Abstract
The stereochemistry of the hexuronic acid residues of the structure of glycosaminoglycans (GAGs) is a key feature that affects their interactions with proteins and other biological functions. Electron based tandem mass spectrometry methods, in particular electron detachment dissociation (EDD), have been able to distinguish glucuronic (GlcA) from iduronic acid (IdoA) residues in some heparan sulfate tetrasaccharides by producing epimer-specific fragments. Similarly, the relative abundance of glycosidic fragment ions produced by collision induced dissociation (CID) or EDD have been shown to correlate with the type of hexuronic acid present in chondroitin sulfate (CS) GAGs. The present work examines the effect of charge state and the degree of sodium cationization on the CID fragmentation products that can be used to distinguish GlcA and IdoA containing chondroitin sulfate A (CSA) and dermatan sulfate (DS) chains. The cross-ring fragments 2,4An and 0,2Xn formed within the hexuronic acid residues are highly preferential for chains containing GlcA, distinguishing it from IdoA. The diagnostic capability of the fragments requires the selection of a molecular ion and fragment ions with specific ionization characteristics, namely charge state and the number of ionizable protons. The ions with the proper characteristics display diagnostic properties for all the CS and DS chains (dp4-dp10) studied.
Introduction
Chondroitin sulfate (CS) is a type of glycosaminoglycan (GAG) that is responsible for a variety of important biological activities.1, 2 These linear polysaccharides are biosynthesized by addition of alternating residues of N-acetylgalactosamine (GalNAc) and glucuronic acid (GlcA), producing chains with a variety of degrees of polymerization (dp). During their biosynthesis, enzymatic modifications may occur on the differentially elongated chains of CS, such as O-sulfation, and epimerization of GlcA to iduronic acid (IdoA), thus producing highly heterogeneous products.3 Samples extracted from natural sources are thus mixtures of different lengths of CS chains with different sets of modifications within these sequences. Differential sulfation and epimerization of uronic acids leads to a diverse group of compounds differing in length and composition. Even those having the same composition may exhibit a variety of isomeric structures.
CS can be subdivided into different classes that are differentiated by the extent and location of the sulfo modifications and the hexuronic acid C5 stereochemistry. The basic repeating disaccharide unit of CS polysaccharides is N-acetyl-galactosamine (GalNAc) and a uronic acid (GlcA or IdoA). Chondroitin sulfate A (CSA) is O-sulfated at C4 of GalNAc and the uronic acid is mainly GlcA while dermatan sulfate (CSB or DS) have the same amino sugar structure as CSA, but most of the GlcA residues are converted to their epimer form, IdoA.1, 4 Other forms of CS include chondroitin sulfate C (CSC), in which the GalNAc residues are sulfated at the 6-O position, or forms with multiple sulfation sites within the disaccharide unit including 2-O sulfation at the hexuronic acid.1 GAG chains can contain differentially modified domains, with IdoA appearing continuously within a given portion and intermittently in other portions of the intact chain.1, 5
The structural characterization of chondroitin sulfate at the molecular level is required to gain insight regarding the structural motifs associated with protein binding and regulation of biological pathways, which can impact clinical and biomedical applications.6 The details of the composition of CSA/DS are known to have a profound effect on their binding activity. For example, IdoA-rich CSA/DS chains are found in large quantities in specific parts of the brain, and have been found to be critical in neuritogenesis during the brain development of mice and other organisms.7, 8 DS is also found in connective tissue maintenance, is implicated with embryonic development,9, 10 and has been found to suppress blood coagulation by activating heparin cofactor II, a plasma protein which inhibits thrombin.9, 11 Mutations in CSA/DS sulfotransferases lead to abnormal levels of CSA/DS in corneal tissue, causing macular corneal dystrophy.12 CSA/DS chains play critical roles in the central nervous system, regulating both brain development and plasticity. Some pathogens use CS and DS to bind to host cells as well. For example, the malarial parasite P. falciparum cannot attach themselves to mutant CHO cells defective in CS.13 Because of these and many other functions of CS GAGs, knowledge of their molecular structures and how these relate to protein binding will be important to the development of important new therapies.
Various analytical techniques for characterizing CS, including nuclear magnetic resonance (NMR) and mass spectrometry (MS), have been implemented for elucidating GAG structures, including differentiation of hexuronic acid C5 stereochemistry.14-16 The low sample quantities recovered from natural sources makes MS analysis particularly applicable due to its high sensitivity and its capability to analyze mixtures. The production of sufficient glycosidic and cross-ring fragments during tandem mass spectrometry is required to provide detailed molecular level structural information, and there has been considerable research regarding ion activation methods to produce this rich information for GAGs.17-28 The undesired decomposition of sulfo modifications is problematic for GAGs, particularly those with multiple sites of sulfation, but recent advances in ion activation has enabled the acquisition of more detailed structural information from these biomolecules.15, 17, 18, 21-24, 29-34 The combination of glycosidic and cross-ring fragmentation enables the identification of the positions of the sulfo groups within the residue and in some cases the epimeric state of the hexuronic acid.13, 20, 21, 23, 27, 28, 35, 36 Research has shown that epimerization of hexuronic acid and sulfate position along GalNAc residues can affect the number and the intensity of some glycosidic and cross-ring bond cleavages obtained in a given MS/MS experiment.21, 27, 28 Thus, diagnostic ions that can distinguish differentially modified chains can be obtained.
CID is a commonly used approach for ion activation, however, the resulting fragmentation often causes significant losses of SO3 from labile sulfo modifications. Such losses can be reduced through deprotonation of acidic groups during ESI, or by metal cation/hydrogen ion exchange.17, 35, 36 CID has been used in the past to sequence intact chains of CS from the proteoglycan bikunin.37 Different isomers could be identified through the intensity of the glycosidic fragments obtained from CID spectra. Relative abundance of B, X and Y CID ions have been previously used for the characterization of a hexuronic acid's epimeric state in CS GAGs.25, 27, 28, 38 Recently, relative abundance of MSn B and Y fragment ions was used to distinguish between CSA and DS.24 The ratio of the peaks intensities for specific hexuronic acids must be first established from standards, and then applied to mixtures. These ratios might be sensitive to instrument parameters and, thus, are probably not universal. Diagnostic ions that occur for one epimer but not the other would be ideal.
