Abstract
We demonstrate that surface-induced dissociation (SID) coupled with ion mobility-mass spectrometry (IM-MS) is a powerful tool for determining the stoichiometry and quaternary structure of a multi-subunit ribonucleoprotein (RNP) complex assembled in Mg2+. We investigated here Pyrococcus furiosus (Pfu) RNase P, an archaeal RNP that catalyzes tRNA 5′ maturation. Previous step-wise, Mg2+-dependent reconstitutions of Pfu RNase P using its catalytic RNA subunit and two interacting protein cofactor pairs (RPP21•RPP29 and POP5•RPP30) revealed functional RNP intermediates en route to the RNase P enzyme, but provided no information on subunit stoichiometry. Our native MS studies with the proteins alone showed RPP21•RPP29 and (POP5•RPP30)2 complexes, but indicated a 1:1 composition for all subunits when either or both protein complexes bind the cognate RNA. These results highlight the utility of SID and IM-MS in resolving conformational heterogeneity and yielding insights on RNP assembly.
Keywords: RNase P, native mass spectrometry, stoichiometry, RNA-protein complex, surface-induced dissociation, ion mobility
Nano-electrospray ionization-based native mass spectrometry (MS) has been used to determine the stoichiometry, structure and interactions of several non-covalent protein assemblies, including large viral capsids (106 Da range)[1], but its use has been restricted to only a few ribonucleoprotein (RNP) complexes assembled in the absence of divalent cations.[1c, 2] The limited progress with RNPs is partly due to the weak signal intensity and poor resolution caused by ionization suppression[3] and peak broadening,[4] respectively; both are inevitable consequences of non-specific RNP-attachment of non-volatile cations (e.g., Mg2+), which are required for the assembly of many cellular RNPs. Formation of analyte ions in native MS is dependent on volatile buffers (e.g., ammonium acetate),[5] which may not be ideal for forming the functionally relevant RNP complex. Collectively, these factors lead to complicated spectra with several different Mg2+-associated RNA/RNP ions of similar m/z. To overcome these challenges, it is critical to isolate low abundance ionic species and identify their subunit make-up. Here, we demonstrate how tandem MS (MS/MS) and ion mobility (IM), which have already provided information about the architecture and dynamics of protein complexes,[1a-e, 1g-i, 6] can also be used to resolve RNP heterogeneity and to determine the stoichiometry of archaeal RNase P, a multi-subunit RNP.
In all organisms, RNase P is an essential enzyme that cleaves the 5′-leader of precursor tRNAs (pre-tRNAs) in a Mg2+-dependent manner to yield functional mature tRNAs.[7] While an RNA-free, protein-based form of RNase P has been reported in several eukaryotes,[8] the RNP form is found in all domains of life. The RNP form comprises a catalytic RNase P RNA (RPR) and a variable number of RNase P proteins (RPPs) depending on the source: one in Bacteria, up to five in Archaea and as many as ten in Eukarya.[7, 9] A common ancestry for all RPRs is evident in their highly conserved sequences and structural elements, but sequence similarity for RPPs is shared only between Archaea and Eukarya. While eukaryotic RNase P has not been reconstituted in vitro, RNase P from several archaea has been assembled from recombinant RPR and RPPs.[10] High-resolution structures of archaeal RPPs have also been determined.[9a, 11] These advances make archaeal RNase P an attractive experimental model for understanding how multiple proteins modulate the structure, dynamics and function of an RNA in a large RNP, while serving as a tractable surrogate for its eukaryotic relative.
In our biochemical studies on RNase P from the archaeon Pyrococcus furiosus (Pfu), we determined that addition of the four RPPs to the RPR enhances its kcat/KM by 4250-fold and that the RPPs function as binary complexes (RPP21•RPP29 and POP5•RPP30).[10d] The assembly of the RPR with each binary complex is functional, albeit only at higher concentrations of Mg2+ (120 instead of 30 mM) and with a lower kcat/KM than the RNP reconstituted with both pairs.[10a, 10d] These findings were presaged and subsequently confirmed by genetic and structural studies. First, yeast two-hybrid analysis of Methanothermobacter thermautotrophicus and Pyrococcus horikoshii (Pho) RPPs uncovered strong interactions between RPP21 and RPP29 and between POP5 and RPP30.[12] Second, RPP21 and RPP29 were found to interact as a heterodimer by X-ray crystallography and NMR.[11a, 11d] Third, the crystal structure of POP5 and RPP30 from Pho revealed a heterotetramer with a POP5 dimer placed between two RPP30 monomers.[11b] Dynamic light scattering, NMR line widths and hydrodynamic measurements corroborated a similar (POP5•RPP30)2 assembly in Pfu.[11c, 13] Although comparable studies were not performed on the binary RPPs complexed with the RPR, these results led to some speculations, like the idea of Pho RNase P being made up of two copies each of the RPR and RPPs, centered around a (POP5•RPP30)2 assembly flanked symmetrically by two copies of RPP21•RPP29.[11b]
While isotope-coded tags, two-dimensional gel electropohoresis followed by fluorescent staining, metabolic labeling (SILAC), chemical labeling (iTRAQ) and label-free spectral counts have been employed in MS-based studies to establish the protein composition and stoichiometry in stage-specific spliceosomal RNP complexes,[14] here we have used SID and IM-MS to simultaneously determine the RNA and protein stoichiometry of Pfu RNase P. In IM-MS, ions are driven by an electric field to pass through a drift tube filled with bath gas, and those with large collisional cross sections and low charge states are slowed down by the bath gas.[1h, 1i, 6b, 6c, 15] IM can thus assist in resolving complicated spectra because it can separate ions with the same m/z but different drift times based on differences in charge states, sizes and shapes. Ion source activation and collision-induced dissociation (CID) have been used to remove solvents, salts and detergents attached to protein ions in the gas phase;[16] we have also employed this collision-induced cleaning technique to remove Mg2+ attached non-specifically to RNAs and RNPs. Because Mg2+ can stabilize protein and RNP complexes[17] and create structures that are difficult for CID to break apart, surface-induced dissociation (SID) was used to dissociate RNPs in the presence of Mg2+. SID deposits high energy into the ions through a single collision event with a surface[18] to reveal dissociation pathways with high-energy barriers, which are likely for RNP disassembly. In tandem with IM, SID can be employed to dissociate a putative RNP complex, thereby confirming its identity and revealing substructures. Together, these MS techniques allowed us to determine a 1:1 composition for all subunits and discern a possible assembly pathway of Pfu RNase P, reconstituted in the presence of Mg2+ from recombinant RPR, RPP21•RPP29 and POP5•RPP30. (See Supplement for comment on the fifth archaeal RPP.)
