Abstract
Inosine monophosphate dehydrogenase (IMPDH) is a pivotal enzyme in the de novo pathway of guanine nucleotide biosynthesis. Inhibitors of this enzyme decrease intracellular guanine nucleotide levels by 50-80% and have potential as anti-neoplastic agents. Both mycophenolic acid (MPA) and AVN-944 are highly specific inhibitors of IMPDH that cause cell cycle arrest or apoptosis in lymphocytes and leukemic cell lines. We have examined the mechanisms by which these two agents cause cytotoxicity. Both MPA and AVN-944 inhibit the growth of K562 cells, and induce apoptosis in Raji B and CCRF-CEM T cells. Both compounds strikingly inhibit RNA synthesis within 2 h of exposure. Depletion of guanine nucleotides by MPA and AVN-944 also causes an early and near-complete reduction in levels of the 45S precursor rRNA synthesis and the concomitant translocation of nucleolar proteins including nucleolin, nucleophosmin, and nucleostemin from the nucleolus to the nucleoplasm. This efflux correlates temporally with the sustained induction of p53 in cell lines with wild type p53. We conclude that inhibition of IMPDH causes a primary reduction in rRNA synthesis and secondary nucleolar disruption and efflux of nucleolar proteins that most likely mediate cell cycle arrest or apoptosis. The ability of AVN-944 to induce apoptosis in a number of leukemic cell lines supports its potential utility in the treatment of hematologic malignancies.
Keywords: IMPDH, RNA polymerase I, AVN-944, GTP depletion, Nucleolar protein, ribosomal RNA
Introduction
IMPDH is the rate limiting enzyme for the de novo synthesis of guanine nucleotides that are critical for cell proliferation 1. Inhibitors of IMPDH such as mycophenolate mofeteil (Cellcept), a prodrug of mycophenolic acid (MPA), have been used in organ and stem cell transplantation and in autoimmune diseases as highly effective immunosuppressive agents 2. Since IMPDH inhibitors induce cell cycle arrest and apoptosis in a number of neoplastic cell lines, they also have considerable potential as antineoplastic agents, both alone and in combination with other cytotoxic drugs. A new and potent IMPDH inhibitor, AVN-944, has been developed based on the structural analysis of the active site of the enzyme and is a selective, noncompetitive inhibitor of the enzyme directed against both of the two human IMPDH isoforms (I and II; Ki 6–10 nM) 1. AVN-944, in contrast to MPA, does not undergo glucuronidation and is currently in Phase I clinical trials for the treatment of refractory hematologic malignancies.
The potential mechanisms by which IMPDH inhibitors inhibit the proliferation of normal lymphocytes and tumor cell lines have undergone extensive investigation 3-7. These studies have been aided by the fact that the selectivity of these agents for IMPDH can be demonstrated by reversing the observed biologic effects through the addition of guanosine or guanine, both of which replete guanine nucleotide pools through the hypoxanthine guanine phosphoribosyl transferase (HPRT) salvage pathway 2. Recent studies have demonstrated that guanine nucleotide depletion leads to apoptosis and concomitant inhibition of signaling in IL-3 dependent murine myeloid cell lines through effects on both the Ras-MAPK and mTOR pathways 8,9. Other studies have demonstrated that the induction of apoptosis in multiple myeloma cell lines occurs through both caspase-dependent 10 and caspase-independent 7 apoptotic mechanisms. Despite a large number of such empiric observations on potential downstream targets, the upstream sensing event that detects depletion of guanine nucleotides and initially triggers cell cycle arrest or apoptotic cell death has not been elucidated.
We and others have noted that depletion of guanine nucleotides induces the translocation of several nucleolar proteins from the nucleolus into the nucleus 11,12, suggesting a GTP-dependent event that may have consequences for cellular proliferation. We have therefore asked whether the effects of MPA and AVN944 may be mediated by changes in the localization of these nucleolar proteins. As a result of these studies, we suggest that the well-defined growth-inhibitory and pro-apoptotic effects of IMPDH inhibition on malignant cells may be mediated, at least in part, through a primary inhibition of ribosomal RNA synthesis and a secondary disruptive effect on nucleolar organization.
Materials and Methods
Cell culture conditions and reagents used
U2OS, K562, Raji, and CCRF-CEM cells were grown in Dulbecco's Modified Eagle (DMEM) or RPMI 1640 medium supplemented with 10% fetal bovine serum and 100 U/ml of penicillin and streptomycin. The cells were kept at 37°C in an atmosphere containing 5% CO2. Uridine, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was obtained from Sigma (Sigma-Chemical Co., USA). Uridine ([5,6-3H], 35-50 Ci/mmol) was purchased from ICN Biomedicals (Costa Mesa, Calif., USA). Actinomycin-D was obtained from Calbiochem (La Jolla, CA, USA). 4′,6-diamino-2-phenylindol (DAPI) was purchased from Molecular Probes, Inc ( Eugene, OR, USA). Anti-cmyc tag monoclonal antibody (mAb) (9E10) and anti-nucleolin monoclonal antibody (mAb) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Rabbit polyclonal anti-NPM was from Cell Signaling Technology Inc. (Beverly, MA, USA), rabbit polyclonal anti-PAF53 was from BD PharMingen (Lexington, KY, USA), anti-ARF mouse monoclonal antibody (P02) was purchased from NeoMarkers (Fremont, CA, USA). Fluorescein (FITC)-conjugated donkey anti-rabbit F(ab')2 and rhodamine-conjugated goat anti-mouse secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA, USA).
Total RNA Extraction and cDNA Synthesis
Total RNA was extracted from U2OS cells with Trizol reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instruction. Two micrograms of total RNA were used for first-strand cDNA synthesis using oligo (dT) primers and SuperScript II RNase H− Reverse Transcriptase (SuperScript™ First-Strand Synthesis System for RT-PCR, Invitrogen).