Electron based ion activation methods are promising for locating sites of sulfo modification and for assigning the uronic acid (GlcA versus IdoA) in GAG chains.20, 23, 35 Electron detachment dissociation (EDD) produces a relatively high number of useful product ions with minimal loss of sulfo groups, and it has been found to produce unique fragment ions that can distinguish between GlcA and IdoA in modestly-sulfated heparan sulfate tetrasaccharides.21 EDD, electron induced dissociation (EID), and negative electron transfer dissociation (NETD) combined with multivariate analysis have also been applied in distinguishing chondroitin and dermatan sulfate GAGs.19 The effect of chain length and sodiation level on the EDD spectra of GAGs has also been investigated.20, 23, 35
Most of the previous work investigated molecular ions in which the number of ionized acidic groups was equal to the number of sulfate groups within the molecules.25, 30 This is significant, as it has been shown that if a sulfo group is protonated, it is much more susceptible to decomposition.28 In most instances, CID of these precursor ions does not produce large number of cross-ring cleavages. Recently, we have shown that ionizing all the acidic groups (carboxyl as well as sulfo) within highly sulfated GAGs lead to the production of significantly more useful product ions, including cross-ring fragments.17, 18 The current work investigates cross-ring fragment ions formed during this process. This work shows that CID of molecular ions of CSA and DS with a single free proton produces abundant glycosidic and cross-ring fragments (in hexuronic acid residues) ions. These cross-ring daughter ions are found to be useful in distinguishing GlcA and IdoA residues with a single stage of MS/MS. Production of these ions is sensitive to the precursor charge state and the number of ionized species within the GAG chain.
Methods
CSA and DS Preparation
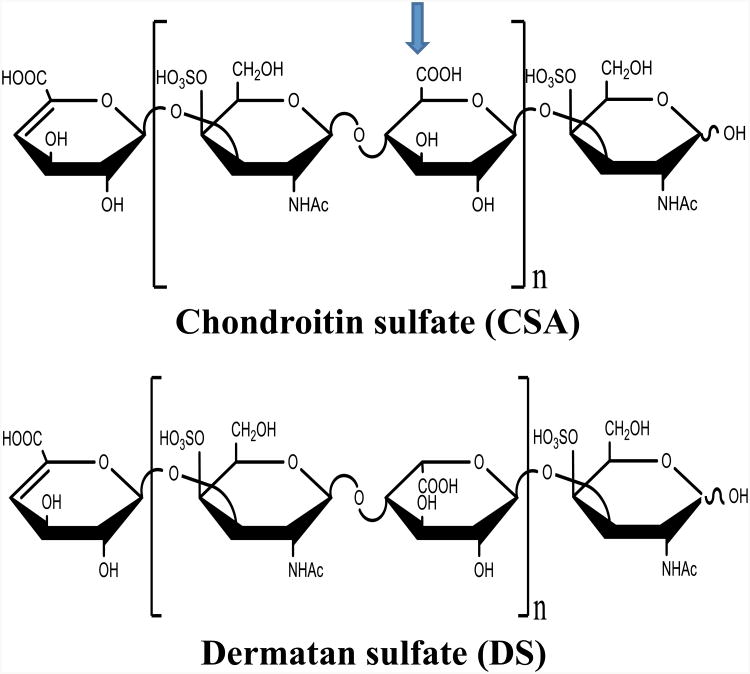
CSA and DS oligosaccharides were independently prepared by partial enzymatic depolymerization. CSA was depolymerized from bovine trachea chondroitin sulfate A (Celsus Laboratories, Cincinnati, OH) while DS used in this work originated from porcine intestinal mucosa dermatan sulfate (Celsus Laboratories, Cincinnati, OH). Chondroitin ABC lyase from Proteus vulgaris, EC 4.2.2.4 (Seikagaku, Japan) was used to incubate 20 mg/mL solution of each sample in 50 mM Tris HCL/60 mM sodium acetate buffer, pH 8 at 37° C. When the UV absorbance at 232 nm indicated 50% complete, the digestion mixture was heated for 3 min at 100° C. Ultra-filtration was carried out using a 5000 MWCO membrane to remove the enzyme and the high-molecular-weight oligosaccharide. Concentrate the remaining oligosaccharide mixture, rotary evaporation was used and then fractionated by low pressure GPC on a Bio-Gen p10 (Bio-Rad, Richmond, CA) column. The oligosaccharide fractions were desalted by GPC on a Bio-Gel P2 column and freeze dried.39 Strong anion exchange high-pressure liquid chromatography (SAX-HPLC) on a semi-preparative SAX S5 Spherisorb column (Waters Corp, Milford, MA) was used for further purification of the oligosaccharides. The resulting SAX-HPLC with over 90% oligosaccharides fractions were collected, desalted by GPC and then freeze-died. The dried solid was reconstituted in water and purified one more time using SAX-HPLC. Only the oligosaccharides within the top 30% of the chromatogram peak was collected, desalted and freeze-dried. Oligomer concentration in the solution was determined by measuring the absorbance at 232 nm (e=3800 M−1 cm−1). The final oligosaccharide fractions were characterized by PAGE, ESI-MS, and high-field NMR spectroscopy.40 The general structures for the molecules used are shown in Figure 1.
Figure 1.
General structures for CSA and DS oligosaccharides. The value of n for the molecules analyzed is 1, 2, 3 and 4 for dp4, 6, 8, and 10 respectively.
Mass Spectrometry Analysis
The experiments were carried out using a 9.4 Bruker Apex Ultra Qh-FTICR instrument (Billerica, MA), with electrospray ionization operating in negative ionization mode. The sample concentrations were 0.05-0.1 mg/mL in 50:50 MeOH:H2O. Oligomers were introduced individually at the rate of 2 uL/min. Between 0.5-1.0 mM NaOH in the spray solution was used to vary the degree of sodiation of the analytes and varying the degree of ionization. Ions of interest were isolated in a mass selective quadrupole using 3 Da isolation window and CID experiments of CSA/DS dp4-dp10 molecular ions with different level of Na+/H+ exchange were performed in the collision cell external to the high magnetic field region. The intensity of the precursor ion was maintained above the fragment ions during CID experiments. The fragment ions were assigned using high resolution accurate mass measurement assisted with Glycoworkbench software.41 Wolff and Amster annotation23 derived from Domon and Costello nomenclature42 was used to designate the identified product ions.
Samples were run in triplicates acquired more than one month apart. The error bars in the graphs represent the standard error of those three measurements. Relative intensity calculations were obtained by dividing the intensity of the diagnostic product ion by the total ion current in the spectrum excluding the precursor.
PCA Analysis
Principal component analysis (PCA) was used to visualize the differences between the spectra of epimeric compounds. PCA was performed using MATLAB (MathWorks, Natick, MA, USA) and the PLS toolbox (Eigenvector Research, Inc., Wenatchee, WA, USA). Five spectra were obtained for each CS and DS oligosaccharide and ion assignment was carried out and the intensity of at least 40 of the assigned fragment ions was used for PCA analysis. Before the PCA analysis, the intensity of the ions was normalized using the base peak of the background spectrum of each respective oligosaccharide chain. A PCA matrix was created in an excel spreadsheet with each sample spectrum represented in a row while the intensity of the particular ions was entered in a given column. The data was mean-centered and cross-validated during the analysis.