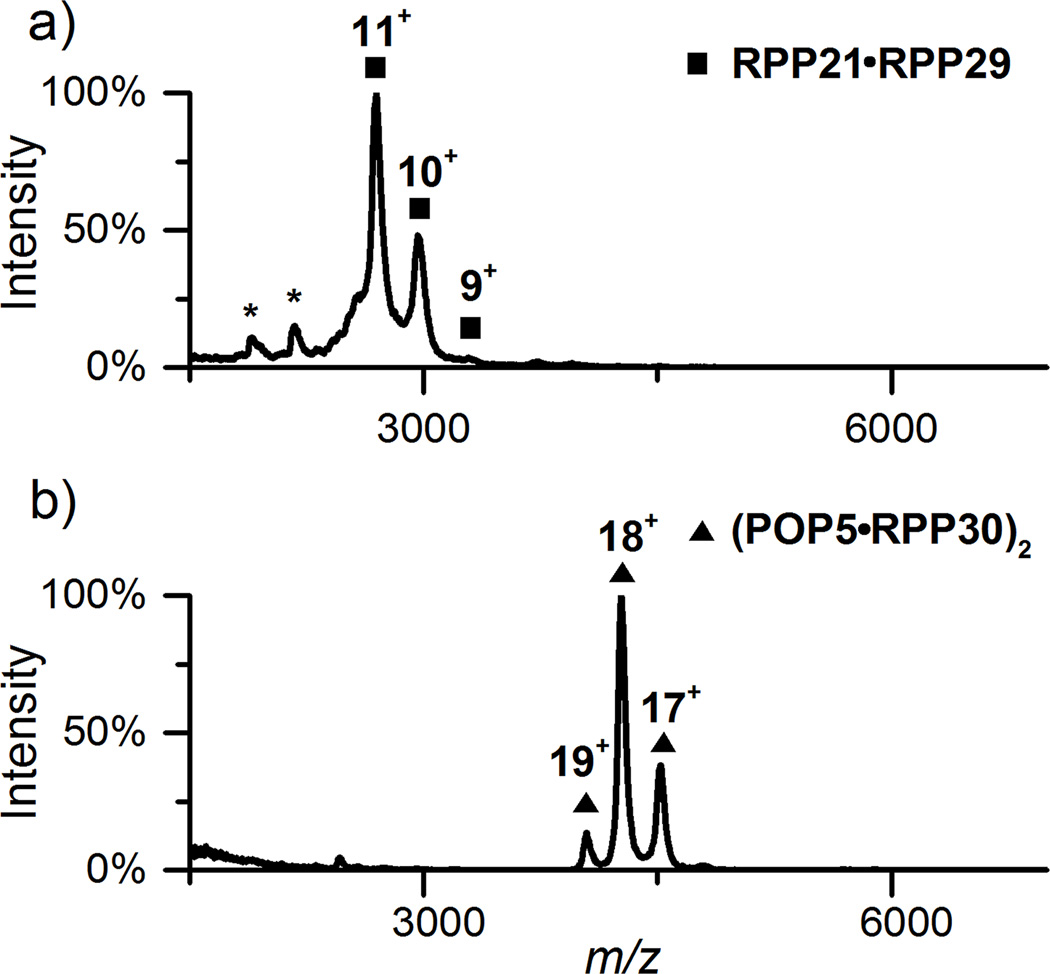
To ascertain whether the structures of the RPP complexes observed previously by X-ray crystallography and NMR are preserved in the gas phase, we performed native MS on Pfu RPP21•RPP29 and POP5•RPP30. The spectra indeed confirm an RPP21•RPP29 heterodimer and a (POP5•RPP30)2 heterotetramer in 500 mM NH4OAc (Figure 1) as well as in 100 mM and 800 mM (not shown), and provide a reference for their RPR-bound states in the experiments described below.
Figure 1.
Native MS spectra of Pfu RPP21•RPP29 heterodimer (a) and (POP5•RPP30)2 heterotetramer (b) in 500 mM NH4OAc. In each spectrum, the dominant peaks and charge states are indicated. Asterisks indicate the 7+ and 8+ states of RPP29, a small amount likely generated from the binary complex during storage or analysis. See Table S-1 for predicted and observed masses.
A major challenge in determining the stoichiometry of RNP complexes such as archaeal RNase P is the optimization of experimental conditions for MS that also closely reflect the native functional state. We focused our initial attention on Pfu RPR reconstituted with all four RPPs, a complex that exhibits maximum pre-tRNA cleavage activity in 50 mM Tris-HCl/HEPES-KOH (pH 7.5-8), 800 mM NH4OAc and 30 mM MgCl2.[10a, 10d] Because Tris/HEPES and 30 mM Mg2+ complicate MS experiments, we assembled Pfu RPR, RPP21•RPP29 and POP5•RPP30 in 800 mM NH4OAc and systematically explored a lower Mg(OAc)2 range (2 to 10 mM). In addition, we found that careful optimization of the acceleration voltage during collision-induced cleaning was critical to achieve distinguishable RNP signals (Figure S-1).
