Abstract
Specific gene expression in oocytes and its surrounding cumulus (CC) and granulosa (GC) cells is needed for successful folliculogenesis and oocyte maturation. The aim of the present study was to compare genome-wide gene expression and biological functions of human GC and CC. Individual GC and CC were derived from 37 women undergoing IVF procedures. Gene expression analysis was performed using microarrays, followed by a meta-analysis. Results were validated using quantitative real-time PCR. There were 6029 differentially expressed genes (q < 10−4); of which 650 genes had a log2 FC ≥ 2. After the meta-analysis there were 3156 genes differentially expressed. Among these there were genes that have previously not been reported in human somatic follicular cells, like prokineticin 2 (PROK2), higher expressed in GC, and pregnancy up-regulated nonubiquitous CaM kinase (PNCK), higher expressed in CC. Pathways like inflammatory response and angiogenesis were enriched in GC, whereas in CC, cell differentiation and multicellular organismal development were among enriched pathways. In conclusion, transcriptomes of GC and CC as well as biological functions, are distinctive for each cell subpopulation. By describing novel genes like PROK2 and PNCK, expressed in GC and CC, we upgraded the existing data on human follicular biology.
Introduction
Precise and timely coordination between oocyte maturation and the process of somatic cell proliferation and differentiation is crucial for the proper development of the ovarian follicle [1]. Granulosa cells (GC) differentiate into two functionally distinct groups during ovarian follicle growth: cumulus cells (CC) that are in physical contact with the oocyte, and mural GC, which line the wall of the antral follicle [2]. Bidirectional communication between the oocyte and its surrounding somatic cells through paracrine signalling and gap junctions is required during folliculogenesis for the development of follicular compartments, oocyte maturation and competence acquisition [3].
In order to secure its own optimal growth and maturation [4], the oocyte regulates the differentiation, proliferation, apoptosis and luteinisation of GC and CC by secretion of paracrine factors such as growth differentiation factor 9 (GDF-9) and bone morphogenetic protein 15 (BMP-15) [5]. GDF-9 and BMP-15 activate signalling pathways and induce GC gene expression which leads to a functional change of these cells necessary for oocyte development [6].
On the other hand, somatic follicular cells provide support for oocyte growth, resumption of meiosis, nuclear and cytoplasmic maturation and control oocyte's transcriptional activity through the activation of different signalling pathways [7]. For example, in mice, GC derived Kit ligand (KitL) has been shown to stimulate oocyte growth via activation of phosphatidylinositol 3-kinase (PI3K) pathway [8]. Furthermore, binding of KitL to its receptor Kit on the oocyte activates Ras, Raf, mitogen activated protein kinase (MAPK) and protein kinase B (PKB/Akt)—apoptosis related signalling pathways [9].
Identification of key molecules and signalling pathways within GC and CC is necessary to gain insight of how the oocyte acquires developmental competence during its growth within the follicle and for uncovering molecular regulators of this process. This could help optimize in vitro techniques for oocyte maturation. There have been some studies conducted on human GC and CC in order to provide information on cell-type specific transcriptome [10, 11, 12]. These studies identified specific enriched biological processes in GC (cell cycle, immune response, cell proliferation, steroidogenesis) and CC (cell signalling, protein degradation, angiogenesis). However, our knowledge of the key genes and pathways of human folliculogenesis and oocyte competence acquisition is still incomplete. Identification of specific genes and functions of human GC and CC would help in understanding the functional specifics of each follicular compartment and its role in the process of human folliculogenesis.
The aim of the present study was to identify differentially expressed genes and analyse biological processes in human GC and CC obtained during IVF procedures.
Materials and Methods
Patient population and IVF procedure
Thirty-seven (37) IVF patients were included in this study. All patients were less than 35 years old, had a body mass index (BMI) between 17 and 26 kg/m2, were enrolled in the IVF procedure due to tubal cause of infertility and had their first or second IVF procedure. Their partner’s spermiogram was normal according to the WHO criteria [13]. The study was approved by the National Medical Ethics Committee of the Republic of Slovenia and written informed consent was obtained from all women prior to participation.
All patients were treated with recombinant FSH (Puregon; Schering Plough, New Jersey, USA) and GnRH antagonist cetrorelix acetate (Cetrotide; Asta Medica AG, Frankfurt, Germany). Vaginal ultrasound examination was performed to monitor follicular development. Final follicular maturation was induced by administering 10,000 IU of human chorionic gonadotropin (hCG) (Pregnyl; N.V. Organon, Oss, the Netherlands) when at least three follicles were ≥17 mm. Ultrasound-guided transvaginal puncture was performed 34–36 h later. Each follicle was aspirated separately. After aspiration, cumulus-oocyte complexes (COC) were removed from the follicular fluid. Cumulus cells of each oocyte were removed immediately. Classical IVF was used for oocyte fertilization. Embryo transfer was performed on day 5 after oocyte retrieval. The GC and CC samples in this study were all derived from follicles containing mature, MII, oocytes.
Study design
We first performed genome-wide gene expression analysis using microarrays on 34 individual GC and 30 individual CC samples derived from 21 women. The next step included quantitative real-time PCR (qPCR) validation performed on 19 individual GC and 19 individual CC samples derived from 16 women that were newly included in the study. Data from two previous studies, where comparison of human GC and CC gene expression profiles was performed, were used for the meta-analysis for the identification of consistent differentially expressed genes.
Samples collection and preparation
Immediately after oocyte retrieval, CC of each oocyte were mechanically removed by a needle and a glass denudation pipette (Swemed, Göteborg, Sweden), washed in phosphate-buffered saline (PBS), snap frozen in liquid nitrogen and stored at -80°C until RNA isolation.
For GC purification, Dynabeads CD45 Magnetic Beads (Invitrogen Dynal AS, Oslo, Norway) were used for the depletion of CD45+ cells according to the manufacturer's protocol with some adjustments. The beads were used to separate leukocyte CD45+ cells from GC to exclude the risk of contamination [14]. Follicular fluids were centrifuged at 800x g at 4°C for 10 minutes. Follicular fluids were then snap frozen in liquid nitrogen and stored at -80°C. The sediment was resuspended in 2 ml of lysis solution (0.15 M NH4Cl) to remove red blood cells, incubated for 10 minutes and centrifuged at 600x g at 4°C for 10 minutes. The supernatant was discarded and the sediment resuspended in 1 ml PBS with 0.1% bovine serum albumin (BSA) and 0.002 M ethylenediaminetetraacetic acid (EDTA). The suspension was added to the tubes containing 50 μl of Dynabeads CD45 and incubated at 4°C for 30 minutes with gentle tilting and rotation of the mixer. The tubes were pre-coated with 2 ml of Roswell Park Memorial Institute medium (RPMI) with 1% fetal calf serum (FCS) for 5 minutes. After incubation, the samples were diluted with PBS with 0.1% BSA and 0.002 M EDTA to 4 ml and placed in a magnet for 10 minutes. The supernatant was transferred to a new pre-coated tube and centrifuged at 800x g at 4°C for 15 minutes. The supernatant was discarded and sediment resuspended in 1 ml of PBS. The samples were then centrifuged at 2000 x g for 5 minutes, supernatant discharged and sediment snap frozen in liquid nitrogen and stored at -80°C. The purity of the samples was tested with flow cytometry. The samples were prepared from a GC suspension before isolation and compared to cell suspension after CD45+ depletion. The contamination with leucocytes was investigated using antibodies CD45 conjugated with Fluorescin Isothiocyanate (FITC) and CD14 Phycoerythrin (PE) (both from BD Bioscience, San Jose, USA). Samples were washed and resuspended in PBS with 1% BSA and incubated with antibodies for 20 minutes. The purity of the samples was characterized by Forward Scatter (FSC) versus Side Scatter (SSC) followed by gating on CD45 and CD14. Data were collected on FACSCanto flow cytometer (BD Bioscience, San Jose, USA) and expression of various markers was assessed using FlowJo analysis software (TreeStarInc, Ashland, USA).
Before depletion of CD45+ cells there were, in average, 14.8% of granulocytes, 9.7% of lymphocytes, 9.7% of monocytes and 66.9% of GC present. After the depletion, the proportions of leukocytes were significantly lower. In average, there were 4.7% of granulocytes, 1.8% of monocytes, 1.7% of lymphocytes and 91.8% of GC. Thus, we have achieved a sufficient purity of GC samples, as it has been established that a gene expression profile of a more than 75% pure sample was indistinguishable from a 100% pure sample [15].
RNA extraction and cDNA synthesis
TRI reagent (Sigma—Aldrich, St.Louis, USA) was used for RNA extraction according to a slightly modified manufacturer’s protocol. Due to a small sample volume, glycogen was used as a carrier to increase RNA yield. Individual GC and CC samples were homogenized in 500 μL TRI reagent supplemented with 125 μg of glycogen (Ambion, Austin, USA). After 2 min incubation at room temperature, 100 μL of chloroform was added and the sample was vortexed vigorously. RNA was precipitated from the aqueous phase with isopropanol and collected after 15 min of centrifugation at 12,000x g and 4°C. RNA pellet was washed three times with 75% ethanol, dried and dissolved in 15 μL of RNAse free water. The integrity of the RNA samples was assessed with an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, USA) and the total RNA quantity was measured with a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, USA). SuperScript Vilo reverse transcriptase (Invitrogen, Carlsbad, USA) was used for cDNA synthesis from 150 ng RNA according to the manufacturer’s instructions.
Microarray analysis
Microarray profiling of global gene expression on RNA samples from GC and CC was performed using Agilent SurePrint G3 Human gene expression 8x60K two-color microarrays (Agilent eArray design identifier: 028004), with 42.405 unique probes targeting for most of the RefSeq mRNA sequences. The experimental design comprised of co-hybridization of each tested sample with common universal reference RNA sample (Universal Human Reference RNA from Agilent). The labeling procedure was performed using Agilent’s two-color Low Input Quick Amp Labeling Kit (two-color) according to manufacturers’ instructions, labeling test samples with Cy3 and reference samples with Cy5 dye.