Results and Discussion
CSA and DS GAGs differ by the stereochemistry of C5 in their hexuronic acid residues, a feature that plays a key role in their biological function. Although considerable effort has been made to develop methods to distinguish CSA and DS stereoisomers using tandem mass spectrometry, most of published work on these biomolecules investigated molecular ions with the number of ionized acidic groups equal to the number of sulfate groups within the chains. As the sulfate groups are considerably more acidic than carboxyl groups, and are ionized in aqueous solution, it is easy to generate Mn− (where n is equal to the number of sulfate modifications) by electrospray ionization. The molecular ions of CSA and DS often produce ions of identical mass-to-charge, even for different chain lengths, as the charge generally increases in direct proportion to degree of polymerization. Generally, CID of these molecular ions produce B and Y glycosidic fragments, but relatively few cross-ring fragments, thus reducing the extent of structural information that can be derived.24 Use of higher charge state precursors or derivatized CSA/DS molecules eliminate the isobaric nature of these molecules enabling tandem mass spectrometry of mixtures of different chain length of the oligomers.30 Use of higher charge state precursor ions also increases the number of product ions observed. The diagnostic value of the ring fragments obtained from glucuronic acid (GlcA) and iduronic acid (IdoA) residues was investigated for CSA versus DS chains with a degree of polymerization from (dp) 4 to 10.
Diagnostic Ion Criteria
The number of ionized acidic groups in the precursor ion of a GAG is known to play a significant role in controlling the diagnostic value of the product ions 43. Although several precursor ions that vary in the number of ionized groups (charge state and Na+/H+ exchange) were found to produce 2,4A and 0,2X uronic acid fragment ions that exhibit reproducible intensity differences between CSA and DS, the precursor ions that produced all the diagnostic ions were found to have particular characteristics. The best choice for a precursor ion is one in which the charge state was higher than the number of sulfate groups within the chain by one, and also one in which the molecular ion contained a single free acidic hydrogen. Such a precursor was found to produce both A and X diagnostic fragment ions. These results suggest that a single mobile proton plays an important role in the formation of these significant cross-ring fragment products.
Furthermore, even for the molecular ions with the specific characteristics mentioned above, only product ions with a specific charge and sodium level combinations were diagnostic. Diagnostic 2,4An ions had all the acidic groups ionized except one. Whenever any of these ions were produced, regardless of the precursor ion from which they were derived, they were found to be diagnostic. In most cases there were no such products from molecular ions lacking Na+, and their prevalence and diagnostic qualities increased with the number of Na+ present in the molecular ion. Fully ionized precursor ions generally produced fewer fragment ions including the ones that were diagnostic, consistent with the observation that a single mobile proton is necessary for the production of 2,4A and 0,2X products.
Table 1 shows the abundance of 2,4An fragments fitting these criteria for CSA and DS oligomers of dp 4-10. For these samples, the number of ionizable groups is equal to the dp value, and the number of sulfo groups is equal to one-half of the dp value. The appropriate precursor is therefore one whose charge state is (1+dp/2) and that has lost (dp-1) protons, either through ionization or by Na+/H+ exchange. In all cases, the 2,4A3 is less diagnostic than the longer fragment ions, with only a 2-4 times difference in intensity for GlcA versus IdoA. The longer fragments showed a much higher preference for producing a 2,4An fragment for GlcA versus IdoA, and in the case of dp 10, this fragment is only observed for GlcA when n>3.
Table 1.
Diagnostic ion intensities for CSA and DS dp4-dp10. The intensities are reported as percentages of the total ion intensity of the tandem mass spectrum excluding the precursor ion intensity. The ratio column compares the intensity of diagnostic ions for DS to those from CSA. The protons column lists the number of ionizable protons in the product ions.
| dp | Precursor ion | 2,4An | protons | CSA | DS | Ratio |
|---|---|---|---|---|---|---|
| 4 | [M-3H]3− | 2,4A3 | 1 | 17.1 ± 0.7 | 4.7 ± 0.2 | 0.28 |
| 6 | [M-5H+Na]4− | 2,4A3 | 1 | 2.02 ± 0.06 | 0.96 ± 0.04 | 0.48 |
| [2,4A5+Na]2− | 1 | 6.8 ± 0.7 | 0.32 ± 0.02 | 0.05 | ||
| 8 | [M-7H+2Na]5− | 2,4A3 | 1 | 1.17 ± 0.03 | 0.32 ± 0.03 | 0.28 |
| [2,4A5+Na]2− | 1 | 0.64 ± 0.04 | 0.08 ± 0.003 | 0.13 | ||
| 2,4A5+2Na | 1 | 0.11 ± 0.01 | 0.00 | 0.00 | ||
| [2,4A7+2Na]3− | 1 | 2.1 ± 0.2 | 0.07 ± 0.005 | 0.03 | ||
| 10 | [M-9H+3Na]6− | 2,4A3 | 1 | 0.66 ± 0.06 | 0.24 ± 0.01 | 0.36 |
| [2,4A5+Na]2− | 1 | 0.17 ± 0.02 | 0.00 | 0.00 | ||
| [2,4A7+2Na]3− | 1 | 0.29 ± 0.02 | 0.00 | 0.00 | ||
| [2,4A7+3Na]2− | 1 | 0.26 ± 0.01 | 0.00 | 0.00 | ||
| [2,4A9+3Na]4− | 1 | 0.63 ± 0.10 | 0.00 | 0.00 |
For the 0,2Xn fragments that are found to be diagnostic, the ions were either fully ionized or contained a single free acidic proton, especially for longer chains. The ions with a single free proton are also accompanied by another peak resulting from loss of SO3 that is also diagnostic, as will be shown below, in the description of the individual oligomers.
dp4 CSA and DS
CSA/DS tetrasaccharide epimers were the shortest chains that were investigated in this study. They have a total of four acidic groups (2 sulfate and 2 carboxyl groups). In all the CS/DS oligomers described in this study, the non-reducing end hexuronic acid residue is a Δ-uronic acid, that is, it has a double bond between C4 and C5, a result of the enzymatic process that produced these oligomers from the much longer CS or DS material from with they were derived. For this reason, there is only one GlcA or IdoA residue to assign in the dp4 oligomers. The precursor ion with the aforementioned diagnostic characteristics is [M-3H]3−, which is not sodiated. The MS/MS spectrum obtained for the two epimers is shown in Figure 2. For CSA, the most intense product ions are C2/Z2 (isobaric species that are indistinguishable in the tandem mass spectra due to formation of a Δ-uronic acid at the nonreducing end of the Z-fragment), 2,4A3, and Y1−, shown in green in the figure. DS dp4 produces the ions B2−, Y32−, and 0,2X32− as the most intense products, and these are highlighted in red. The differences in intensity of the ions resulting from CSA and those of DS is due to hexuronic acid residue C5 stereochemistry, as that is the only structural difference between the two chains.