Generation of expression constructs
The resulting first-strand cDNA was used for PCR amplification with high fidelity pfx DNA polymerase (Invitrogen). The primers used for TIF-IA and EGFP-PAF53 are listed in table 1. The italicized bases are the restriction enzyme sites (EGFP-PAF53: Xho I and EcoR1; TIF-IA: Bam H1 and Xho I) with three or four extra bases added at each end to promote efficient digestion by the restriction enzymes.
Table 1.
| Gene | Primer sequence | Product (bp) |
|---|---|---|
| TIF-IA | 5'- ttcaggatccaaatggcggcaccgctgcttcacacg -3' (forward) 5'- catgctcgagtcagaggggactgggttgcatgtacaac -3' (reverse) |
1970 |
| EGFP-PAF53 | 5'- agatctcgagatggcggcggaggtgttgccgagtgcg -3' (forward) 5'- gcagaattcgctaggtaatcttcctccgctttgccaggc-3' (reverse) |
1260 |
| 18S rRNA to ITS (between 18S rRNA and 5.8S) |
5'- ttggatggtttagtgaggc-3' (forward) 5'- cacgcccttctttctctctc-3' (reverse) |
394 |
| GAPDH | 5'- ggtgaaggtcggagtcaacg -3' (forward) 5'- caaagttgtcatggatgacc -3' (reverse) |
500 |
The TIF-IA and PAF53 PCR products were subcloned into the Bam H1 and Xho I site of myc-pcDNA 3 (myc-pcDNA3) and the Xho I and EcoR1 of p-EGFP-C3 (BD, biotech), respectively, and the resulting plasmids were sequenced. The EGFP-PAF53 fusion constructs and myc-tagged TIF-IA were introduced into U2OS cells by Superfect transfection (Qiagen) according to the manufacturer's instruction.
Immunocytochemistry
U2OS and 293 T cells were grown on coverslips in 24-well plates in DMEM complete medium for 12 h and were then treated with MPA (2 μM), AVN-944 (1μM), Actinomycin D (5 nM or 500 nM), or vehicle control for the time indicated. The cells were fixed in 4% phosphate-buffered saline (PBS)-paraformaldehyde solution for 20 min followed by permeabilization with 0.1% Triton X-100 in PBS for 15 min. After blocking with 5% bovine serum albumin in PBS for 30 min, coverslips were inverted onto a 30-μl drop of the primary antibodies diluted in PBS containing 0.1% Triton-X-100 and 5% BSA (1:50 for NPM, nucleolin, ARF and nucleostemin; 1: 200 for myc). After 1 h incubation with antibody and subsequent washing with PBS × 3 (10 min each), coverslips were inverted again onto 30-μl drops of the secondary antibodies diluted in PBS containing 5% BSA and 0.1% Triton-X 100 ( rhodaminecongugated secondary antibody (1:300); fluorescein isothiocyanate-conjugated secondary antibody (1:50)). After washing, coverslips were stained with DAPI (300 nM) for 5 minutes, and washed with PBS × 3 (5 min. each), mounted in Gel Mount IM Aqueous Mounting Medium (Sigma-Chemical Co., USA) and examined by fluorescent microscopy.
Determination of 45S precursor rRNA by semiquantitative RT-PCR
Total RNAs were extracted as described above. Two micrograms of total RNA were used for first-strand cDNA synthesis using random primers and SuperScript II RNase H− Reverse Transcriptase (SuperScript™ First-Strand Synthesis System for RT-PCR, Invitrogen). The 45S rRNA precursor fragment was then amplified with pfx DNA polymerase (Invitrogen). PCR reactions were carried out in volumes of 50 μL containing 2 μL cDNA, 2 μM forward and reverse primers, 5.0 μL 10× Accuprime Pfx Reaction Mix from the Accuprime Pfx DNA Polymerase Kit, 1 μL Accuprime Taq DNA polymerase from the Accuprime Taq DNA Polymerase System. The two step PCR was first run at 95°C for 2 min, followed by 35 cycles (95°C for 15 s; 68°C for 40 s), and a final extension step (68°C, 2min). GAPDH fragment was amplified with Tag DNA polymerase (promega). The PCR was first run at 94°C for 2 min, followed by 30 cycles (94°C for 30 s; 56°C for 30 s, and 72°C for 30 s), and a final extension step (72°C, 7min). The primer pairs used for 45 S rRNA precursor and GAPDH amplification are listed in table 1. PCR products were cloned into the TOPO blunt cloning vector (Invitrogen), and the resulting clones were randomly picked and sequenced using M13 forward and reverse primers.
Measurement of cell proliferation by MTT assay
The effects of AVN-944 and MPA on proliferation of CCRF-CEM, K562, and Raji cells were determined using the modified MTT assay 13. Exponentially-growing cells were plated in 0.5 ml aliquots of growth medium into 48-well plates at 2×104 cells per well and then incubated with various concentrations of AVN-944, MPA, or vehicle alone. After 96 h incubation, cytotoxicity assays were performed by the modified MTT method. All samples were tested in triplicate. Values represent the means of triplicate wells and the S.E. of the mean was less than 10%.
Measurement of RNA Synthesis
To measure [3H]-uridine incorporation into RNA, exponentially-growing U2OS cells and CCRF-CEM cells were plated in growth medium at 1×105 cells/ml. After incubation in the absence or presence of MPA (2 μM) or actinomycin-D (5, 50, and 500 nM), cells were labeled for 30 min after addition of [3H]-uridine (5 μCi/ml). Cells were washed three times with ice-cold PBS, and total RNA was isolated using Trizol reagent (GIBCO BRL). Total RNAs were quantitated and 10 μg RNA dissolved in RNA-free water. Radioactivity was determined by the addition of 6 ml of scintillation fluid followed by liquid scintillation counting. Triplicate samples were analyzed for each treatment.
