Abstract
Purpose
To study the association of serum levels of inflammatory mediators and angiogenic factors with genetic polymorphism in Pakistani age-related macular degeneration (AMD) patients.
Methods
This was a cross-sectional and case-control study that included 90 AMD patients diagnosed through slit-lamp examination, fundoscopy, and ocular coherence tomography. For reference and comparison purposes, 100 healthy age-matched subjects (controls) were also recruited. IL-6, IL-8, VEGF, and CRP levels were estimated in the serum samples of patients and control subjects. Using restriction fragment length polymorphism, single nucleotide polymorphisms were studied in IL-6 (rs1800795, rs1800796, rs1800797), IL-8 (rs4073, rs2227306, rs2227543), VEGF (rs3025039, rs699947), and CRP genes (rs1205, rs1130864). Since the data were obtained from a sample population, the Box–Cox transformation algorithm was applied to reduce heterogeneity of error. Multivariate analyses of variance (M-ANOVA) were applied on the transformed data to investigate the association of serum levels of IL-6, IL-8, VEGF, and CRP with AMD. Genotype and allele frequencies were compared through χ2 tests applying Hardy–Weinberg equilibrium. The serum concentrations of IL-6 and IL-8, VEGF, and CRP between homozygotes and heterozygotes were compared through one-way ANOVA. Significance level was p<0.05.
Results
Compared to control subjects, serum IL-6 (p<0.0001), IL-8 (p<0.0001), VEGF (p<0.0001), and CRP (p<0.0001) levels were significantly elevated in the AMD patients. For rs1800795, patients with the GG genotype showed significantly raised levels of IL-6 compared to those with GC and CC genotypes (p<0.0001). Serum IL-8 levels were significantly higher in patients with the GG genotype compared to the GC and CC genotypes for the single nucleotide polymorphism (SNP) rs2227543 (p<0.002). Similarly, significantly higher VEGF levels were detected for genotype TT for rs3025039 SNP (p<0.038). However, no significant alteration in serum CRP levels was detected in hetero- or homozygotes for rs1205 and rs1130864 SNPs.
Conclusions
Serum IL-6, IL-8, and VEGF levels are substantially increased in AMD, and the levels coincide with polymorphism in the respective gene. No such relationship appears to exist with regard to SNPs of CRP.
Introduction
Age-related macular degeneration (AMD) is an ophthalmic disease that is characterized by early subclinical changes in choroidal and retinal pigment epithelium. The rationale for progression of AMD leading to “geographic atrophy” or “choroidal neovascularization” remains enigmatic. Knowledge of the pathogenesis and causal factors of the disease is therefore needed. AMD appears to be a devastating disease because of the dramatic loss of the central visual field that ultimately affects quality of life [1].
Being a normal process of life, aging acts as a major determinant of AMD; however several other factors, including oxidative stress and a genetic tendency, also contribute toward the pathogenesis of AMD [2]. Pathologically, AMD is considered to be an inflammatory disease [3] because as the disease progresses, macrophages and giant cells accumulate near the drusen, subretinal neovascular membrane, and Bruch’s membrane. Microglial cells then get involved in the inflammatory process; they support the phagocytosis of debris by secreting chemokines and cytokines to create neurotoxic milieu, all resulting in the progression of AMD pathogenesis [4].
The single nucleotide polymorphism (SNP) of many inflammatory factors has been studied in AMD patients. These include toll-like receptor 3, complement factor H, CX3C chemokine receptor 1 (CX3CR1), and interleukin 8 (IL-8) [4]. The variation in cytokine production at the individual level is influenced by genetic polymorphism in the respective gene(s). Consequently, it becomes difficult to consider that AMD is a single-gene disorder. Since the genes associated with AMD show a high degree of variability and interference [5], it seems logical to investigate these variations and determine the role of inflammatory factors in the pathology of AMD. IL-8, which is a member of the chemokine family, is a multifunctional cytokine and is involved in both chronic and acute inflammation [6]. Due to its pro-angiogenic properties, IL-8 appears to be involved in choroidal neovascularization (CNV), a clinical and pathologic complication associated with AMD. It has been shown that new blood vessels that grow during AMD express both VEGF and IL-8 [7]. IL-6 is another cytokine with numerous functions ranging from regulation of the immune response to acute-phase reactions and hematopoiesis [8]. Under normal healthy conditions, the balance between anti-angiogenic and angiogenic growth factors prevents the growth of anomalous blood vessels, but CNV arises as soon as an imbalance occurs between the two [9].
Reduced oxygen levels in the retina stimulate VEGF production in many cell types, including endothelial cells, Muller cells, RPE cells, and pericytes [10]. Elevated VEGF levels in the retina promote neovascularization [11], whereas an inhibition of VEGF downregulates CNV [12]. Bhutto et al. [13] showed that in eyes affected with AMD, VEGF levels are elevated in the posterior segment before CNV occurs, but the exact mechanism of VEGF release/stimulation in CNV is not clear.
C-reactive protein (CRP), which is another inflammatory marker, is a known risk factor for cardiovascular and peripheral arterial disease [14,15]. Hypertension and diabetes mellitus are systemic disorders that increase the risk of vascular abnormalities in the retina. Inflammation and endothelial cell dysfunction are therefore suggested to play significant roles in the development of retinopathy [16]. Increased levels of CRP in the serum greatly increase the risk of inflammatory disorders [17]. Studies on aged African Blacks and young White populations showed that plasma CRP levels are related to genetic make-up [18,19]. In this context, the CRP gene haplotype strongly influences plasma CRP levels in an individual [20,21], and alleles associated with elevated plasma CRP levels may confer an increased risk of AMD.
We hypothesized that gene polymorphism is an important determinant in the pathogenesis of AMD. The aims of the current study were to determine the levels of IL-6, IL-8, VEGF, and CRP in the sera of AMD patients and to investigate their association with the SNPs of respective genes. Using the standard restriction fragment length polymorphism (RFLP) technique, we analyzed three SNPs of interleukins genes: IL-6 (rs1800795, rs1800796, rs1800797) and IL-8 (rs4073, rs2227306, rs2227543); and two SNPs of VEGF genes: (rs3025039, rs699947) and CRP (rs1205, rs113086).
Methods
Ethics
The study conformed to the tenets of the Declaration of Helsinki [22] and was formally approved by the Bioethics Committee of Quaid-i-Azam University, Islamabad, Pakistan (Protocol# BEC-FBS-QAU-06) and the Ethics Committee of Al-Shifa Trust Eye Hospital, Rawalpindi, Pakistan. Written informed consent was obtained from each subject after narrating the study procedures.
