Abstract
IMP dehydrogenase (IMPDH) and GMP reductase (GMPR) belong to the same structural family, share a common set of catalytic residues and bind the same ligands. The structural and mechanistic features that determine reaction outcome in the IMPDH/GMPR family have not been identified. Here, we show that the GMPR reaction utilizes the same intermediate E-XMP* as IMPDH, but this intermediate reacts with ammonia instead of water. A single crystal structure of human GMPR type 2 with IMP and NADPH fortuitously captures three different states, each of which mimic a distinct step in the catalytic cycle of GMPR. The cofactor is found in two conformations, an "in" conformation poised for hydride transfer, and an "out" conformation where the cofactor is 6 Å from IMP. Mutagenesis, substrate/cofactor analog experiments demonstrate that the “out” conformation is required for the deamination of GMP. Remarkably, the cofactor is part of the catalytic machinery activating ammonia.
Understanding the chemical and structural basis of enzyme catalysis remains a central challenge in biochemistry. Although enzyme function is often presumed from protein sequence 1, subtle differences in sequence, and hence in structure, can profoundly change reaction outcomes 2, 3. The inability to reliably predict the consequences of mutations also limits enzyme redesign. A deeper understanding of how enzymes with similar structures and catalytic machinery promote different chemical transformations is required.
This predicament is vividly illustrated with (β/α)8 barrel proteins, also known as TIM barrels after the founding structure of triose phosphate isomerase. This fold is the most common among enzyme structures 4–6. The current SCOP and CATH databases list 33 and 31 homologous superfamilies with this topology, respectively 7, 8. The (β/α)8 barrel is also the most versatile protein fold, catalyzing isomerization, condensation, phosphotransfer, hydride transfer and over 20 other reactions 9. The remarkable ability of the (β/α)8 barrel to morph into catalysts with both novel substrate and reaction specificities creates a particularly difficult problem for the determination of function from sequence and structure.
Gene duplication is believed to propel the diversification of (β/α)8 barrel functions by allowing one gene copy to diverge constraint-free while the other copy maintains essential functions (2, 10–13). Several models for the evolution of new activities have been proposed, differing in the timing of the gene duplication, the emergence of new function and the application of selective pressure, and circumstances where each model can operate are easily envisioned. The "escape from adaptive conflict" model is particularly attractive for the evolution of new enzyme activities. In this model, ancestral enzymes were promiscuous generalists capable of catalyzing several different reactions. Duplication relieves the constraint of maintaining the entire reaction repertoire and allows the emergence of specialist enzymes that catalyze smaller sets of reactions more efficiently 14.
The (β/α)8 barrel proteins IMPDH and GMPR present a challenge for the "generalist to specialist" paradigm of enzyme evolution 2. These enzymes have ~30% sequence identity, bind the same ligands with similar affinities and catalyze similar reactions, but with opposing metabolic consequences (Figure 1A; 15). IMPDH catalyzes the conversion of IMP into XMP with the concomitant reduction of NAD+; this is the first committed step in guanine nucleotide biosynthesis. GMPR catalyzes the reduction of GMP to IMP and ammonia with the concomitant oxidation of NADPH. IMPDH is responsible for the expansion of the guanine nucleotide pool required for proliferation 16. In contrast, the guanine nucleotide pool shrinks during differentiation as a result of the expression of GMPR. Thus promiscuous activities would appear to be problematic for these two enzymes – a GMPR activity would be deleterious in IMPDH, and likewise an IMPDH reaction would be unwelcome in GMPR.
Figure 1.
The reactions catalyzed by IMPDH and GMPR. (a) Purine nucleotide interconversions. (b) The mechanism of the IMPDH reaction.
The fact that these promiscuous activities are not prominent features of both enzyme reactions is even more remarkable from a mechanistic perspective. Both reactions are believed to utilize the same thioimidate intermediate (E-XMP*), but this intermediate partitions very differently on the two enzymes. In IMPDH, E-XMP* is formed by the oxidation of IMP and is hydrolyzed to XMP (Figure 1B). This reaction is well characterized 15. The catalytic Cys310, assisted by the Thr312/Glu421 dyad, attacks C2 of IMP and hydride is rapidly transferred to NAD+ (Streptococcus pyogenes IMPDH numbering). Structures of ternary complexes mimicking E•IMP•NAD+ find the cofactor and nucleotide optimally aligned for this reaction, with C4 of the nicotinamide stacked against C2 of the purine 17–20. NADH departs and a mobile flap moves into the vacant dinucleotide site, carrying Arg406/Tyr407 into the active site 21. Arg406 acts as the base to activate water, assisted by Thr312 and Glu421 22–24. Thus IMPDH cleverly solves the problem of how to catalyze two different chemical transformations by rearranging one side of the active site. As expected, Cys310, Thr312 and Arg406/Tyr407 are completely conserved among IMPDHs. However, Glu421 is often substituted with Gln, with little consequence to the overall reaction 20, 24.