For the analysis of 45S pre-rRNA synthesis, K562 cells were treated with 2 μM MPA or 1 μM AVN-944 for 1.5, 3.5, or 7.5 h, respectively, and then metabolically labeled with 2.0 μCi/ml [3H]-uridine ( 36 Ci/mmol, Amersham Bioscience, Piscataway, NJ) in complete medium for 30 min and washed three times with ice cold PBS. RNA was extracted with Trizol (Invitrogen, Carlsbad, CA), and 15 μg of total RNA loaded onto a 1% agarose gel containing formaldehyde. The RNAs were electrophoretically separated and transferred to Nytran SuPerCharge TurboBlotter membrane (Whatman, Sanford, ME). Dried membranes were treated with EN3HANCE (Perkin Elmer Life Sciences, Boston, MA) and subjected to autoradiography at −70° C with an intensifying screen for 4 days.
Immunoprecipitation, SDS–PAGE and immunoblotting
Cells were treated with MPA, actinomycin-D, or the DMSO vehicle for the time indicated. Cells were rinsed twice with phosphate-buffered saline (PBS) and lysed in a buffer containing 20 mM Tris-HCl (pH 7.5), 137 mM NaCl, 1% Triton X-100, 10% glycerol, 2 mM EDTA, 150 μM Na3VO4, 0.25mM phenylmethysulfonyl fluoride (PMSF), 5 μg/ml leupeptin, 10 nM microcystin LR. The lysate was sonicated, and 20 μg protein from total cell lysis subjected to SDS–PAGE and transferred onto polyvinylidene difluoride membrane (Immobilon-P; Millipore, Bedford, MA, USA). Specific antigens were probed using the corresponding primary antibody, followed by horseradish peroxidase conjugated secondary antibody. Western blots were visualized using enhanced chemiluminescence (ECL) (Chemiluminescence Reagent Plus, Perkin-Elmer Life Sciences, Boston, MA, USA).
Results
Induction of cytotoxicity by AVN-944 and MPA
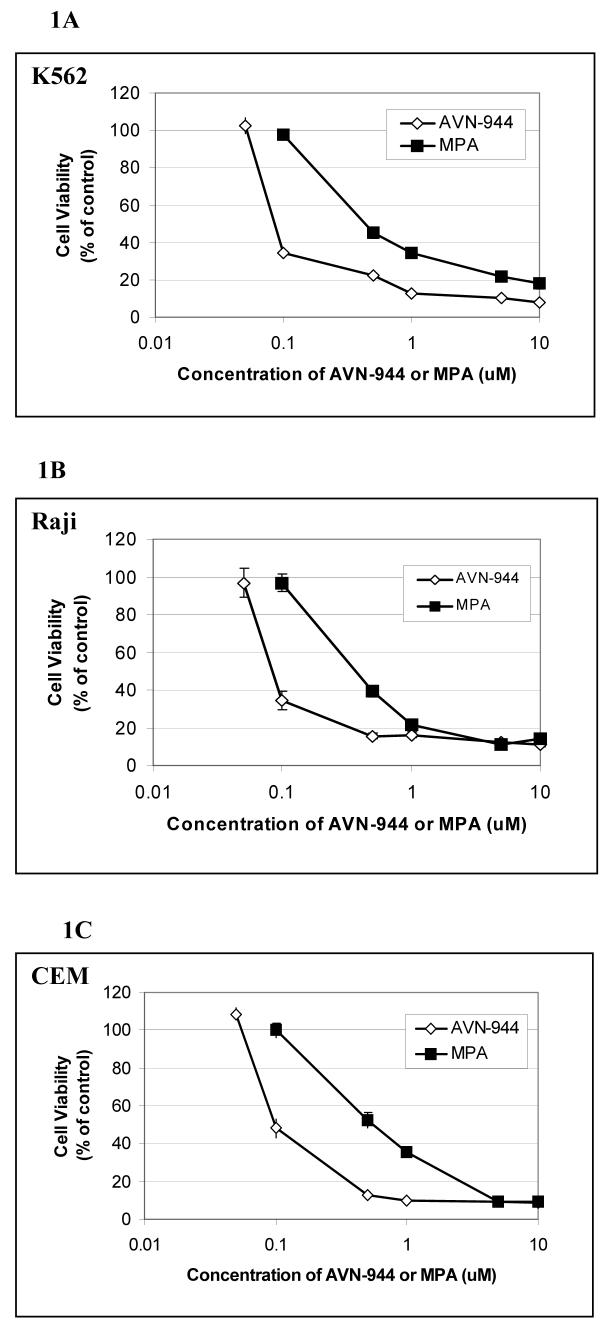
We examined the relative effects of the two IMPDH inhibitors on growth of the human myeloid progenitor K562 cell line, the Burkitt lymphoma Raji cell line (B cell origin), and the human T cell leukemia CCRF-CEM cell line. As shown in Fig. 1, the growth of all three cell lines was inhibited by both compounds in a dose-dependent manner, with AVN-944 being at least 2-fold more potent than MPA. The IC50s ranged from 0.13 to 0.16 μM for AVN-944 and from 0.40 to 0.58 μM for MPA. Growth inhibition was completely reversible with guanosine for both agents (not shown), as has been demonstrated in numerous previous studies 8,14. Apoptosis as measured by Annexin V staining was induced in Raji and CCRF-CEM cell lines, whereas K562 cells underwent erythroid differentiation (data not shown).
Fig. 1. Inhibition of cell growth by MPA and AVN-944 in human leukemia K562 cells, Raji, and CCRF-CEM cells.
K562 (A), Raji (B), and CCRF-CEM (C) cells were exposed to 1-10 μM MPA or 1-10 μM AVN-944 for 96 h. Cytotoxicity assays were performed by the modified MTT method. All assays were performed in triplicate. All values represent the mean +/− SEM.