Study design and subjects
This was a cross-sectional and case-control study. Detailed proformas were designed to record anthropometric parameters and the clinical history of the disease. A total of 3,911 patients who presented for ophthalmic complications at the Al-Shifa Trust Eye Hospital over a period of about 1 year were screened, and the presence of AMD was diagnosed after slit-lamp examination (C-153 Topcon SL-3D; Santa Clara, CA), ocular coherence tomography (OCT; Zeiss, Oberkochen, Germany), and fluorescent fundus (FFA) done through retinal camera TRC50EX using digital imaging system, IMAGEnet 2000, (Topcon). Inclusion criteria for patients were subjects of both genders aged ≥50 years, nondiabetic, and normotensive with no complication(s) of the eye other than AMD. Subjects with arthritis, diabetes, cancer, end-stage renal disease, arteriosclerosis, and/or using antimetabolite or anti-inflammatory therapy were excluded from the study. Consequently, only 59 males and 31 females who fulfilled the inclusion criteria were selected to participate. Control subjects met the same inclusion criteria and underwent detailed examination for confirmation of healthy ocular conditions. The identity of the healthy subjects and AMD patients was confirmed during the interviews and through certificates of residence as to whether they were Pakistanis by birth. Patients and subjects whose identity as Pakistanis could not be confirmed were omitted from the study.
Blood sampling
Cubital blood (6 ml) was collected for estimation of serum IL-6, IL-8, VEGF, CRP levels, and DNA extraction. We collected blood samples in gel tubes, prepared the sera, and stored them at −20 °C. To avoid long-term storage, serum samples were analyzed after every 20 samples.
Determination of IL-6, IL-8, VEGF, and CRP Levels
Commercially available ELISA kits were used for the estimation of IL-6 and IL-8 (Immunotech, Marseille Cedex, France), VEGF (Cusabio, Wuhan, Hubei, China), and CRP (Amgenix, San Jose, CA). Analyses were performed in triplicate on a fully automated microplate reader with washer (KHB-ST 360; Shanghai, China) following the manufacturer’s instructions. Briefly, for IL-6, 100 µl of standard and serum samples were added to the wells of a 96-well plate precoated with monoclonal antibody. After incubation for 2 h at 18-25 °C, the plate was washed thoroughly with washing solution (PBS + 0.05% Tween 20) and 200 µl of enzyme-conjugated horse reddish protein (HRP) streptavidin substrate was added. The plate was incubated for 30 min in dark at 18-25 °C, after which stop solution (2M H2SO4) was added and optical density (OD) was recorded on an ELISA plate reader at 405 nm wavelength. A standard curve was plotted from the ODs observed for the standard solutions (0.00–1000 pg/ml), and the concentration of IL-6 was calculated from the standard curve. The analytic range for the IL-6 ELISA kit was 3 pg/ml to 150 ng/ml. The intra-assay coefficient of variation (CV) ranged between 1.6% and 6.8%; the inter-assay CV ranged between 7.9% and 14.6%.
For estimation of serum IL-8, 150 µl of standard or sample was added per well and incubated for 2 h at 18–25 °C while shaking. After thoroughly washing the plate, 50 μl of biotinylated antibody and 100 μl of streptavidin-horseradish peroxidase (HRP) conjugate were added and incubated for 30 min at 18–25 °C. Then, 50 μl of stop solution was added, and absorbance was recorded at 450 nm by an ELISA plate reader. A standard curve was plotted from the ODs observed for the standard solutions (0.00–2000 pg/ml), and the concentration of IL-8 was calculated from the standard curve. The kit was sensitive to measure 8 pg/ml. Intra-assay CV ranged between 2.3% and 5.5%, whereas inter-assay CV ranged between 7.6% and 10.1%.
For the measurement of serum VEGF levels, samples were diluted 1:2 with distilled water and 100 μl of standard or diluted samples was added to the wells of precoated plate. Samples were incubated for 2 h at 37 °C, after which the contents were removed. Then, 100 µl of biotin antibody solution was added, and the plate was incubated for 1 h at 37 °C. The plate was then washed thoroughly, and 100 μl of HRP-avidin solution was added. Samples in the plate were incubated again for 1 h at 37 °C and followed by another washing. Ninety microliters of tetramethylbenzidine (TMB) solution was added and incubated for about 15–30 min at 37 °C; 50 μl stop solution (2M H2SO4) was then added to each well. Absorbance was recorded immediately on a microplate reader at 450 nm and 570 nm. The reading at 570 nm was subtracted from the 450-nm reading, and a standard curve was plotted from the readings of the standards. VEGF concentration was calculated through interpolation of the calibration curve. The unknown concentration in the serum samples was calculated from the standard curve and multiplied by the dilution factor. The detection range of the kit was 31.25–2000 pg/ml. The intra-assay CV was <8%, while the inter-assay CV was <10%.
For the estimation of CRP serum concentration, samples were diluted to 1:100 with distilled water. To each well of the plate, 50 µl of standard or sample was added, followed by 50 µl of high sensitivity C - reactive protein (hsCRP) sample diluents. The plate was shaken thoroughly for 30 min and washed thoroughly. Then, 100 µl of CRP enzyme conjugate reagent was added to each well, mixed thoroughly for 30 s, and incubated for 30 min. The plate was washed again, and 100 µl of TMB reagent was added to each well and mixed for about 5 s. Incubation at 18-22 °C was performed for 30 min, after which a stop solution (2M H2SO4) was added that changed the color from blue to yellow. Absorbance was recorded at 450 nm as above. A standard curve was plotted, and the concentration was calculated by multiplying the obtained value by the dilution factor. The detectable range was 1–100 ng/ml. The intra-assay CV ranged between 3.68% and7.67% CV, while the inter-assay CV ranged between 6.04% and 9.82%.
DNA extraction
For DNA extraction, cubital blood was collected in tubes containing potassium ethylene diamine tetra acetic acid (K3.EDTA) (Becton Dickinson and Company, BD, Franklin Lakes, NJ). Whole blood genomic DNA extraction was performed by using the standard organic method of DNA extraction [23]. PCRs were run using the specific primer sets designed for each SNP: IL-6 (rs1800795, rs1800796, rs1800797); IL-8 (rs4073, rs2227306, rs2227543); VEGF (rs3025039, rs699947); and CRP (rs1205, rs113086; Table 1).
Table 1. Primer sequence and PCR optimization conditions with restriction enzymes.