Surprisingly little is known about the mechanism of GMPR; even the presence of E-XMP* has yet to be demonstrated. Most of the key catalytic residues of IMPDH are also found in GMPR; the Cys, Thr and Glu are strictly conserved (Cys186, Thr188 and Glu289 in Escherichia coli GMPR numbering). Mutation of any one of these residues compromises enzymatic function in vivo 24, 25. Presumably these residues have similar functions in GMPR as in IMPDH, i.e., Cys186 reacts to form E-XMP* and Thr188/Glu289 are involved in the activation of the leaving group instead of water. However, no counterpart exists for the Arg/Tyr dyad of IMPDH, so some of the catalytic machinery remains to be identified in GMPR. Curiously, steady-state kinetic investigations indicate that the ternary E•GMP•NADPH complex forms before reaction proceeds 26–28. Thus the deamination reaction must occur in the presence of NADPH. This observation creates a mechanistic conundrum: how is the leaving group activated if NADPH is stacked against GMP?
Here we show that the GMPR reaction involves the same E-XMP* intermediate as IMPDH. However, where the IMPDH reaction uses two different protein conformations to catalyze the two different chemical transformations, GMPR uses two different cofactor conformations. Where E-XMP* reacts with water in IMPDH, it reacts exclusively with ammonia in GMPR. Contrary to previous reports, we find that GMPR can catalyze the synthesis of GMP from IMP, ammonia and NADP+ with sufficient efficiency to support bacterial proliferation. These findings provide new insight into the determinants of reaction specificity in the IMPDH/GMPR family and suggest that the metabolic role of GMPR should be revised to acknowledge its role in GMP synthesis.
RESULTS
GMPR and IMPDH are not promiscuous
The structural and presumed mechanistic similarity of GMPR and IMPDH, as well as the close resemblance of their substrates, suggested that cofactor specificity/availability might determine their respective metabolic roles. Cellular concentrations of NADPH exceed those of NADP+, so that anabolic reactions such as GMPR usually utilize this cofactor. Conversely, NAD+ is more abundant than NADH, and therefore the cofactor of choice for catabolic reactions. Therefore we tested the ability of GMPR to catalyze the conversion of IMP to XMP in the presence of NADP+. No reaction was detected with E. coli GMPR (EcGMPR; less than 0.01% of the activity of IMPDH). Similarly, no reaction could be detected when GMP and NADH were incubated with IMPDH (less than 0.1% of the activity of EcGMPR). Therefore, cofactor selectivity cannot account for the metabolic roles of GMPR and IMPDH. Thus, despite their close structural similarity, these two enzymes are very specialized.
Ligand affinity was investigated to further assess the similarity of the GMPR and IMPDH active sites (Supplementary Results, Supplementary Table 1 compares our values to a compilation of literature values). The value of Km for GMP was 3.2 ± 0.5 µM in EcGMPR, which was similar to the affinity of GMP for IMPDH. IMP and XMP were competitive inhibitors of EcGMPR with values of Ki = 49 ± 14 µM and 28 ± 5 µM, respectively. These affinities were also comparable to those observed with IMPDH. Thus the affinity of substrates and products does not determine the reaction specificity of GMPR and IMPDH.