Effect of GTP depletion on the translocation of nucleolar proteins
To determine the effects of IMPDH inhibition on the localization of nucleolar proteins, U2OS osteosarcoma cells were treated with MPA or AVN-944 in the absence or presence of guanosine. This adherent cell line was chosen because changes in the nucleolar architecture are most easily documented. As shown in Fig. 2A, nucleolin, nucleophosmin and nucleostemin had all translocated from the nucleolus to nucleoplasm after 24 h of MPA treatment. Nucleolar protein translocation was completely prevented by guanosine. AVN-944 induced the translocation of these nucleolar proteins after 4 h of treatment, while MPA took approximately 8 h to have a similar effect (Fig. 2B). Similar data were obtained with the MCF-7 cells (data not shown).
Fig. 2. Effect of MPA and AVN-944 on the localization of nucleophosmin, nucleolin, and nucleostemin and reversibility by guanosine.

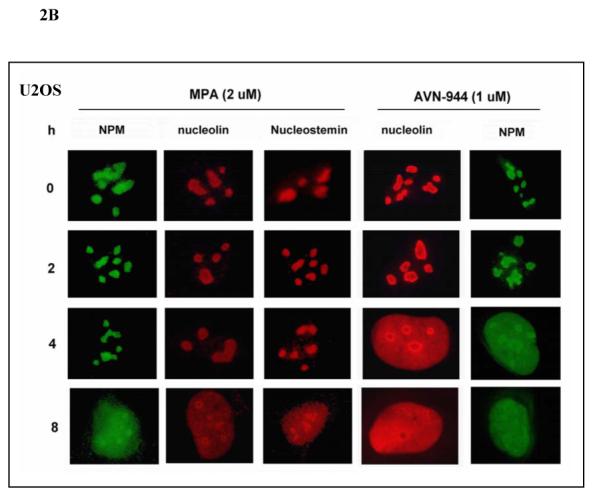
(A) U2OS cells were treated with 2 μM MPA in the absence or presence of 100 μM guanosine for 24 h. (B) U2OS cells were treated with 2 μM MPA or 1 μM AVN-944 for 2 h, 4 h, and 8 h. Cells were then fixed, permeabilized, stained for nucleolin, nucleostemin, or NPM, as described in Materials and Methods, and observed using fluorescent microscopy.
Effects of IMPDH inhibition on p53 activation and ARF expression
The translocation of nucleophosmin (NPM) and nucleostemin, as well as of other nucleolar proteins such as L11 and L23, has been shown to be involved in the activation of p5315-18. In the case of NPM, there is evidence that the tumor suppressor ARF is released from binding to NPM in the nucleolus and preferentially binds to p53, preventing its degradation through the MDM2 ubiquitin ligase pathway 19. Nucleostemin, on the other hand, appears to have a direct inhibitory effect on cellular proliferation both dependent 20 and independent 21 of p53. To determine whether p53 is induced as a correlate of the translocation of NPM and nucleostemin, the effect of IMPDH inhibition on p53 expression was examined in U2OS cells and MCF-7 breast cancer cells known to express wild-type p53. Marked induction of p53 occurred at 1 μM AVN-944 after 4 h and the effect was sustained up to 24 h (Fig. 3A, 3C). Immunostaining of ARF demonstrated that nucleolar ARF was detectable in 293T cells but undetectable in U2OS cells and MCF-7 cells (data not shown). Therefore, 293T cells were used to ask whether ARF was translocated as one potential mechanism for the activation of p53. As shown in Fig. 3B, 1 μM AVN-944 induced ARF translocation in this cell line at 4 h with nearly complete translocation occurring at 8 h.
Fig. 3. Effects of AVN-944 on p53 and ARF.

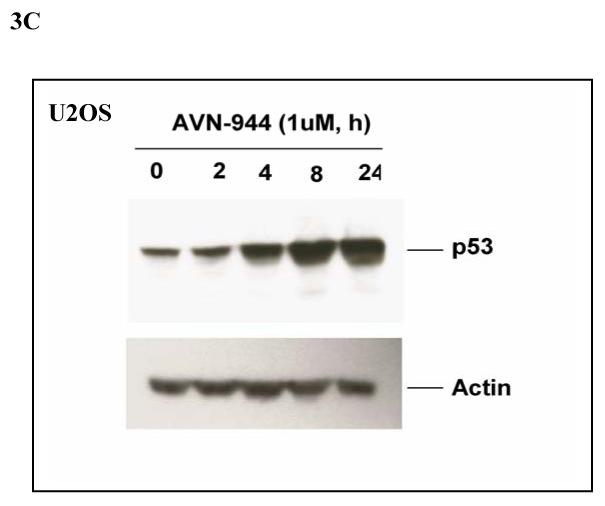
MCF-7 cells (A) or 293 T cells (B) were treated with AVN-944 at the time and concentration indicated. Cells were then fixed, permeabilized, and stained for p53 and ARF as described in the Materials and Methods. (C) U2OS cells were treated with AVN-944 at the time and concentration indicated, and lysed as described in Materials and Methods. 30 μg of total protein were loaded for immunoblot analysis for p53.