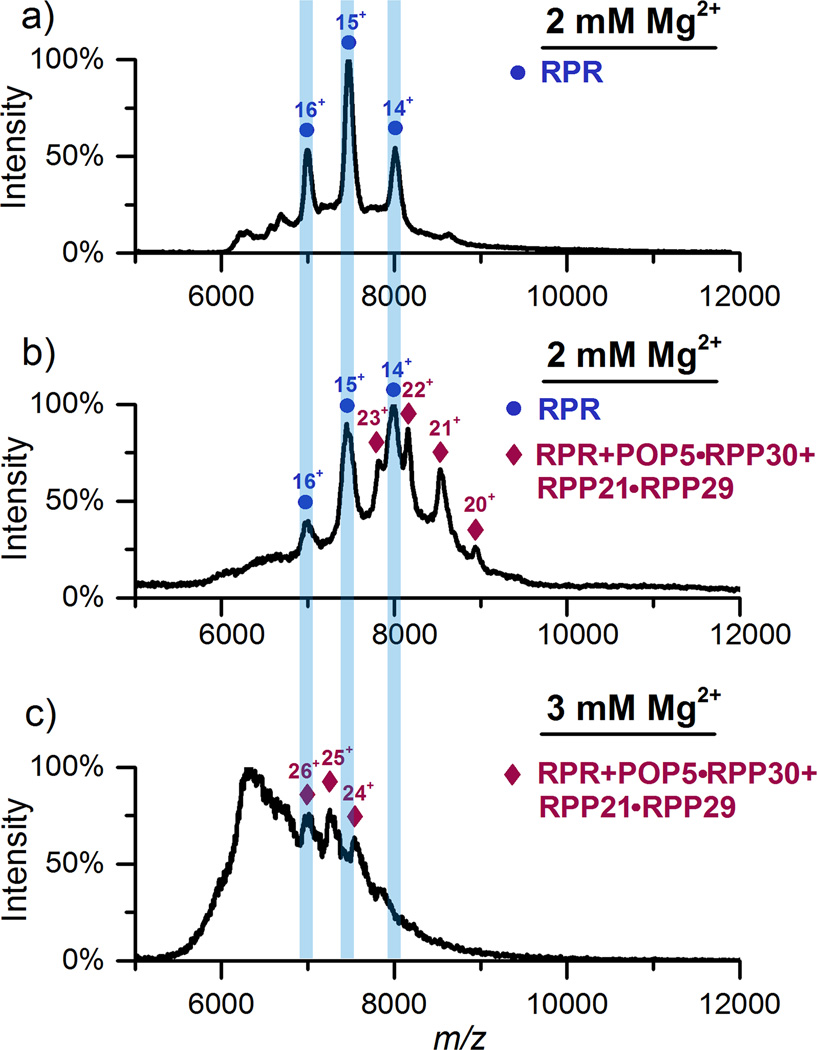
We empirically determined that assembling Pfu RPR, RPP21•RPP29 and POP5•RPP30 in 800 mM NH4OAc and ≤ 3 mM Mg(OAc)2 yielded the best MS data (Figure 2), though we have no evidence of protein aggregation at ≥ 3 mM Mg2+. While the overall RNP stoichiometry and oligomeric state remains the same with either 2 or 3 mM Mg(OAc)2, the data as expected are superior with the former. Figure 2a shows the spectrum for the RPR in 2 mM Mg(OAc)2, observed in the positive-ion mode due to bound Mg2+. Heterogeneous Mg2+ complexation in the RPR can also cause peak broadening, leading to the high background signal in the m/z range (6800 to 8200) of the RPR spectrum. When the RPR was assembled with the four proteins in 2 mM Mg(OAc)2, two series of peaks were observed (Figure 2b). The first set of peaks corresponds to the RPR, mirroring the charge states observed with the RPR alone (Figure 2a), while the second matches an RPR bound to one copy of each RPP. Although of poorer spectral quality, the 4-RPP Pfu RNase P complex in 3 mM Mg(OAc)2 also shows a 1:1 composition for all subunits (Figure 2c), an unexpected but informative result given the (POP5•RPP30)2 heterotetrameric structure observed in the absence of the RPR (Figure 1).
Figure 2.
a) Pfu RPR in 800 mM NH4OAc and 2 mM Mg(OAc)2, observed in positive-ion mode. b) RPR, RPP21•RPP29 and POP5•RPP30 in 800 mM NH4OAc and 2 mM Mg(OAc)2. Peaks corresponding to RPR alone (blue bars) and RPR with one copy of each of the four RPPs are indicated. c) Same as (b) except with 3 mM Mg(OAc)2. Collision-induced cleaning in the first collision cell was optimized to improve peak quality in each case [70 V (a); 130 V (b); 90 V (c); collision energy is V x charge state]. See Table S-1 for predicted and observed masses.
