Abstract
Recently, the kelch-like protein 3 (KLHL3)–Cullin3 complex was identified as an E3 ubiquitin ligase for with no lysine (WNK) kinases, and the impaired ubiquitination of WNK4 causes pseudohypoaldosteronism type II (PHAII), a hereditary hypertensive disease. However, the involvement of WNK kinase regulation by ubiquitination in situations other than PHAII has not been identified. Previously, we identified the WNK3–STE20/SPS1-related proline/alanine-rich kinase–Na/K/Cl cotransporter isoform 1 phosphorylation cascade in vascular smooth muscle cells and found that it constitutes an important mechanism of vascular constriction by angiotensin II (AngII). In this study, we investigated the involvement of KLHL proteins in AngII-induced WNK3 activation of vascular smooth muscle cells. In the mouse aorta and mouse vascular smooth muscle (MOVAS) cells, KLHL3 was not expressed, but KLHL2, the closest homolog of KLHL3, was expressed. Salt depletion and acute infusion of AngII decreased KLHL2 and increased WNK3 levels in the mouse aorta. Notably, the AngII-induced changes in KLHL2 and WNK3 expression occurred within minutes in MOVAS cells. Results of KLHL2 overexpression and knockdown experiments in MOVAS cells confirmed that KLHL2 is the major regulator of WNK3 protein abundance. The AngII-induced decrease in KLHL2 was not caused by decreased transcription but increased autophagy-mediated degradation. Furthermore, knockdown of sequestosome 1/p62 prevented the decrease in KLHL2, suggesting that the mechanism of KLHL2 autophagy could be selective autophagy mediated by sequestosome 1/p62. Thus, we identified a novel component of signal transduction in AngII-induced vascular contraction that could be a promising drug target.
Keywords: hypertension, vascular, angiotensin
Recently, the kelch-like protein 3 (KLHL3) and Cullin3 (Cul3) were identified as the genes responsible for a hereditary hypertensive disease—pseudohypoaldosteronism type II (PHAII).1 Because the with no lysine (WNK) kinases (WNK1 and WNK4) had already been identified as the responsible genes for PHAII and the KLHL proteins were known to serve as substrate adaptors of Cul3-based E3 ubiquitin ligase,1–3 we speculated and recently showed that KLHL3 functions as an E3 ligase with Cul3 for WNK4 and that the impaired ubiquitination of WNK4 and its subsequent increase within the cell stimulates the downstream OSR1/STE20/SPS1-related proline/alanine-rich kinase (SPAK)–NaCl cotransporter signaling and causes PHAII.4 In addition to WNK4, WNK1 and other WNK kinases (WNK2 and WNK3) have been identified as substrates of KLHL3–Cul3 E3 ligase, because KLHL3 can bind to all WNKs in a highly conserved domain (acidic domain).5 Furthermore, we recently reported that KLHL2 possesses a kelch repeat domain (WNK binding domain) highly similar to that of KLHL3 and that KLHL2–Cul3 also functions as an E3 ligase for all WNK kinases.6 These data strongly suggest that any combination of KLHL2/KLHL3 with WNK1/WNK2/WNK3/WNK4 could occur in various types of cells and that KLHL-mediated regulation of WNK kinase abundance by ubiquitination could be involved in the mechanisms of WNK kinase regulation under various pathophysiologic conditions other than PHAII. To date, we have identified several upstream regulators for WNK kinases, including hormonal factors, such as aldosterone, angiotensin II (AngII), insulin, and vasopressin.7–11 However, the mechanisms of signal transduction from these factors to WNK kinases have not necessarily been clarified. We speculated that one of these regulations could be mediated by the ubiquitination of WNKs by KLHL2 and/or KLHL3.
We recently identified WNK3–SPAK–Na/K/Cl cotransporter isoform 1 (NKCC1) signaling in mouse arteries, which plays an important role in the regulation of vascular smooth muscle cell contractions with AngII.12 Influx of Cl− through NKCC1 changes membrane depolarization and leads to voltage-gated Ca channel activation, which causes the elevation of peripheral resistance.13 SPAK regulates NKCC1 activity by its phosphorylation in its amino terminal domain,14 and SPAK is regulated by the phosphorylation by WNK3 as well as WNK1.15 Although we have clarified that WNK3 is necessary for SPAK–NKCC1 activation by AngII using WNK3 knockout mice, little is known about how AngII activates WNK3–SPAK–NKCC1 signaling.
In this study, we identified that KLHL2 and not KLHL3 was expressed in mouse aorta and vascular smooth muscle cells. We also showed that AngII rapidly decreased KLHL2 protein abundance, thereby causing the increase in WNK3 and its downstream activation. Interestingly, the rapid decrease in KLHL2 by AngII was mediated by autophagy-mediated degradation of KLHL2, which could be a novel mechanism in KLHL and WNK regulations as well as AngII-induced signal transduction.