Effect of IMPDH inhibition on RNA synthesis
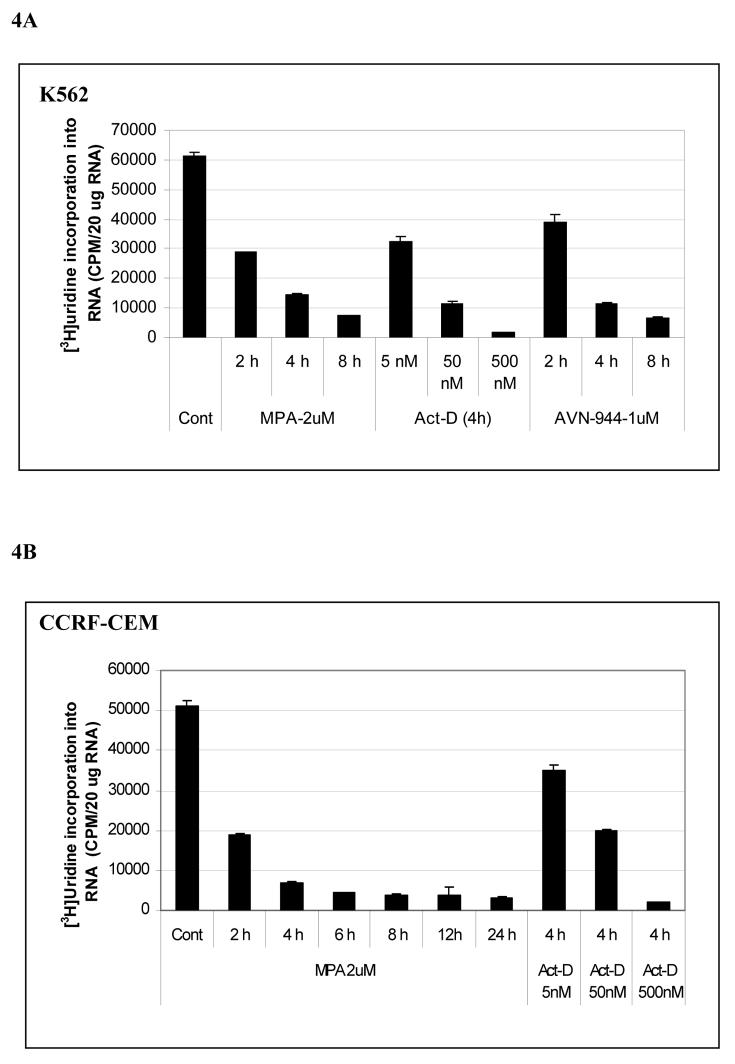
We next sought to determine whether GTP depletion had a direct effect on RNA synthesis as determined by the incorporation of [3H] uridine into purified total cellular RNA. Maximum inhibition of the incorporation of [3H] uridine incorporation occurred at 2 μM MPA or 1 μM AVN-944 in both K562 and CCRF-CEM cells, with more than 80% inhibition at 4 h and sustained inhibition lasting for 24 h (Fig. 4A, 4B). Similar inhibition was seen with 500 nM Act-D, a dose that has been shown to inhibit RNA polymerases I and II 22. Moreover, the total RNA and precursor rRNA synthesis was markedly inhibited by MPA at 2 h in U2OS cells , and was almost completely abolished at 4 h, an effect that was sustained for up to 24 h (data not shown). Since Pol I-directed rRNA synthesis accounts for the majority of total RNA synthesis, we next asked whether 45 S rRNA precursor synthesis was impaired using semiquantitative RT-PCR. The primer pairs used amplify a region extending from the 3' end of 18 S rRNA to the internal transcribed sequence (ITS) located between 18 S rRNA and 5.8 S rRNA, indicative of the amount of 45S precursor rRNA (Fig. 4C). Precursor rRNA synthesis was markedly inhibited by MPA at 2 h and 4 h in CCRF-CEM cells and K562 cells, respectively, and inhibition was sustained up to 8 h (Fig. 4C). In order to rule out an effect of IMPDH inhibition on RNA processing, 15 μg [H3]-uridine labeled RNA from cells treated with either MPA or AVN-944 were run on an agarose gel containing formaldehyde, transferred, and autoradiographed. As shown in Fig. 4D, the 47S/45S precursor form, the mature rRNA (28S, 18S, and 5.8S), and 32S intermediate comprise the majority of the radioactivity in untreated K562 cells. A decrease in newly synthesized rRNA was observed with both drugs at 2 h, and an absolute decline occurred at 4 h. Total 28S and 18S ribosomal RNAs were only mildly decreased under these conditions.
Fig. 4. Effects of MPA, AVN-944, and Actinomycin D on total RNA synthesis and prerRNA synthesis in K562 and CCRF-CEM cells.


K562 cells were treated with MPA, AVN-944, or actinomycin-D at the concentrations and times indicated and [3H]-uridine incorporation into RNA was measured (A, B); the effect of MPA on the expression of pre-rRNA levels was determined by RT-PCR (C) as described in Materials and Methods. Primers amplified the 394 bp fragment extending from the 3' end of 18 S rRNA to the internal transcribed sequence (ITS) located between 18 S rRNA and 5.8 S rRNA of the intact pre-rRNA (shown as model figure, C). Analysis of rRNA processing (D). Extracted total [3H]-uridine-labeled RNAs from untreated K562 cells and cells treated with 2 μM MPA or 1 μM AVN-944 were separated on 1% agarose gel containing formaldehyde, transferred to membrane, and detected by autoradiography as described in Materials and Methods. The bottom panel shows 28S and 18S ribosomal RNA after transfer.
Co-translocation of PAF53 and TIF-IA to the nucleolar periphery
To further investigate the possible involvement of the RNA Polymerase I complex in the inhibition of rRNA synthesis, the EGFP-tagged Pol I subunit PAF53 and myc-tagged TIF-IA, a Pol I-associated initiation factor, were generated by RT-PCR from U2OS cells. The effects of MPA or AVN-944 treatment on the localization of myc-TIF-IA and EGFP-PAF53 transfected into U2OS cells were followed over time using fluorescent microscopy. As shown in Fig. 5A, EGFP-PAF53 was predominantly localized in the nucleolus, whereas TIF-IA was distributed throughout the nucleus in untreated cells. Co-transfection of myc-tagged TIF-IA and EGFP-tagged PAF53 demonstrated that both TIF-IA and PAF53 co-translocated to the identical nucleolar cap region following MPA treatment (Fig. 5A). Similar co-translocation of TIF-IA and PAF53 to the nucleolar periphery was induced by Act-D at both 5 nM and 500 nM concentrations (Fig. 5B and data not shown). The co-translocation of PAF53 and TIF-IA, two important components of the Pol I complex, correlated temporally with the rapid and potent inhibition of pre-rRNA synthesis.