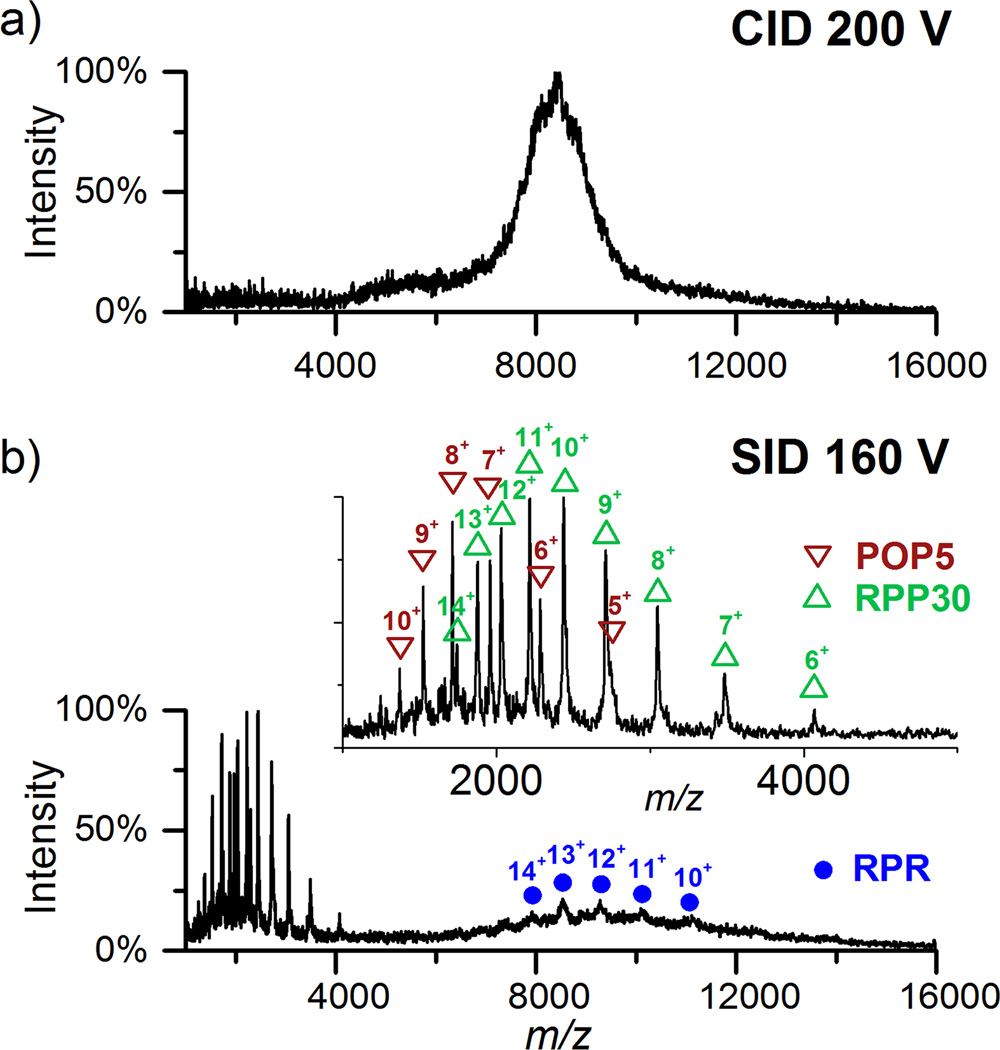
To further confirm the composition of the RNP complex (Figure 2b), CID and SID were employed. Because the signal intensity of the RNP complexes is too low to select a particular peak for dissociation, we chose to dissociate the complexes in the high m/z range (>6400). While even the highest CID acceleration voltage (200 V) did not dissociate the complex (Figure 3a), SID produced the RPR, POP5 and RPP30 fragments, validating their presence in the parental RNP (Figure 3b). By subtractive analysis, one can also deduce the presence of RPP21 and RPP29 in the parental ion: the mass of the complex minus the mass sum of RPR+POP5•RPP30 equals the mass of RPP21•RPP29 (Table S-1). These data highlight the previously unproven ability of SID in dissociating Mg2+-stabilized RNP complexes.
Figure 3.
CID and SID of ions in the high m/z range (>6400) present in the assembly of RPR, RPP21•RPP29 and POP5•RPP30 in 800 mM NH4OAc and 2 mM Mg(OAc)2. a) CID at 200 V (4000 to 4600 eV), the highest acceleration voltage possible with the instrument used, did not dissociate the RNP complex effectively. b) SID of precursor (whole charge state envelope) at 160 V (3200-3680 eV) successfully dissociated the RNP complex to produce POP5 and RPP30 monomers as well as the RPR (in the m/z range 8000-11000).