| Gene variants | rs number | Primer sequence | uncut | cut | Restriction enzymes | Annealing temperature (˚C) | |
|---|---|---|---|---|---|---|---|
| IL-6 174 G > C |
1800795 |
Forward: 5' TGACTTCAGCTTTACTCTTTGT3' |
198 |
G: 198 |
SfaN1 |
56.0 |
|
| Reverse: 5'CTGATTGGAAACCTTATTAAG3' |
C:144 |
||||||
| IL-6 572 C > G |
1800796 |
Forward: 5'GCAAAGTCCTCACTGGGAGGA3' |
296 |
C: 296 |
BsrB1 |
64.0 |
|
| Reverse: 5'TCTGACTCCATCGCAGCCC3' |
G: 202 |
||||||
| IL-6 597 A > G |
1800797 |
Forward: 5'GGAGTCACACACTCCACCTG3' |
419 |
G: 419 |
Fok1 |
64.0 |
|
| Reverse: 5'AGCAGAACCACTCTTCCTTTACTT3' |
A: 362 |
||||||
| IL-8 251 A > T |
4073 |
Forward: 5′TCATCCATGATCTTGTTCTAA3' |
542 |
T: 542 |
Mfe1 |
60.0 |
|
| Reverse: 5′GGAAAACGCTGTAGGTCAGA3' |
A: 449 |
||||||
| IL-8 781 C > T |
2227306 |
Forward: 5′CTCTAACTCTTTATATAGGAATT3' |
203 |
T: 203 |
EcoR1 |
56.0 |
|
| Reverse: 5′GATTGATTTTATCAACAGGCA3' |
C: 185 |
||||||
| IL-8 1633 C > T |
2227543 |
Forward: 5′CTGATGGAAGAGAGCTCTGT3' |
397 |
T: 397 |
NlaIII |
60.0 |
|
| Reverse: 5′TGTTAGAAATGCTCTATATTCTC3' |
C: 233 |
||||||
| VEGF 936 C > T |
3025039 |
Forward 5ꞌ AAGGAAGAGGAGACTCTGCGC3ꞌ |
198 |
T:198 |
HpyCH4 III |
58.0 |
|
| Reverse 5ꞌ TATGTGGGTGGGTGTGTCTACAG3ꞌ |
C:112 |
||||||
| VEGF 2578 A > C |
699947 |
Forward 5ꞌ GGATGGGGCTGACTAGGTAAGC3ꞌ |
324 |
C:324 |
BglII |
55 .0 |
|
| Reverse 5ꞌ AGCCCCCTTTTCCTCCAAC3ꞌ |
A:202 |
||||||
| CRP 1846 C > T |
1205 |
Forward 5ꞌ GGAGTGAGACATCTTCTTG3ꞌ |
227 |
A:227 |
HpyCH4 III |
50.0 |
|
| Reverse 5ꞌ CTTATAGACCTGGGCAGT3ꞌ |
G:131 |
||||||
| CRP 1444 G > A | 1130864 | Forward 5ꞌ AGCTCGTTAACTATGCTGGG3ꞌ |
181 |
C:181 |
HpyCH4 III | 59.0 | |
| Reverse 5ꞌ CTTCTCAGCTCTTGCCTTAT3ꞌ | T:156 | ||||||
PCR and restriction fragment length polymorphism (RFLP)
Unless otherwise indicated, all reagents were obtained from Invitrogen (Paisley, UK). For each sample, PCR was performed with a final volume of 20 μl. This mixture contained 40 ng genomic DNA, 2 μl of PCR buffer without Mg2+, 1 μl of 25 mM MgCl2, 0.2 μl (5 U/μl) Taq DNA polymerase, 1 μl of 2 mM dNTPs, and 1 μl each of 20 nM forward and reverse primers (e-oligos, Gene Link, Inc, New York, NY). The final volume was adjusted to 20 μl with PCR-grade water.
PCR was performed in a thermal cycler (Thermo electron Corporation, Millford). PCR conditions were the following: denaturation at 94 °C for 5 min with 1 cycle, denaturation at 94 °C for 45 s, annealing for 45 s at the temperature specific for each primer set (Table 1), and extension at 72 °C for 1 min. The number of cycles at step 2 was 35. In step 3, the final extension was performed at 72 °C for 10 min. The PCR product was mixed with 5 U of restriction enzyme and the reaction buffer as per the manufacturer’s instructions. The cuttable and uncuttable products are shown in Table 1. The mixture was incubated for 16–18 h at 37 °C and, 5 μl of product after the enzyme reaction was loaded onto 3% agarose gel containing ethidium bromide. A 100-bp DNA ladder (Invitrogen) was run alongside to compare the sizes of fragments.
DNA sequencing
Sequencing was carried out through the standard method of Sanger DNA sequencing. For each SNP, purified PCR products were loaded onto an ABI 3130 genetic analyzer, 3013 (Applied Biosystems, Life Technology, Thermo electron Corporation, Millford, CA). The results of sequencing were read, and the SNP genotypes were validated.
Statistical analysis
Data were transformed using XLSTAT (version 2015, Belmont, MA). Statistical analyses were performed with Statistical Product and Service Solutions (SPSS) software (version 18.0; IBM, Chicago, IL), and graphs were drawn using GraphPad Prism software (GraphPad Prism Version 5.00 for Windows; San Diego, CA). The data were transformed using the method described by Box and Cox [24] for reducing heterogeneity of error that permitted the assumption of equal variance to be met. The data were transformed using the method as described by Box and Cox for reducing heterogeneity of error that permitted the assumption of equal variance to be met. Box and Cox transformation provided an algorithm through which the optimal value of the transformation parameter λ was selected by the method of maximum likelihood. Transformed data were subjected to multivariate analysis of variance (M-ANOVA) for statistical evaluation of the association of serum IL-6, IL-8, CRP, and VEGF levels with AMD and gender of the subject. One-way ANOVA was applied to evaluate the association of heterozygosity and homozygosity of studied SNPs with the serum levels of each protein. The expected Hardy–Weinberg frequencies for the alleles and genotypes were calculated, and any deviation of data from Hardy–Weinberg equilibrium was tested through χ2 analysis. For each SNP, allele frequencies are given in the results section. The probability value was p<0.05.
Results
Anthropomorphic history and diagnostics
Diagnostic symptoms for AMD included small or large drusen, sub retinal CNV, hemorrhages, edema, geographic atrophy and RPE atrophy. Of the AMD patients, 65.5% were males and 34.5% were females. Of the 100 control subjects, 55 were males and were 45 females. There was no significant difference as regards age in any of the groups (p<0.448). Diagnostic characteristics of the AMD patients are presented in Table 2.
Table 2. Number of subjects, age, gender and diagnostic parameters in control subjects and AMD patients.
| Characteristics | Control | AMD |
|---|---|---|
| N |
100 |
90 |
| Males |
55 |
63 |
| Females |
45 |
27 |
| Age (median ± SE) |
63.0 ± 0.84 |
70.0 ± 1.07 |
| Wet AMD |
NA |
59 |
| Dry AMD |
NA |
31 |
| Maculopathy Pattern |
NA |
32 |
| Exudative Pattern |
NA |
40 |
| Atrophic Pattern | NA | 18 |
NA: not applicable
Serum levels of IL-6, IL-8, VEGF and CRP
The ANOVA for λ= 0.413 that yielded the lowest error sums of squares was used for hypothesis testing. M-ANOVA showed that serum levels of IL-6, IL-8, VEGF and CRP were significantly elevated in AMD patients (p<0.0001; Wilk’s λ = 0.064; F= 278.7); however, these levels were not found to be associated with gender (p<0.560; Wilk’s λ = 0.997; F= 0.463). The levels of all these factors were found significantly associated with the type of AMD that is wet or dry (p<0.0001; Wilk’s λ = 0.018; F= 147.406). IL-6 and IL-8 levels were significantly increased in patients with dry AMD (p<0.0001), while serum VEGF and CRP levels were significantly elevated in patients with wet AMD (p<0.0001; Figure 1A-D). There was no significant difference in the serum levels of inflammatory factors with respect to age groups (p>0.705; Wilk’s λ = 0.983; F=0.631; Table 3). No significant change was observed in the concentration of IL-6, IL-8, VEGF, and CRP with respect to the pattern of AMD (p<0.302; Wilk’s λ = 0.882; F=1.202; Table 4).