Figure 2.
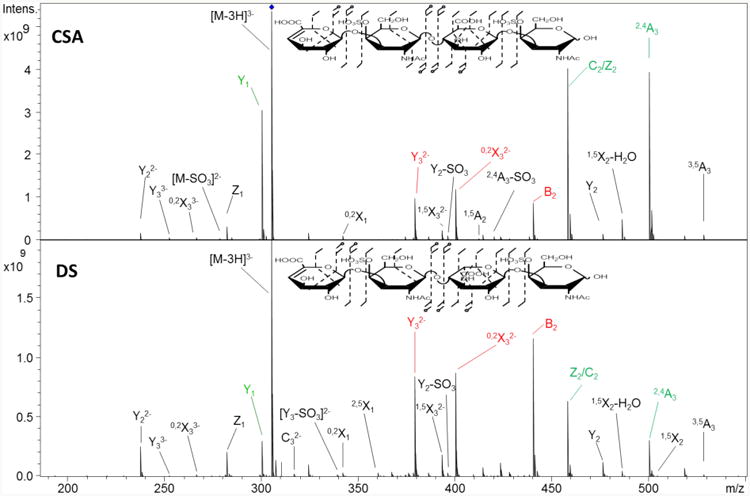
The CID tandem mass spectra for dp4 CSA and DS chains displaying differences in ion abundance and distributions. Product ions are singly-charged unless otherwise indicated in the annotation. The ions highlighted in red have been found to be diagnostic for this chain by other researchers. The fragment ions (shown in green) were found in this work and they are highly diagnostic for CSA and DS. Relative abundance of ion pair B2 and C2/Z2 also differ between the two chains.
The statistical differences between the CSA and DS tandem mass spectra were evaluated by PCA, which clearly distinguishes the CSA and DS CID mass spectra, and assigned over 99% of the differences to a single principal component (supplementary data, S1.) The loadings plot for this analysis (supplementary data, S2), identifies the peaks that are the largest contributors to the differences between the tandem mass spectra of the epimers. One fragment ion that appears here as a diagnostic peak, and also appears for all of the oligomers in this study, is the uronic acid cross-ring fragment, 2,4A3, whose fractional abundance is approximately four times less intense in DS compared to CSA for the dp4 chains, Table 1. (Fractional intensity is the abundance of a peak divided by the sum of the intensities of all other product ions.)
Some of the other distinguishing ions have been observed in the previous work on doubly deprotonated precursors, [M-2H]2− and they follow a similar pattern for this precursor.19, 27 For instance Y1 is more intense in CSA than in DS. Other previously identified diagnostic ions were also present; these include Z1, Y32−, and 0,2X32− although their intensity differences are not very pronounced. The molecular ion investigated here reveals other ions, such as B2, that are more intense in DS than in CSA. Another useful detail that can be used to compare the two isomers is the B2 intensity, which is lower compared to the Z2/C2 fragment ions (these have identical compositions, and cannot be distinguished by their m/z values) in CSA, while in the DS the relative intensity ratio is reversed, as seen in Figure 2d. This can be useful in quick, non-quantitative identification of contamination of a CSA or DS sample.
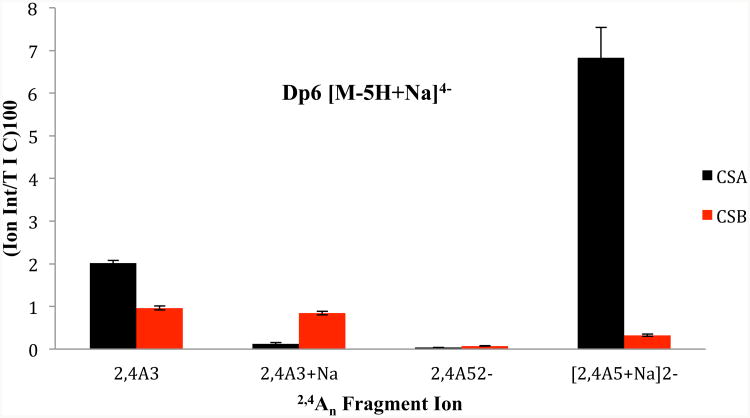
Dp6 CSA and DS
The hexasaccharides have a Δ-uronic acid at the non-reducing end, and two uronic acids with C5 stereochemistry. The precursor ion that is diagnostic is [M-5H+Na]4−, and its CID spectrum is shown in Figure 3. The fragment ion distribution is similar to that observed for dp4. The second most intense fragment in both spectra is [2,4A5+Na]2−, a fragment occurring within the hexuronic acid near the reducing end and that distinguishes GlcA from IdoA (Table 1, Figures 3 and 4). For DS, the notable diagnostic ions include B3+Na, [B4+Na]2−, B2, and C2+Na/Z2+Na. We also observe ions whose intensity differences were previously reported to be diagnostic of GlcA versus IdoA, specifically Y32−, [Y5+Na]3−, [0,2X5+Na]3−.27 PCA of the two mass spectra (CSA versus DS) show that the two sets of spectra resolve from each other with a single principal component for 99.9% of the differences (supplemental data, S3). The loadings plot (S4) shows that many of the ions that differentiate the two sets of spectra are cross-ring fragments, 2,4An and 0,2Xn.
Figure 3.
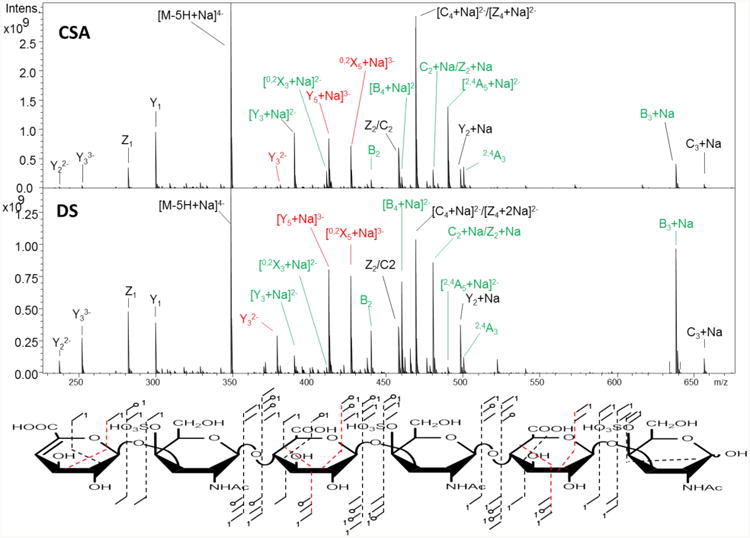
CID tandem mass spectra for dp6 CSA and DS showing ions that differ in intensity between the two chains. Only the ions with intensity differences between the two chains are annotated in the spectra but all the fragment ions found in the two spectra are indicated in the structure below the spectra with 2,4A and 0,2X ions appearing in red. The ions that are shown in red in the spectra have been reported by others in previous studies before, but are found to be less diagnostic of C5 stereochemistry than the ions highlighted in green.