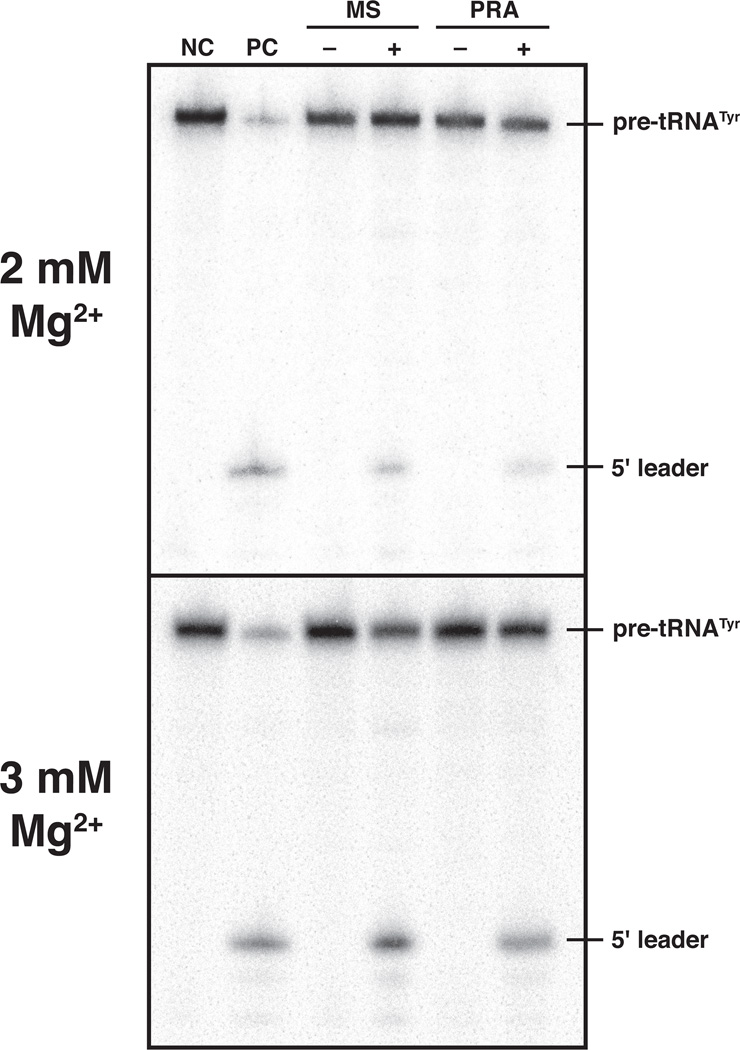
We next investigated whether the 4-RPP Pfu RNase P complex is functional under the conditions employed for native MS. Because the 2 and 3 mM Mg(OAc)2 used in the MS studies are well below the reported optimum of 30 mM,[10d] we used single-turnover conditions (i.e., [E]>>[S]) to facilitate a qualitative comparison. Whether we assembled Pfu RPR, RPP21•RPP29 and POP5•RPP30 in 800 mM NH4OAc and 2 or 3 mM Mg(OAc)2 (Figure 4, MS lanes) or in 50 mM HEPES-KOH (pH 8), 800 mM NH4OAc and 2 or 3 mM MgCl2 (Figure 4, PRA lanes), the activity is similar and shows the expected substrate cleavage specificity; without the RPPs, the RPR is not active in 2 or 3 mM Mg2+ (data not shown). These results indicate that a functional Pfu RNase P is assembled under conditions identical to those used in the MS studies.
Figure 4.
Comparison of the pre-tRNATyr processing activity at 55°C of the 4-RPP Pfu RNase P complex reconstituted in vitro under conditions employed for native MS (MS) and those previously optimized for function in vitro (PRA). [10a, 10d] NC, a negative control with pre-tRNATyr alone; PC, a positive control with in vitro reconstituted Escherichia coli RNase P and pre-tRNATyr; – and + are parallel reactions performed without and with Pfu RNase P, respectively. See supplement for additional details.
Having determined the subunit stoichiometry of the 4-RPP Pfu RNase P complex, we next explored assembly intermediates en route to this final RNP. Although the RPR is active without its cognate RPPs in 500 mM Mg2+, its activity is significantly increased by RPP21•RPP29 or POP5•RPP30 at lower substrate and Mg2+ concentrations. That RPR+RPP21•RPP29 and RPR+POP5•RPP30 constitute minimal functional complexes is evident from their kcat/KM values being, respectively, 8- to 80-fold higher than the RPR alone and 600- to 50-fold lower than the RPR with both binary RPPs.[10d] These partial complexes exhibited highest activity in 50 mM Tris-HCl/HEPES-KOH (pH 7.5-8), 100 mM NH4OAc and 120 mM MgCl2. Here also we determined the lowest Mg2+ concentration that would permit assembly. Based on the premise that most of the 120 mM Mg2+ used in the activity assays served as counterions to aid the native RNA/RNP fold, we proportionately increased NH4OAc from 100 to 500 mM to compensate for the decreased ionic strength associated with lowering the Mg2+ concentration.
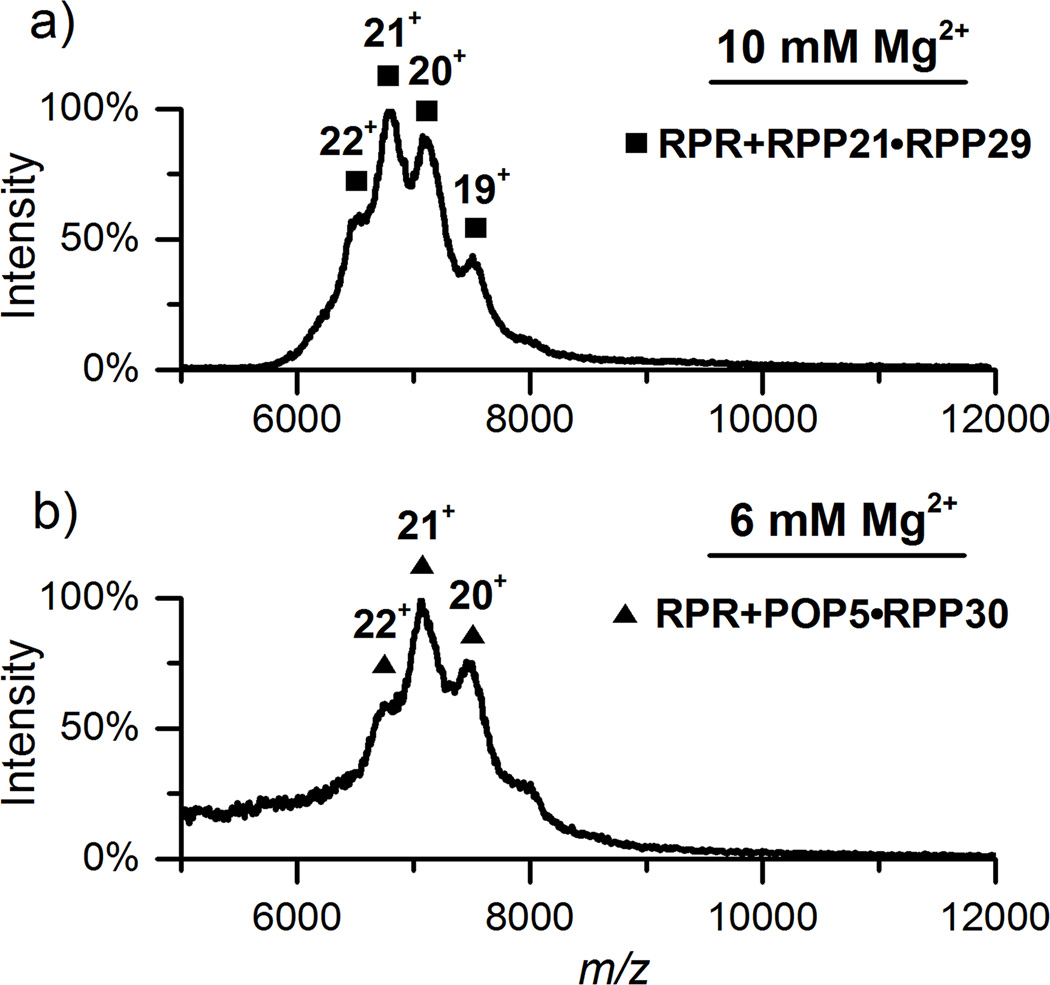
We then determined the stoichiometry of Pfu RPR+RPP21•RPP29 and RPR+POP5•RPP30 complexes in 500 mM NH4OAc with varying Mg(OAc)2 concentrations. In 10 mM Mg(OAc)2RPP21•RPP29 binds the RPR to generate a complex with a mass consistent with either RPR+RPP21•RPP29, RPR+(RPP21)2 or RPR+(RPP29)2 because the masses of RPP21 and RPP29 differ by only 0.7 kDa (Figure 5a; Table S-1). Of these, the first complex is the most likely based on the strong interaction between RPP21 and RPP29 as revealed in high-resolution NMR and crystal structures,[11a, 11d] and the activation of the RPR by RPP21•RPP29, but not by RPP21 or RPP29.[10d] We attribute the 2.7-kDa difference between our experimentally determined value and the predicted mass (Table S-1) to severe peak broadening caused by the 10 mM Mg(OAc)2 required for generating this RNP complex. Even with optimization of collision-induced cleaning voltage, exceeding a certain Mg2+ threshold clearly poses a problem for MS analysis.
Figure 5.