Results
KLHL2 and Not KLHL3 Is Expressed in the Mouse Aorta and Mouse Vascular Smooth Muscle Cells
We have reported that KLHL2 is the closest homolog to KLHL3 among >40 members of the KLHL proteins and that KLHL2 could interact with all WNK kinases, including WNK3.6 Furthermore, WNK3 showed the highest binding affinity with KLHL2 and KLHL3 among the WNK kinases, suggesting that WNK3 as well as WNK4 could be regulated by KLHL2 or KLHL3 in its signaling cascade.6 To investigate the role of KLHL2 and KLHL3 in WNK3 signaling in vascular smooth muscle cells, we first examined whether KLHL2 and KLHL3 were expressed in the mouse aorta and mouse vascular smooth muscle (MOVAS) cells by RT-PCR. As shown in Figure 1A, KLHL2 was expressed in the mouse aorta and MOVAS cells; however, KLHL3 mRNA was not detected. In the immunoblots of aorta and MOVAS cells, the anti-KLHL2/KLHL3 antibody detected double bands around 65 kD, whereas the KLHL3-specific antibody did not detect any signal (Figure 1B). The KLHL2 knockdown by siRNA decreased both bands detected by the anti-KLHL2/KLHL3 antibody (Figure 1C). On the basis of these data, we concluded that the double bands detected by the anti-KLHL2/KLHL3 antibody in the mouse aorta and MOVAS cells were KLHL2. The lower band may be the major KLHL2 band that corresponds to cloned KLHL2 cDNA because the overexpressed KLHL2 had a 3XFLAG tag (Figure 1B). Although the nature of the upper band remains to be determined, we can exclude the possibility of protein modifications by phosphorylation, glycosylation, and neddylation by the experiments using phosphatase, glycosidase, and MLN4924 (data not shown). With these results, we focused on KLHL2 in WNK3 regulation in the mouse aorta and MOVAS cells.
Figure 1.
KLHL2 and not KLHL3 is expressed in mouse aorta and MOVAS cells. (A) Expression of KLHL2 and KLHL3 in the mouse aorta and MOVAS cells was investigated by RT-PCR. KLHL2 was expressed in the mouse aorta and MOVAS cells, although KLHL3 mRNA was not detected. (B) Immunoblots of KLHL2 and KLHL3 in the mouse aorta and MOVAS cells. 3XFLAG-tagged KLHL2 and KLHL3 that were overexpressed in MOVAS cells were used as controls. The anti-KLHL2/KLHL3 antibody recognized both 3XFLAG-tagged KLHL2 and KLHL3 and detected double bands around 65 kD in the aorta and MOVAS cells. The anti-KLHL3 antibody recognized only 3XFLAG KLHL3 but detected no band in the aorta and MOVAS cells. (C) Immunoblot with the anti-KLHL2/KLHL3 antibody in KLHL2 knockdown MOVAS cells. The KLHL2 knockdown by siRNA decreased both bands. GAPDH, glyceraldehyde-3-phosphate dehydrogenase
Salt Depletion and Acute AngII Infusion Decrease KLHL2 and Increase WNK3 in the Mouse Aorta
We have reported that the regulation of the WNK3–SPAK–NKCC1 phosphorylation cascade by AngII in arteries is important for the maintenance of BP in cases of salt depletion.12 However, how AngII regulated WNK3 remains unclear. Previously, we could not detect WNK3 protein in the mouse aorta with any WNK3 antibodies that were available at that time. However, we have recently improved the method of detecting the WNK3 protein in tiny mouse aortic tissues (Supplemental Figure 1). As shown in Figure 2, A and B, a high-salt diet and a low-salt diet decreased and increased WNK3 protein levels in mouse aortas, respectively. In contrast, KLHL2 was increased and decreased by a high-salt diet and a low-salt diet, respectively. As shown in Figure 2, C and D, the WNK3 protein level in the aortas of mice that were infused with AngII was increased and that of KLHL2 was decreased. These results indicate that salt intakes and AngII regulated WNK3 and KLHL2 protein levels in a reciprocal manner.
Figure 2.
Dietary salt and acute AngII infusion decrease KLHL2 and increase WNK3 in mouse aorta. (A) Representative immunoblots and (B) densitometry analyses (n=6) of KLHL2 and WNK3 in aortas from mice fed high-, normal-, and low-salt diets. KLHL2 in mouse aorta was decreased by a low-salt diet and increased by a high-salt diet. WNK3 in the mouse aorta was increased by a low-salt diet and decreased by a high-salt diet. (C) Representative immunoblots and (D) densitometry analyses (n=4) of KLHL2 and WNK3 in aortas from mice at 30 minutes after AngII infusion. Acute AngII infusion decreased KLHL2 and increased WNK3. **P<0.01.
AngII Rapidly Regulates KLHL2 and WNK3 Protein Levels and SPAK–NKCC1 Phosphorylation Cascade in MOVAS Cells
To investigate whether the regulation of WNK3 and KLHL2 by AngII that was observed in the aorta was, indeed, involved in the regulation of WNK3–SPAK–NKCC1 signaling, we performed cell culture studies using MOVAS cells. As shown in Figure 3, AngII rapidly decreased the KLHL2 protein levels in minutes, and WNK3, phosphorylated SPAK, and phosphorylated NKCC1 levels were also increased concomitantly. RT-PCR (Figure 4) revealed that both salt depletion in the mouse aorta and AngII treatment in MOVAS cells did not affect WNK3 mRNA levels, indicating that both regulatory increases in the WNK3 protein were not caused by the increased transcription of WNK3. Therefore, we then investigated the involvement of KLHL2 in the rapid regulation of WNK3 protein levels by AngII.
Figure 3.
AngII rapidly decreases KLHL2 and increases WNK3–SPAK–NKCC1 phosphorylation cascade in MOVAS cells. (A) Representative immunoblots and (B) densitometry analyses (n=5) of KLHL2, WNK3, phosphorylated SPAK (pSPAK), and phosphorylated NKCC1 (pNKCC1) in MOVAS cells treated by AngII. In 1 minute after AngII treatment, KLHL2 was decreased, and the WNK3–SPAK–NKCC1 phosphorylation cascade was activated. After 10 minutes, the tendency was more prominent. *P<0.05; **P<0.01.
Figure 4.
WNK3 mRNA levels in mouse aortas and MOVAS cells do not change with both salt intake and AngII. (A) WNK3 mRNA levels in mouse aortas under high-, regular-, and low-salt diets analyzed by quantitative RT-PCR. WNK3 mRNA levels in mouse aortas did not change with salt intake (n=11). (B) WNK3 mRNA levels of AngII-treated MOVAS cells were analyzed by quantitative RT-PCR. WNK3 mRNA levels in MOVAS did not increase with AngII treatment (n=3).