Microarray slides were scanned using Agilent High Resolution Microarray Scanner System after hybridization, using the manufacturer’s recommended scanning settings. Subsequently, microarray features were extracted using Agilent Feature Extraction (FE) software v10.7.3.1. Background was subtracted using background de-trending algorithm implemented in FE software, features with non-uniform fluorescence profiles were removed from further analyses and linear lowess intra-array normalization was performed to correct for the presence of dye bias. The consistency of log ratio values was inspected using manufacturers provided spike-in probes and reproducibility was evaluated by investigating the extent of variance in the population of replicated probes on the array. The microarray data are available at the Gene Expression Omnibus (GEO) public repository (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE55654.
Statistical analysis of microarray data
All the steps described in this section, including post-processing, statistical comparisons and classification performance estimations were performed using packages from Bioconductor v2.8 project in R statistical environment version 2.13.1.
Processed fluorescence values obtained by feature extraction were imported and inspected for presence of missing values and other irregularities. Probe annotations were obtained from the Agilent’s eArray service (earray.chem.agilent.com). Prior to the calculation of Cy3/Cy5 log2 ratios, fluorescence values were offset by 100 units to prevent an undesirable increase in log2 ratio variance in the population of features with low fluorescence values. Quality assessment of microarray hybridizations was performed by inspecting the signal distribution in box and MA plots. Principal component analysis was done to identify the presence of possible batch and other confounding effects.
Statistical comparisons of expression values were evaluated using moderated t-test approach implemented in limma [16]. Significance values and log2 fold-change were calculated and afterwards, p-values were controlled for multiple testing effect by the method described by Benjamini and Hochberg [17] and implemented into the limma workflow.
Meta-analysis
Two datasets that analysed global gene expression profiles in human GC and CC samples with accession numbers GSE18559 [10] and E-MEXP-3641 [11], together with our own data. were included in the meta-analysis. Raw data from microarray experiments were obtained from Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih.gov/geo/) [18] and from ArrayExpress repository (www.ebi.ac.uk/arrayexpress/).
Raw datasets were put through quality control steps implemented in arrayQualityMetrics package, followed by intra-array quantile and inter-array loess normalization, where necessary. Afterwards, intensities of probe replicates targeting specific transcripts were median averaged and only values for transcripts interrogated in all three studies were retained through further meta-analysis steps.
Differential expression of genes across all three studies was calculated using meta-analysis algorithms implemented in the RankProd package [19]. RankProd uses a non-parametric approach that selects genes displaying consistently highly ranked across different microarray studies and is therefore a feasible meta-analytic tool across a variety of microarray studies originating from different laboratories and performed under differing conditions [19]. We utilized RPadvance function, with origin parameter set to account for data originating from three different sources. P values were calculated using permutation approach (1000 permutations) and predicted false positive rates estimated across permutation cycles [20].
Gene ontology and IPA analysis
Gene ontology (GO) analysis was performed to define enriched, cell specific GO profiles using Gene Codis [21]. We uploaded our two sets of genes: one set of 524 genes that were up-regulated in GC and one set of 126 that were up-regulated in CC. All of the selected genes had a log2 fold difference > ± 2 and q-value < 10−4 so they were considered significantly differentially expressed. Up-regulation of genes in GC population, as mentioned in this manuscript, refers to down-regulation of genes in CC and vice versa.
Our list of significantly differentially expressed genes (log2 FC > ±2, q-value < 10−4) was subjected to Ingenuity Pathway Analysis (IPA) Software (Ingenuity Systems Inc, Redwood City, USA) for network enrichment analysis in each cell subpopulation.
Quantitative real-time PCR analysis
We performed validation of microarray data results using qPCR on an independent set of GC and CC samples. The genes selected for validation were angiotensin I converting enzyme 2 (ACE2 ), dual specificity phosphatase 6 (DUSP6), fibrinogen gamma chain (FGG), HtrA serine peptidase 1 (HTRA1), interleukin 1, beta (IL1B), pregnancy up-regulated nonubiquitous CaM kinase (PNCK), prokineticin 2 (PROK2), ryanodine receptor 2 (cardiac) (RYR2).
Pre-designed TaqMan Gene Expression assays (Applied Biosystems, Foster City, CA, USA) were used for mRNA quantification. All reactions were performed in triplicates in 96-well formats in a 20 μl final volume. The measurements were performed on ABI Prism 7000 Sequence detection system (Applied Biosystems, Foster City, USA). Thermal cycling conditions were as follows: an initial step at 50°C for 2 min, denaturation at 95°C for 10 min, amplification for 40 cycles at 95°C for 15s and at 60°C for 1 min. The threshold cycle (Ct) values were then determined for each assay and normalized to internal control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) that was co-ran with each sample. Differences in gene expression were calculated using the delta-delta Ct method, as previously described by Livak and Schmittgen [22]. The significance of the mean relative gene expression level differences was calculated using two-sample t-distribution test, differences were considered significant at α < 0.05.
Results
Global differential gene expression
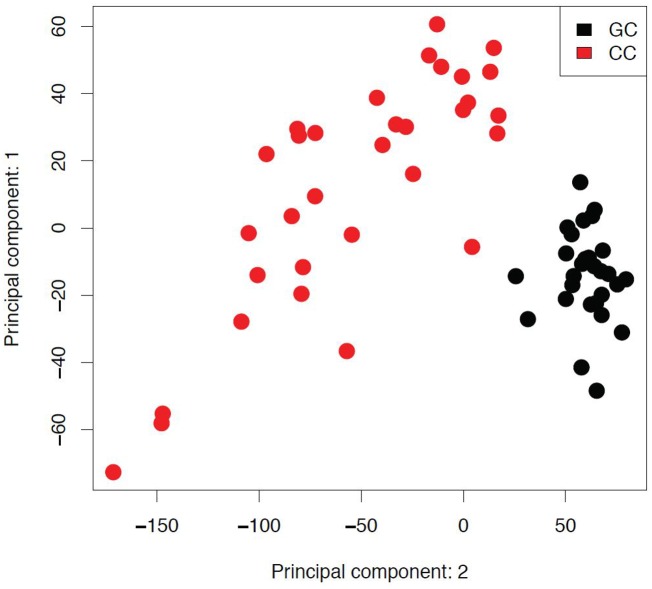
The comparison of global gene expression profiles of GC and CC showed cell type specific profiles. Complete transcriptional profiles of the two cell populations were subjected to principal component analysis (PCA); first two principal variance components clearly separated expression profiles of GC and CC populations (Fig 1).
Fig 1. Principal component analysis between granulosa and cumulus cells.
The axes of the plot represent first two main components of variance, obtained by performing principal component analysis (PCA) on global expression profiles. GC- granulosa cells; CC- cumulus cells.
There were 6029 genes significantly (q < 10−4) differentially expressed between GC and CC in our study, with 650 having a log2 fold change ≥ 2, with 524 genes higher expressed in GC and 126 genes higher expressed in CC. The lists of top 100 differentially expressed genes are represented in Tables 1 and 2 for GC and CC, respectively. The full list of differentially expressed genes is represented in S1 Table.
Table 1. The list of top 100 genes up-regulated in GC.