a) RPR and RPP21•RPP29 in 500 mM NH4OAc and 10 mM Mg(OAc)2. b) RPR and POP5•RPP30 in 500 mM NH4OAc and 6 mM Mg(OAc)2. In both a and b, the dominant peaks correspond to the RPR bound to one copy of the respective binary RPP complex. Collision-induced cleaning was used to partially remove non-specific adducts, providing better resolved peaks with optimal collision voltage at 110 V.
POP5•RPP30 binds the RPR in 6 mM Mg(OAc)2 to form RPR+POP5•RPP30 (Figure 5b), but it forms a (POP5•RPP30)2 heterotetramer under identical conditions without RPR (data not shown). The abrupt structural change of POP5•RPP30 in the presence of the RPR with or without RPP21•RPP29 motivated us to examine possible intermediates. After considerable effort, we found that when POP5•RPP30 was assembled with RPR in 500 mM NH4OAc and 4 mM Mg(OAc)2, IM could be used to extract minor signals corresponding to RPR+(POP5•RPP30)2 from background noise and other interference (Figure S-2b). However, when the Mg(OAc)2 concentration was increased from 4 to 6 mM, which is more favorable for RNase P activity, RPR+(POP5•RPP30)2 disappears and only RPR+POP5•RPP30 is observed (Figures 5b and S-2d); even with a two-fold excess of RPPs over RPR, we failed to observe RPR+(POP5•RPP30)2 at 6 mM Mg(OAc)2 (data not shown). Although present, the significance of the low-abundance, RNA-bound heterotetramer at 4 mM Mg(OAc)2 is unclear. These data suggest that tight RPR binding is concomitant with disruption of this heterotetramer (Figure S-3).
Based on functional assays, we had proposed that assembly of the Pfu RPR with RPP21•RPP29 and POP5• RPP30 entails binding of either RPP complex to the RPR followed by binding of the other.[10d] Under the conditions used for MS, RPP21•RPP29 requires at least 10 mM Mg2+, while POP5•RPP30 binds the RPR in as little as 6 mM Mg2+. These findings suggest that POP5•RPP30 may be the first to assemble with the RPR in cellular Mg2+ concentration (< 2 mM).[19]
Although the stoichiometry and oligomeric state of the archaeal/eukaryal RNase P have not been previously determined experimentally, the expectation that RPPs might be present in multiple copies, in contrast to our finding of single copies, was based on at least three lines of evidence. First, in the absence of RPR, crystallographic and NMR studies revealed a (POP5•RPP30)2 heterotetramer[11b, 11c, 13] (now confirmed in the gas phase; Figure 1). Although data were not shown, deletion of a loop in Pho POP5 renders it defective in homodimerization and abolishes Pho RNase P activity;[11b] because this mutant could only form POP5•RPP30 and not (POP5• RPP30)2, tetramer formation was inferred as essential for activity. However, a defect in this mutant’s ability to bind RPR could also impair its activity, an untested premise. Second, an EM study on yeast RNase P and MRP (a sister RNP enzyme to RNase P that requires most of the RPPs for function) suggests the presence of two copies of RPP30, one bound to POP5 and another to RPP14 (a POP5 homolog).[20] Finally, staining of native preparations of yeast RNase P and MRP suggests greater amounts of some RPPs than others,[21] though the bias of gel stains for some protein compositions imparts uncertainty to this argument. In contrast, the mass determined by native MS, which is free of the above caveats, provides evidence for only one RPR and one of each RPP in both 4- and 2-RPP Pfu RNase P complexes, the most definitive stoichiometry of an active RNase P to date.
