Letter to the Editor
Up to 20% of patients with aplastic anemia (AA) who do not undergo allogeneic hematopoietic stem cell transplantation (HSCT) will ultimately develop myelodysplastic syndrome (MDS) or acute myelogenous leukemia (AML). (1) Distinguishing which patients will evolve to MDS could help guide decision-making between immunosuppresive therapy (IST) and HSCT, but risk factors for malignant evolution remain poorly defined. Recent studies utilizing single nucleotide polymorphism (SNP) arrays have identified somatic alterations in 15-20% of patients with AA. (2, 3) However, the unbiased identification of variants by either SNP arrays or traditional Sanger sequencing typically requires an allele fraction >25% of the involved specimen. Because the progression from AA to MDS may occur over years or even decades, subclones that ultimately evolve into MDS could be present at very low fractions at the time of AA diagnosis.
Sanger sequencing of specimens from patients with AA has not identified recurrent alterations, suggesting that they are either not present or exist within subclones below the level of detection with this method. Next-generation sequencing to a high depth of coverage offers the potential to identify subclonal mutations with sensitivity proportional to the depth of sequencing. We developed a platform for exon capture followed by deep sequencing (4) of 219 genes (Supplementary Table 1) that are recurrently mutated in hematologic malignancies (see Supplementary Methods). Patients with AA and available blood or bone marrow were identified by searching databases of stored specimens at the Dana-Farber Cancer Institute and Brigham and Women’s Hospital. All studies were performed under the approval of the Institutional Review Board of the Dana-Farber Cancer Institute/Harvard Cancer Center.
We identified 39 patients with AA and banked bone marrow or peripheral blood specimens (Table 1). None of the patients had abnormalities detected by metaphase karyotyping or fluorescence in situ hybridization of bone marrow specimens. Severe AA was defined as a hypocellular bone marrow for age and at least two of the following in the peripheral blood: platelet count < 20,000/mm3, a corrected reticulocyte count < 1%, and/or absolute neutrophil count (ANC) < 500/mm3. Patients with peripheral blood ANC < 200/mm3 were categorized as very severe AA. Four patients had a paroxysmal nocturnal hemoglobinuria (PNH) clone identified at >5% based on loss of CD55/CD59 expression by flow cytometry, although none had clinical manifestations of PNH. Six additional patients had pre-existing hematologic disorders (Table 1).
Table 1.
Clinical characteristics of 39 patients with aplastic anemia, stratified by the presence or absence of mutations. Where indicated, fewer than 39 patients were evaluable.
| Characteristic | Total cohort (n = 39) | No mutation (n = 30) | Mutation present (n = 9) | P value |
|---|---|---|---|---|
|
| ||||
| Sex - n (%) | ||||
| Male | 22 (56) | 18 (60) | 4 (44) | 0.46 |
| Female | 17 (44) | 12 (40) | 5 (56) | |
|
| ||||
| Age at diagnosis, yr | ||||