Figure 4.
The graph compares the relative intensity of 2,4An fragments obtained from dp6 CSA and DS with the standard error bar from triplicate runs. Several 2,4An ions are found in the mass spectra, but only 2,4A3 and [2,4A5+Na]2− had the diagnostic qualities discussed in the text. The 2,4A3 fragment ion shows less of a difference between the two uronic acid epimers than other 2,4An ions closer to the reducing end, namely the [2,4A5+Na]2− ion, a feature that is found for the mass spectra of all the chain lengths studied here.
The dp6 diagnostic precursor produces cross-ring fragments in each uronic acid residue that are diagnostic of the uronic acid stereochemistry. [2,4A5+Na]2− has an abundance in DS that is 17 times lower than in CSA, and cross-ring fragment 2,4A3 has half the intensity in DS than in CSA, Table 1. These data, and other shown below suggest that the closer the 2,4An fragment is to the reducing end, the more diagnostic it appears to be, that is, the larger the difference in intensity of this cross ring fragment between CSA and DS. Figure 4 compares the behavior of the 2,4An product ions that fit the requirement of having one ionizable proton, versus those with more or fewer such protons. The fragment ion, [2,4A3+Na]−, has no ionizable protons, and is actually more abundant for IdoA than for GlcA (i.e. DS versus CSA). The fragment ion, 2,4A52−, has two ionizable protons, and it is found in both the CSA and DS dp6 tandem mass spectra, but at very low intensity.
Dp8 CSA and DS
The dp8 precursor ion [M-7H+2Na]5− fits the criteria for producing diagnostic ions, and yields mass spectra with slightly different distributions of ions from dp4 and dp6 (mass spectra are found in supplemental data, S5). The most intense peak corresponds to the isobaric glycosidic fragment ions [Z6+2Na]3−/[C6+2Na]3−. The next most intense fragments are from glycosidic B and Y fragments. The data from the mass spectra were analyzed by PCA, and CSA segregates from DS with two principal components that account for 79% and 20%, respectively, of the differences between the spectra (supplemental data, S6). The loadings plot (S7) finds several peaks that contribute to the differences, including glycosidic and cross-ring cleavages. 2,4An peaks and 0,2Xn peaks are significant contributors. There are cross-ring fragment ions exhibiting diagnostic characteristics in every uronic acid except the non-reducing end residue, and their intensities are compared graphically in Figure 5a. The diagnostic species are 2,4A3−, [2,4A5+Na]2−, [2,4A5+2Na]− and [2,4A7+2Na]3−, all of which contain a single ionizable proton. The ratio of these fragment ions in DS and CSA spectra can be found in Table 1. Consistent with the finding for dp6, the closer these cross ring fragments are to the reducing end, the larger the difference revealed by the ratio of intensities for IdoA versus GlcA. As expected, the other 2,4An cross-ring fragment ions that do not contain a single ionizable proton do not show diagnostic qualities, as can be seen in Figure 5. For instance the [2,4A3+Na]− ion, with no ionizable protons, shows the opposite behavior to the other diagnostic ions, that is, it is more intense for DS than for CSA. The [2,4A5+2Na]2− ion also has no ionizable protons, and does not follow the trend of the diagnostic ions, while the [2,4A5+Na]2− ion has one ionizable proton, and is diagnostic of the presence of GlcA versus IdoA. Figure 5b shows an expansion of the mass scale highlighting the region where these cross-ring products are found, emphasizing the utility of selecting the appropriate product ions for assigning uronic acid stereochemistry.
Figure 5.

(a) A graph of the relative intensities of 2,4An CID fragments obtained from the two dp8 chains. The ions indicated with an arrow have the diagnostic qualities discussed in the text. (b). Mass scale expansion highlighting the 2,4An fragments obtained in CSA and DS with [2,4A7 +2Na]3− almost exclusively in CSA. (c). A structure with the fragment ions obtained from [M-7H+2Na]5− precursor. 2,4A and 0,2X fragments are shown in red within the structure.
Dp 10 CSA and DS
CSA and DS dp10 were the longest GAG oligomers analyzed in this study. The precursor ion that fit the requirement for charge state and having a single ionizable proton, and that produced peaks diagnostic of the uronic acid stereochemistry is [M-9H+3Na]6−. As with dp8, the majority of intense fragments are not the ones resulting from cross-ring cleavages but instead B and Y ions (supplemental data, S8). The most intense peak is assigned as [C8+3Na]4−/[Z8+3Na]4−. Although there are plenty of 2,4An and 0,2Xn fragment ions in the hexuronic acid residues of both CSA and DS, there are extremely few ring fragments occurring at the amino sugar for this chain. The annotated structure of dp10 CSA shows only one fragment ion at the reducing end glucosamine residue, Figure 6c.
Figure 6.
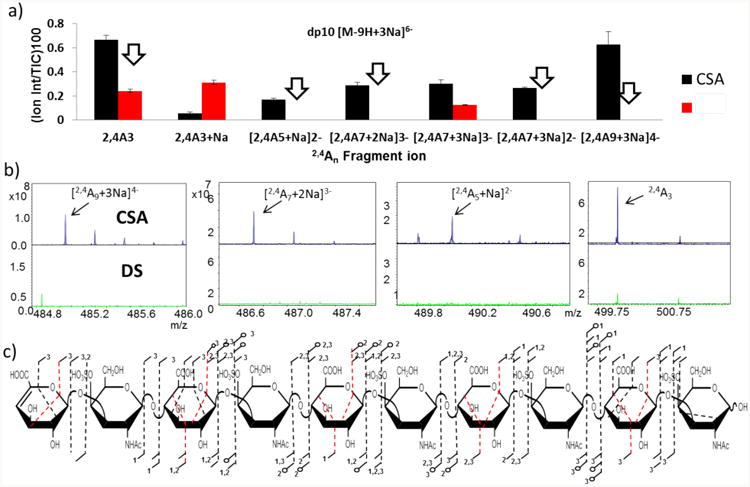
(a). The graph shows the 2,4An ions obtained from the dp10 CSA and DS chains with the ions with the arrows indicating ions with the diagnostic characteristics discussed in the text. With the exception of the 2,4A3, these crossring appeared only in the tandem mass spectrum of CSA and were absent for DS. (b).The zoom in of some of these particular diagnostic ions is shown below the graph. (c). Structure with fragment ions obtained from the diagnostic precursor ion with the 2,4A and 0,2X fragments shown in red.