Although we have previously shown using WNK3 knockout mice that the AngII-induced activation of SPAK–NKCC1 signaling in the aorta was totally dependent on WNK3,12 we also investigated the involvement of WNK1 and WNK4 in this regulation. However, WNK4 mRNA and protein were not detected in the mouse aorta and MOVAS cells (Supplemental Figure 2A). Therefore, whether WNK4 undergoes similar regulation by AngII remains to be determined. In contrast, the WNK1 protein was detected in the mouse aorta and MOVAS cells, and similar to WNK3, it was decreased by a high-salt diet and increased by a low-salt diet and AngII infusion (Supplemental Figure 2B). However, the AngII-induced phosphorylation of SPAK and NKCC1 was diminished in WNK3 knockdown MOVAS cells but not WNK1 knockdown cells (Supplemental Figure 3), confirming the dominant role of WNK3 in AngII-induced SPAK–NKCC1 signaling in the aorta and vascular smooth muscle cells.
KLHL2 Regulates WNK3 Protein Levels in MOVAS Cells
To investigate whether the rapid regulation of WNK3 protein abundance was caused by KLHL2, we performed knockdown and overexpression experiments for KLHL2 in MOVAS cells. As shown in Figure 5, WNK3 was increased in KLHL2 knockdown cells, and the phosphorylation of SPAK and NKCC1 was also increased. However, KLHL2 overexpression decreased WNK3 protein levels and the phosphorylation of SPAK and NKCC1; however, Cul3 overexpression alone did not affect them. The inactivation of the WNK3–SPAK–NKCC1 cascade was more evident in both KLHL2- and Cul3-overexpressing cells (Figure 6), suggesting the importance of the complex formation of KLHL2–Cul3 as an E3 ligase in WNK3 regulation.6 Thus, we performed a Cul3 knockdown experiment. As shown in Figure 7, Cul3 knockdown increased WNK3 protein under basal and AngII-stimulated conditions, showing the involvement of Cul3 in WNK3 degradation. Interestingly, KLHL2 protein was also increased by Cul3 knockdown, suggesting that KLHL2 protein may be regulated by Cul3-based E3 ligase as shown for KLHL3.5 However, such an increase of KLHL2 did not lead to a reduction in WNK3 protein under the Cul3 knockdown condition, clearly suggesting that Cul3 is necessary for KLHL2-mediated WNK3 degradation. AngII could still decrease KLHL2 under the Cul3 knockdown condition (Figure 7), suggesting that the increased degradation of KLHL2 by AngII may be independent of Cul3.
Figure 5.
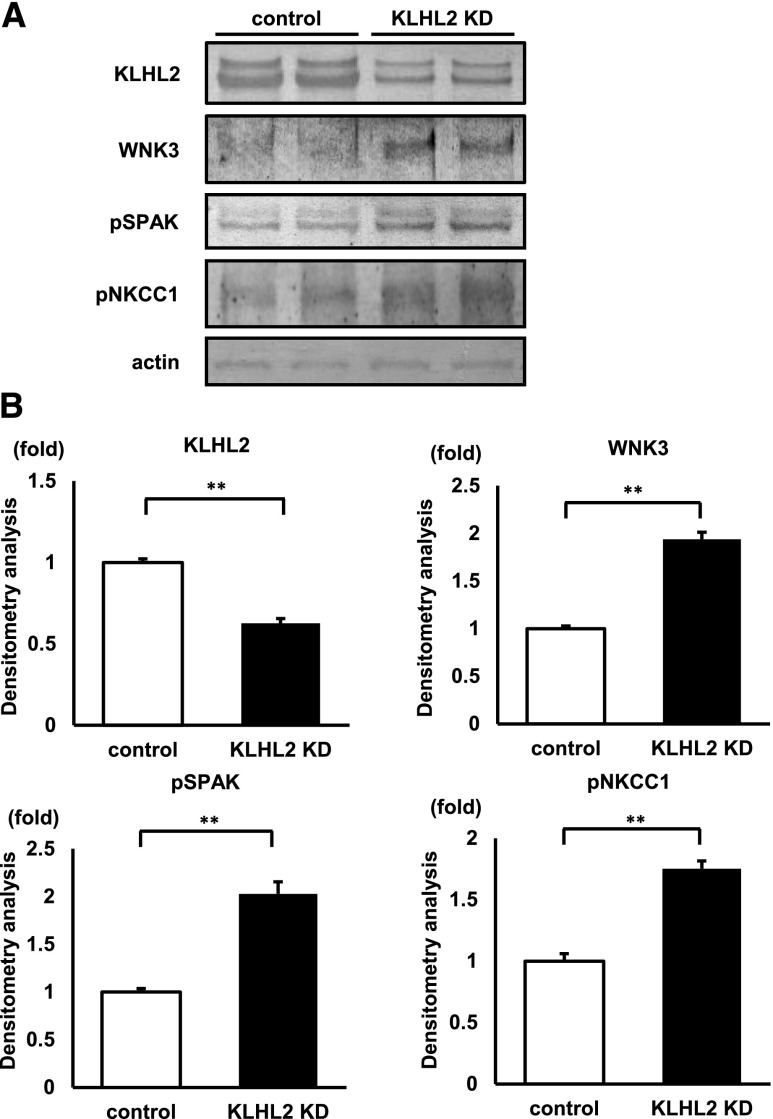
KLHL2 knockdown (KD) activates WNK3–SPAK–NKCC1 phosphorylation cascade in MOVAS cells. (A) Representative immunoblots and (B) densitometry analyses (n=14) of KLHL2, WNK3, phosphorylated SPAK (pSPAK), and phosphorylated NKCC1 (pNKCC1) in KLHL2 KD MOVAS cells. KLHL2 KD increased WNK3 expression and phosphorylation of SPAK and NKCC1. **P<0.01.