Figure 1.
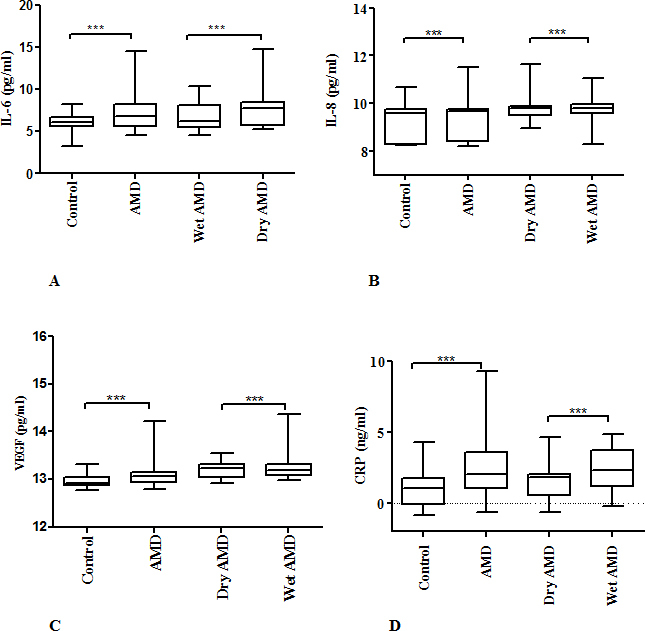
The box-and-whisker plot represents the mean and minimum to maximum range of IL-6, IL-8, VEGF and CRP levels in transformed data. Serum levels were measured in 100 control subjects and 90 AMD patients. Among AMD patients 63 had wet type and 27 had dry type AMD. The serum levels of (A) IL-6, (B) IL-8, (C) VEGF and (D) CRP were significantly elevated in AMD patients compared to control subjects (p<0.0001). The levels of (A) IL-6 and (B) IL-8 were significantly elevated in dry AMD patients compared to wet AMD patients. The levels of (C) VEGF and (D) CRP were significantly elevated in wet AMD patients compared to dry AMD patients (p<0.0001).
Table 3. Comparison of levels of IL-6 (pg/ml), IL-8 (pg/ml), VEGF (pg/ml) and CRP (ng/ml) with respect to each age group.
| Age group | Group | N | IL-6 | IL-8 | VEGF | CRP |
|---|---|---|---|---|---|---|
| 50-59 |
Control |
45 |
8.55±0.37 |
13.03±0.15 |
16.96±0.57 |
4.042±0.75 |
| |
AMD |
15 |
11.18±0.80 |
12.82±0.22 |
19.38±0.63 |
1.646±0.32 |
| 60-69 |
Control |
30 |
10.10±0.39 |
12.69±1.11 |
16.82±0.69 |
4.91±0.99 |
| |
AMD |
33 |
9.73±0.45 |
13.61±0.21 |
17.35±3.87 |
4.34±0.82 |
| 70-79 |
Control |
21 |
9.117±0.48 |
12.38±0.22 |
19.18±0.56 |
1.947±0.77 |
| |
AMD |
29 |
10.54±0.43 |
13.31±0.18 |
19.14±0.51 |
3.62±0.61 |
| 80-89 |
Control |
4 |
9.866±1.24 |
12.65±0.57 |
18.07±1.90 |
4.25±2.75 |
| |
AMD |
10 |
9.51±0.89 |
13.50±0.26 |
17.45±1.22 |
4.79±1.27 |
| 90-99 |
Control |
0 |
none |
none |
none |
none |
| |
AMD |
2 |
17.72±5.35 |
14.62±1.60 |
16.24±3.87 |
-0.15±0.34 |
| ≥100 |
Control |
0 |
none |
none |
none |
none |
| AMD | 1 | 12.346 | 12.37 | 22.46 | 2.797 |
none: means no subject in these age groups could be found in the control group
Table 4. Levels of IL-6 (pg/ml), IL-8 (pg/ml), VEGF (pg/ml) and CRP (ng/ml) with respect to pattern of AMD.
| Protein | N | IL-6 | IL-8 | VEGF | CRP |
|---|---|---|---|---|---|
| Maculopathy |
32 |
11.71±0.94 |
13.73±0.81 |
17.03±0.97 |
3.90±0.91 |
| Exudative |
40 |
9.83±0.40 |
13.30±0.56 |
18.81±0.54 |
4.26±0.62 |
| Atrophic |
18 |
10.94±0.47 |
12.94±0.59 |
18.79±0.60 |
2.83±0.61 |
| p Value | - | 0.058 | 0.284 | 0.249 | 0.280 |
Allele and genotype frequencies of IL-6
Among SNPs of IL-6, the frequency of the CC genotype for SNP rs1800795 (p<0.002; odds ratio [OR] = 0.4981; confidence interval [CI] 0.2422 – 1.0243) was significantly increased compared to other genotypes. The frequency of allele C increased significantly (p<0.0001; OR= 3.7096; CI 2.2357 – 6.1551) in AMD patients compared to control subjects. For SNP rs1800796, the difference in genotype frequency between the control subjects and AMD patients was not significant (p<0.7958; OR= 1.7024; CI 0.9555 – 3.033). There was no significant difference in the allele frequency between control group and AMD patients (p<0.8957; OR = 1.0279; CI 0.6829 – 1.5472). When SNP rs1800797 was compared to other genotypes, the frequency of genotype GA was significantly reduced (p<0.0001; OR= 0.1499; CI 0.0626-0.3588). Furthermore, the frequency of allele A was significantly reduced (p<0.0001; OR= 0.1843; CI 0.0798-0.4258) in AMD patients when compared with healthy controls. The serum concentration of IL-6 was significantly higher in AMD patients with genotype GG compared to those with genotypes GC or CC for SNP rs1800795 (p<0.0001; F= 8.801). No significant change was observed in serum IL-6 concentration with respect to genotype in other SNPs of IL-6 (Table 5).