Two nucleoside drugs, ribavirin and mizoribine, target IMPDH via their monophosphate forms. The x-ray crystal structures of the ribavirin monophosphate and mizoribine monophosphate complexes mimic the features of E-XMP*, and the the mizoribine monophosphate complex mimicks the transition state for the hydrolysis reaction 15. These observations suggest that the affinity of these ligands would provide some insights into the stability and reactivity of E-XMP*. Ribavirin monophosphate was a relatively weak inhibitor of EcGMPR (Ki = 98 ± 25 µM) compared to IMPDHs (Ki = 0.1–0.6 µM 15). In contrast, mizoribine monophosphate was a potent inhibitor of both GMPR and IMPDH (Ki = 0.015 ± 0.003 µM for EcGMPR versus 0.0005–0.008 µM for IMPDHs 15). This observation suggests that GMPR is a previously unrecognized target of the immunosuppressive drug mizoribine. Taken together, these observations suggested that GMPR and IMPDH have very similar interactions with substrates and products.
Mechanism of the GMPR reaction
The reaction of EcGMPR was characterized in detail in order to investigate the reaction specificity of the IMPDH/GMPR family. As expected, an intersecting line pattern was observed in the Lineweaver-Burk plot (Supplementary Figure 1), indicating that no reaction occurs until both GMP and cofactor are bound to the enzyme (Km = 10.1 ± 0.8 µM for NADPH; kcat values are shown in Table 1). NMR analysis revealed that deuterium was transferred from 4S-[2H]-NADPH, but not from 4R-[2H]-NADPH (Supplementary Figure 2). The hydride is also transferred to the 4S position of the cofactor during the IMPDH reaction 15. Surprisingly, unlike IMPDH, a Vm isotope effect of 2.2 ± 0.1 was observed when 4S-[2H]-NADPH is the cofactor, indicating that the hydride transfer reaction was at least partially rate-limiting in GMPR (similar steady state kinetics and isotope effect experiments were reported while this work was under review 28). These observations demonstrated that the substrate and cofactor are aligned similarly on both GMPR and IMPDH, but the kinetics of the hydride transfer reaction are very different.
Table 1.
Values of kcat for EcGMPR reactions.
| Parameter | kcat (s−1) | |||
|---|---|---|---|---|
| wild-type | C186A | T188A | E289Q | |
| GMP + NADPH | 0.35 ± 0.01 | ≤ 0.0001 b | (8.8 ± 0.6)×10−4 b | (3.8 ± 0.3)×10−4 b,c |
| 2-Cl-IMP + NADPH | 0.40 ± 0.01 | ≤ 0.0001 d | 0.021 ± 0.001 d | 0.027 ± 0.001 d |
| GMP + APADPH | < 0.0008 | n.d. | n.d. | n.d. |
| 2-Cl-IMP + APADPH | 0.08 ± 0.01 | n.d. | n.d. | n.d. |
Reactions were performed in 75 mM Tris, pH 7.8, 100 mM KCl, 1 mM DTT, and 1 mM EDTA at 25°C. NADPH consumption was monitored as described in Methods. Values are the average and standard error of at least two independent experiments. n.d., no data.
These reactions were performed at saturating concentrations of GMP (1 mM) and NADPH (1 mM).
A complete turnover was not observed.
These assays were conducted at saturating concentrations of 2-Cl-IMP (0.8 mM) and NADPH (1 mM).
Detection of E-XMP*
Although the GMPR reaction is presumed to proceed via E-XMP*, this mechanism had not been experimentally verified. Since hydride transfer was slow, E-XMP* should accumulate during the steady-state phase of the reaction. When the reaction was performed with 8-[14C]-GMP and NADPH, radioactivity co-precipitated with enzyme (Supplementary Figure 3). No radioactivity was associated with the enzyme at the completion of the reaction, as expected if a transient intermediate was formed. Approximately 15–20% of the enzyme accumulated as E-XMP* during the steady state. Similar amounts of radioactivity were also trapped in the presence of NADP+. Mass spectrometry revealed that a new peak was formed in the presence of NADP+ that was 347 Da heavier than the apoenzyme, as expected for E-XMP* (Figure 2). Also as expected, no E-XMP* formed when GMPR was incubated with GMP in the absence of NADPH and NADP+ (Figure 2). Therefore the deamination reaction only occurs in the presence of cofactor, although the redox state of the cofactor was not important.
Figure 2.
Formation of E-XMP* during the GMPR reaction. ESI mass spectra of EcGMPR incubated with GMP alone (bottom trace) and with GMP and NADP+ (top trace). Black asterisks (*) indicate EcGMPR that has likely undergone oxidation. EcGMPR observed: 37665 Da (theoretical: 37665 Da). E-XMP* observed: 38012 Da (theoretical: 38010 Da).
