;13:565. doi: 10.1007/s10544-011-9527-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao W, Cui CH, Bose S, Guo D, Shen C, Wong WP, Halvorsen K, Farokhzad OC, Teo GSL, Phillips JA, Dorfman DM, Karnik R, Karp JM. Proc. Natl. Acad. Sci. U. S. A. 2012;109:19626. doi: 10.1073/pnas.1211234109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Gottert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. J. Am. Chem. Soc. 2008;130:8633. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dharmasiri U, Njoroge SK, Witek MA, Adebiyi MG, Kamande JW, Hupert ML, Barany F, Soper SA. Anal. Chem. 2011;83:2301. doi: 10.1021/ac103172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamande JW, Hupert ML, Witek MA, Wang H, Torphy RJ, Dharmasiri U, Njoroge SK, Jackson JM, Aufforth RD, Snavely A, Yeh JJ, Soper SA. Anal. Chem. 2013;85:9092. doi: 10.1021/ac401720k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheng W, Ogunwobi OO, Chen T, Zhang J, George TJ, Liu C, Fan ZH. Lab Chip. 2014;14:89. doi: 10.1039/c3lc51017d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wirkner M, Alonso JM, Maus V, Salierno M, Lee TT, García AJ, del Campo A. Adv. Mater. 2011;23:3907. doi: 10.1002/adma.201100925. [DOI] [PubMed] [Google Scholar]
- 18.Ikehata H, Ono T. J. Radiat. Res. 2011;52:115. doi: 10.1269/jrr.10175. [DOI] [PubMed] [Google Scholar]
- 19.Situma C, Wang Y, Hupert M, Soper SA. Abstr. Pap. Am. Chem. Soc. 2004;228:U118. [Google Scholar]
- 20.Situma C, Wang Y, Hupert M, Barany F, McCarley RL, Soper SA. Anal. Biochem. 2005;340:123. doi: 10.1016/j.ab.2005.01.044. [DOI] [PubMed] [Google Scholar]
- 21.Soper SA, Hashimoto M, Situma C, Murphy MC, McCarley RL, Cheng YW, Barany F. Methods. 2005;37:103. doi: 10.1016/j.ymeth.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Situma C, Hashimoto M, Soper SA. Biomol. Eng. 2006;23:213. doi: 10.1016/j.bioeng.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Situma C, Moehring AJ, Noor MAF, Soper SA. Anal. Biochem. 2007;363:35. doi: 10.1016/j.ab.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindahl T. Annu. Rev. Biochem. 1982;51:61. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- 25.Melamede RJ, Hatahet Z, Kow YW, Ide H, Wallace SS. Biochemistry. 1994;33:1255. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- 26.Jackson JM, Witek MA, Hupert ML, Brady C, Pullagurla S, Kamande J, Aufforth RD, Tignanelli CJ, Torphy RJ, Yeh JJ, Soper SA. Lab Chip. 2014;14:106. doi: 10.1039/c3lc50618e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tinland B, Pluen A, Sturm J, Weill G. Macromolecules. 1997;30:5763. [Google Scholar]
- 28.Steel AB, Levicky RL, Herne TM, Tarlov MJ. Biophys. J. 2000;79:975. doi: 10.1016/S0006-3495(00)76351-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng SH, Ng MHL, Lau KM, Liu HSY, Chan JCW, Hui ABY, Lo KW, Jiang H, Hou J, Chu RW, Wong WS, Chan PHN, Ng HK. Blood. 2007;109:2089. doi: 10.1182/blood-2006-04-018770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.