Table 5. Genotype and allele frequencies in control and AMD patients for IL-6 SNPs.
| Genotype | t test |
Control (N=100) |
AMD (N=90) |
Alleles | Allele frequency reported in Pakistan (%) [25] |
Control |
AMD |
p value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| p value | F value | p value | F value | HWE allele frequency | HWE allele frequency | |||||
| rs1800795 |
|
|||||||||
| GG |
0.0001*** |
0.105 |
2.707 |
0.0001*** |
8.801 |
G |
87 |
0.865 (86.5%) |
0.633 (63.3%) |
0.0001*** |
| GC |
0.916 |
C |
13 |
0.135 (13.5%) |
0.367 (36.7%) |
|||||
| CC |
NA |
|
||||||||
|
rs1800796 |
|
|||||||||
| GG |
0.010* |
0.514 |
0.671 |
0.293 |
1.254 |
G |
67 |
0.59 (59%) |
0.583 (58.3%) |
0.896 |
| GC |
0.0001*** |
C |
33 |
0.41 (41%) |
0.417 (41.7%) |
|||||
| CC |
0.044* |
|
||||||||
|
rs1800797 |
|
|||||||||
| GG |
0.0001*** |
3.154 | 0.081 | 0.254 | 0.616 | G |
86 |
0.82 (82%) |
0.96 (96%) |
0.0001*** |
| GA |
0.733 |
A |
14 |
0.18 (18%) |
0.04 (4%) |
|||||
| AA | NA | |||||||||
HWE: Hardy Weinberg Equilibrium; NA: none of the subject had this genotype therefore t test was not available; *p < 0.05; ***p < 0.0001
Allele and genotype frequencies of IL-8
In case of IL-8, the difference in genotype frequency for SNP rs4073 between control and AMD patients was not significant (p<0.4239; OR=1.301; CI 0.7347-2.3039). Allele frequency was not significantly different between control subjects and AMD patients (p<0.2366; OR= 1.1635; CI 0.7703 – 1.7575). For SNP rs2227306, there was no statistically significant difference in the genotype frequency between AMD patients and control subjects (p<0.0547; OR= 0.7101; CI 0.371 – 1.3592). The frequency of allele T was significantly lower than allele C in AMD patients (p<0.0001; OR= 0.5193; CI 0.3133-0.8608). For SNP rs2227543, the frequency of genotype TT was significantly higher in AMD patients compared to control subjects (p<0.0371; OR= 0.8906; CI 0.5034-1.5756). The frequency of allele T was significantly higher in AMD patients compared to the control group (p<0.0372; OR= 1.5529; CI 1.0257-2.3511). For SNP rs2227543, serum IL-8 levels were significantly higher in AMD patients with genotype TT compared to those with the CC or CT genotypes (p<0.002; F= 6.80). No significant change in serum IL-8 levels was observed with respect to other SNPs (Table 6).
Table 6. Genotype and allele frequencies in control and AMD patients for IL-8 SNPs.
| Genotype | t test |
Control (N=100) |
AMD (N=90) |
Alleles | Allele frequency reported in Pakistan (%) [25] |
Control |
AMD |
p value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| p value | F value | p value | F value | HWE allele frequency | HWE allele frequency | |||||
|
rs4073 |
|
|||||||||
| TT |
0.0001*** |
0.368 |
1.015 |
0.340 |
1.099 |
T |
64 |
0.625 (62.5%) |
0.589 (58.9%) |
0.2366 |
| AT |
0.007** |
A |
36 |
0.375 (37.5%) |
0.411 (41.1%) |
|||||
| AA |
0.146 |
|
||||||||
|
rs2227306 |
|
|||||||||
| CC |
0.0001*** |
0.125 |
2.148 |
0.951 |
0.050 |
C |
74 |
0.73 (73%) |
0.84 (84%) |
0.0001*** |
| CT |
0.0001*** |
T |
26 |
0.27 (27%) |
0.16 (16%) |
|||||
| TT |
0.137 |
|
||||||||
|
rs2227543 |
|
|||||||||
| CC |
0.0001*** |
0.710 | 0.495 | 0.002** | 6.80 | C |
68 |
0.66 (66%) |
0.555 (55.5%) |
0.0372 |
| CT |
0.0001*** |
T |
32 |
0.34 (34%) |
0.445 (45.5%) |
|||||
| TT | 0.833 | |||||||||
HWE: Hardy Weinberg Equilibrium; **p < 0.01; ***p < 0.0001
Allele and genotype frequencies of VEGF
In the SNPs of VEGF, for rs3025039, the genotype frequency in AMD patients was not significantly different compared to control subjects (p<0.5124; OR= 0.899; CI 0.3544 – 2.2807). There was no significant difference in allelic frequency between AMD patients and control subjects (p<0.310; OR= 1.3956; CI 0.7329 – 2.6577). For the gene variant rs699947, the genotype frequency between AMD patients and control subjects was not significantly different (p<0.052; OR= 1.0114; CI 0.4079 – 2.5078). The frequency of allele A was significantly higher in AMD patients compared to the control subjects (p<0.0363; OR=1.5539; CI 1.0283-2.3482). Serum VEGF levels were significantly elevated in AMD patients with TT genotype for gene variant rs3025039 compared to patients with CC or CT genotypes (p < 0.038). For gene variant rs699947, no significant change in serum VEGF concentration was observed between the control subjects and AMD patients with respect to the genotype (Table 7).
Table 7. Genotype and allele frequencies in control and AMD patients for VEGF SNPs.
| Genotype | t test |
Control (N=100) |
AMD (N=90) |
Alleles | Allele frequency reported in Pakistan (%) [25] |
Control |
AMD |
p value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| p value | F value | p value | F value | HWE allele frequency | HWE allele frequency | |||||
|
rs3025039 |
|
|||||||||
| CC |
0.801 |
0.62 |
0.481 |
0.038* |
3.441 |
C |
93 |
0.905 (90.5%) |
0.872 (87.2%) |
0.310 |
| CT |
0.258 |
T |
7 |
0.095 (9.5%) |
0.128 (12.8%) |
|||||
| TT |
0.342 |
|
||||||||
|
rs699947 |
|
|||||||||
| CC |
0.349 |
0.755 | 0.283 | 0.938 | 0.396 | C |
59 |
0.65 (65%) |
0.54 (54%) |
0.036 |
| CA |
0.134 |
A |
41 |
0.35 (35%) |
0.46 (46%) |
|||||
| AA | 0.295 | |||||||||
HWE: Hardy Weinberg Equilibrium; *p < 0.05
Allele and genotype frequencies of CRP
In the gene variants of CRP, genotype frequency for SNP rs1205 between control subjects and AMD patients was not significantly different (p<0.6849; OR= 0.4779; CI 0.2398 – 0.9524). No significant difference was found in allele frequency between AMD patients and control subjects (p<0.342; OR= 1.107; CI 0.7376 – 1.6614). For SNP rs1130864, there was no significant difference in genotype frequency between control subjects and AMD patients (p<0.2411; OR= 1.0338; CI 0.5739 – 1.8623). No significant difference was observed in allele frequency between controls subjects and AMD patients (p<0.1560; OR= 1.4043; CI 0.8784 – 2.2452). Likewise, no significant difference in serum CRP levels was observed between heterozygous or homozygous AMD patients (p<0.618; F= 0.484) and control subjects for rs1205 variant (p<0.233; F=1.487). The CRP levels were not significantly different in heterozygous and homozygous individuals for gene variant rs1130864 in AMD patients (p<0.534; F= 0.633) or healthy subjects (p<0.171; F= 1.810; Table 8). For the above SNPs, allele frequencies reported previously [25] are also given in Table 5- Table 8 for comparison purpose.