Taken together, our study illustrates the value of collision-induced cleaning and native tandem SID coupled with IM-MS for improved peak resolution, confirmation of subunit composition, and parsing conformational heterogeneity of a Mg2+-containing RNA-protein complex. It is a valuable structural biology tool that can provide insights into the structure and dynamics of large RNA-protein complexes, which might be refractory to other high-resolution methods.
Supplementary Material
Footnotes
Funding support from the NSF (MCB-0843543 and DBI-0923551) and NIH (RO1 GM067807) is gratefully acknowledged.
References
- 1.a) Hernández H, Robinson CV. Nat. Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]; b) Benesch JLP, Ruotolo BT, Simmons DA, Robinson CV. Chem. Rev. 2007;107:3544–3567. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]; c) Kaddis CS, Lomeli SH, Yin S, Berhane B, Apostol MI, Kickhoefer VA, Rome LH, Loo JA. J. Am. Soc. Mass Spectrom. 2007;18:1206–1216. doi: 10.1016/j.jasms.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kaddis CS, Loo JA. Anal. Chem. 2007;79:1778–1784. doi: 10.1021/ac071878c. [DOI] [PubMed] [Google Scholar]; e) Heck AJR. Nat. Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]; f) Heck AJR, van den Heuvel RHH. Mass Spectrom. Rev. 2004;23:368–389. doi: 10.1002/mas.10081. [DOI] [PubMed] [Google Scholar]; g) Pace CN, Treviño S, Prabhakaran E, Scholtz JM. Philos. Trans. R. Soc. B. 2004;359:1225–1235. doi: 10.1098/rstb.2004.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Ruotolo BT, Benesch JLP, Sandercock AM, Hyung S-J, Robinson CV. Nat. Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]; i) Uetrecht C, Watts NR, Stahl SJ, Wingfield PT, Steven AC, Heck AJR. Phys. Chem. Chem. Phys. 2010;12:13368–13371. doi: 10.1039/c0cp00692k. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Shoemaker GK, van Duijn E, Crawford SE, Uetrecht C, Baclayon M, Roos WH, Wuite GJL, Estes MK, Prasad BVV, Heck AJR. Mol. Cell. Proteomics. 2010;9:1742–1751. doi: 10.1074/mcp.M900620-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]; k) Uetrecht C, Barbu IM, Shoemaker GK, van Duijn E, Heck AJR. Nat. Chem. 2011;3:126–132. doi: 10.1038/nchem.947. [DOI] [PubMed] [Google Scholar]; l) Uetrecht C, Heck AJR. Angew. Chem. Intl. Ed. Engl. 2011;50:8248–8262. doi: 10.1002/anie.201008120. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2011;123:8398–8413. [Google Scholar]
- 2.a) Callaghan AJ, Grossmann JG, Redko YU, Ilag LL, Moncrieffe MC, Symmons MF, Robinson CV, McDowall KJ, Luisi BF. Biochemistry. 2003;42:13848–13855. doi: 10.1021/bi0351099. [DOI] [PubMed] [Google Scholar]; b) Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Science. 2005;310:1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]; c) Akashi S, Watanabe M, Heddle JG, Unzai S, Park S-Y, Tame JRH. Anal. Chem. 2009;81:2218–2226. doi: 10.1021/ac802354j. [DOI] [PubMed] [Google Scholar]
- 3.a) King R, Bonfiglio R, Fernandez-Metzler C, Miller-Stein C, Olah T. J. Am. Soc. Mass Spectrom. 2000;11:942–950. doi: 10.1016/S1044-0305(00)00163-X. [DOI] [PubMed] [Google Scholar]; b) Annesley TM. Clin. Chem. 2003;49:1041–1044. doi: 10.1373/49.7.1041. [DOI] [PubMed] [Google Scholar]
- 4.a) Crain PF, McCloskey JA. Curr. Opin. Biotechnol. 1998;9:25–34. doi: 10.1016/s0958-1669(98)80080-3. [DOI] [PubMed] [Google Scholar]; b) Pan J, Xu K, Yang X, Choy W-Y, Konermann L. Anal. Chem. 2009;81:5008–5015. doi: 10.1021/ac900423x. [DOI] [PubMed] [Google Scholar]
- 5.Verkerk UH, Kebarle P. J. Am. Soc. Mass Spectrom. 2005;16:1325–1341. doi: 10.1016/j.jasms.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 6.a) Hall Z, Robinson CV. J. Am. Soc. Mass Spectrom. 2012;23:1161–1168. doi: 10.1007/s13361-012-0393-z. [DOI] [PubMed] [Google Scholar]; b) Jurneczko E, Barran PE. Analyst. 2011;136:20–28. doi: 10.1039/c0an00373e. [DOI] [PubMed] [Google Scholar]; c) Borsdorf H, Mayer T, Zarejousheghani M, Eiceman GA. Appl. Spectrosc. Rev. 2011;46:472–521. [Google Scholar]
- 7.Lai LB, Cho I-M, Chen W-Y, Gopalan V. In: Ribonuclease P. Liu F, Altman S, editors. New York, NY: Springer; 2010. pp. 153–172. [Google Scholar]