| Median (range) | 34.8 (4 - 65.7) | 34.5 (4 – 65.7) | 44.9 (6 – 58) | 0.85 |
|
| ||||
| Age at sequencing, yr | ||||
| Median (range) | 37.4 (17.4 – 66.3) | 35.2 (20.8 - 66.3) | 44.9 (17.7 – 58.7) | 0.53 |
|
| ||||
| Years with AA at sequencing, yr | ||||
| Median (range) | 0.5 (0 - 24.8) | 0.46 (0 - 24.8) | 0.67 (0 – 19.3) | 0.21 |
|
| ||||
| White blood cell count, 109/L | ||||
| Median (range) | 2.0 (0.3 – 5.8) | 2.1 (0.3 – 5.8) | 1.6 (0.6 – 3.6) | 0.55 |
|
| ||||
| Absolute neutrophil count, /mm3 | ||||
| Median (range) | 474 (0 – 2560) | 665 (0 – 2560) | 470 (0 – 1760) | 0.46 |
|
| ||||
| Platelets, 109/L | ||||
| Median (range) | 16 (4 – 151) | 15 (4 – 151) | 17 (5 – 51) | 0.56 |
|
| ||||
| Hemoglobin, g/dL | ||||
| Median (range) | 8.9 (3.9 – 12.7) | 8.8 (3.9 – 12.7) | 9.4 (6.8 – 11.8) | 0.52 |
|
| ||||
| Corrected reticulocyte count, % | ||||
| Median (range) | 0.57 (0.04 – 0.97) | 0.59 (0.04 – 0.97) | 0.38 (0.12 – 0.62) | 0.28 |
|
| ||||
| Marrow cellularity - n (%) (n=38 evaluable) | ||||
| <10% | 16 (41) | 13 (43) | 3 (33) | 0.72 |
| 10 - 20% | 19 (49) | 15 (50) | 4 (44) | |
| 21 - 30% | 2 (5) | 1 (3) | 1 (11) | |
| >30% | 1 (3) | 1 (3) | - | |
|
| ||||
| AA severity - n (%) (n=35 evaluable) | ||||
| Moderate | 6 (15) | 5 (17) | 1 (11) | 0.89 |
| Severe | 20 (51) | 17 (57) | 3 (33) | |
| Very severe | 9 (23) | 7 (23) | 2 (22) | |
|
| ||||
| PNH clone - n (%) (n=31 evaluable) | ||||
| Absent | 27 (69) | 22 (73) | 5 (56) | 0.21 |
| Present | 4 (10) | 2 (7) | 2 (22) | |
|
| ||||
| Pre-existing hematologic disorder - n (%) | ||||
| Absent | 33 (85) | 25 (83) | 8 (89) | 1.00 |
| Present | 6 (15) | 5 (17) | 1 (11) | |
| Dyskeratosis congenita | 3 | 3 | - | |
| Fanconi anemia | 1 | 1 | - | |
| Pure red cell aplasia | 1 | 1 | - | |
| Schwachman-Diamond Syndrome | 1 | - | 1 | |
|
| ||||
| IST prior to sequencing - n (%) | ||||
| Yes | 19 (49) | 12 (40) | 7 (78) | 0.06 |
| No | 20 (51) | 18 (60) | 2 (22) | |
Abbreviations: PNH, Paroxysmal nocturnal hemoglobinuria; IST. Immunosuppressive therapy.
Paired-end sequencing was performed to mean depth >450 haploid coverage. Among the 39 patients, we identified 9 that each harbored a single non-dbSNP coding variant (Figure 1A). Seven of the mutations were present in allele fractions less than 10%. All 9 mutations were confirmed to be present by Sanger sequencing (if present in ≥20% of called reads) or by mass spectrometry-based genotyping (if present in <20% of called reads) and 7 were confirmed to be somatic by Sanger sequencing of either germline material or CD3-positive T-cells. For the remaining two mutations (BCR g.2365393C>T and ASXL1 g.31023451delCATT), neither germline material nor CD3-positive T-cells were available, but both mutations were present in <10% of reads, strongly suggesting that they were somatically acquired. In univariate analysis (Table 1), harboring a mutation was not associated with any particular demographic factor or clinical factor, although 7 (36.8%) of 19 patients who received IST prior to sample acquisition had mutations detected, compared with only 2 (10%) of 20 patients who were previously untreated (p=0.06 by two-sided Fisher’s exact test).
Figure 1.