Table 8. Genotype and allele frequencies in control and AMD patients for CRP SNPs.
| Genotype | t test |
Control (N=100) |
AMD (N=90) |
Alleles | Allele frequency reported in Pakistan (%) [25] |
Control |
AMD |
p value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| p value | F value | p value | F value | HWE allele frequency | HWE allele frequency | |||||
|
rs1205 |
|
|||||||||
| CC |
0.856 |
0.233 |
1.487 |
0.618 |
0.484 |
C |
75 |
0.575 (57.5%) |
0.55 (55%) |
0.342 |
| CT |
0.0001*** |
T |
25 |
0.425 (42.5) |
0.45 (45%) |
|||||
| TT |
0.123 |
|
||||||||
|
rs1130864 |
|
|||||||||
| AA |
0.001** |
0.171 | 1.810 | 0.534 | 0.633 | G |
72 |
0.79 (79%) |
0.72 (72%) |
0.156 |
| AG |
0.002** |
A |
28 |
0.21 (21%) |
0.28 (28%) |
|||||
| GG | 0.905 | |||||||||
HWE: Hardy Weinberg Equilibrium; **p < 0.01; *** p < 0.0001
Discussion
We found significant elevations in serum IL-6, IL-8, VEGF, and CRP levels in our AMD patients compared to healthy controls. Although, gene variants for IL-6, IL-8, and VEGF showed a significant association with the incidence of AMD, we did not observe any significant association between gene variants of CRP and the risk of AMD. This appears to coincide with the previously available epidemiologic, genetic, and pathologic data that support the view that inflammatory processes contribute to the initiation and progression of AMD [2,3,26]. Although plasma is preferable for estimating the above parameters [27], our patients presented and were diagnosed at different time intervals over the year and their samples were preserved at −20 °C. Consequently, sera were used for analyses instead of plasma.
There was no statistical difference in the ages of control subjects and AMD patients, supporting the comparability of the two groups. The unusual finding of our study is that the number of patients with wet-type AMD was greater than the dry type. This is in contrast to previous data showing that nearly four-fifths of all AMD cases present with dry AMD, which is characterized by early stage AMD without exudation or scarring of retinal tissue [26]. When compared with other populations of the world [28], this deviation in our sample population is possibly because dry AMD progresses slowly; patients perceive this visual loss as age related and so report it less frequently. It is expected that further large-scale studies on the general Pakistani population would confirm the proportion of patients with dry AMD.
In the present cohort of AMD patients, elevated levels of interleukins were indicative of the involvement of inflammatory processes in the pathology of AMD. As increased serum concentrations of pro-inflammatory cytokines, such as IL-6 and IL-8, are related to the production of reactive oxygen species, which further leads to the production of such cytokines, the positive feedback not only promotes inflammatory processes but also enhances vascularization, contributing ultimately to progression of the disease [29]. Our study revealed that patients with the CC genotype had lower serum IL-6 levels compared to those with the GG genotype. Allele C for SNP rs1800795 has previously been associated with reduced IL-6 levels [30]. The functional C/G transversion in SNP rs1800795 modulates binding of transcription factors, such as GATA1. The relationship between this genotype and IL-6 serum levels is complex due to the involvement of environmental interactions [31].
Importantly, our study implies that the presence of allele A for gene variant rs1800797 appears to reduce the risk of AMD as its frequency was higher in control subjects than in AMD patients. This SNP is present in the promoter region of the IL-6 gene. Variation in this region is related to the functional alteration of the protein product that ultimately leads to occurrence of the disease [32]. Our study reveals that the presence of allele T in SNP rs2227306 and allele C in SNP rs2227543 for the IL-8 gene is related to the risk of AMD as frequencies of these alleles were found to be higher in AMD patients compared to normal subjects. At the biochemical level, a significant elevation in serum IL-8 levels in AMD patients suggests that the SNPs under investigation are functional variants of the gene. A study in a Taiwan Chinese population examined 14 candidate SNPs of five interleukin genes and found that for SNP 2227306, allele T had a significant association with wet-type AMD, but for IL-6, no specific SNP was identified as a risk factor for AMD [5]. Thus, AMD seems to be the result of a linkage between many different gene loci related to the risk of disease.
We found serum VEGF concentration to be significantly increased in AMD patients compared to control subjects. Moreover, patients with wet-type AMD showed significantly higher VEGF levels compared to patients with dry-type AMD. VEGF is an important angiogenesis regulator that is overexpressed in retinal tissues of AMD patients. One of the key features of wet AMD is angiogenesis, which leads to neovascularization and ultimately blindness in the elderly population. Previous studies have provided evidence that VEGF is a major mediator of angiogenesis and vascular leakage in wet-type AMD [33].
Altered levels of VEGF gene expression due to polymorphism contributes to alteration in protein production [34]. In the same AMD patients, the frequency of allele A in VEGF gene variant rs699947 was found to be significantly higher compared to age-matched control subjects, while no significant change in allele or gene frequency was noticeable for gene variant rs3025039. It is known that SNP rs699947 is present in the promoter region, while SNP rs3025039 lies in the exon [35]. A significant association between AMD and allele T for SNP rs3025039 has been reported by Lin et al. [36] in the Chinese population, yet this association was not found in other studies performed on Caucasian [37] and Tunisian [38] populations.
We found that CRP levels were significantly elevated in AMD patients compared to control subjects. During chronic low-level inflammation, CRP levels are slightly raised as documented in a subset of aged individuals [39]. Meta-analysis has previously indicated a link between CRP and AMD, with elevated CRP levels being related to a twofold likelihood of late-onset AMD [40]; however, we could not find any association between serum CRP levels and its respective SNPs. One possibility is that CRP has some direct pathophysiologic role in AMD. This role is expected to be mediated through its capacity to induce complement activation via an unknown alternative pathway and contribute to tissue damage through several complement-mediated mechanisms [41]. In the present investigation, no significant change as related to CRP gene variants was observed. We studied SNPs rs1205 and rs1130864, which are present in the untranslated region of the CRP gene. Although our study provides evidence that raised serum CRP levels are associated with AMD, this might not be solely due to genetic variation. Kim et al. [42] acquired similar results regarding the CRP gene in wet AMD patients. Genetic susceptibility of an individual in complex diseases such as AMD is the result of the cumulative effect of several predictive alleles studied with full haplotype information rather than a single allelic variation [43]. The polymorphisms in genes of several cytokines influence variation in cytokine production via gene transcription [44].
In conclusion, the present study provides evidence that elevated serum IL-6, IL-8, VEGF, and CRP levels are substantially increased in AMD. The IL-6 gene variants rs1800795 and rs1800797, IL-8 gene variants rs2227306, rs2227543 and VEGF gene variant rs699947 are involved in the AMD pathogenesis. However, the gene variants studied for CRP did not show any association with the disease. The data support the role of inflammation in AMD pathogenesis but do not provide specific insights as regards molecular mechanisms contributing to the disease.
The number of patients and controls in our study was relatively few due to very stringent inclusion criteria; thus, the findings do not represent the whole population of the country. Further functional and genomic studies are required to confirm our findings with studies performed on larger and more diverse samples from the same population so that involvement of inflammatory and angiogenic factors in the pathogenesis of AMD can be delineated.