| Gene Symbol | Gene name | Absolute array intensity (log2A) | logFC(CC/GC) | q value |
|---|---|---|---|---|
| ADAM8 | ADAM metallopeptidase domain 8 | 11.4 | -3.30 | 3.99E-15 |
| AIF1 | allograft inflammatory factor 1 | 12.3 | -3.44 | 2.28E-13 |
| ALAS2 | aminolevulinate, delta-, synthase 2 | 12.7 | -3.56 | 1.09E-12 |
| APBB1IP | amyloid beta | 10.4 | -3.56 | 1.78E-15 |
| APOBEC3A | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A | 7.3 | -3.29 | 3.00E-12 |
| APOBR | apolipoprotein B receptor | 7.9 | -3.30 | 1.71E-16 |
| AQP9 | aquaporin 9 | 8.3 | -3.61 | 7.09E-15 |
| ARHGAP30 | Rho GTPase activating protein 30 | 7.7 | -3.59 | 1.78E-16 |
| ARHGDIB | Rho GDP dissociation inhibitor | 13.5 | -3.58 | 2.76E-14 |
| BCL2A1 | BCL2-related protein A1 | 9.6 | -3.52 | 2.57E-13 |
| C16orf54 | chromosome 16 open reading frame 54 | 8.7 | -4.03 | 4.33E-16 |
| C5AR1 | complement component 5a receptor 1 | 8.1 | -4.03 | 1.27E-17 |
| CAMP | cathelicidin antimicrobial peptide | 7.4 | -3.26 | 3.62E-13 |
| CASP1 | caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | 9.4 | -3.47 | 2.04E-15 |
| CCL3 | chemokine (C-C motif) ligand 3 | 10.3 | -3.22 | 4.68E-11 |
| CCL5 | chemokine (C-C motif) ligand 5 | 8.3 | -3.47 | 1.65E-15 |
| CCR1 | chemokine (C-C motif) receptor 1 | 9.7 | -3.45 | 2.21E-12 |
| CCR7 | chemokine (C-C motif) receptor 7 | 8.2 | -3.47 | 9.49E-18 |
| CD247 | CD247 molecule | 6.4 | -3.28 | 3.01E-17 |
| CD300A | CD300a molecule | 7.9 | -3.20 | 8.23E-15 |
| CD52 | CD52 molecule | 11.4 | -3.31 | 8.27E-15 |
| CD53 | CD53 molecule | 10.2 | -3.25 | 1.48E-14 |
| CD97 | CD97 molecule | 12.1 | -3.48 | 7.57E-17 |
| CMTM2 | CKLF-like MARVEL transmembrane domain containing 2 | 7.6 | -3.50 | 8.96E-15 |
| CX3CR1 | chemokine (C-X3-C motif) receptor 1 | 9.9 | -3.49 | 4.10E-15 |
| CYTH4 | cytohesin 4 | 6.4 | -3.28 | 1.40E-16 |
| CYTIP | cytohesin 1 interacting protein | 9.4 | -3.39 | 5.52E-14 |
| DEFA3 | defensin, alpha 3, neutrophil-specific | 8.5 | -4.01 | 2.00E-13 |
| ENST00000390547 | immunoglobulin heavy constant alpha 1 [Source:HGNC Symbol;Acc:5478] [ENST00000390547] | 9.1 | -3.23 | 2.60E-12 |
| EVI2B | ecotropic viral integration site 2B | 8.8 | -3.72 | 3.31E-14 |
| FAM65B | family with sequence similarity 65, member B | 7.3 | -3.36 | 1.17E-11 |
| FCAR | Fc fragment of IgA | 6.9 | -3.36 | 1.09E-16 |
| FCGR2A | Fc fragment of IgG, low affinity IIa, receptor | 10.0 | -3.37 | 5.60E-15 |
| FCGR2A | Fc fragment of IgG, low affinity IIa, receptor (CD32) | 10.0 | -3.50 | 4.91E-14 |
| FGD3 | FYVE, RhoGEF and PH domain containing 3 | 9.5 | -3.38 | 4.21E-18 |
| FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 9.8 | -3.65 | 1.27E-18 |
| FPR2 | formyl peptide receptor 2 | 7.4 | -3.40 | 1.68E-13 |
| GABRP | gamma-aminobutyric acid | 8.8 | -3.85 | 6.37E-15 |
| GIMAP4 | GTPase, IMAP family member 4 | 7.5 | -3.27 | 2.32E-13 |
| GLT1D1 | glycosyltransferase 1 domain containing 1 | 8.7 | -3.31 | 9.42E-14 |
| GMFG | glia maturation factor, gamma | 12.6 | -3.72 | 4.86E-15 |
| GZMA | granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) | 7.7 | -3.70 | 5.46E-16 |
| GZMB | granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) | 8.6 | -3.45 | 8.26E-18 |
| HBA2 | hemoglobin, alpha 2 | 16.5 | -4.89 | 2.77E-14 |
| HBM | hemoglobin, mu | 9.5 | -3.56 | 4.72E-16 |
| HBQ1 | hemoglobin, theta 1 | 11.7 | -4.11 | 1.87E-15 |
| HCAR3 | hydroxycarboxylic acid receptor 3 | 9.1 | -3.42 | 6.30E-14 |
| HCK | hemopoietic cell kinase | 7.7 | -3.26 | 6.18E-16 |
| HCLS1 | hematopoietic cell-specific Lyn substrate 1 | 12.3 | -3.73 | 1.16E-16 |
| IL18RAP | interleukin 18 receptor accessory protein | 7.7 | -3.42 | 1.55E-13 |
| IL1B | interleukin 1, beta | 9.3 | -3.67 | 4.64E-15 |
| IL2RB | interleukin 2 receptor, beta | 7.8 | -3.31 | 2.40E-18 |
| INPP5D | inositol polyphosphate-5-phosphatase | 10.5 | -3.66 | 3.37E-17 |
| ITGAL | integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) | 8.3 | -3.27 | 8.87E-17 |
| ITGB2 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | 7.1 | -3.67 | 6.33E-16 |
| KLF2 | Kruppel-like factor 2 (lung) | 10.6 | -4.05 | 1.54E-18 |
| KLRB1 | killer cell lectin-like receptor subfamily B, member 1 | 7.3 | -3.42 | 1.04E-14 |
| KRT1 | keratin 1 | 8.2 | -3.42 | 9.50E-09 |
| LAPTM5 | lysosomal protein transmembrane 5 | 10.0 | -3.39 | 1.80E-13 |
| LCP1 | lymphocyte cytosolic protein 1 (L-plastin) | 12.6 | -3.43 | 1.71E-13 |
| lincRNA:chr1:205404902–205417627_F | lincRNA:chr1:205404902–205417627 forward strand | 7.5 | -3.38 | 6.18E-14 |
| lincRNA:chr10:17250419–17261819_F | lincRNA:chr10:17250419–17261819 forward strand | 8.7 | -3.55 | 2.86E-15 |
| lincRNA:chr15:52209683–52223508_R | lincRNA:chr15:52209683–52223508 reverse strand | 7.9 | -3.37 | 1.42E-10 |
| lincRNA:chr17:29887612–29925137_F | lincRNA:chr17:29887612–29925137 forward strand | 7.7 | -3.72 | 1.80E-13 |
| lincRNA:chr17:73598283–73599500_F | lincRNA:chr17:73598283–73599500 forward strand | 10.4 | -3.73 | 1.17E-12 |
| lincRNA:chr4:769425–775573_F | lincRNA:chr4:769425–775573 forward strand | 11.6 | -3.76 | 8.00E-14 |
| lincRNA:chr5:12625075–12747025_F | lincRNA:chr5:12625075–12747025 forward strand | 11.2 | -3.98 | 4.90E-15 |
| LOC100128348 | cDNA FLJ46249 fis, clone TESTI4021377 | 6.1 | -3.25 | 1.26E-18 |
| LOC100133286 | uncharacterized LOC100133286 | 9.2 | -3.29 | 6.98E-13 |
| LOC100652730 | Homo sapiens hypothetical LOC100652730 (LOC100652730), miscRNA | 7.0 | -3.21 | 3.98E-12 |
| LSP1 | lymphocyte-specific protein 1 | 10.4 | -3.42 | 1.82E-17 |
| LTB | lymphotoxin beta (TNF superfamily, member 3) | 9.1 | -4.01 | 6.09E-18 |
| LYN | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | 10.6 | -3.59 | 9.89E-15 |
| MBP | myelin basic protein | 6.3 | -3.40 | 1.06E-13 |
| MNDA | myeloid cell nuclear differentiation antigen | 8.1 | -3.35 | 1.62E-11 |
| MPEG1 | macrophage expressed 1 | 7.6 | -3.21 | 8.07E-15 |
| NCF1 | neutrophil cytosolic factor 1 | 8.5 | -3.21 | 1.89E-17 |
| NCF2 | neutrophil cytosolic factor 2 | 8.7 | -3.44 | 2.04E-15 |
| NFAM1 | NFAT activating protein with ITAM motif 1 | 8.4 | -3.44 | 1.54E-16 |
| NFE2 | nuclear factor (erythroid-derived 2) | 11.8 | -3.55 | 1.07E-13 |
| PLEK | Pleckstrin | 7.8 | -3.40 | 2.19E-15 |
| PREX1 | phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 | 11.2 | -3.49 | 3.03E-15 |
| PROK2 | prokineticin 2 | 8.2 | -4.29 | 5.90E-14 |
| PTPRC | protein tyrosine phosphatase, receptor type, C | 10.5 | -3.38 | 3.77E-11 |
| PTPRC | tyrosine phosphatase, receptor type, C | 10.5 | -3.63 | 4.10E-16 |
| RASSF5 | Ras association (RalGDS/AF-6) domain family member 5 | 6.9 | -3.24 | 3.81E-15 |
| RGL4 | ral guanine nucleotide dissociation stimulator-like 4 | 8.0 | -3.42 | 5.30E-15 |
| S100A12 | S100 calcium binding protein A12 | 9.3 | -3.68 | 6.60E-14 |
| S100A4 | S100 calcium binding protein A4 | 13.1 | -3.32 | 1.44E-14 |
| S1PR4 | sphingosine-1-phosphate receptor 4 | 9.8 | -3.77 | 1.61E-16 |
| SECTM1 | secreted and transmembrane 1 | 10.8 | -3.59 | 3.72E-16 |
| SORL1 | sortilin-related receptor, L(DLR class) A repeats containing | 12.4 | -3.47 | 8.10E-13 |
| SPI1 | spleen focus forming virus (SFFV) proviral integration oncogene spi1 | 9.3 | -3.22 | 2.42E-19 |
| TAGAP | T-cell activation RhoGTPase activating protein | 7.6 | -3.81 | 6.17E-17 |
| TBC1D10C | TBC1 domain family, member 10C | 10.3 | -3.39 | 8.67E-16 |
| TLR1 | toll-like receptor 1 | 8.3 | -3.36 | 2.16E-12 |
| TNFSF10 | tumor necrosis factor (ligand) superfamily, member 10 | 8.6 | -3.43 | 1.18E-11 |
| TRIM58 | tripartite motif containing 58 | 9.6 | -3.36 | 4.32E-11 |
| TTTY16 | testis-specific transcript, Y-linked 16 (non-protein coding) | 9.0 | -4.03 | 1.06E-14 |
| VWA1 | von Willebrand factor A domain containing 1 | 11.1 | -3.45 | 6.99E-14 |
Table 2. The list of top 100 genes up-regulated in CC.