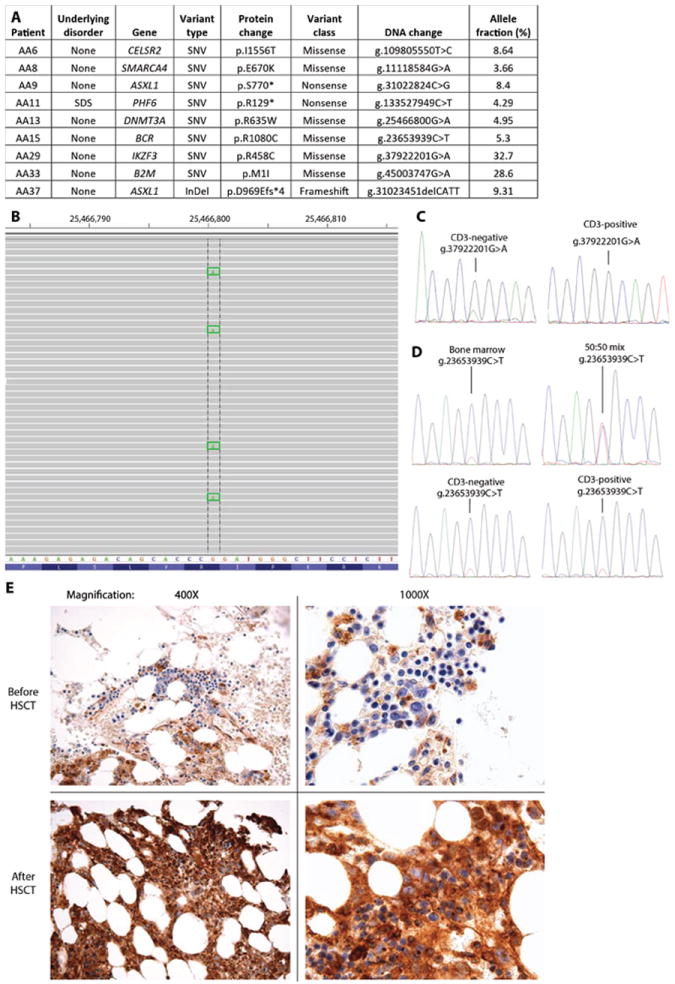
A. Mutations identified in 9 of 39 cases of aplastic anemia. Abbreviations: SNV, single nucleotide variant; InDel, insertion/deletion; ND, not determined. InDel notation signifies amino acid change at position of deletion, and frameshift with premature stop codon after four amino acids. B. Snapshot of sequencing reads from patient AA13 that overlap DNMT3A g.25466800 showing multiple mutant calls. C. Sanger sequencing of CD3-negative and CD3-positive sorted populations from specimen AA29 identifies a mutant IKZF3 g.37922201G>A allele only in the CD3-negative population. D. Sanger sequencing identifies the BCR g.2365393C>T mutation at a low frequency in unsorted, CD3-positive and CD3-negative populations from AA15. In contrast, a 50:50 mix of wild-type and mutant DNA results in equal representation of both alleles by Sanger sequencing. E. Immunohistochemistry demonstrates complete loss of B2M staining in hematopoietic elements from a bone marrow specimen obtained prior to allogeneic HSCT (top) and essentially ubiquitous membrane staining in a specimen from after allogeneic HSCT (bottom). Two different magnifications are shown.
We also sequenced 4 additional patients with bone marrow disorders who did not meet criteria for AA. There were no mutations recovered from a patient with pure red cell aplasia, a patient with transient idiopathic cytopenias or a patient with Diamond-Blackfan anemia. A patient who was initially diagnosed with AA but was found to have monosomy 7 in 10% of interphase cells by FISH harbored a DNMT3A V372F mutation in 3.5% of reads (Supplementary Figure 1).
Among the patients with AA, one harbored a DNMT3A R635W mutation previously described in AML (Figure 1, Supplementary Figure 2) and two cases harbored premature truncation mutations in ASXL1 (Supplementary Figure 3). All of these mutations were present in <10% of reads, which was confirmed by mass spectrometry-based genotyping (Supplementary Figures 2, 3). To our knowledge, this demonstrates for the first time that sequencing to significant depth can distinguish patients with AA who harbor subclonal mutations associated with MDS and AML (5).