Acknowledgments
The authors thank all the participants of the study, the patients and the control subjects. The research was supported by the funds provided by Higher Education Commission (HEC), Islamabad, Pakistan under the program “access to scientific instrumentation” and a graduate student scholarship grant (PIN No.1061414Bm044) by the HEC to the principal author. Authors declare that they have no conflict of interest. We are grateful to Prof. Dr. Wajid Ali Khan, Chief Consultant and Prof. Dr. Nadeem Qureshi, Head Vitreo-retina Department and all the residents and staff of Al-Shifa Trust Eye hospital for their cooperation during the screening of the patients, to the Director NORI and Mrs. Shahnaz Murtaza, Head Diagnostics, NORI, Islamabad, for extending laboratory facilities to carry out ELISA. We are thankful to Dr. Kiran Afshan, Assistant Professor, Department of Animal Sciences, Quaid-i-Azam University, Islamabad, Pakistan for her advice on statistical analyses.
References
- 1.Grisanti S, Tatar O. The role of vascular endothelial growth factor and other endogenous interplayers in age-related macular degeneration. Prog Retin Eye Res. 2008;27:372–90. doi: 10.1016/j.preteyeres.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Klein R, Cruickshanks KJ, Nash SD, Krantz EM, Nieto FJ, Huang GH, Pankow JS, Klein BE. The prevalence of age-related macular degeneration and associated risk factors. Arch Ophthalmol. 2010;128:750–8. doi: 10.1001/archophthalmol.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hageman GS, Luthert PJ, Chong NV, Johnson LV, Anderson DH, Mullins RF. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch's membrane interface in aging and age-related macular degeneration. Prog Retin Eye Res. 2001;20:705–32. doi: 10.1016/s1350-9462(01)00010-6. [DOI] [PubMed] [Google Scholar]
- 4.Gupta N, Brown KE, Milam AH. Activated microglia in human retinitis pigmentosa, late-onset retinal degeneration, and age-related macular degeneration. Exp Eye Res. 2003;76:463–71. doi: 10.1016/s0014-4835(02)00332-9. [DOI] [PubMed] [Google Scholar]
- 5.Tsai YY, Lin JM, Wan L, Lin HJ, Tsai Y, Lee CC, Tsai CH, Tsai FJ, Tseng SH. Interleukin gene polymorphisms in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49:693–8. doi: 10.1167/iovs.07-0125. [DOI] [PubMed] [Google Scholar]
- 6.Tong Y, Liao J, Zhang Y, Zhou J, Zhang H. Mao M. LOC387715/HTRA1 gene polymorphisms and susceptibility to age-related macular degeneration: A HuGE review and meta-analysis. Mol Vis. 2010;16:1958–81. [PMC free article] [PubMed] [Google Scholar]
- 7.Heinzmann A, Ahlert I, Kurz T, Berner R, Deichmann KA. Association study suggests opposite effects of polymorphisms within< i> IL8</i> on bronchial asthma and respiratory syncytial virus bronchiolitis. J Allergy Clin Immunol. 2004;114:671–6. doi: 10.1016/j.jaci.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 8.Kalayoglu MV, Bula D, Arroyo J, Gragoudas ES, D’Amico D, Miller JW. Identification of Chlamydia pneumoniae within human choroidal neovascular membranes secondary to age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol. 2005;243:1080–90. doi: 10.1007/s00417-005-1169-y. [DOI] [PubMed] [Google Scholar]
- 9.Kauffmann DJ, Van Meurs JC, Mertens DA, Peperkamp E, Master C, Gerritsen ME. Cytokines in vitreous humor: interleukin-6 is elevated in proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci. 1994;35:900–6. [PubMed] [Google Scholar]
- 10.Zhou J, Pham L, Zhang N, He S, Gamulescu MA, Spee C, Ryan SJ, Hinton DR. Neutrophils promote experimental choroidal neovascularization. Mol Vis. 2005;11:414–24. [PubMed] [Google Scholar]
- 11.Aiello LP, Northrup JM, Keyt BA, Takagi H, Iwamoto MA. Hypoxic regulation of vascular endothelial growth factor in retinal cells. Arch Ophthalmol. 1995;113:1538–44. doi: 10.1001/archopht.1995.01100120068012. [DOI] [PubMed] [Google Scholar]
- 12.Pierce EA, Avery RL, Foley ED, Aiello LP, Smith LE. Vascular endothelial growth factor/vascular permeability factor expression in a mouse model of retinal neovascularization. Proc Natl Acad Sci USA. 1995;92:905–9. doi: 10.1073/pnas.92.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhutto IA, McLeod DS, Hasegawa T, Kim SY, Merges C, Tong P, Lutty GA. Pigment epithelium-derived factor (PEDF) and vascular endothelial growth factor (VEGF) in aged human choroid and eyes with age-related macular degeneration. Exp Eye Res. 2006;82:99–110. doi: 10.1016/j.exer.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson GS, Pierce EA, Rook SL, Foley E, Webb R, Smith LE. Oligodeoxynucleotides inhibit retinal neovascularization in a murine model of proliferative retinopathy. Proc Natl Acad Sci USA. 1996;93:4851–6. doi: 10.1073/pnas.93.10.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simundic AM. New insights in the pathophysiology of inflammation. Biochem Med. 2011;21:243–4. doi: 10.11613/bm.2011.033. [DOI] [PubMed] [Google Scholar]
- 16.Thiruvagounder M, Khan S, Sheriff DS. The prevalence of metabolic syndrome in a local population in India. Biochem Med. 2010;20:249–52. [Google Scholar]
- 17.Colak E, Jaković NK, Žorić L, Radosavljević A. Stanković, Singh NM. The association of lipoprotein parameters and C-reactive protein in patients with age-related macular degeneration. Ophthalmic Res. 2011;46:125–32. doi: 10.1159/000323815. [DOI] [PubMed] [Google Scholar]
- 18.Kikuchi M, Nakamura M, Ishikawa K, Suzuki T, Nishihara H, Yamakoshi T, Nishio K, Taki K, Niwa T, Hamajima N, Terasaki H. C-reactive protein levels in patients with polypoidal choroidal vasculopathy and patients with neovascular age-related macular degeneration. Ophthalmology. 2007;114:1722–7. doi: 10.1016/j.ophtha.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 19.Pankow JS, Folsom AR, Cushman M, Borecki IB, Hopkins PN, Eckfeldt JH, Tracy RP. Familial and genetic determinants of systemic markers of inflammation: the NHLBI family heart study. Atherosclerosis. 2001;154:681–9. doi: 10.1016/s0021-9150(00)00586-4. [DOI] [PubMed] [Google Scholar]
- 20.Vickers MA, Green FR, Terry C, Mayosi BM, Julier C, Lathrop M, Ratcliffe PJ, Watkins HC, Keavney B. Genotype at a promoter polymorphism of the interleukin-6 gene is associated with baseline levels of plasma C-reactive protein. Cardiovasc Res. 2002;53:1029–34. doi: 10.1016/s0008-6363(01)00534-x. [DOI] [PubMed] [Google Scholar]