| Gene Symbol | Gene name | Absolute array intensity (log2A) | logFC(CC/GC) | q value |
|---|---|---|---|---|
| ACE2 | angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 | 8.7 | 3.39 | 3.04E-20 |
| ACOXL | acyl-CoA oxidase-like | 8.3 | 3.12 | 2.36E-23 |
| ACTA1 | actin, alpha 1, skeletal muscle | 7.9 | 2.71 | 5.34E-14 |
| ADAMTS4 | ADAM metallopeptidase with thrombospondin type 1 motif, 4 | 6.1 | 2.57 | 2.17E-18 |
| ALPK2 | alpha-kinase 2 | 7.7 | 2.37 | 2.26E-18 |
| AMH | anti-Mullerian hormone | 8.8 | 3.15 | 1.80E-19 |
| BEX1 | brain expressed, X-linked 1 | 13.2 | 3.23 | 8.97E-23 |
| BMPER | BMP binding endothelial regulator | 6.0 | 3.18 | 3.15E-26 |
| BUB1 | budding uninhibited by benzimidazoles 1 homolog | 9.7 | 2.26 | 2.51E-19 |
| C1orf141 | chromosome 1 open reading frame 141 | 7.4 | 2.44 | 2.13E-16 |
| C2CD4B | C2 calcium-dependent domain containing 4B | 7.1 | 2.64 | 7.96E-18 |
| CACNA1C | calcium channel, voltage-dependent, L type, alpha 1C subunit | 9.9 | 2.33 | 2.31E-16 |
| CBLN1 | cerebellin 1 precursor | 6.9 | 2.51 | 4.18E-13 |
| CDH11 | cadherin 11, type 2, OB-cadherin | 9.7 | 2.24 | 1.76E-13 |
| CHGA | chromogranin A (parathyroid secretory protein 1) | 7.4 | 2.57 | 3.04E-24 |
| CHGB | chromogranin B (secretogranin 1) | 8.5 | 2.23 | 8.80E-14 |
| CIB4 | calcium and integrin binding family member 4 | 7.2 | 2.87 | 3.69E-19 |
| CILP | cartilage intermediate layer protein, nucleotide pyrophosphohydrolase | 8.4 | 3.09 | 1.33E-17 |
| CLSTN2 | calsyntenin 2 | 8.3 | 2.26 | 2.26E-12 |
| CORO2A | coronin, actin binding protein, 2A | 9.3 | 2.60 | 1.25E-22 |
| CTSK | cathepsin K | 10.9 | 2.77 | 1.97E-16 |
| CYP19A1 | cytochrome P450, family 19, subfamily A, polypeptide 1 | 7.2 | 2.32 | 4.76E-13 |
| CYP19A1 | cytochrome P450, family 19, subfamily A, polypeptide 1 | 7.2 | 2.26 | 7.06E-18 |
| DAPL1 | death associated protein-like 1 | 7.0 | 3.51 | 1.69E-12 |
| DAPL1 | death associated protein-like 1 | 7.0 | 2.72 | 1.35E-12 |
| DHH | desert hedgehog | 7.7 | 2.39 | 2.44E-18 |
| DKFZp451A211 | ref|PREDICTED: Homo sapiens DKFZp451A211 protein (DKFZp451A211), mRNA [XM_003403663] | 7.7 | 2.46 | 3.22E-19 |
| DOK5 | docking protein 5 | 8.7 | 2.65 | 4.18E-18 |
| DPYSL4 | dihydropyrimidinase-like 4 | 9.7 | 2.34 | 1.65E-12 |
| DUOXA2 | dual oxidase maturation factor 2 | 6.0 | 2.60 | 1.32E-13 |
| E2F7 | E2F transcription factor 7 | 11.1 | 3.43 | 1.07E-16 |
| E2F7 | E2F transcription factor 7 | 11.1 | 2.25 | 3.17E-07 |
| EPHB1 | EPH receptor B1 | 7.1 | 2.40 | 1.97E-16 |
| F3 | coagulation factor III (thromboplastin, tissue factor) | 9.8 | 2.33 | 5.27E-15 |
| FABP3 | fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) | 7.9 | 2.31 | 8.86E-18 |
| FAM110C | family with sequence similarity 110, member C | 7.3 | 2.46 | 3.33E-13 |
| FAM150B | family with sequence similarity 150, member B | 6.9 | 2.92 | 9.90E-18 |
| FAM150B | family with sequence similarity 150, member B | 6.9 | 2.75 | 6.21E-17 |
| FAM189A1 | family with sequence similarity 189, member A1 | 8.4 | 2.26 | 3.52E-14 |
| FGF11 | fibroblast growth factor 11 | 7.8 | 2.42 | 1.26E-18 |
| FN1 | fibronectin 1 | 10.4 | 2.51 | 2.14E-14 |
| FOXG1 | forkhead box G1 | 8.2 | 3.30 | 5.27E-14 |
| GABBR2 | gamma-aminobutyric acid (GABA) B receptor, 2 | 7.8 | 2.46 | 2.75E-12 |
| GAL | galanin prepropeptide | 12.2 | 2.92 | 9.22E-15 |
| GAP43 | growth associated protein 43 | 7.4 | 2.36 | 7.87E-22 |
| GDF6 | growth differentiation factor 6 | 8.7 | 3.36 | 1.80E-19 |
| GJA5 | gap junction protein, alpha 5 | 6.8 | 2.35 | 1.24E-16 |
| GRIK3 | glutamate receptor, ionotropic, kainate 3 | 7.2 | 2.76 | 6.85E-13 |
| HTRA1 | HtrA serine peptidase 1 | 14.0 | 3.79 | 2.99E-20 |
| IGFBP5 | insulin-like growth factor binding protein 5 | 11.3 | 3.53 | 3.37E-23 |
| ISM1 | isthmin 1 homolog | 6.9 | 2.56 | 7.96E-18 |
| KCNK3 | potassium channel, subfamily K, member 3 | 8.0 | 2.66 | 5.64E-20 |
| KCNT1 | potassium channel, subfamily T, member 1 | 7.2 | 2.25 | 1.41E-15 |
| KRTAP13-1 | keratin associated protein 13–1 | 7.1 | 2.84 | 9.16E-26 |
| KRTAP13-2 | keratin associated protein 13–2 | 7.5 | 2.57 | 1.01E-20 |
| LEFTY2 | eft-right determination factor 2 | 9.3 | 2.41 | 2.43E-13 |
| LIMS2 | LIM and senescent cell antigen-like domains 2 | 6.9 | 2.54 | 2.88E-20 |
| LOC100506189 | ref|PREDICTED: Homo sapiens hypothetical LOC100506189 (LOC100506189), miscRNA [XR_108925] | 7.3 | 2.43 | 1.14E-15 |
| LOC100506189 | ref|PREDICTED: Homo sapiens hypothetical LOC100506189 (LOC100506189), miscRNA [XR_108925] | 7.3 | 2.41 | 4.79E-12 |
| LOC100506189 | ref|PREDICTED: Homo sapiens hypothetical LOC100506189 (LOC100506189), miscRNA [XR_108925] | 7.3 | 2.39 | 4.79E-15 |
| LRAT | lecithin retinol acyltransferase (phosphatidylcholine—retinol O-acyltransferase) | 7.9 | 2.78 | 5.40E-15 |
| LTBP1 | latent transforming growth factor beta binding protein 1 | 9.5 | 2.25 | 1.19E-16 |
| MT1DP | metallothionein 1D, pseudogene (MT1DP), transcript variant 1 | 6.9 | 2.66 | 4.77E-17 |
| NOS2 | nitric oxide synthase 2, inducible | 7.3 | 2.64 | 2.22E-15 |
| NUDT10 | nudix (nucleoside diphosphate linked moiety X)-type motif 10 | 8.6 | 2.35 | 3.58E-13 |
| PCK1 | phosphoenolpyruvate carboxykinase 1 | 8.6 | 3.22 | 1.69E-16 |
| PCYT1B | phosphate cytidylyltransferase 1, choline, beta | 7.5 | 2.53 | 1.24E-17 |
| PDLIM3 | PDZ and LIM domain 3 | 6.2 | 2.51 | 1.80E-19 |
| PDZD2 | PDZ domain containing 2 | 8.7 | 2.22 | 9.38E-15 |
| PLCXD3 | phosphatidylinositol-specific phospholipase C, X domain containing 3 | 7.5 | 2.32 | 2.71E-14 |
| PLOD2 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | 11.8 | 2.50 | 1.01E-18 |
| PNCK | pregnancy up-regulated non-ubiquitously expressed CaM kinase | 8.0 | 3.35 | 3.88E-22 |
| PPP1R14C | protein phosphatase 1, regulatory (inhibitor) subunit 14C | 7.8 | 2.45 | 2.14E-15 |
| PRB1 | proline-rich protein BstNI subfamily 1 | 7.9 | 2.23 | 1.24E-12 |
| PRB2 | proline-rich protein BstNI subfamily 2 | 8.3 | 3.44 | 3.12E-14 |
| PRB4 | proline-rich protein BstNI subfamily 4 | 10.2 | 2.24 | 4.59E-12 |
| RHOBTB3 | Rho-related BTB domain containing 3 | 11.3 | 2.78 | 9.13E-18 |
| RTN4RL1 | reticulon 4 receptor-like 1 | 8.8 | 2.37 | 3.85E-20 |
| RYR2 | ryanodine receptor 2 | 7.6 | 2.85 | 6.30E-25 |
| SCG2 | secretogranin II | 8.4 | 3.28 | 9.42E-22 |
| SEMA5B | sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B | 6.9 | 2.34 | 7.90E-11 |
| SERPINA3 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 | 11.4 | 2.27 | 1.36E-15 |
| SERPINA5 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 | 9.4 | 2.46 | 1.66E-17 |
| SLC15A1 | solute carrier family 15 (oligopeptide transporter), member 1 | 7.1 | 2.91 | 3.11E-18 |
| SLC28A3 | solute carrier family 28 (sodium-coupled nucleoside transporter), member 3 | 7.1 | 2.50 | 2.39E-17 |
| SMOC2 | SPARC related modular calcium binding 2 | 10.3 | 2.73 | 4.09E-19 |
| SMOC2 | SPARC related modular calcium binding 2 | 10.3 | 2.45 | 6.25E-20 |
| SPON2 | spondin 2, extracellular matrix protein | 13.5 | 2.42 | 4.77E-15 |
| ST6GAL2 | ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 | 6.5 | 3.24 | 3.87E-24 |
| SYNDIG1 | synapse differentiation inducing 1 | 8.7 | 3.75 | 5.40E-25 |
| TAC3 | tachykinin 3 | 9.5 | 2.80 | 1.48E-22 |
| TMEM114 | transmembrane protein 114 | 7.1 | 3.00 | 4.84E-13 |
| TMEM132C | transmembrane protein 132C | 8.0 | 3.09 | 5.64E-20 |
| TNC | tenascin C | 11.2 | 3.16 | 1.78E-19 |
| TTBK1 | tau tubulin kinase 1 | 5.9 | 2.26 | 1.06E-27 |
| ULBP1 | UL16 binding protein 1 | 9.6 | 3.99 | 4.90E-23 |
| VWC2 | von Willebrand factor C domain containing 2 | 6.7 | 2.25 | 1.81E-11 |
| WISP1 | WNT1 inducible signaling pathway protein 1 | 6.8 | 2.23 | 2.26E-18 |
| WNT3A | wingless-type MMTV integration site family, member 3A | 8.1 | 2.72 | 4.46E-17 |
| XLOC_l2_007928 | lincRNA (XLOC_l2_007928), lincRNA [TCONS_l2_00014464] | 11.2 | 2.73 | 2.55E-16 |
Meta-analysis showed there were 3156 genes consistently significantly (q < 10−4) differentially expressed between GC and CC, with 1596 showing higher expression in GC and 1560 in CC. A high concordance was found between the results of our and previous two studies in terms of the direction of expression of differentially expressed genes, as well as the selection of the highest significantly differentially expressed genes. The list of 500 most differentially expressed genes across the three studies is presented in S2 and S3 Tables for GC and CC, respectively.