We sorted CD3-positive and CD3-negative populations to determine whether mutations we recovered from unselected cells are present in one or both compartments. Both IKZF3 g.37922201G>A and ASXL1 g.31023451delCATT were only recovered from CD3-negative populations (Figure 1C). In contrast, BCR g.2365393C>T and CELSR2 g.109805550T>C were present in both CD3-positive and CD3-negative populations at similar frequencies (Figure 1D, Supplementary Figure 4). Thus, some but not all subclonal mutations recovered from patients with AA are present within CD3-positive T-cells.
The ASXL1 g.31023451delCATT mutation in patient AA37 was present in 9.3% of reads from a bone marrow specimen obtained 9 months after the initial diagnosis of AA. To determine whether the mutation was present at an earlier timepoint or other mutations arose and then were lost, we sequenced another bone marrow specimen obtained at the time of diagnosis (i.e., 9 months prior) using the same platform. No mutations were identified in this earlier specimen with the standard calling algorithm. However, visual inspection of the reads at ASXL1 identified the g.31023451delCATT in 1 of 624 reads (0.16%) (Supplementary Figure 5), suggesting that the mutant allele expanded approximately 50-fold over 9 months of observation.
In one patient, we identified an M1I mutation in the start codon of β2-microglobulin (B2M), the invariant subunit of the major histocompatibility complex (MHC) class I. Two years after this patient was diagnosed with AA, the patient had a complete and sustained response after treatment with antithymocyte globulin and cyclosporine. The patient relapsed four years later, underwent allogeneic HSCT three years after that, and was in complete remission one year later. The sequenced specimen was obtained after relapse and prior to HSCT.
Peptides presented by MHC class I are recognized by CD8+ cytotoxic T-cells, leading to target cell destruction. B2M is required for MHC class I surface expression and CD8+ T cell function. (6) B2M is deleted or mutated in 29% of diffuse large B-cell lymphomas (DLBCL) and the most common mutation (5 of 25 cases with mutations) was at the start codon (7), exactly as observed in our patient with AA (Figure 1E). Immunohistochemistry for β2-microglobulin expression in bone marrow obtained from the sequenced specimen demonstrated loss of B2M expression within hematopoietic cells, with preserved staining in surrounding non-hematopoietic cells (Figure 1E). In comparison, a bone marrow specimen isolated 4 months after allogeneic HSCT demonstrated nearly universal membrane staining within hematopoietic and stromal cells (Figure 1E).
If the B2M mutation results in complete loss of MHC class I, the cells should become susceptible to natural killer (NK) cell-mediated cytotoxicity. Of note, DLBCLs also frequently harbor loss-of-function mutations in CD58, which results in evasion from NK cell cytolysis. (7) CD58 was included in our sequencing panel (Supplementary Table 1), but no mutations were recovered in CD58 in any cases. Therefore the B2M-mutated clone may have additional mechanisms of NK cell escape not evident by our targeted sequencing approach.
These results demonstrate that a subset of patients with AA harbor relatively rare clones that contain pathogenic mutations, including premature stop and frame-shift mutations in ASXL1 and a previously described variant in DNMT3A. The paucicellular nature of bone marrow specimens from patients with AA (e.g. Figure 1E) suggests that a significant fraction of the cells sequenced in these specimens may be non-hematopoietic, and thus lack the mutation. As such, the true fraction of hematopoietic cells harboring the mutation may be significantly higher than suggested by the allele burden. Most importantly, nearly all of the mutations we recovered would not have been detected by other methods, including unbiased Sanger sequencing. Whether these mutated clones are long-lived, capable of initiating malignant transformation, or otherwise of consequence remains to be determined.
Whole genome sequencing of hematopoietic stem cells from normal individuals has identified a high burden of age-associated mutations. (8) In addition, alterations associated with hematologic malignancies (e.g. BCL2-IGH (9, 10)) can be recovered from a large fraction of the general population. Together, these findings reinforce the point that the presence of mutations within specimens from patients with AA does not prove that those mutations are pathogenic. Further studies are needed to determine whether the recovery of subclonal mutations predicts for either the response to immunosuppressive therapy or malignant evolution, and thus would serve as a valuable assay to guide therapy.