Fig. 5. MPA and actinomycin-D induce a distinct co-translocation of EGFP-tagged PAF53 (Pol I subunit) and myc-tagged TIF-IA to the nucleolar cap.

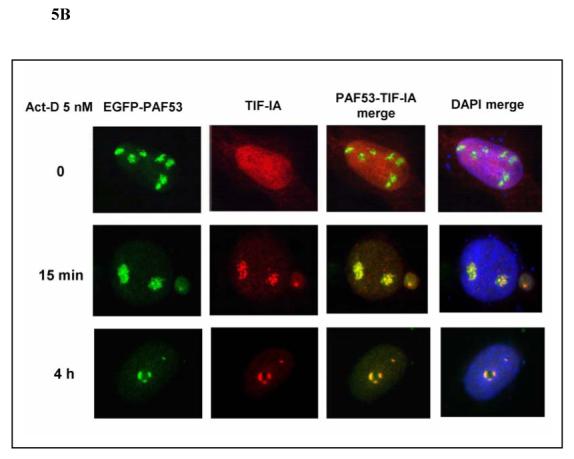
PAF53 cDNA subcloned into EGFP-C3 and TIF-IA cDNA subcloned into pcDNA3 were transfected into U2OS cells. 24 h after transfection, cells were treated with MPA (A) or actinomycin-D (B) at the concentrations and for the times indicated. The myc-tagged TIF-IA was fixed, permealized, and stained as described in the Materials and Methods, and observed using fluorescent microscopy.
Translocation of nucleolar proteins induced by actinomycin-D
U2OS cells were treated with actinomycin-D at concentrations of 5 nM, a dose that specifically inhibits RNA polymerase I 23, and 500 nM, a dose that inhibits both pol I and Pol II 22,23. As shown in Fig. 6, nucleolin, NPM, and nucleostemin move from the nucleolus to the nucleus in response to 5 nM actinomycin D for 4 h and to an even greater extent with 500 nM actinomycin-D. These data indicate that IMPDH inhibitors that cause depletion of guanine nucleotides have a direct effect on nucleolar protein localization that is similar to that caused actinomycin D, a well-known inhibitor of rRNA synthesis. They strongly suggest that inhibition of rRNA synthesis followed by nucleolar disruption is a primary event leading to the cellular cytotoxicity caused by MPA or AVN-944 treatment.
Fig. 6. Actinomycin-D induces translocation of NPM, nucleolin, and nucleostemin.

U2OS cells were treated with actinomycin-D at 5 nM and 500 nM. Cells were then fixed, permealized, and stained for NPM, nucleolin, and nucleostemin as described in the Materials and Methods, and observed using fluorescent microscopy.
Discussion
The nucleolus is a dynamic nuclear sub-compartment that regulates a great number of cellular functions that include the transcription and processing of pre-rRNA and ribosome subunit assembly 24,25, and maturation of non-nucleolar RNAs or RNPs 26,27. Other more general functions that have been attributed to the nucleolus include regulation of telomerase function, regulation of the cell cycle, tumor suppressor activities, cell stress sensing and signaling 28,29. The initial step in nucleolar formation is ribosomal gene transcription, which is mediated by RNA Polymerase I (Pol I) and its associated transcription factors: UBF (upstream-binding factor), SL1 (selectivity factor) and TIF-IA (transcription initiation factor IA) 24,25. Inhibition of Pol I activity by actinomycin D and genetic inactivation of the Pol I-associated transcription factor TIF-IA both lead to diminished pre-rRNA synthesis and a secondary disruption of nucleolar structure as manifest by the efflux of nucleolar proteins into the nucleus 22,30,31. Hence, nucleolar integrity is entirely dependent on ongoing rRNA synthesis.
Our results have demonstrated that depletion of GTP through inhibition of IMPDH in a number of different cell lines leads to inhibition of pre-rRNA synthesis as measured both by the incorporation of [H3]-uridine into precursor RNA and by quantitation of precursor rRNA using the polymerase chain reaction. It has recently been shown that nucleostemin, a nucleolar protein thought to be preferentially expressed in stem cells and in cancer cells 20, is targeted to or retained in the nucleolus through the binding of GTP and that lowering of GTP concentrations through IMPDH inhibition allows repartitioning of the protein into the nucleoplasm 20. Similarly, nucleophosmin, a protein lacking a GTP-binding motif, was shown a number of years ago to have GTP-dependent nucleolar retention 11. As a result of these observations, it has been postulated that a GTP-driven cycle may be important for nucleolar organization 32. Our data support a more fundamental explanation, namely that GTP depletion affects nucleolar protein localization through its effect on rRNA synthesis. We have used actinomycin D to determine whether its known ability to inhibit pre-rRNA synthesis causes an efflux of nucleolar proteins that is temporally and quantitatively similar to that seen with GTP depletion. Indeed, actinomycin D at 5 nM (and more potently at 500 nM) causes an egress of nucleostemin, nucleolin, and nucleophosmin into the nucleoplasm at 4 h, as do both MPA and AVN944. We conclude from these results that IMPDH inhibition and the consequent GTP depletion primarily inhibit rRNA synthesis. Whether this inhibition results from a limitation of GTP as a substrate for Pol I enzymatic activity, from its effect on essential components of Pol I activity such as TIF-1A and PAF53, which relocate to the nucleolar periphery where they are known to be inactive 27,33,34, or from an as yet undiscovered mechanism remains to be determined. However, a major result of inhibiting Pol I-directed rRNA synthesis is nucleolar disruption and the efflux of nucleolar proteins into the nucleoplasm.