Gene ontology and IPA analysis
To determine significantly (p < 0.01) overrepresented biological functions in each cell population the list of differentially expressed genes in our study (524 up-regulated in GC, 126 up-regulated in CC) was subjected to GO analysis. Functions such as immune response, inflammatory response and chemotaxis were among the overrepresented in the GC compartment (Table 3). In the CC compartment protein binding, cell adhesion and multicellular organismal development were among overrepresented (Table 4).
Table 3. Top enriched biological functions in GC according to gene ontology analysis.
Hyp_c: hypergeometric distribution used for p value calculation.
| Items | Items details | Hyp_c |
|---|---|---|
| GO:0006954 | Inflammatory response | 5.00E-09 |
| GO:0006935 | Chemotaxis | 2.93E-06 |
| GO:0006955 | Immune response | 5.55E-06 |
Table 4. Top enriched biological functions in CC according to gene ontology analysis.
Hyp_c: hypergeometric distribution used for p value calculation.
| Items | Items details | Hyp_c |
|---|---|---|
| GO:0005515 | Protein binding | 0.01 |
| GO:0007155 | Cell adhesion | 0.03 |
| GO:0007275 | Multicellular organismal development | 0.03 |
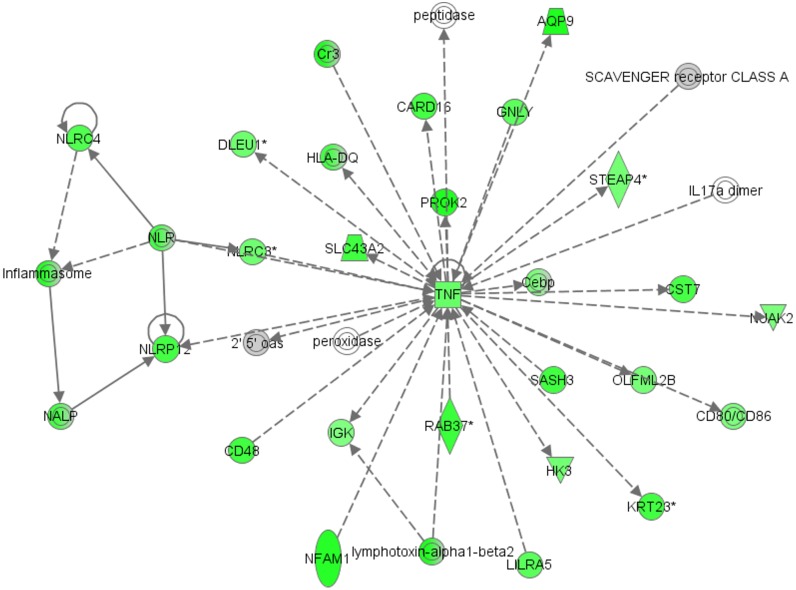
IPA Software was used for network enrichment analysis of differentially expressed genes in each cell subpopulation. Top network enriched in the GC compartment was connected with hematological system development and function, humoral immune response and cellular movement (Enrichment score: 32) (Fig 2). Thirty genes of this network were highly expressed in GC in our study; among them being tumor necrosis factor (TNF) gene, which is at the central position of the network.
Fig 2. Top network enriched in granulosa cells (Enrichment score: 32) generated with Ingenuity Pathway Analysis (IPA).
The network is associated with hematological system development and function, cancer and cellular movement. Up-regulated genes in our study are marked in green colour; the colour intensity of the nodes indicates the degree of up-regulation. Transcripts in grey were not differentially expressed. Genes are represented as nodes, and the biological relationship between two nodes is represented as a line: the plain line indicates direct interaction, the dashed line indicates indirect interaction; the line without arrowhead indicates binding only, the line finishing with a vertical line indicates inhibition; the line with an arrowhead indicates ‘acts on’.
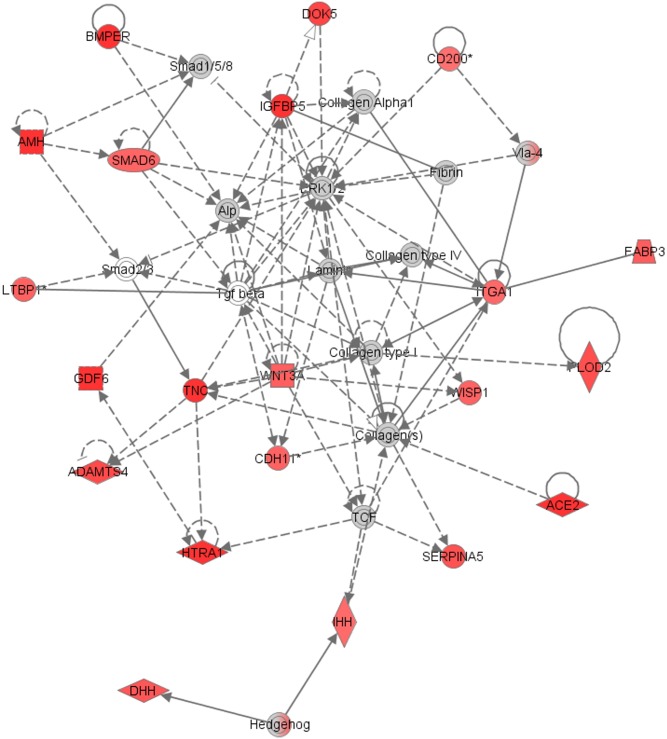
In the CC compartment, the top enriched network was connected with cellular development, skeletal and muscular system development and function and tissue development (Enrichment score: 41) (Fig 3). There were 21 genes of this network highly expressed in CC in our study. The network includes insulin-like growth factor binding protein 5 (IGFBP5) and tenascin C (TNC) genes with functions in extracellular matrix.
Fig 3. Top network enriched in cumulus cells (Enrichment score: 41) generated with Ingenuity Pathway Analysis (IPA).
The network is associated with cellular development, skeletal and muscular system development and function and tissue development. Up-regulated genes in our study are marked in red colour; the colour intensity of the nodes indicates the degree of up-regulation. Transcripts in grey were not differentially expressed. Genes are represented as nodes, and the biological relationship between two nodes is represented as a line: the plain line indicates direct interaction, the dashed line indicates indirect interaction; the line without arrowhead indicates binding only, the line finishing with a vertical line indicates inhibition; the line with an arrowhead indicates ‘acts on’.
Validation of differential expression by quantitative real-time PCR
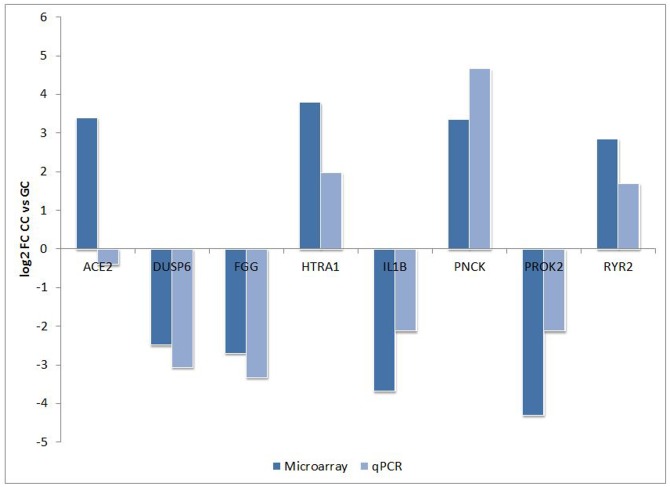
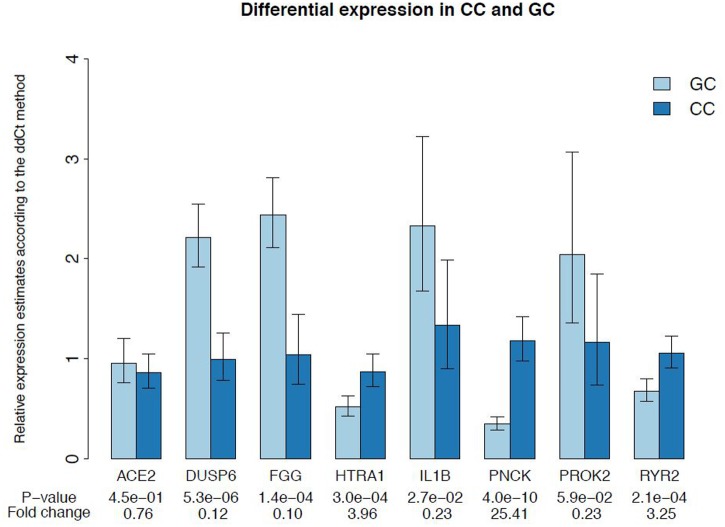
Microarray data from our study were validated using qPCR. Expression levels of 8 genes were checked on 19 individual GC and 19 individual CC samples derived from 16 newly included women. According to the microarray data, genes DUSP6, FGG, IL1B and PROK2 showed higher expression in GC, whereas ACE2, HTRA1, PNCK and RYR2 showed higher expression in CC. Results of the qPCR analysis were in agreement with microarray data for 7 out of 8 selected genes (Fig 4). According to qPCR data, the expression of these 7 genes significantly differed between GC and CC samples (Fig 5). For ACE2 gene, the qPCR analysis failed to confirm the differential expression from the microarray data.
Fig 4. Validation of microarray results by qPCR in additional 19 GC and 19 CC samples, derived from sixteen women.