The 2,4An fragments for the dp 10 are highly diagnostic, in that most appear only in the tandem mass spectrum of CSA, and are absent for DS, Table 1. Figure 6b presents mass scale expansions of the regions where the diagnostic 2,4An fragments appear. Only the 2,4A3 shows a peak in the DS spectrum, but it is three times less intense in the later spectrum. In contrast, 2,4An fragment ions that have more or fewer than a single ionizable proton are not diagnostic of the uronic acid stereochemistry, as shown in Figure 6a for [2,4A3+Na]− (with no ionizable protons) and [2,4A7+3Na]3− (also with no ionizable protons.)
0,2Xn fragments with no ionizable protons as well as ones with a single ionizable proton are also found to have diagnostic characteristics in the mass spectra of both dp8 and dp10. Figure 7 compares the intensities of several such peaks in the tandem mass spectra of CSA and DS. Many 0,2Xn ions show much higher abundance for GlcA rather than IdoA. Some of these appear in the mass spectra of both oligomers, such as [0,2X3+2Na]−, with no ionizable proton, [0,2X5+2Na]2−, with one ionizable proton, and [0,2X5+2Na]3−, with no ionizable proton. The shortest cross-ring fragment, 0,2X1, with no ionizable proton, was found to be less diagnostic of uronic acid stereochemistry than the longer fragment ions, similar to observation for 2,4An fragment ions. Ions with more than one free acidic proton, such as [0,2X7+2Na]3− (two ionizable protons), from dp10, were not diagnostic.
Figure 7.

Graphs showing the 0,2X fragments found in the CID tandem mass spectra of dp8 and dp10 CSA and DS chains. Product ions with 0 or 1 ionizable protons were found to be diagnostic with few exceptions.
Conclusions
The data presented above show that it is possible to observe significant differences between the CID mass spectra of CSA and DS, specifically by examining the abundance of cross-ring fragmentation in the uronic acid residues. The diagnostic value of the cross-ring products is related to the number of ionizable protons in the selected precursor as well as in the product ions. For 2,4An ions, both the precursor and product must contain a single ionizable proton for the fragment ion to have diagnostic value. Addition of NaOH to the spray solution helps to obtain higher charge states and to remove most of the ionizable protons from the precursor ion by Na+/H+ exchange. 2,4An and 0,2Xn ions were present in all the spectra taken from dp4-dp10 CS but 0,2Xn were found to be less diagnostic of the uronic acid type for dp4 and dp6. This is the first indication that unique CID fragments can distinguish between CSA and DS chains. This approach shows great potential for providing detailed structural information for GAG oligomers using a method of ion activation that is widely available on a number of mass spectrometry platforms.
Supplementary Material
S1. Score Plot for dp4 CSA/DS [M-3H]3−
S2. Loadings Plot for dp4 CSA/DS [M-3H]3−
S3. Score Plot for dp6 CSA/DS [M-5H+Na]4−
S4. Loadings Plot for dp6 CSA/DS [M-5H+Na]4−
S5. Dp8 CID spectra
S6. Score Plot for dp8 CSA/DS [M-7H+2Na]5−
S7. Loadings Plot for dp8 CSA/DS [M-7H+2Na]5−
S8. Dp10 CSDS
S9. Score Plot for dp10 CSA/DS [M-9H+3Na]6−
S10. Loadings Plot for dp10 CSA/DS [M-9H+3Na]6−
Acknowledgments
MJK and IJA acknowledge generous financial support from the National Institutes of Health, grant P41 GM103390.
References
- 1.Sugahara K, Mikami T, Uyama T, Mizuguchi S, Nomura K, Kitagawa H. Recent Advances in the Structural Biology of Chondroitin Sulfate and Dermatan Sulfate. Curr Opin Struct Biol. 2003;13:612. doi: 10.1016/j.sbi.2003.09.011. doi: http://dx.doi.org/http://dx.doi.org/10.1016/j.sbi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Gandhi NS, Mancera RL. The Structure of Glycosaminoglycans and Their Interactions with Proteins. Chem Biol Drug Des. 2008;72:455. doi: 10.1111/j.1747-0285.2008.00741.x. doi: http://dx.doi.org/10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 3.Silbert JE, Sugumaran G. Biosynthesis of Chondroitin/Dermatan Sulfate. IUBMB Life. 2002;54:177. doi: 10.1080/15216540214923. doi: http://dx.doi.org/10.1080/15216540214923. [DOI] [PubMed] [Google Scholar]
- 4.Sisu E, Flangea C, Serb A, Zamfir AD. Modern Developments in Mass Spectrometry of Chondroitin and Dermatan Sulfate Glycosaminoglycans. Amino Acids. 2011;41:235. doi: 10.1007/s00726-010-0682-4. doi: http://dx.doi.org/10.1007/s00726-010-0682-4. [DOI] [PubMed] [Google Scholar]
- 5.Akatsu C, Mizumoto S, Kaneiwa T, Maccarana M, Malmstrom A, Yamada S, Sugahara K. Dermatan Sulfate Epimerase 2 Is the Predominant Isozyme in the Formation of the Chondroitin Sulfate/Dermatan Sulfate Hybrid Structure in Postnatal Developing Mouse Brain. Glycobiology. 2011;21:565. doi: 10.1093/glycob/cwq208. doi: http://dx.doi.org/10.1093/glycob/cwq208. [DOI] [PubMed] [Google Scholar]
- 6.Shriver Z, Liu D, Sasisekharan R. Emerging Views of Heparan Sulfate Glycosaminoglycan Structure/Activity Relationships Modulating Dynamic Biological Functions. Trends Cardiovasc Med. 2002;12:71. doi: 10.1016/s1050-1738(01)00150-5. [DOI] [PubMed] [Google Scholar]
- 7.Mitsunaga C, Mikami T, Mizumoto S, Fukuda J, Sugahara K. Chondroitin Sulfate/Dermatan Sulfate Hybrid Chains in the Development of Cerebellum - Spatiotemporal Regulation of the Expression of Critical Disulfated Disaccharides by Specific Sulfotransferases. J Biol Chem. 2006;281:18942. doi: 10.1074/jbc.M510870200. doi: http://dx.doi.org/10.1074/jbc.M510870200. [DOI] [PubMed] [Google Scholar]