Further support for this conclusion comes from recent data on the conditional genetic inactivation of TIF-IA, a transcription initiation factor required for Pol I activity 31. Cre-mediated depletion of TIF-IA leads to nucleolar disruption, cell cycle arrest and p53 up-regulation in murine embryonic fibroblasts. The explanation for the activation of p53 in these cells is the disruption of the p53-MDM2 complex through the release of ribosomal proteins (in this case, L11) from the nucleolus, resulting in p53 stabilization and cell cycle arrest. Alternatively, it has been shown in other systems that the release of the tumor suppressor ARF from its binding to NPM in the nucleolus allows ARF to displace MDM2 from p53, thus preventing its ubiquitination and destruction 35,36. Our results demonstrate that p53 is indeed activated as a consequence of IMPDH inhibition in cell lines containing unmutated p53. Indeed, ribonucleotide depletion has been implicated as one of the many causes of p53 activation since the early experiments of Linke et al. 37, although the underlying mechanism has not been demonstrated to our knowledge. Our results suggest the following overall sequence of events: depletion of guanine nucleotides through inhibition of de novo synthesis from IMP decreases the synthesis of precursor rRNA which in turn leads to the disruption of nucleolar organization, the efflux of nucleolar proteins into the nucleus, and p53 activation.
We have also shown in 293T cells expressing ARF that its nucleolar localization is disrupted as a result of GTP depletion. It is important to note that neither endogenous ARF expression nor wild type p53 are essential for the growth inhibitory or apoptotic effects of GTP depletion, since ARF expression was only detected in 293 T cells that have p53 inactivation and since the three hematopoietic cell lines studied all have mutated p53 (K562 -insertion-frame shift mutation; CCRF-CEM -missense mutation; and Raji -missense mutation) 38. The pro-apoptotic and growth inhibitory effects of GTP depletion in p53-null cells underscores the existence of alternative effects of nucleolar protein efflux (e.g. nucleostemin and other ribosomal proteins such as L11) on other downstream pathways that have not yet been fully elucidated. Similarly, the absence of ARF expression in U2OS and MCF-7 cells clearly implicates additional mechanisms of p53 activation, as has been suggested in a previous study 31.
Since disruption of ribosomal biogenesis and nucleolar organization appear to be increasingly important as potential therapeutic targets in cancer 33,39,40, we suggest that the new, more potent IMPDH inhibitors may have benefit in the treatment of hematologic malignancies. It will be of considerable interest to examine the nucleolar architecture in the leukemic cells of patients treated with IMPDH inhibitors and to determine whether nucleolar disruption is predictive of a therapeutic response.
Acknowledgments
This work was supported by NIH grant 5R01-CA064192 to BSM and by a Translational Research Award to BSM from the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict-of-interest disclosure: B.S.M. is an unpaid consultant to Avalon Pharmaceuticals and the remaining authors declare no competing financial interests.
References
- 1.Zimmermann AG, Gu JJ, Laliberte J, et al. Inosine-5′-monophosphate dehydrogenase: regulation of expression and role in cellular proliferation and T lymphocyte activation. Prog Nucleic Acid Res Mol Biol. 1998;61:181–209. doi: 10.1016/s0079-6603(08)60827-2. [DOI] [PubMed] [Google Scholar]
- 2.Allison AC, Eugui EM. Mechanisms of action of mycophenolate mofetil in preventing acute and chronic allograft rejection. Transplantation. 2005;80:S181–190. doi: 10.1097/01.tp.0000186390.10150.66. [DOI] [PubMed] [Google Scholar]
- 3.Laliberte J, Yee A, Xiong Y, et al. Effects of guanine nucleotide depletion on cell cycle progression in human T lymphocytes. Blood. 1998;91:2896–2904. [PubMed] [Google Scholar]
- 4.Heinschink A, Raab M, Daxecker H, et al. In vitro effects of mycophenolic acid on cell cycle and activation of human lymphocytes. Clin Chim Acta. 2000;300:23–28. doi: 10.1016/s0009-8981(00)00297-7. [DOI] [PubMed] [Google Scholar]
- 5.Yalowitz JA, Pankiewicz K, Patterson SE, et al. Cytotoxicity and cellular differentiation activity of methylenebis(phosphonate) analogs of tiazofurin and mycophenolic acid adenine dinucleotide in human cancer cell lines. Cancer Lett. 2002;181:31–38. doi: 10.1016/s0304-3835(02)00045-9. [DOI] [PubMed] [Google Scholar]
- 6.Greupink R, Bakker HI, Reker-Smit C, et al. Studies on the targeted delivery of the antifibrogenic compound mycophenolic acid to the hepatic stellate cell. J Hepatol. 2005;43:884–892. doi: 10.1016/j.jhep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 7.Ishitsuka K, Hideshima T, Hamasaki M, et al. Novel inosine monophosphate dehydrogenase inhibitor VX-944 induces apoptosis in multiple myeloma cells primarily via caspase-independent AIF/Endo G pathway. Oncogene. 2005;24:5888–5896. doi: 10.1038/sj.onc.1208739. [DOI] [PubMed] [Google Scholar]
- 8.Gu JJ, Gathy K, Santiago L, et al. Induction of apoptosis in IL-3-dependent hematopoietic cell lines by guanine nucleotide depletion. Blood. 2003;101:4958–4965. doi: 10.1182/blood-2002-08-2547. [DOI] [PubMed] [Google Scholar]
- 9.Gu JJ, Santiago L, Mitchell BS. Synergy between imatinib and mycophenolic acid in inducing apoptosis in cell lines expressing Bcr-Abl. Blood. 2005;105:3270–3277. doi: 10.1182/blood-2004-10-3864. [DOI] [PubMed] [Google Scholar]