For all genes, except ACE2, there was a statistically significant difference in expression between CC and GC. Data are presented as mean log2 fold change (FC) between expression in CC and GC. Blue bars represent log2 FC as measured by microarray; red bars represent log2 FC as measured by qPCR.
Fig 5. qPCR validation of microarray data.
The results represent mean relative mRNA expression of the selected genes ± SEM.
Discussion
In the present study differential gene expression between human GC and CC surrounding mature oocytes was analysed. Transcriptome profiling of individual human GC and CC samples revealed two distinct cell subpopulations with 7033 genes significantly (q < 10−4) differentially expressed. Out of these, 650 genes had at least a 2-fold log2 difference of expression.
As the number of study samples included in microarray studies is generally much lower than the number of tested variables (genes), testing large number of variables on a small number of study samples can result in false-positive and false-negative detections. This effect can contribute to low reproducibility of results between the studies performed by different research groups [23]. Combining datasets from several experiments where same condition has been investigated can increase the statistical power for detection of biologically relevant genes [24]. For this reason, we decided to perform a meta-analysis in order to maximize the potential for the discovery of consistently differentially expressed genes between human GC and CC. There were 3156 genes that showed consistent significant differential expression between GC and CC after meta-analysis. Among these, there were genes that have not yet been reported to be expressed in human somatic follicular cells like PROK2 and PNCK.
Differentially expressed genes
The list of significantly differentially expressed genes derived from our study, as well as from the meta-analysis, contained genes that have not yet been described in human GC and CC, like PROK2 and PNCK. qPCR analysis confirmed differential expression of these two genes.
The PROK2 gene was over-expressed in GC. Prokineticin 1 and 2 (PROK1 and 2) are multifunctional proteins that were first described in the gastrointestinal tract, where they induce contractions of smooth muscle fibres [25]. They exert their effects through G-protein coupled receptors, prokineticin receptor 1 and 2 (PROKR1 and PROKR2) [26]. PROK2 is mainly expressed in the central nervous system and nonsteroidogenic cells of the testes [27]. It has been established that PROK2 system is involved in angiogenesis [27], control of circadian rhythms [28] and sexual maturation [29]. PROK2 was shown to be expressed in the olfactory bulb of mice, where it is involved in neurogenesis and morphogenesis. Homozygous PROK2 and PROKR2 knockout mice show hypoplasia of the olfactory bulb [30] and atrophy of the reproductive system (testis, ovary, uterus, vagina, mammary gland) [31]. Defects of the PROK2 pathway in humans affect the neuroendocrine control of reproduction by causing hypogonadotropic hypogonadism due to the GnRH deficiency, which is caused by a reduction in the number of GnRH neurons in the preoptic region of the hypothalamus [32]. Furthermore, inactivating mutations of PROK2 and PROKR2 have been discovered in 2–7% of patients with Kallmann syndrome, who suffer from anosmia and hypogonadotropic hypogonadism [33]. According to the IPA analysis in our study, PROK2 gene was a part of the top enriched network connected to hematological system development and function, cancer and cellular movement. The development of perifollicular vascularization during folliculogenesis is of fundamental importance for the nourishment of the growing follicle and thus, development of a mature and competent oocyte [34]. Vascular endothelial growth factor (VEGF) is an important regulator of this process [35]. In the bovine corpus luteum (CL), PROK1 stimulates the expression of VEGF and this finding implies that it has an indirect role in the process of angiogenesis in the CL [36]. Expression of PROK2 in GC might imply that this gene also has a role in angiogenesis during folliculogenesis. Furthermore, PROK2 in the enriched IPA network was indirectly related to the tumor necrosis factor (TNF) gene at the central position. During mammalian ovulation, TNF causes apoptosis of ovarian surface epithelium and breakdown of the extracellular matrix in the follicle wall [37]. By this the oocyte can be released from the follicle. The connection of PROK2 with TNF might thus indicate that PROK2 is involved in the process of releasing the cumulus-oocyte complex from the follicle at the time of ovulation.
Another gene that has not been previously described in somatic cells of ovarian follicles and showed higher expression on our set of CC samples and after meta-analysis was PNCK. PNCK is a calmodulin kinase I isoform [38]. PNCK has a role in the murine central nervous system development, as its mRNA and protein are present there during embryonic development [39]. In adult rats, it is also expressed in the breast, uterus, heart and stomach [40]. It has been shown that PNCK mRNA over-expression causes epidermal growth factor receptor (EGFR) protein degradation leading to inhibition of EGF-induced mitogen-activated protein kinase (MAPK) activation [41]. After the LH surge, EGF-like factors (amphiregulin, epiregulin, β-cellulin) act on EGFR to activate Ras-MAPK3/1 pathway in GC and CC; this activation is crucial for the CC expansion, final oocyte maturation and GC luteinization [42]. In our study, only CC samples that surrounded mature oocytes were used for analyses; therefore, we presume that PNCK expression in CC after LH/hCG triggering oocyte maturation perhaps maintains inactivation of Ras-MAPK3/1 pathway, as the effects of this pathway are no longer needed with CC expansion and oocyte maturation being complete.
Among consistently differentially expressed genes that have already been described in human GC and CC there were also FGG and RYR2. Gene FGG that codes fibrinogen was expressed higher in GC. This finding is in accordance with a study of Parrot et al. [43], where higher expression of FGG was found in equine GC and the presence of FSH increased its expression. Fibrinogen is crucial for blood coagulation and tissue reparation, and its presence in the ovary is probably necessary for restoration of tissue integrity after follicle rupture at ovulation. Gene RYR2, a regulator of intracellular Ca++ concentrations, showed higher expression in CC. Positive correlation of RYR2 expression with expression of genes involved in cell to cell communication in CC has been described [11]. Our findings further support the assumption that intercellular signaling in CC is dependent upon gap junction proteins as well as Ca++ signaling.
Interestingly, we also identified a number of long non-coding RNA (lncRNA) transcripts as differentially expressed, when comparing GC and CC expression profiles. LncRNAs are transcripts of >200 nucleotides without known protein coding function [44]. They play a role in regulating gene expression at the level of chromatin modifications, transcription and post-transcriptional processing [45]. In human CC, differential expression of several lncRNAs was found between mature and germinal vesicle oocytes [46] and between high and poor-quality embryos [47]. These results together with the results of our study show that lncRNAs play a role in the development of oocytes. It is possible that lncRNAs expression participates in driving the differentiation of GC and CC and further studies are required to characterize their biological functions during the process of oocyte maturation.
Biological functions enriched in GC
The 524 up-regulated genes in GC represent biological functions such as inflammatory and immune response, and these results are consistent with previous studies [10, 11]. Ovulation is considered to be a process similar to inflammation due to the rupture of surface epithelium followed by a healing process [48]. After the release of the oocyte from the ovary the coagulation cascade is activated through invasion of leucocytes and cytokine release, that are needed for tissue healing [49]. Among the genes involved in the inflammatory process that were significantly up-regulated in GC in our study was IL1B. IL1 system is composed of ligands IL1A and IL1B [50], receptors IL1R1 and IL1R2 [51] and a receptor antagonist IL1RA [52]. In human cultured GC IL1B was shown to regulate plasminogen activator activity and steroidogenesis [53]. In our study IL1B, IL1RA and IL1R2 were up-regulated in GC, IL1A expression was undetectable and IL1R1 was up-regulated in CC. These findings are in accordance with the study of Martoriati and Gérard [54], where equine GC showed the same expression of IL1 system genes. They confirm the proposed involvement of the IL1 system in the events regulating oocyte maturation and ovulation [55] (Gérard et al., 2004). The higher expression of IL1R1 in CC probably indicates that IL1B exerts its effects on CC through this receptor. In the present study, IL6 and IL8 were also up-regulated in GC. This finding is in accordance with previous studies, where it has been established that IL1 induces expression of pro-inflammatory genes (IL6, IL8) at the site of the follicle rupture [56]. Cytokines IL6 and IL8 induce cellular proliferation which is needed for the healing of the ovarian surface epithelium [57]. Furthermore, IL6 has an important role in ovulation process as it is involved in regulation of meitoic maturation, CC expansion and CL formation [58].
The group of 'toll-like' receptors (TLR1,2, 4, 6, 7, 8), that has a role in the innate immune response in somatic cells of ovarian follicles [59], was found to be up-regulated in GC in our study. Our results are in line with a previous study that compared transcriptomes of GC and CC [10]. In mouse CC, TLR genes activate the expression of several signalling molecules (IL6, PTGS2, PTX3) that have a known role in CC expansion [60], cumulus-oocyte complex oviductal migration [61] and fertilization [62]. In hen GC, treatment with LH led to an increase in TLR's mRNA in pre-ovulatory follicles [59]. In the study of Woods et al. [63] it was shown that TLR signaling pathway influences hen GC steroidogenesis and apoptosis in relation to follicle size. Additionally, it has been suggested that TLR's function as a detection system to initiate tissue regeneration after injury [64]. According to these findings, we may presume that TLR signaling pathway also modulates human folliculogenesis. Furthermore, the high expression of TLR's in GC of pre-ovulatory follicles might support the suggestion that they are important in the healing process at the sight of the follicle rupture after ovulation. Additional studies are needed to determine the effects of TLR mediated signaling in GC during folliculogenesis.
Biological functions enriched in CC
The 126 significantly up-regulated genes in CC represent biological functions as multicellular organismal development and cell differentiation. Genes IGFBP5 and TNC were among the genes involved in signal transduction. IGFBP5 is a binding protein for insulin-like growth factors 1 and 2 (IGF1, IGF2), that are known to stimulate extracellular matrix production [65] and regulate steroidogenesis [66] during folliculogenesis. IGFBP proteins control the bioactivity of IGFs and controlled access to IGF1 is crucial for the proper coordination of early oocyte and follicular development [67]. In the present study, IGFBP5 expression was increased in CC and IGF1 expression was increased in GC. These results are in line with the study of Ingman et al. [68] where treatment of CC culture media with IGF1 increased extracellular matrix IGFBP5 by 2.5-fold. Our results probably indicate that in pre-ovulatory follicles, bioavailability of IGF1 protein derived from GC is regulated by IGFBP5 expression in CC.