Figure 6.
KLHL2 overexpression downregulates the WNK3–SPAK–NKCC1 phosphorylation cascade in MOVAS cells. (A) Representative immunoblots and (B) densitometry analyses (n=4) of KLHL2, WNK3, phosphorylated SPAK (pSPAK), and phosphorylated NKCC1 (pNKCC1) in Cul3- and KLHL2-overexpressing MOVAS cells. KLHL2 overexpression alone significantly decreased WNK3 expression and the phosphorylation of SPAK and NKCC1, and the coexpression of KLHL2 with Cul3 further downregulated the signal cascade; however, Cul3 overexpression alone did not affect the cascade. *P<0.05; **P<0.01.
Figure 7.
Cul3 knockdown (KD) increases WNK3 protein. (A) Representative immunoblots and (B) densitometry analyses (n=4) of Cul3, KLHL2, and WNK3 in Cul3 KD MOVAS cells. Cul3 KD increased WNK3 protein in both AngII(−) and AngII(+) conditions, suggesting the involvement of Cul3 in WNK3 degradation. AngII could still decrease KLHL2 in Cul3 KD MOVAS cells, suggesting a Cul3-independent degradation mechanism of KLHL2 by AngII. **P<0.01.
Thus, AngII decreased KLHL2 protein by a Cul3-independent mechanism, and the decreased KLHL2–Cul3 E3 ligase increased WNK3 abundance, which activated the downstream SPAK–NKCC1 signaling.
The Rapid Decrease in KLHL2 by AngII Is Caused by Autophagy-Mediated Degradation
Because KLHL2 was identified as the major regulator of WNK3 in MOVAS cells, we then investigated the mechanism of how the KLHL2 level was rapidly decreased by AngII. KLHL2 mRNA levels in MOVAS cells were not decreased with AngII treatment (Figure 8A). Furthermore, as shown in Figure 3, the decrease in KLHL2 induced by AngII was very rapid and occurred within minutes. Taken together, these data clearly indicate that the decrease in KLHL2 by AngII may be mediated by the activation of KLHL2 degradation.
Figure 8.
KLHL2 is degraded by the autophagy mechanism in MOVAS cells. (A) KLHL2 mRNA levels in AngII-treated MOVAS cells were analyzed by quantitative RT-PCR. KLHL2 mRNA in MOVAS cells did not decrease with AngII treatment (n=14). (B) Representative immunoblots and (C) densitometry analyses (n=6) of KLHL2 in MOVAS cells treated with chloroquine, a lysosomal degradation inhibitor. AngII did not decrease KLHL2 when MOVAS cells were treated with chloroquine. (D) Representative immunoblots and (E) densitometry analyses (n=6) of KLHL2 in MOVAS cells treated with epoxomicin, a proteasome inhibitor. AngII decreased KLHL2 in MOVAS cells treated with epoxomicin. **P<0.01.
To confirm this hypothesis and identify the degradation mechanism, we performed the experiments using protein degradation inhibitors. As shown in Figure 8, B and C and Supplemental Figure 4, chloroquine, an autophagy inhibitor, inhibited the decrease in KLHL2 by AngII treatment. In addition, other types of autophagy inhibitors (bafilomycin A1 and 3-MA) showed similar effects. However, epoxomicin, a potent proteasome inhibitor, did not inhibit the decrease in KLHL2 by AngII (Figure 8, D and E). These results indicate that the KLHL2 decrease induced by AngII was caused by the activated degradation of KLHL2 through autophagy.
Selective Autophagy Mediated by p62 Degrades KLHL2 on AngII Stimulation
Kelch-like ECH-associated protein 1 (Keap1), a member of the KLHL family (KLHL19), forms E3 ligase with Cul3 for NF E2-related factor 2, a novel regulator of the antioxidant response.16 Recently, there have been some reports showing that Keap1 is degraded by selective autophagy mediated by sequestosome1/p62.17,18 The experiments using protein degradation inhibitors strongly suggested that the KLHL2 degradation by AngII was not mediated by the proteasome system but by autophagy. Thus, we examined whether a mechanism similar to p62-mediated Keap1 degradation may be functioning on AngII-induced KLHL2 degradation by autophagy. We performed knockdown experiments of p62 in MOVAS cells. As shown in Figure 9, compared with the control cells, KLHL2 was increased in the p62 knockdown cells. Furthermore, p62 knockdown inhibited the decrease in KLHL2 by AngII stimulation. In another vascular smooth muscle cell line (rat vascular smooth muscle cells [SV40LT–SMC]), the same behavior was confirmed (Supplemental Figure 5). From these results, it was shown that the decrease in KLHL2 by AngII was mediated by selective autophagy facilitated by p62.
Figure 9.
The decrease in KLHL2 by AngII is mediated by selective autophagy mediated by p62. (A) Representative immunoblots and (B) densitometry analyses (n=4) of p62 and KLHL2 in p62 knockdown (KD) MOVAS cells. p62 KD increased KLHL2 expression and inhibited the decrease in KLHL2 by AngII. *P<0.01; **P<0.05.