- 21.Carlson CS, Aldred SF, Lee PK, Tracy RP, Schwartz SM, Rieder M, Liu K, Williams OD, Iribarren C, Lewis EC, Fornage M, Boerwinkle E, Gross M, Jaquish C, Nickerson DA, Myers RM, Siscovick DS, Reiner AP. Polymorphisms within the C - reactive protein (CRP) promoter region are associated with plasma CRP levels. Am J Hum Genet. 2005;77:64. doi: 10.1086/431366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rennie D. Disclosure to the reader of institutional review board approval and informed consent. JAMA. 1997;277:922–3. [PubMed] [Google Scholar]
- 23.Sambrook J, Russell DW. Molecular Cloning. A laboratory manual. 3rd Edition. Volume 1. Cold Spring Harbour Laboratory Press, New York, USA. 2001. [Google Scholar]
- 24.Box GEP, Cox DR. An analysis of transformations. J R Stat Soc, B. 1964;26:211–52. [Google Scholar]
- 25.Kersey PJ, Allen JE, Christensen M, Davis P, Falin LJ, Grabmueller C, Hughes DS, Humphrey J, Kerthornou A, Khobova J, Langridge N, McDowall MD, Matheswari U, Maslen G, Nuhn M, Ong CK, Paulini M, Pedro H, Toneva I, Tuli MA, Walts B, Williams G, Wilson D, Youens-Clar K, Monaco MK, Stein J, Wei X, Ware D, Bosler DM, Howe KL, Kulesha E, Lawson D, Staines DM. Ensembl Genomes 2013: scaling up access to genome-wide data. Nucleic Acids Res. 2014;42(D1):D546–52. doi: 10.1093/nar/gkt979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seddon JM, Gensler G, Rosner B. C-Reactive Protein and CFH, ARMS2 / HTRA1 gene variants are independently associated with risk of macular degeneration. Ophthalmology. 2010;117:1560–6. doi: 10.1016/j.ophtha.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banks RE, Forbes MA, Kinsey SE, Stanley A, Ingham E, Walters C, Selby PJ. Release of the angiogenic cytokine vascular endothelial growth factor (VEGF) from platelets: significance for VEGF measurements and cancer biology. Br J Cancer. 1998;77:956–64. doi: 10.1038/bjc.1998.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dewan A, Liu M, Hartman S, Zhang SSM, Liu DT, Zhao C, Tam PO, Chan WM, Lam DS, Snyder M, Barnstable C, Pang CP, Hoh J. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–92. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 29.Owen CG, Fletcher AE, Donoghue M, Rudnicka AR. How big the burden of visual loss caused by age related macular degeneration in the United Kingdom? Br J Ophthalmol. 2003;87:312–7. doi: 10.1136/bjo.87.3.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding X, Patel M, Chan CC. Molecular pathology of age-related macular degeneration. Prog Retin Eye Res. 2009;28:1–18. doi: 10.1016/j.preteyeres.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szmitko PE, Wang CH, Weisel RD, de Almeida JR, Anderson TJ, Verma S. New markers of inflammation and endothelial cell activation. Circulation. 2003;108:1917–1923. doi: 10.1161/01.CIR.0000089190.95415.9F. [DOI] [PubMed] [Google Scholar]
- 32.Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, Woo P. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Sawada T, Sawada O, Saishin Y, Liu P, Ohji M. Serum and plasma vascular endothelial growth factor concentrations before and after intravitreal injection of aflibercept or ranibizumab for age-related macular degeneration. Am J Ophthalmol. 2014;158:738–44. doi: 10.1016/j.ajo.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Ferrara N, Damico L, Shams N, Lowman H, Kim R. Development of ranibizumab, an anti–vascular endothelial growth factor antigen binding fragment, as therapy for neovascular age-related macular degeneration. Retina. 2006;26:859–70. doi: 10.1097/01.iae.0000242842.14624.e7. [DOI] [PubMed] [Google Scholar]
- 35.Jain L, Vargo CA, Danesi R, Sissung TM, Price DK, Venzon D, Venitz J, Figg WD. The role of vascular endothelial growth factor SNPs as predictive and prognostic markers for major solid tumors. Mol Cancer Ther. 2009;8:2496–508. doi: 10.1158/1535-7163.MCT-09-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin JM, Wan L, Tsai YY, Lin HJ, Tsai Y, Lee CC, Tsai CH, Tseng SH, Tsai FJ. Vascular endothelial growth factor gene polymorphisms in age-related macular degeneration. Am J Ophthalmol. 2008;145:1045–51. doi: 10.1016/j.ajo.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 37.Huang C, Xu Y, Li X, Wang W. Vascular endothelial growth factor A polymorphisms and age-related macular degeneration: a systematic review and meta-analysis. Mol Vis. 2013;19:1211. [PMC free article] [PubMed] [Google Scholar]
- 38.Habibi I, Sfar I, Chebil A, Kort F, Bouraoui R, Jendoubi-Ayed S, Makhlouf M, Abdallah TB, El Matri L, Gorgi Y. Vascular endothelial growth factor genetic polymorphisms and susceptibility to age-related macular degeneration in Tunisian population. Biomarker Research. 2014;2:15. doi: 10.1186/2050-7771-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaumberg DA, Christen WG, Kozlowski P, Miller DT, Ridker PM, Zee RY. A prospective assessment of the Y402H variant in complement factor H, genetic variants in C-reactive protein, and risk of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2006;47:2336–40. doi: 10.1167/iovs.05-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higgins GT, Wang JH, Dockery P, Cleary PE, Redmond HP. Induction of angiogenic cytokine expression in cultured RPE by ingestion of oxidized photoreceptor outer segments. Invest Ophthalmol Vis Sci. 2003;44:1775–82. doi: 10.1167/iovs.02-0742. [DOI] [PubMed] [Google Scholar]
- 41.Du Clos TW. Function of C-reactive protein. Ann Med. 2000;32:274–8. doi: 10.3109/07853890009011772. [DOI] [PubMed] [Google Scholar]
- 42.Kim IK, Ji F, Morrison MA, Adams S, Zhang Q, Lane AM, Capone A, Dryja TP, Ott J, Miller JW, DeAngelis MM. Comprehensive analysis of CRP, CFH Y402H and environmental risk factors on risk of neovascular age-related macular degeneration. Mol Vis. 2008;14:1487. [PMC free article] [PubMed] [Google Scholar]
- 43.Szalai AJ, Wu J, Lange EM, McCrory MA, Langefeld CD, Williams A, Zakharkin SO, George V, Allison DB, Cooper GS, Xie F, Fan Z, Edberg JC, Kimberly RP. Single-nucleotide polymorphisms in the C-reactive protein (CRP) gene promoter that affect transcription factor binding, alter transcriptional activity and associate with differences in baseline serum CRP level. J Mol Med. 2005;83:440–7. doi: 10.1007/s00109-005-0658-0. [DOI] [PubMed] [Google Scholar]
- 44.Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]