TNC is an important glycoprotein of the extracellular matrix [69]. It is involved in cellular functions as adhesion, migration, embryonic development, wound healing and tumour metastasis [70]. It was shown to be overexpressed in the stroma of malignant ovarian tumours and it was proposed to be involved in the process of tumour invasion [71]. In ovarian cancer cell line expressions of transforming growth factor beta 1 (TGFB1) and TNC are significantly related [72]. Furthermore, TGFB1 induces secretion of TNC from ovarian fibroblasts [68]. During folliculogenesis TGFB1 enables CC hyaluronic acid production and expansion, regulates CC steroidogenesis and promotes GC proliferation [73, 74, 75]. TNC was significantly higher expressed in CC after our microarray analysis as well as after the meta-analysis. Over-expression of TNC in CC might thus indicate that TNC is one of the genes through which TGFB1 exerts its effects during the process of oocyte maturation. Evidently, if this process is pathological, it is related to ovarian tumour invasion. The precise role of TNC in ovarian folliculogenesis and the possible relation with TGFB1 will need to be determined in future studies.
In conclusion, with this study we upgraded the existing data on differentially expressed genes between GC and CC by description of PROK2 and PNCK genes whose expression has, to the best of our knowledge, not yet been reported in human GC and CC. These genes could be involved in any number of the processes regulating folliculogenesis. Further research is needed to elucidate their potential physiological roles. Pathway enrichment and IPA analyses suggested that GC are involved in angiogenesis and ovarian healing process after follicle rupture, whereas pathways enriched in CC are connected with intercellular communication and extracellular matrix formation. Identification of genes and pathways that contribute to the development of mature and competent oocytes has the potential to improve oocyte in vitro maturation procedures.
Supporting Information
(DOC)
(DOC)
(DOC)
(DOCX)
(DOCX)
Data Availability
The microarray data are available at the Gene Expression Omnibus (GEO) public repository (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE55654.
Funding Statement
This work was funded by the Slovenian Research Agency (https://www.arrs.gov.si/en/novo.asp), grant number P3-0326. Tanja Burnik Papler is funded as a young researcher by the Slovenian Research Agency. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Moley KH, Schreiber JR. Ovarian follicular growth, ovulation and atresia. Endocrine, paracrine and autocrine regulation. Adv Exp Med Biol. 1995;77: 103–119. [DOI] [PubMed] [Google Scholar]
- 2. Greenwald GS, Terranova PF. Follicular selection and its control In: Knobil E, Neill JD, eds. The Physiology of Reproduction. New York: Raven Press; 1988; 387–445. [Google Scholar]
- 3. Kidder GM, Vanderhyden BC. Bidirectional communication between oocytes and follicle cells: ensuring oocyte developmental competence. Can J Physiol Pharmacol. 2010;88: 399–413. 10.1139/y10-009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knight PG, Glister C. Focus on TGF-b Signalling: TGF-b superfamily members and ovarian follicle development. Reproduction. 2006; 132: 191–206. [DOI] [PubMed] [Google Scholar]
- 5. Gilchrist RB, Lane M, Thompson JG. Oocyte-secreted factors: regulators of cumulus cell function and oocyte quality. Hum Reprod Update. 2008;14: 159–177. 10.1093/humupd/dmm040 [DOI] [PubMed] [Google Scholar]
- 6. Li Q, McKenzie LJ, Matzuk MM. Revisiting oocyte-somatic cell interactions: in search of novel intrafollicular predictors and regulators of oocyte developmental competence. Mol Hum Reprod. 2008;14: 673–678. 10.1093/molehr/gan064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Binelli M, Murphy BD. Coordinated regulation of follicle development by germ and somatic cells. Rep Fertil Dev. 2010;22: 1–12. [DOI] [PubMed] [Google Scholar]
- 8. Thomas FH, Ethier JF, Shimasaki S, Vanderhyden BC. Follicle-stimulating hormone regulates oocyte growth by modulation of expression of oocyte and granulosa cell factors. Endocrinology. 2005;146: 941–949. [DOI] [PubMed] [Google Scholar]
- 9. Wang JM, Chao JR, Chen W, Kuo ML, Yen JJ, Yang-Yen HF. The antiapoptotic gene mcl-1 is upregulated by the phosphatidylinositol 3'-kinase/factor complex containing CREB. Mol Cell Biol. 1999;19: 6195–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kõks S, Velthut A, Sarapik A, Altmäe S, Reinmaa E, Schalkwyk LC, et al. The differential transcriptome and ontology profiles of floating and cumulus granulosa cells in stimulated human antral follicles. Mol Hum Reprod. 2010;16: 229–240. 10.1093/molehr/gap103 [DOI] [PubMed] [Google Scholar]
- 11. Grøndahl ML, Andersen CY, Bogstad J, Borgbo T, Boujida VH, Borup R. Specific genes are selectively expressed between cumulus and granulosa cells from individual human pre-ovulatory follicles. Mol Hum Reprod. 2012;18: 572–584. 10.1093/molehr/gas035 [DOI] [PubMed] [Google Scholar]
- 12. Bonnet A, Cabau C, Bouchez O, Sarry J, Marsaud N, Foissac S, et al. An overview of gene expression dynamics during early ovarian folliculogenesis: specificity of follicular compartments and bi-directional dialog. BMC Genomics. 2013;9: 904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cooper TG, Noonan E, von Eckardstein S, Auger J, Baker HW, Hermann M, et al. World Health Organization reference values for human semen characteristics. Hum Reprod Update. 2010;16: 231–245. 10.1093/humupd/dmp048 [DOI] [PubMed] [Google Scholar]
- 14. Assou S, Haouzi D, De Vos J, Hamamah S. Human cumulus cells as biomarkers for embryo and pregnancy outcomes. Mol Hum Rep. 2010;16: 531–538. [DOI] [PubMed] [Google Scholar]
- 15. Szaniszlo P, Wang N, Sinha M, Reece LM, Van Hook JW, Luxon BA, et al. Getting the right cells to the array: Gene expression microarray analysis of cell mixtures and sorted cells. Cytometry A. 2004;59: 191–202. [DOI] [PubMed] [Google Scholar]
- 16. Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004: Article3 [DOI] [PubMed] [Google Scholar]
- 17. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc: Series B (Statistical Methodology). 1995; 289–300. [Google Scholar]
- 18. Sean D, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23: 1846–1847. [DOI] [PubMed] [Google Scholar]
- 19. Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22: 2825–2827. [DOI] [PubMed] [Google Scholar]
- 20. Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19: 368–375. [DOI] [PubMed] [Google Scholar]
- 21. Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, et al. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37: W317–322. 10.1093/nar/gkp416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25: 402–408. [DOI] [PubMed] [Google Scholar]
- 23. Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 2006;7: 55–65. [DOI] [PubMed] [Google Scholar]
- 24. Ghosh D, Barette TR, Rhodes D, Chinnaiyan AM. Statistical issues and methods for meta-analysis of microarray data: a case study in prostate cancer. Funct Integr Genomics. 2003;3: 180–188. [DOI] [PubMed] [Google Scholar]
- 25. Li M, Bullock CM, Knauer DJ, Ehlert FJ, Zhou QY. Identification of two prokineticin cDNAs: Recombinant proteins potently contract gastrointestinal smith muscle. Mol Pharmacol. 2001;59: 692–698. [DOI] [PubMed] [Google Scholar]
- 26. Lin R. Characterization of endocrine gland-derived vascular endothelial growth factor signaling in adrenal cortex capillary endothelial cells. J Biol Chem. 2002;277: 8724–8729. [DOI] [PubMed] [Google Scholar]
- 27. Le Couter J, Lin R, Tejada M, Frantz G, Peale F, Hillan KJ, et al. The endocrine-gland-derived VEGF homologue Bv8 promotes angiogenesis in the testis: Localization of Bv8 receptors to endothelial cells. Proc Natl Acad Sci USA. 2003;100: 2685–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou QY, Cheng MY. Prokineticin 2 and circadian clock output. FEBS J. 2005;272: 5703–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Balasubramanian R, Plummer L, Sidis Y, Pitteloud N, Martin C,Zhou QY, et al. The puzzles of the prokineticin 2 pathway in human reproduction. Mol Cell Endocrinol. 2011;346: 44–50. 10.1016/j.mce.2011.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ng KL, Li JD, Cheng MY, Leslie FM, Lee AG, Zhou QY.Dependence of olfactory bulb neurogenesis on prokineticin 2 signaling. Science. 2005;308: 1923–1927. [DOI] [PubMed] [Google Scholar]
- 31. Matsumoto S. Abnormal development of the olfactory bulb and reproductive system in mice lacking prokineticin receptor PKR2. Proc Natl Acad Sci USA. 2006;103: 4140–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pitteloud N, Zhang C, Pignatelli D, Li JD, Raivio T, Cole LW, et al. Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA. 2007;104: 17447–17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dode C, Teixeira L, Levilliers J, Fouveaut C, Bouchard P, Kottler ML, et al. Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genetics. 2006;2: e175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Suzuki T, Sasano H, Takaya R, Fukaya T, Yajima ANagura H. Cyclic changes of vasculature and vascular phenotypes in normal human ovaries. Hum Rep. 1998;13: 953–959. [DOI] [PubMed] [Google Scholar]
- 35. Lam PM, Haines C. Vascular endothelial growth factor plays more than an angiogenic role in the female reproductive system. Fertil Steril. 2005;84: 1775–1778. [DOI] [PubMed] [Google Scholar]