Discussion
In this study, we uncovered a new AngII signaling pathway in vascular smooth muscle cells for the control of vascular tone and BP. We had previously found that dietary salt intake and AngII regulate the WNK3–SPAK–NKCC1 phosphorylation cascade in MOVAS cells.12 However, how AngII regulated this cascade remains unclear. We have now shown that AngII invoked p62-mediated selective autophagic degradation of KLHL2 acting as E3 ligase with Cul3 for WNK3 and that the reduction of KLHL2 led to the increase in WNK3 protein abundance, which activated the downstream phosphorylation signaling of SPAK–NKCC1 (Figure 10).
Figure 10.
The mechanism of vasoconstriction mediated AngII–KLHL2–WNK3 signaling. In vascular smooth muscle cells, WNK3 is degraded by KLHL2–CUL3-mediated ubiquitination. With AngII stimulation, KLHL2 is degraded by selective autophagy by p62, which leads to the activation of the WNK3–SPAK–NKCC1 phosphorylation cascade and vasoconstriction. AT1R, angiotensin II type-I receptor; P, phosphorylation.
KLHL2 and KLHL3 are known to be involved in the degradation of WNK1/WNK2/WNK3/WNK4. Each of them forms a complex with Cul3 and acts as E3 ligase on WNK1/WNK2/WNK3/WNK4.4–6,19,20 The KLHL3–Cul3 complex acts as an E3 ubiquitin ligase for WNK kinases, and KLHL3 mutations have been reported to cause PHAII through the impaired ubiquitination of WNK4.4 However, the involvement of WNK kinase regulation by ubiquitination in the situation other than PHAII has not been identified. This may be the first report that a KLHL protein is, indeed, involved in the known physiologic regulation of the WNK signal cascade.
We showed that the AngII-induced WNK3 protein increase in MOVAS was surprisingly rapid, which was accompanied by the same rapid decrease in KLHL2. Knockdown and overexpression of KLHL2 were, indeed, able to regulate WNK3 protein abundance in MOVAS; this indicated that AngII-induced WNK3 signaling was mediated by the rapid regulatory decrease in the KLHL2 protein. In signal transduction, the regulation of key molecules by protein degradation may be a good mechanism that can promptly adapt to external stimuli, because there is no need for transcription and translation. The mechanism clarified in this study involves the regulation of degradation of two molecules (i.e., KLHL2 and WNK3).
Autophagy is initially identified as a catabolic mechanism that protects against cellular starvation through the degradation of cellular components in lysosomes. In addition to such a classic role, autophagy has been shown to be involved in the regulation of numerous biologic processes, including signal transduction.21 There have been some reports that showed the link between AngII and autophagy. Pan et al.22 reported that AngII induced the downregulation of microRNA-30, which increased beclin-1 and induced excessive autophagy in cardiac myocytes. In the podocytes of the kidney, AngII elicited reactive oxygen species, which increased LC3-II and beclin-1 and induced autophagosomes.23 Recently, in vascular smooth muscle cells, AngII also increased the autophagy levels through the activation of the AT1 receptor.24 However, all of these reports showed that AngII could induce bulk autophagy but not selective autophagy mediated by p62; p62 is known as a key protein that can induce selective autophagy. In this study, it was first shown that KLHL2 was degraded by selective autophagy mediated by p62. In this respect, although additional information on how AngII regulated p62-mediated KLHL2 degradation remains to be determined, this report may describe the first selective autophagy mediated by AngII, which could be an important switching mechanism of signal transduction.
In this study, we clarified how AngII activated the WNK3–SPAK–NKCC1 phosphorylation cascade in MOVAS cells. This is the first demonstration that a KLHL protein physiologically regulates the WNK signal cascade and that AngII regulates its downstream signaling by autophagy. Thus, our study identified a novel component of signal transduction in AngII-induced vascular smooth muscle cell contraction, which could be a promising drug target for hypertension.
Concise Methods
Dietary Salt and Drug Infusion Study Protocols
For experiments examining the effects of dietary salt intake, C57BL/6J mice were fed a high-salt diet (4% NaCl [wt/wt]), normal diet (0.9% NaCl [wt/wt]), or low-salt diet (0.01% NaCl [wt/wt]) for 1 week. All foods were obtained from Oriental Yeast Co., Ltd.
For acute AngII infusion, we implanted the infusion tube into the right external jugular vein 5 days before the administration of AngII and infused AngII at a dose of 25 μg/g intravenously. The mice were then euthanized 30 minutes after AngII administration as reported previously.8,12
Plasmids
Mouse KLHL2 cDNA was isolated by RT-PCR using mouse kidney mRNA from a C57BL/6J mouse. Sequences of the amplification primers used were as follows: KLHL2 sense, 5′-ATGGAGACGCCGCCACTGCCTCCCGC-3′; KLHL2 antisense, 5′-TCATAACGGTTTATCAATAACTGTGACC-3′. The mouse KLHL3 cDNA clone was purchased from Origene. Mouse KLHL2 and KLHL3 cDNA was cloned into 3XFLAG-CMV10 vector (Sigma-Aldrich).
RT-PCR
Total RNA from the mouse aorta, mouse kidney, and MOVAS cells was extracted using TRIzol reagent (Invitrogen). The total RNA was reverse transcribed using Omniscript RT (Qiagen). Sequences of RT-PCR primers used were as follows: KLHL2 sense, 5′-TTCTTAACCTCGGCATC-3′; KLHL2 antisense, 5′-AACTCCTGTCTTACGTCCTT-3′; KLHL3 sense, 5′-GGACCAACACTTGGAAGCAG-3′; and KLHL3 antisense, 5′-AGTGCTCATGTTGGTGGG-3′. The WNK3 and WNK4 primers have been described previously.12,25 The primers for GAPDH were purchased from TAKARA BIO.