- 8.Bao XF, Nishimura S, Mikami T, Yamada S, Itoh N, Sugahara K. Chondroitin Sulfate/Dermatan Sulfate Hybrid Chains from Embryonic Pig Brain, Which Contain a Higher Proportion of L-Iduronic Acid Than Those from Adult Pig Brain, Exhibit Neuritogenic and Growth Factor Binding Activities. J Biol Chem. 2004;279:9765. doi: 10.1074/jbc.M310877200. doi: http://dx.doi.org/10.1074/jbc.M310877200. [DOI] [PubMed] [Google Scholar]
- 9.Dundar M, Muller T, Zhang Q, Pan J, Steinmann B, Vodopiutz J, Gruber R, Sonoda T, Krabichler B, Utermann G, Baenziger JU, Zhang LJ, Janecke AR. Loss of Dermatan-4-Sulfotransferase 1 Function Results in Adducted Thumb-Clubfoot Syndrome. Am J Hum Genet. 2009;85:873. doi: 10.1016/j.ajhg.2009.11.010. doi: http://dx.doi.org/10.1016/j.ajhg.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou S, Maccarana M, Min TH, Strate I, Pera EM. The Secreted Serine Protease Xhtra1 Stimulates Long-Range Fgf Signaling in the Early Xenopus Embryo. Dev Cell. 2007;13:226. doi: 10.1016/j.devcel.2007.07.001. doi: http://dx.doi.org/http://dx.doi.org/10.1016/j.devcel.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Vicente CP, He L, Pavão MS, Tollefsen DM. Antithrombotic Activity of Dermatan Sulfate in Heparin Cofactor Ii-Deficient Mice. Blood. 2004;104:3965. doi: 10.1182/blood-2004-02-0598. doi: http://dx.doi.org/10.1182/blood-2004-02-0598. [DOI] [PubMed] [Google Scholar]
- 12.Plaas AH, West LA, Thonar EJ, Karcioglu ZA, Smith CJ, Klintworth GK, Hascall VC. Altered Fine Structures of Corneal and Skeletal Keratan Sulfate and Chondroitin/Dermatan Sulfate in Macular Corneal Dystrophy. J Biol Chem. 2001;276:39788. doi: 10.1074/jbc.M103227200. doi: http://dx.doi.org/10.1074/jbc.M103227200. [DOI] [PubMed] [Google Scholar]
- 13.Yamada S, Sugahara K. Potential Therapeutic Application of Chondroitin Sulfate/Dermatan Sulfate. Curr Drug Disc Technol. 2008;5:289. doi: 10.2174/157016308786733564. doi: http://dx.doi.org/10.2174/157016308786733564. [DOI] [PubMed] [Google Scholar]
- 14.Sudo M, Sato K, Chaidedgumjorn A, Toyoda H, Toida T, Imanari T. 1h Nuclear Magnetic Resonance Spectroscopic Analysis for Determination of Glucuronic and Iduronic Acids in Dermatan Sulfate, Heparin, and Heparan Sulfate. Anal Biochem. 2001;297:42. doi: 10.1006/abio.2001.5296. doi: http://dx.doi.org/10.1006/abio.2001.5296. [DOI] [PubMed] [Google Scholar]
- 15.Casu B, Torri G. Structural Characterization of Low Molecular Weight Heparins. Semin Thromb Hemost. 1999;25 Suppl 3:17. [PubMed] [Google Scholar]
- 16.Scott JE, Heatley F. Detection of Secondary Structure in Glycosaminoglycans Via the H N.M.R. Signal of the Acetamido Nh Group. Biochem J. 1982;207:139. doi: 10.1042/bj2070139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kailemia MJ, Li L, Ly M, Linhardt RJ, Amster IJ. Complete Mass Spectral Characterization of a Synthetic Ultralow-Molecular-Weight Heparin Using Collision-Induced Dissociation. Anal Chem. 2012;84:5475. doi: 10.1021/ac3015824. doi: http://dx.doi.org/10.1021/ac3015824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kailemia MJ, Li L, Xu Y, Liu J, Linhardt RJ, Amster IJ. Structurally Informative Tandem Mass Spectrometry of Highly Sulfated Natural and Chemoenzymatically Synthesized Heparin and Heparan Sulfate Glycosaminoglycans. Mol Cell Proteomics. 2013;12:979. doi: 10.1074/mcp.M112.026880. doi: http://dx.doi.org/10.1074/mcp.M112.026880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leach FE, III, Ly M, Laremore TN, Wolff JJ, Perlow J, Linhardt RJ, Amster IJ. Hexuronic Acid Stereochemistry Determination in Chondroitin Sulfate Glycosaminoglycan Oligosaccharides by Electron Detachment Dissociation. J Am Soc Mass Spectrom. 2012;23:1488. doi: 10.1007/s13361-012-0428-5. doi: http://dx.doi.org/10.1007/s13361-012-0428-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolff JJ, Amster IJ, Chi L, Linhardt RJ. Electron Detachment Dissociation of Glycosaminoglycan Tetrasaccharides. J Am Soc Mass Spectrom. 2007;18:234. doi: 10.1016/j.jasms.2006.09.020. doi: http://dx.doi.org/10.1016/j.jasms.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolff JJ, Chi L, Linhardt RJ, Amster IJ. Distinguishing Glucuronic from Iduronic Acid in Glycosaminoglycan Tetrasaccharides by Using Electron Detachment Dissociation. Anal Chem. 2007;79:2015. doi: 10.1021/ac061636x. doi: http://dx.doi.org/10.1021/ac061636x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolff JJ, Laremore TN, Busch AM, Linhardt RJ, Amster IJ. Influence of Charge State and Sodium Cationization on the Electron Detachment Dissociation and Infrared Multiphoton Dissociation of Glycosaminoglycan Oligosaccharides. J Am Soc Mass Spectrom. 2008;19:790. doi: 10.1016/j.jasms.2008.03.010. doi: http://dx.doi.org/10.1016/j.jasms.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolff JJ, Laremore TN, Busch AM, Linhardt RJ, Amster IJ. Electron Detachment Dissociation of Dermatan Sulfate Oligosaccharides. J Am Soc Mass Spectrom. 2008;19:294. doi: 10.1016/j.jasms.2007.10.007. doi: http://dx.doi.org/10.1016/j.jasms.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bielik AM, Zaia J. Multistage Tandem Mass Spectrometry of Chondroitin Sulfate and Dermatan Sulfate. Int J Mass Spectrom. 2011;305:131. doi: 10.1016/j.ijms.2010.10.017. doi: http://dx.doi.org/http://dx.doi.org/10.1016/j.ijms.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller MJC, Costello CE, Malmström A, Zaia J. A Tandem Mass Spectrometric Approach to Determination of Chondroitin/Dermatan Sulfate Oligosaccharide Glycoforms. Glycobiology. 2006;16:502. doi: 10.1093/glycob/cwj093. doi: http://dx.doi.org/10.1093/glycob/cwj093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saad O, Leary J. Delineating Mechanisms of Dissociation for Isomeric Heparin Disaccharides Using Isotope Labeling and Ion Trap Tandem Mass Spectrometry. J Am Soc Mass Spectrom. 2004;15:1274. doi: 10.1016/j.jasms.2004.05.008. doi: http://dx.doi.org/10.1016/j.jasms.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Zaia J, Li XQ, Chan SY, Costello CE. Tandem Mass Spectrometric Strategies for Determination of Sulfation Positions and Uronic Acid Epimerization in Chondroitin Sulfate Oligosaccharides. J Am Soc Mass Spectrom. 2003;14:1270. doi: 10.1016/S1044-0305(03)00541-5. doi: http://dx.doi.org/10.1016/s1044-0305(03)00541-5. [DOI] [PubMed] [Google Scholar]