- 36. Kisliouk T, Podlovni H, Spanel-Borowski K, Ovadia O, Zhou QY, Meidan R. Prokineticins (endocrine gland-derived vascular endothelial growth factor and BV8) in the bovine ovary: expression and role as mitogens and survival factors for corpus luteum-derived endothelial cells. Endocrinology. 2005;146: 3950–3958. [DOI] [PubMed] [Google Scholar]
- 37. Murdoch WJ, Lund SA. Prostaglandin-independent anovulatory mechanism of indomethacin action: inhibition of tumor necrosis factor alpha-induced sheep ovarian cell apoptosis. Biol Reprod. 1999;61: 1655–1659. [DOI] [PubMed] [Google Scholar]
- 38. Naito Y, Watanabe Y, Yokokura H, Sugita R, Nishio M, Hidaka H. Isoform-specific activation and structural diversity of calmodulin kinase I. J Biol Chem. 1997;272: 32704–32708. [DOI] [PubMed] [Google Scholar]
- 39. Jusuf AA, Rina S, Sakagami H, Terashima T. Expression of Ca2+/calmodulin-dependent protein kinase (CaMK) I beta 2 in developing rat CNS. Neuroscience. 2002;109: 407–420. [DOI] [PubMed] [Google Scholar]
- 40. Gardner HP, Rajan JV, Ha SI, Copeland NG, Gilbert DJ, Jenkins NA, et al. Cloning, characterization, and chromosomal localization of Pnck, a Ca2+/calmodulin-dependent protein kinase. Genomics. 2000;63: 279–288. [DOI] [PubMed] [Google Scholar]
- 41. Deb TB, Coticchia CM, Barndt R, Zuo H, Dickson RB, Johnson MD. Pregnancy upregulated nonubiquitous calmodulin kinase induces ligand-independent EGFR degradation. Am J Physiol Cell Physiol. 2008;295: C365–C377. 10.1152/ajpcell.00449.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fan HY, Liu Z, Shimada M, Sterneck E, Johnson PF, Hedrick SM, et al. MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science. 2009;324: 938–941. 10.1126/science.1171396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Parrott JA, Whaley PD, Skinner MK. Extrahepatic expression of fibrinogen by granulosa cells: potential role in ovulation. Endocrinology. 1993;133: 1645–1649. [DOI] [PubMed] [Google Scholar]
- 44. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81: 145–166. 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136: 629–641. 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 46. Yerushalmi GM, Salmon-Divon M, Yung Y, Maman E, Kedem A, Ophir L, et al. Characterization of the human cumulus cell transcriptome during final follicular maturation and ovulation. Mol Hum Reprod. 2014;20: 719–735. 10.1093/molehr/gau031 [DOI] [PubMed] [Google Scholar]
- 47. Xu XF, Li J, Cao YX, Chen DW, Zhang ZG, He XJ, et al. Differential Expression of Long Noncoding RNAs in Human Cumulus Cells Related to Embryo Developmental Potential: A Microarray Analysis. Reprod Sci. 2015;22: 672–678. 10.1177/1933719114561562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Espey LL. Current status of the hypothesis that mammalian ovulation is comparable to an inflammatory reaction. Biol Reprod. 1994;50: 233–238. [DOI] [PubMed] [Google Scholar]
- 49. Fleming JS, Beaugie CR, Haviv I, Chenevix-Trench G, Tan OL. Incessant ovulation, inflammation and epithelial ovarian carcinogenesis: revisiting old hypotheses. Mol Cell Endocrinol. 2006;247: 4–21. [DOI] [PubMed] [Google Scholar]
- 50. Dinarello CA. The interleukin-1 family: 10 years of discovery. FASEB J. 1994;8: 1314–1325. [PubMed] [Google Scholar]
- 51. Sims JE, Dower SK. Interleukin-1 receptors. Eur Cytokine Netw. 1991;5: 539–546. [PubMed] [Google Scholar]
- 52. Arend WP. Interleukin 1 receptor antagonist: A new member of the interleukin 1 family. J Clin Invest. 1991;88: 1445–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Piquette GN, Simon C, el Danasouri I, Frances A, Polan ML. Gene regulation of interleukin-1 beta, interleukin-1 receptor type I, and plasminogen activator inhibitor-1 and -2 in human granulosa-luteal cells. Fertil Steril. 1994;62: 760–770. [DOI] [PubMed] [Google Scholar]
- 54. Martoriati A, Gérard N. Interleukin-1 (IL-1) system gene expression in granulosa cells: kinetics during terminal preovulatory follicle maturation in the mare. Reprod Biol Endocrinol. 2003;6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gérard N, Caillaud M, Martoriati A, Goudet G, Lalmanach AC. The interleukin-1 system and female reproduction. J Endocrinol. 2004;180: 203–212. [DOI] [PubMed] [Google Scholar]
- 56. Rae MT, Niven D, Ross A, Forster T, Lathe R, Critchley HO, et al. Steroid signalling in human ovarian surface epithelial cells: the response to interleukin-1alpha determined by microarray analysis. J Endocrinol. 2004;183: 19–28. [DOI] [PubMed] [Google Scholar]
- 57. Auersperg N, Wong AS, Choi KC, Kang SK, Leung PC. Ovarian surface epithelium: biology, endocrinology, and pathology. Endocr Rev. 2001;22: 255–288. [DOI] [PubMed] [Google Scholar]
- 58. Kawasaki F, Kawano Y, Kosay Hasan Z, Narahara H, Miyakawa I. The clinical role of interleukin-6 and interleukin-6 soluble receptor in human follicular fluids. Clin Exp Med. 2003;3: 27–31. [DOI] [PubMed] [Google Scholar]
- 59. Shimada M, Hernandez-Gonzalez I, Gonzalez-Robanya I, Richards JS. Induced expression of pattern recognition receptors in cumulus oocyte complexes: novel evidence for innate immune-like functions during ovulation. Mol Endocrinol. 2006;20: 3228–3239. [DOI] [PubMed] [Google Scholar]
- 60. Liu Z, Shimada M, Richards JS. The involvement of the toll-like receptor family in ovulation. J Assist Reprod Gen. 2008;25: 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Salustri A, Garlanda C, Hirsch E, De Acetis M, Maccagno A, Bottazzi B, et al. PTX3 plays a key role in the organization of the cumulus oophorus extracellular matrix and in in vivo fertilization. Development. 2004;131: 1577–1586. [DOI] [PubMed] [Google Scholar]
- 62. Shimada M, Yanai Y, Okazaki T, Noma N, Kawashima I, Mori T, et al. Hyaluronan fragments generated by sperm-secreted hyaluronidase stimulate cytokine/chemokine production via the TLR2 and TLR4 pathway in cumulus cells of ovulated COCs, which may enhance fertilization. Development. 2008;135: 2001–2011. 10.1242/dev.020461 [DOI] [PubMed] [Google Scholar]
- 63. Woods DC, Schorey JS, Johnson AL. Toll-like receptor signaling in hen ovarian granulosa cells is dependent on stage of follicle maturation. Reproduction. 2009;137: 987–996. 10.1530/REP-08-0320 [DOI] [PubMed] [Google Scholar]
- 64. Zhang Z, Schluesener HJ. Mammalian toll-like receptors: from endogenous ligands to tissue regeneration. Cell Mol Life Sci. 2006;63: 2901–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pricci F, Pugliese G, Romano G, Romeo G, Locuratolo N, Pugliese F, et al. Insulin-like growth factors I and II stimulate extracellular matrix production in human glomerular mesangial cells. Comparison with transforming growth factor-beta. Endocrinol. 1996;137: 879–885. [DOI] [PubMed] [Google Scholar]
- 66. Giudice LC. Insulin-like growth factors and ovarian development. Endocr Rev. 1992;13: 641–669. [DOI] [PubMed] [Google Scholar]
- 67. Monget P, Bondy C. Importance of the IGF system in early folliculogenesis. Mol Cell Endocrinol. 2000;163: 89–93. [DOI] [PubMed] [Google Scholar]
- 68. Ingman WV, Owens PC, Armstrong DT. Differential regulation by FSH and IGF-I of extracellular matrix IGFBP-5 in bovine granulosa cells: effect of association with the oocyte. Mol Cell Endocrinol. 2000;164: 53–58. [DOI] [PubMed] [Google Scholar]
- 69. Familiari G, Verlengia C, Nottola SA, Renda T, Micara G, Aragona C, et al. Heterogeneous distribution of fibronectin, tenascin-C, and laminin immunoreactive material in the cumulus-corona cells surrounding mature human oocytes from IVF-ET protocols-evidence that they are composed of different subpopulations: an immunohistochemical study using scanning confocal laser and fluorescence microscopy. Mol Reprod Dev. 1996;43: 392–402. [DOI] [PubMed] [Google Scholar]
- 70. Erickson H, Bourdon M. Tenascin: an extracellular matrix protein prominent in specialized embryonic tissues and tumours. Annu Rev Cell Biol. 1989;5: 71–92. [DOI] [PubMed] [Google Scholar]
- 71. Wilson KE, Langdon SP, Lessells AM, Miller WR. Expression of the extracellular matrix protein tenascin in malignant and benign ovarian tumours. Br J Cancer. 1996;74: 999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wilson KE, Bartlett JM, Miller EP, Smyth JF, Mullen P, Miller WR, et al. Regulation and function of the extracellular matrix protein tenascin-C in ovarian cancer cell lines. Br J Cancer. 1999;80: 685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vanderhyden BC, Macdonald EA, Nagyova E, Dhawan A. Evaluation of members of the TGF beta superfamily as candidates for the oocyte factors that control mouse cumulus expansion and steroidogenesis. Hum Reprod Suppl. 2003;61: 55–70. [PubMed] [Google Scholar]
- 74. Dragovic RA, Ritter LJ, Schulz SJ, Amato F, Armstrong DT, Gilchrist RB. Role of oocyte-secreted growth differentiation factor 9 in the regulation of mouse cumulus expansion. Endocrinol. 2005;146: 2798–2806. [DOI] [PubMed] [Google Scholar]
- 75. Gilchrist RB, Ritter LJ, Myllymaa S, Kaivo-Oja N, Dragovic RA, Hickey TE, et al. Molecular basis of oocyte-paracrine signalling that promotes granulosa cell proliferation. J Cell Sci. 2006;119: 3811–3821. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
(DOC)
(DOCX)
(DOCX)
Data Availability Statement
The microarray data are available at the Gene Expression Omnibus (GEO) public repository (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE55654.