Supplementary Material
Acknowledgments
The authors thank Paul van Hummelen for assistance with sequencing. This research was supported by the Edward P. Evans Foundation (A.Y.), the Conquer Cancer Foundation (A.A.L), Lauri Strauss Leukemia Foundation (A.A.L.), and Leukemia & Lymphoma Society (A.A.L.), the Jock and Bunny Adams Education and Research Fund (J.H.A.), the Stellato Fund (D.M.W.) and a Stand Up To Cancer Innovative Research Grant (D.M.W.).
Footnotes
Author contributions
A.A.L. designed and performed research, collected and interpreted data, and wrote the manuscript. O.O. designed and performed research, interpreted data and performed statistical analysis. N.K. performed research, collected and analyzed data. S.K. performed research, collected and analyzed data. A.Y. designed research. R.E. designed research and interpreted data. N.W. contributed analytical tools. G.A.A. performed statistical analysis and interpreted data. S.J.R. performed research, contributed analytical tools and interpreted data. J.H.A. designed research and wrote the manuscript. D.M.W. designed research, interpreted data and wrote the manuscript.
Conflict of Interest Disclosure
The authors report no relevant conflicts of interest.
References
- 1.Afable MG, 2nd, Tiu RV, Maciejewski JP. Clonal evolution in aplastic anemia. Hematology Am Soc Hematol Educ Program. 2011;2011:90–95. doi: 10.1182/asheducation-2011.1.90. [DOI] [PubMed] [Google Scholar]
- 2.Afable MG, 2nd, Wlodarski M, Makishima H, Shaik M, Sekeres MA, Tiu RV, et al. SNP array-based karyotyping: differences and similarities between aplastic anemia and hypocellular myelodysplastic syndromes. Blood. 2011 Jun 23;117(25):6876–6884. doi: 10.1182/blood-2010-11-314393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katagiri T, Sato-Otsubo A, Kashiwase K, Morishima S, Sato Y, Mori Y, et al. Frequent loss of HLA alleles associated with copy number-neutral 6pLOH in acquired aplastic anemia. Blood. 2011 Dec 15;118(25):6601–6609. doi: 10.1182/blood-2011-07-365189. [DOI] [PubMed] [Google Scholar]
- 4.Wagle N, Berger MF, Davis MJ, Bluemensteil B, Defelice M, Pochanard P, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2(1):82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012 Sep;12(9):599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 6.Zijlstra M, Bix M, Simister NE, Loring JM, Raulet DH, Jaenisch R. Beta 2-microglobulin deficient mice lack CD4-8+ cytolytic T cells. Nature. 1990 Apr 19;344(6268):742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- 7.Challa-Malladi M, Lieu YK, Califano O, Holmes AB, Bhagat G, Murty VV, et al. Combined genetic inactivation of beta2-Microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell. 2011 Dec 13;20(6):728–740. doi: 10.1016/j.ccr.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012 Jul 20;150(2):264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Summers KE, Goff LK, Wilson AG, Gupta RK, Lister TA, Fitzgibbon J. Frequency of the Bcl-2/IgH rearrangement in normal individuals: implications for the monitoring of disease in patients with follicular lymphoma. J Clin Oncol. 2001 Jan 15;19(2):420–424. doi: 10.1200/JCO.2001.19.2.420. [DOI] [PubMed] [Google Scholar]
- 10.Roulland S, Navarro JM, Grenot P, Milili M, Agopian J, Montpellier B, et al. Follicular lymphoma-like B cells in healthy individuals: a novel intermediate step in early lymphomagenesis. J Exp Med. 2006 Oct 30;203(11):2425–2431. doi: 10.1084/jem.20061292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