Cell Culture and Chemicals
We used the MOVAS cell line from mouse aorta (CRL-2797; ATCC) and the SV40LT-SMC Clone HEP-SA cell line from rat aorta (CRL-2018; ATCC) as a model of vascular smooth muscle cells. Both the MOVAS cells and SV40LT-SMC cells were cultured in DMEM supplemented with 10% (vol/vol) FBS, 2 mM l-glutamine, 100 units/ml penicillin, and 0.1 mg/ml streptomycin at 37°C in a humidified 5% CO2 incubator. MOVAS cells that were cultured in six-well dishes were lysed with 150 μl ice-cold lysis buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 50 mM sodium fluoride, 1 mM sodium orthovanadate, 1% Triton X-100, 0.27 M sucrose, 1 mM DTT, and Complete protease inhibitor cocktail; Roche; 1 tablet per 50 ml). In the p62 knockdown experiments, the cells were lysed with RIPA buffer containing 0.25 mM Tris/HCl (pH 8.0), 0.38% EGTA, 0.1 mM EDTA, 0.18% Na orthovanadate, 2.1% NaF, and Complete protease inhibitor cocktail (Roche; 1 tablet per 50 ml). After centrifugation at 12,000×g for 5 minutes at 4°C, the supernatants were denatured for 20 minutes at 60°C with SDS sample buffer (Cosmo Bio) and subjected to SDS-PAGE.
For AngII stimulation experiments, the MOVAS cells were exposed to 5 μM AngII for 10 minutes. For the experiment using autophagy inhibitors, the cells were incubated with 25 μM chloroquine for 24 hours,26 200 nM bafilomycin A1 for 24 hours,27 or 5 mM 3-MA for 24 hours before AngII treatment.26 As a proteasome inhibitor, we used 1 μM epoxomicin for 4 hours.4
Transfection
The MOVAS cells were transfected by the indicated amount of plasmid DNA with Lipofectamine 2000 reagent (Invitrogen). For the KLHL2 knockdown experiments, we used 50 nM cocktails of three duplexes of siRNA for mouse KLHL2 (siTRIO Library; Origene) with Lipofectamine RNAiMAX (Invitrogen). The oligonucleotide sequences of siRNAs were as follows: mouse KLHL2 siTRIO SR417910A, agaauccugagaaacauugcccaCT; SR417910B, cgagcaaagagaguucgaauaaaGG; and SR417910C, ccuuuaaagucaugaacgaacucAG. Experiments were performed at 48 hours after siRNA transfection. For the p62 knockdown experiments, we used 25 nM cocktails of three duplexes of siRNA for mouse p62 (siTRIO library; Origene) with Lipofectamine 2000 (Invitrogen). The oligonucleotide sequences of siRNAs were as follows: mouse p62 siTRIO SR413467A, ggcuccuacagaccaagaauuacGA; SR413467B, ggcuauguccuaugugaaagaugAC; and SR413467C, cgcaucuacauuaaagagaagaaGG. Experiments were performed at 24 hours after siRNA transfection. For the WNK1 or WNK3 knockdown experiments, we used 50 nM cocktails of three duplexes of siRNA for mouse WNK1 or WNK3 (siTRIO library; Origene) with Lipofectamine RNAiMAX (Invitrogen). The oligonucleotide sequences of siRNAs were as follows: mouse WNK1 siTRIO SR423399A, agaagaggcugaaauguuaaaggGT; SR423399B, gccagagccuaauggaauuaguaTT; and SR423399C, ccauucaaauaaaccaaacuugcTT and mouse WNK3 siTRIO SR423205A, gcagaaguucuaauaguuugacgGG; SR423205B, ggaccgcaaauuaacuaaagcugAG; and SR423205C, cgaauaucaagcauguauaccaaGT. Experiments were performed at 24 hours after siRNA transfection. For the Cul3 knockdown experiments, we used 100 nM cocktails of three duplexes of siRNA for mouse Cul3 (Santa Cruz Biotechnology) with Lipofectamine 2000 (Invitrogen). The oligonucleotide sequences of siRNAs were as follows: mouse Cul3 sc-35131A sense, ccaagcacaugaagacuauTT; sc-35131B sense, gaaggaauguuuagggauaTT; and sc-35131C sense, gacagaaaguagaugaugaTT. Experiments were performed at 36 hours after siRNA transfection. As a negative control, siTRIO negative control siRNA (Origene) was used.
Immunoblotting
For the protein lysate of the mouse aorta, the thoracic aorta was carefully isolated and immediately frozen with liquid nitrogen. After being crushed with a mortar, the aorta was lysed with 150 μl lysis buffer, as reported previously, followed by centrifugation at 6000×g at 4°C.12,28 The supernatant (120 μl) was then denatured at 60°C for 20 minutes.
The primary antibodies used in this study were rabbit anti-KLHL2/KLHL3 antibody (Santa Cruz Biotechnology), anti-KLHL3 antibody (Proteintech), sheep anti-WNK3 antibody (S346C),29,30 rabbit antiphosphorylated SPAK antibody,31 rabbit antiphosphorylated NKCC1 (T206) antibody,32 rabbit antiactin antibody (Cell Signaling Technology), anti-T7 antibody (EMD Millipore), anti-Cul3 antibody (BETHYL), anti-WNK1 antibody (A301–516A; BETHYL), anti-WNK4 antibody,33 and rabbit anti-p62 antibody (Cell Signaling Technology). Alkaline phosphatase-conjugated anti-IgG antibodies (Promega) were used as secondary antibodies for immunoblotting. WesternBlue (Promega) was used for the development of immunoblots. The relative intensities of the immunoblot bands were determined by densitometry with ImageJ software.
Statistical Analyses
Statistical significance was evaluated using an unpaired t test. All data were expressed as means±SEMs. When more than three groups were compared, one-way ANOVA with Fischer’s post hoc test was used. P values<0.05 were considered to indicate statistical significance.