- 28.Zaia J, McClellan JE, Costello CE. Tandem Mass Spectrometric Determination of the 4s/6s Sulfation Sequence in Chondroitin Sulfate Oligosaccharides. Anal Chem. 2001;73:6030. doi: 10.1021/ac015577t. doi: http://dx.doi.org/10.1021/ac015577t. [DOI] [PubMed] [Google Scholar]
- 29.Wolff JJ, Amster IJ, Chi L, Linhardt RJ. Electron Detachment Dissociation of Glycosaminoglycan Tetrasaccharides. J Am Soc Mass Spectrom. 2007;18:234. doi: 10.1016/j.jasms.2006.09.020. doi: http://dx.doi.org/10.1016/j.jasms.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaia J, Li XQ, Chan SY, Costello CE. Tandem Mass Spectrometric Strategies for Determination of Sulfation Positions and Uronic Acid Epimerization in Chondroitin Sulfate Oligosaccharides. J Am Soc Mass Spectrom. 2003;14:1270. doi: 10.1016/S1044-0305(03)00541-5. doi: http://dx.doi.org/10.1016/s1044-0305(03)00541-5. [DOI] [PubMed] [Google Scholar]
- 31.Zaia J, Miller MJC, Seymour JL, Costello CE. The Role of Mobile Protons in Negative Ion Cid of Oligosaccharides. J Am Soc Mass Spectrom. 2007;18:952. doi: 10.1016/j.jasms.2007.01.016. doi: http://dx.doi.org/10.1016/j.jasms.2007.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zamfir A, Seidler DG, Kresse H, Peter-Katalinić J. Structural Characterization of Chondroitin/Dermatan Sulfate Oligosaccharides from Bovine Aorta by Capillary Electrophoresis and Electrospray Ionization Quadrupole Time-of-Flight Tandem Mass Spectrometry. Rapid Commun Mass Spectrom. 2002;16:2015. doi: 10.1002/rcm.820. doi: http://dx.doi.org/10.1002/rcm.820. [DOI] [PubMed] [Google Scholar]
- 33.Zamfir A, Seidler DG, Kresse H, Peter-Katalinić J. Structural Investigation of Chondroitin/Dermatan Sulfate Oligosaccharides from Human Skin Fibroblast Decorin. Glycobiology. 2003;13:733. doi: 10.1093/glycob/cwg086. doi: http://dx.doi.org/10.1093/glycob/cwg086. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Z, Ogden S, Leary JA. Linkage Position Determination in Oligosaccharides: Mass Spectrometry (Ms/Ms) Study of Lithium-Cationized Carbohydrates. J Org Chem. 1990;55:5444. doi: http://dx.doi.org/10.1021/jo00307a011. [Google Scholar]
- 35.Wolff JJ, Laremore TN, Busch AM, Linhardt RJ, Amster IJ. Influence of Charge State and Sodium Cationization on the Electron Detachment Dissociation and Infrared Multiphoton Dissociation of Glycosaminoglycan Oligosaccharides. J Am Soc Mass Spectrom. 2008;19:790. doi: 10.1016/j.jasms.2008.03.010. doi: http://dx.doi.org/10.1016/j.jasms.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zamfir A, Seidler DG, Kresse H, Peter-Katalinic J. Structural Investigation of Chondroitin/Dermatan Sulfate Oligosaccharides from Human Skin Fibroblast Decorin. Glycobiology. 2003;13:733. doi: 10.1093/glycob/cwg086. doi: http://dx.doi.org/10.1093/glycob/cwg086. [DOI] [PubMed] [Google Scholar]
- 37.Ly M, Leach FE, III, Laremore TN, Toida T, Amster IJ, Linhardt RJ. The Proteoglycan Bikunin Has a Defined Sequence. Nat Chem Biol. 2011;7:827. doi: 10.1038/nchembio.673. doi: http://dx.doi.org/10.1038/nchembio.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hitchcock AM, Costello CE, Zaia J. Glycoform Quantification of Chondroitin/Dermatan Sulfate Using a Liquid Chromatography-Tandem Mass Spectrometry Platform. Biochemistry. 2006;45:2350. doi: 10.1021/bi052100t. doi: http://dx.doi.org/10.1021/bi052100t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pervin A, Gallo C, Jandik KA, Han XJ, Linhardt RJ. Preparation and Structural Characterization of Large Heparin-Derived Oligosaccharides. Glycobiology. 1995;5:83. doi: 10.1093/glycob/5.1.83. [DOI] [PubMed] [Google Scholar]
- 40.Muñoz E, Xu D, Avci F, Kemp M, Liu J, Linhardt RJ. Enzymatic Synthesis of Heparin Related Polysaccharides on Sensor Chips: Rapid Screening of Heparin–Protein Interactions. Biochem Biophys Res Commun. 2006;339:597. doi: 10.1016/j.bbrc.2005.11.051. doi: http://dx.doi.org/10.1016/j.bbrc.2005.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ceroni A, Maass K, Geyer H, Geyer R, Dell A, Haslam SM. Glycoworkbench: A Tool for the Computer-Assisted Annotation of Mass Spectra of Glycans†. J Proteome Res. 2008;7:1650. doi: 10.1021/pr7008252. doi: http://dx.doi.org/10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- 42.Domon B, Costello CE. A Systematic Nomenclature for Carbohydrate Fragmentations in Fab-Ms Ms Spectra of Glycoconjugates. Glycoconjugate J. 1988;5:397. doi: http://dx.doi.org/10.1007/Bf01049915. [Google Scholar]
- 43.Kailemia MJ, Li L, Xu Y, Liu J, Linhardt RJ, Amster IJ. Structurally Informative Tandem Mass Spectrometry of Highly Sulfated Natural and Chemoenzymatically Synthesized Heparin and Heparan Sulfate Glycosaminoglycans. Mol Cell Proteomics. 2013;12:979. doi: 10.1074/mcp.M112.026880. doi: http://dx.doi.org/10.1074/mcp.M112.026880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1. Score Plot for dp4 CSA/DS [M-3H]3−
S2. Loadings Plot for dp4 CSA/DS [M-3H]3−
S3. Score Plot for dp6 CSA/DS [M-5H+Na]4−
S4. Loadings Plot for dp6 CSA/DS [M-5H+Na]4−
S5. Dp8 CID spectra
S6. Score Plot for dp8 CSA/DS [M-7H+2Na]5−
S7. Loadings Plot for dp8 CSA/DS [M-7H+2Na]5−
S8. Dp10 CSDS
S9. Score Plot for dp10 CSA/DS [M-9H+3Na]6−
S10. Loadings Plot for dp10 CSA/DS [M-9H+3Na]6−