- 8.Rossmanith W. Biochim. Biophys. Acta Gene Regul. Mech. 2012;1819:1017–1026. doi: 10.1016/j.bbagrm.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Lai LB, Vioque A, Kirsebom LA, Gopalan V. FEBS Lett. 2010;584:287–296. doi: 10.1016/j.febslet.2009.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Walker SC, Engelke DR. Crit. Rev. Biochem. Mol. Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Chen W-Y, Pulukkunat DK, Cho I-M, Tsai H-Y, Gopalan V. Nucleic Acids Res. 2010;38:8316–8327. doi: 10.1093/nar/gkq668. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cho I-M, Lai LB, Susanti D, Mukhopadhyay B, Gopalan V. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14573–14578. doi: 10.1073/pnas.1005556107. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kouzuma Y, Mizoguchi M, Takagi H, Fukuhara H, Tsukamoto M, Numata T, Kimura M. Biochem. Biophys. Res. Commun. 2003;306:666–673. doi: 10.1016/s0006-291x(03)01034-9. [DOI] [PubMed] [Google Scholar]; d) Tsai H-Y, Pulukkunat DK, Woznick WK, Gopalan V. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16147–16152. doi: 10.1073/pnas.0608000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Honda T, Kakuta Y, Kimura K, Saho J, Kimura M. J. Mol. Biol. 2008;384:652–662. doi: 10.1016/j.jmb.2008.09.056. [DOI] [PubMed] [Google Scholar]; b) Kawano S, Nakashima T, Kakuta Y, Tanaka I, Kimura M. J. Mol. Biol. 2006;357:583–591. doi: 10.1016/j.jmb.2005.12.086. [DOI] [PubMed] [Google Scholar]; c) Wilson RC, Bohlen CJ, Foster MP, Bell CE. Proc. Natl. Acad. Sci. U.S.A. 2006;103:873–878. doi: 10.1073/pnas.0508004103. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Xu Y, Amero CD, Pulukkunat DK, Gopalan V, Foster MP. J. Mol. Biol. 2009;393:1043–1055. doi: 10.1016/j.jmb.2009.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Hall TA, Brown JW. Archaea. 2004;1:247–254. doi: 10.1155/2004/743956. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kifusa M, Fukuhara H, Hayashi T, Kimura M. Biosci. Biotechnol. Biochem. 2005;69:1209–1212. doi: 10.1271/bbb.69.1209. [DOI] [PubMed] [Google Scholar]
- 13.Crowe BL, Bohlen CJ, Wilson RC, Gopalan V, Foster MP. Archaea. 2011;2011 doi: 10.1155/2011/891531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Hochleitner EO, Kastner B, Fröhlich T, Schmidt A, Lührmann R, Arnold G, Lottspeich F. J. Biol. Chem. 2005;280:2536–2542. doi: 10.1074/jbc.M409587200. [DOI] [PubMed] [Google Scholar]; b) Agafonov DE, Deckert J, Wolf E, Odenwälder P, Bessonov S, Will CL, Urlaub H, Lührmann R. Mol. Cell. Biol. 2011;31:2667–2682. doi: 10.1128/MCB.05266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Schmidt C, Grønborg M, Deckert J, Bessonov S, Conrad T, Lührmann R, Urlaub H. RNA. 2014;20:406–420. doi: 10.1261/rna.041244.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borsdorf H, Eiceman GA. Appl. Spectrosc. Rev. 2006;41:323–375. [Google Scholar]
- 16.a) Morgner N, Montenegro F, Barrera NP, Robinson CV. J. Mol. Biol. 2012;423:1–13. doi: 10.1016/j.jmb.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Borysik AJ, Robinson CV. Phys. Chem. Chem. Phys. 2012;14:14439–14449. doi: 10.1039/c2cp41687e. [DOI] [PubMed] [Google Scholar]; c) Benesch JLP. J. Am. Soc. Mass Spectrom. 2009;20:341–348. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]; d) McKay AR, Ruotolo BT, Ilag LL, Robinson CV. J. Am. Chem. Soc. 2006;128:11433–11442. doi: 10.1021/ja061468q. [DOI] [PubMed] [Google Scholar]
- 17.a) Han L, Hyung S-J, Ruotolo BT. Angew. Chem. Intl. Ed. Engl. 2012;51:5692–5695. doi: 10.1002/anie.201109127. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2012;124:5790–5793. [Google Scholar]; b) Han L, Hyung S-J, Ruotolo BT. Faraday Discuss. 2013;160:371–388. doi: 10.1039/c2fd20099f. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Han L, Ruotolo BT. Angew. Chem. Intl. Ed. Engl. 2013;52:8329–8332. doi: 10.1002/anie.201301893. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2013;125:8487–8490. [Google Scholar]; d) Liu J, Konermann L. J. Am. Soc. Mass Spectrom. 2014;25:595–603. doi: 10.1007/s13361-013-0814-7. [DOI] [PubMed] [Google Scholar]
- 18.a) Fernandez FM, Wysocki VH, Futrell JH, Laskin J. J. Am. Soc. Mass Spectrom. 2006;17:700–709. doi: 10.1016/j.jasms.2006.01.012. [DOI] [PubMed] [Google Scholar]; b) Wysocki VH, Joyce KE, Jones CM, Beardsley RL. J. Am. Soc. Mass Spectrom. 2008;19:190–208. doi: 10.1016/j.jasms.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wysocki VH, Jones CM, Galhena AS, Blackwell AE. J. Am. Soc. Mass Spectrom. 2008;19:903–913. doi: 10.1016/j.jasms.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Beardsley RL, Jones CM, Galhena AS, Wysocki VH. Anal. Chem. 2009;81:1347–1356. doi: 10.1021/ac801883k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowan JA. In: The Biological Chemistry of Magnesium. Cowan JA, editor. New York, New York: VCH Publishers, Inc.; 1995. pp. 1–23. [Google Scholar]
- 20.Hipp K, Galani K, Batisse C, Prinz S, Bottcher B. Nucleic Acids Res. 2012;40:3275–3288. doi: 10.1093/nar/gkr1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salinas K, Wierzbicki S, Zhou L, Schmitt ME. J. Biol. Chem. 2005;280:11352–11360. doi: 10.1074/jbc.M409568200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.