Disclosures
None.
Acknowledgments
We thank M. Chiga and C. Iijima for help in the experiments. We also thank D. Alessi for provision of antibodies.
This study was supported, in part, by Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (S, A); a Ministry of Education, Culture, Sports, Science and Technology of Japan Grant-in-Aid for Young Scientists (B); a Ministry of Health, Labor, and Welfare Health and Labor Sciences Research Grant; Salt Science Research Foundation Grant 1422; the Takeda Science Foundation; a Banyu Foundation Research Grant; and the Vehicle Racing Commemorative Foundation.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2014070639/-/DCSupplemental.
References
- 1.Boyden LM, Choi M, Choate KA, Nelson-Williams CJ, Farhi A, Toka HR, Tikhonova IR, Bjornson R, Mane SM, Colussi G, Lebel M, Gordon RD, Semmekrot BA, Poujol A, Välimäki MJ, De Ferrari ME, Sanjad SA, Gutkin M, Karet FE, Tucci JR, Stockigt JR, Keppler-Noreuil KM, Porter CC, Anand SK, Whiteford ML, Davis ID, Dewar SB, Bettinelli A, Fadrowski JJ, Belsha CW, Hunley TE, Nelson RD, Trachtman H, Cole TR, Pinsk M, Bockenhauer D, Shenoy M, Vaidyanathan P, Foreman JW, Rasoulpour M, Thameem F, Al-Shahrouri HZ, Radhakrishnan J, Gharavi AG, Goilav B, Lifton RP: Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 482: 98–102, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson FH, Disse-Nicodème S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, Lifton RP: Human hypertension caused by mutations in WNK kinases. Science 293: 1107–1112, 2001 [DOI] [PubMed] [Google Scholar]
- 3.Louis-Dit-Picard H, Barc J, Trujillano D, Miserey-Lenkei S, Bouatia-Naji N, Pylypenko O, Beaurain G, Bonnefond A, Sand O, Simian C, Vidal-Petiot E, Soukaseum C, Mandet C, Broux F, Chabre O, Delahousse M, Esnault V, Fiquet B, Houillier P, Bagnis CI, Koenig J, Konrad M, Landais P, Mourani C, Niaudet P, Probst V, Thauvin C, Unwin RJ, Soroka SD, Ehret G, Ossowski S, Caulfield M, International Consortium for Blood Pressure (ICBP) Bruneval P, Estivill X, Froguel P, Hadchouel J, Schott JJ, Jeunemaitre X: KLHL3 mutations cause familial hyperkalemic hypertension by impairing ion transport in the distal nephron. Nat Genet 44: 456–460, 2012 [DOI] [PubMed] [Google Scholar]
- 4.Wakabayashi M, Mori T, Isobe K, Sohara E, Susa K, Araki Y, Chiga M, Kikuchi E, Nomura N, Mori Y, Matsuo H, Murata T, Nomura S, Asano T, Kawaguchi H, Nonoyama S, Rai T, Sasaki S, Uchida S: Impaired KLHL3-mediated ubiquitination of WNK4 causes human hypertension. Cell Rep 3: 858–868, 2013 [DOI] [PubMed] [Google Scholar]
- 5.Ohta A, Schumacher FR, Mehellou Y, Johnson C, Knebel A, Macartney TJ, Wood NT, Alessi DR, Kurz T: The CUL3-KLHL3 E3 ligase complex mutated in Gordon’s hypertension syndrome interacts with and ubiquitylates WNK isoforms: Disease-causing mutations in KLHL3 and WNK4 disrupt interaction. Biochem J 451: 111–122, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi D, Mori T, Wakabayashi M, Mori Y, Susa K, Zeniya M, Sohara E, Rai T, Sasaki S, Uchida S: KLHL2 interacts with and ubiquitinates WNK kinases. Biochem Biophys Res Commun 437: 457–462, 2013 [DOI] [PubMed] [Google Scholar]
- 7.Chiga M, Rai T, Yang SS, Ohta A, Takizawa T, Sasaki S, Uchida S: Dietary salt regulates the phosphorylation of OSR1/SPAK kinases and the sodium chloride cotransporter through aldosterone. Kidney Int 74: 1403–1409, 2008 [DOI] [PubMed] [Google Scholar]
- 8.Talati G, Ohta A, Rai T, Sohara E, Naito S, Vandewalle A, Sasaki S, Uchida S: Effect of angiotensin II on the WNK-OSR1/SPAK-NCC phosphorylation cascade in cultured mpkDCT cells and in vivo mouse kidney. Biochem Biophys Res Commun 393: 844–848, 2010 [DOI] [PubMed] [Google Scholar]
- 9.Takahashi D, Mori T, Nomura N, Khan MZ, Araki Y, Zeniya M, Sohara E, Rai T, Sasaki S, Uchida S: WNK4 is the major WNK positively regulating NCC in the mouse kidney. Biosci Rep 34: e00107, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishida H, Sohara E, Nomura N, Chiga M, Alessi DR, Rai T, Sasaki S, Uchida S: Phosphatidylinositol 3-kinase/Akt signaling pathway activates the WNK-OSR1/SPAK-NCC phosphorylation cascade in hyperinsulinemic db/db mice. Hypertension 60: 981–990, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saritas T, Borschewski A, McCormick JA, Paliege A, Dathe C, Uchida S, Terker A, Himmerkus N, Bleich M, Demaretz S, Laghmani K, Delpire E, Ellison DH, Bachmann S, Mutig K: SPAK differentially mediates vasopressin effects on sodium cotransporters. J Am Soc Nephrol 24: 407–418, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeniya M, Sohara E, Kita S, Iwamoto T, Susa K, Mori T, Oi K, Chiga M, Takahashi D, Yang SS, Lin SH, Rai T, Sasaki S, Uchida S: Dietary salt intake regulates WNK3-SPAK-NKCC1 phosphorylation cascade in mouse aorta through angiotensin II. Hypertension 62: 872–878, 2013 [DOI] [PubMed] [Google Scholar]