- 10.Takebe N, Cheng X, Fandy TE, et al. IMP dehydrogenase inhibitor mycophenolate mofetil induces caspase-dependent apoptosis and cell cycle inhibition in multiple myeloma cells. Mol Cancer Ther. 2006;5:457–466. doi: 10.1158/1535-7163.MCT-05-0340. [DOI] [PubMed] [Google Scholar]
- 11.Finch RA, Chang DC, Chan PK. GTP gamma S restores nucleophosmin (NPM) localization to nucleoli of GTP-depleted HeLa cells. Mol Cell Biochem. 1995;146:171–178. doi: 10.1007/BF00944610. [DOI] [PubMed] [Google Scholar]
- 12.Tsai RY, McKay RD. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J Cell Biol. 2005;168:179–184. doi: 10.1083/jcb.200409053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carmichael J, DeGraff WG, Gazdar AF, et al. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 1987;47:936–942. [PubMed] [Google Scholar]
- 14.Yalowitz JA, Jayaram HN. Molecular targets of guanine nucleotides in differentiation, proliferation and apoptosis. Anticancer Res. 2000;20:2329–2338. [PubMed] [Google Scholar]
- 15.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. Embo J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai MS, Zeng SX, Jin Y, et al. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5 5-aza-2′-deoxycytidine activates the p53/p21Waf1/Cip1 pathway to inhibit cell proliferation MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. Mol Cell Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai MS, Zeng SX, Jin Y, et al. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enomoto T, Lindstrom MS, Jin A, et al. Essential role of the B23/NPM core domain in regulating ARF binding and B23 stability. J Biol Chem. 2006;281:18463–18472. doi: 10.1074/jbc.M602788200. [DOI] [PubMed] [Google Scholar]
- 19.Lindstrom MS, Zhang Y. B23 and ARF: friends or foes? Cell Biochem Biophys. 2006;46:79–90. doi: 10.1385/CBB:46:1:79. [DOI] [PubMed] [Google Scholar]
- 20.Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beekman C, Nichane M, De Clercq S, et al. Evolutionarily Conserved Role of Nucleostemin (NS): Controlling Proliferation of Stem/Progenitor Cells during Early Vertebrate Development. Mol Cell Biol. 2006 doi: 10.1128/MCB.01183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker M, Baumann C, John S, et al. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 2002;3:1188–1194. doi: 10.1093/embo-reports/kvf244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang S, Deerinck TJ, Ellisman MH, et al. The perinucleolar compartment and transcription. J Cell Biol. 1998;143:35–47. doi: 10.1083/jcb.143.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carmo-Fonseca M, Mendes-Soares L, Campos I. To be or not to be in the nucleolus. Nat Cell Biol. 2000;2:E107–112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- 25.Olson MO. Sensing cellular stress: another new function for the nucleolus? Sci STKE. 2004;2004:pe10. doi: 10.1126/stke.2242004pe10. [DOI] [PubMed] [Google Scholar]
- 26.Thiry M, Lafontaine DL. Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol. 2005;15:194–199. doi: 10.1016/j.tcb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Hernandez-Verdun D. The nucleolus: a model for the organization of nuclear functions. Histochem Cell Biol. 2006;126:135–148. doi: 10.1007/s00418-006-0212-3. [DOI] [PubMed] [Google Scholar]
- 28.Andersen JS, Lam YW, Leung AK, et al. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 29.Raska I, Shaw PJ, Cmarko D. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol. 2006;18:325–334. doi: 10.1016/j.ceb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Chan PK, Bloom DA, Hoang TT. The N-terminal half of NPM dissociates from nucleoli of HeLa cells after anticancer drug treatments. Biochem Biophys Res Commun. 1999;264:305–309. doi: 10.1006/bbrc.1999.1255. [DOI] [PubMed] [Google Scholar]
- 31.Yuan X, Zhou Y, Casanova E, et al. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol Cell. 2005;19:77–87. doi: 10.1016/j.molcel.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 32.Misteli T. Going in GTP cycles in the nucleolus. J Cell Biol. 2005;168:177–178. doi: 10.1083/jcb.200412038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christensen MO, Krokowski RM, Barthelmes HU, et al. Distinct effects of topoisomerase I and RNA polymerase I inhibitors suggest a dual mechanism of nucleolar/nucleoplasmic partitioning of topoisomerase I. J Biol Chem. 2004;279:21873–21882. doi: 10.1074/jbc.M400498200. [DOI] [PubMed] [Google Scholar]
- 34.Shav-Tal Y, Blechman J, Darzacq X, et al. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 36.Lindstrom MS, Jin A, Deisenroth C, et al. Cancer-Associated Mutations in MDM2 Zinc Finger Domain Disrupt Ribosomal Protein Interaction and Attenuate MDM2-Induced p53 Degradation Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Mol Cell Biol. 2006;12:175–186. doi: 10.1128/MCB.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linke SP, Clarkin KC, Di Leonardo A, et al. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 38.Hernandez-Boussard T, Rodriguez-Tome P, Montesano R, et al. IARC p53 mutation database: a relational database to compile and analyze p53 mutations in human tumors and cell lines. International Agency for Research on Cancer. Hum Mutat. 1999;14:1–8. doi: 10.1002/(SICI)1098-1004(1999)14:1<1::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 39.Yoshioka N, Wang L, Kishimoto K, et al. A therapeutic target for prostate cancer based on angiogenin-stimulated angiogenesis and cancer cell proliferation. Proc Natl Acad Sci U S A. 2006;103:14519–14524. doi: 10.1073/pnas.0606708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stirpe F, Battelli MG. Ribosome-inactivating proteins: progress and problems. Cell Mol Life Sci. 2006;63:1850–1866. doi: 10.1007/s00018-006-6078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]