- 13.Orlov SN, Koltsova SV, Tremblay J, Baskakov MB, Hamet P: NKCC1 and hypertension: Role in the regulation of vascular smooth muscle contractions and myogenic tone. Ann Med 44[Suppl 1]: S111–S118, 2012 [DOI] [PubMed] [Google Scholar]
- 14.Dowd BF, Forbush B: PASK (proline-alanine-rich STE20-related kinase), a regulatory kinase of the Na-K-Cl cotransporter (NKCC1). J Biol Chem 278: 27347–27353, 2003 [DOI] [PubMed] [Google Scholar]
- 15.Ponce-Coria J, San-Cristobal P, Kahle KT, Vazquez N, Pacheco-Alvarez D, de Los Heros P, Juárez P, Muñoz E, Michel G, Bobadilla NA, Gimenez I, Lifton RP, Hebert SC, Gamba G: Regulation of NKCC2 by a chloride-sensing mechanism involving the WNK3 and SPAK kinases. Proc Natl Acad Sci U S A 105: 8458–8463, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M: Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13: 76–86, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M: The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12: 213–223, 2010 [DOI] [PubMed] [Google Scholar]
- 18.Fan W, Tang Z, Chen D, Moughon D, Ding X, Chen S, Zhu M, Zhong Q: Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy 6: 614–621, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata S, Zhang J, Puthumana J, Stone KL, Lifton RP: Kelch-like 3 and Cullin 3 regulate electrolyte homeostasis via ubiquitination and degradation of WNK4. Proc Natl Acad Sci U S A 110: 7838–7843, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu G, Peng JB: Disease-causing mutations in KLHL3 impair its effect on WNK4 degradation. FEBS Lett 587: 1717–1722, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boya P, Reggiori F, Codogno P: Emerging regulation and functions of autophagy. Nat Cell Biol 15: 713–720, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan W, Zhong Y, Cheng C, Liu B, Wang L, Li A, Xiong L, Liu S: MiR-30-regulated autophagy mediates angiotensin II-induced myocardial hypertrophy. PLoS ONE 8: e53950, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yadav A, Vallabu S, Arora S, Tandon P, Slahan D, Teichberg S, Singhal PC: ANG II promotes autophagy in podocytes. Am J Physiol Cell Physiol 299: C488–C496, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu KY, Wang YP, Wang LH, Jian Y, Zhao XD, Chen JW, Murao K, Zhu W, Dong L, Wang GQ, Zhang GX: Mitochondrial KATP channel involvement in angiotensin II-induced autophagy in vascular smooth muscle cells. Basic Res Cardiol 109: 416, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Reilly M, Marshall E, Macgillivray T, Mittal M, Xue W, Kenyon CJ, Brown RW: Dietary electrolyte-driven responses in the renal WNK kinase pathway in vivo. J Am Soc Nephrol 17: 2402–2413, 2006 [DOI] [PubMed] [Google Scholar]
- 26.Bae SH, Sung SH, Oh SY, Lim JM, Lee SK, Park YN, Lee HE, Kang D, Rhee SG: Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab 17: 73–84, 2013 [DOI] [PubMed] [Google Scholar]
- 27.Myeku N, Figueiredo-Pereira ME: Dynamics of the degradation of ubiquitinated proteins by proteasomes and autophagy: Association with sequestosome 1/p62. J Biol Chem 286: 22426–22440, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Susa K, Kita S, Iwamoto T, Yang SS, Lin SH, Ohta A, Sohara E, Rai T, Sasaki S, Alessi DR, Uchida S: Effect of heterozygous deletion of WNK1 on the WNK-OSR1/ SPAK-NCC/NKCC1/NKCC2 signal cascade in the kidney and blood vessels. Clin Exp Nephrol 16: 530–538, 2012 [DOI] [PubMed] [Google Scholar]
- 29.Thastrup JO, Rafiqi FH, Vitari AC, Pozo-Guisado E, Deak M, Mehellou Y, Alessi DR: SPAK/OSR1 regulate NKCC1 and WNK activity: Analysis of WNK isoform interactions and activation by T-loop trans-autophosphorylation. Biochem J 441: 325–337, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oi K, Sohara E, Rai T, Misawa M, Chiga M, Alessi DR, Sasaki S, Uchida S: A minor role of WNK3 in regulating phosphorylation of renal NKCC2 and NCC co-transporters in vivo. Biol Open 1: 120–127, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sohara E, Rai T, Yang SS, Ohta A, Naito S, Chiga M, Nomura N, Lin SH, Vandewalle A, Ohta E, Sasaki S, Uchida S: Acute insulin stimulation induces phosphorylation of the Na-Cl cotransporter in cultured distal mpkDCT cells and mouse kidney. PLoS ONE 6: e24277, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang SS, Lo YF, Wu CC, Lin SW, Yeh CJ, Chu P, Sytwu HK, Uchida S, Sasaki S, Lin SH: SPAK-knockout mice manifest Gitelman syndrome and impaired vasoconstriction. J Am Soc Nephrol 21: 1868–1877, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohta A, Rai T, Yui N, Chiga M, Yang SS, Lin SH, Sohara E, Sasaki S, Uchida S: Targeted disruption of the Wnk4 gene decreases phosphorylation of Na-Cl cotransporter, increases Na excretion and lowers blood pressure. Hum Mol Genet 18: 3978–3986, 2009 [DOI] [PubMed] [Google Scholar]