The formation E-XMP* from GMP is expected to be energetically uphill, which can account for the small fraction of E-XMP* trapped in the above experiments. Therefore we characterized the reaction of EcGMPR with 2-Cl-IMP, reasoning that the better leaving group would lead to a greater accumulation of E-XMP*. 2-Cl-IMP was a good substrate for GMPR, with comparable values of Km and kcat to GMP (Table 1; Km = 7.8 ± 1.1 µM). The similarity of the values of kcat for the 2-Cl-IMP and GMP reactions was expected given that hydride transfer was rate-limiting in the GMP reaction. When EcGMPR was incubated with 2-Cl-IMP and NADP+, the major peak in mass spectrum shifted from 37,663 to 38,013 Da, indicating the formation of E-XMP* (Supplementary Figure 4). No unmodified EcGMPR was seen in this spectrum, suggesting that nearly complete conversion to E-XMP* had occurred. The mutation of the catalytic Cys186 to Ala inactivated EcGMPR (Table 1; 25), and no mass shift was observed when 2-Cl-IMP and NADP+ were incubated with the mutant enzyme (Supplementary Figure 5). These experiments demonstrated that the GMPR reaction, like the IMPDH reaction, proceeded via E-XMP*.
Structure overview
To gain insight into the structural basis of reaction specificity in GMPR, we solved the structures of human GMPR type 2 (hGMPR2) in a binary complex with IMP at 2.15 Å resolution and in an E•IMP•NADPH ternary complex at 1.7 Å resolution (Supplementary Table 2). The sequences of hGMPR2 and EcGMPR are 69% identical overall, and the active site residues are completely conserved. Both crystals contained two tetramers in the asymmetric unit (tetramer 1, subunits A-D; tetramer 2, subunits E-H). The monomers had the expected (β/α)8 barrel structure (Supplementary Figure 6). Together with a previously reported structure of the E•GMP complex of hGMPR2 25, these complexes revealed that GMPR undergoes a very different set of conformational rearrangements during the catalytic cycle than were observed in IMPDH.
The eight monomers of the E•IMP crystal had nearly identical conformations, with the exception of residues 25–28, which were disordered in monomers B, C, D and G and had varying conformations in A, E, F and H. All eight monomers contained IMP, and the residues that interacted with IMP had very similar conformations in each active site. IMP was bound in the loops after β6, β 7 and β 8, in a manner very similar to the interaction of GMP with hGMPR2. The ribose of IMP was in the 2’-endo conformation and both the 2’- and 3’- oxygens were hydrogen bonded to Asp219, while the purine ring formed hydrogen bonds to the main chain NHs of Met269, Ser270 and Glu289 as well as with the side chain of Ser270. Nearly identical interactions are found in the E•IMP complex of IMPDH (Figure 3).
Figure 3.
Structure of the IMP/GMP binding site in IMPDH and GMPR. Residues within 4 Å of mononucleotide are shown. Hydrogen bonds are depicted by cyan lines. (a) IMP bound in the active site of S. pyogenes IMPDH (PDB accession number 1ZFJ 49) (dark magenta). (b) GMP bound in the active site of hGMPR2 (2A7R 25) (sea green). (c) IMP in the active site of the binary hGMPR2 complex (steel blue). This figure was rendered with Chimera 50.
However, the E•IMP complex displayed several significant differences relative to the E•GMP complex of hGMPR2. The catalytic machinery appeared poised for the reaction: the thiol of Cys186 was only 2.5 ± 0.1 Å from C2 of the purine ring, and the Thr188/Glu289 dyad was positioned for proton transfer. In contrast, the catalytic machinery is disabled in E•GMP complex: Cys186 is 3.9 Å from C2 in the E•GMP complex and the Glu289 is pointed away from Thr188 25, which explains why the deamination of GMP did not occur in the absence of NAPDH or NADP+. Importantly, residues 279–286 were disordered in all the E•IMP monomers. This finding also contrasts with the E•GMP complex, where Arg286 extends across guanine to form a hydrogen bond with the 2'OH of GMP, providing another impediment for the deamination of GMP 25. This segment must move for a reaction to occur at the C2 position of GMP.
The structures of the ternary complexes
Remarkably, the E•IMP•NADPH crystal captured three different ligand states representing three distinct steps in the catalytic cycle of GMPR. One monomer, G, contained only IMP; this structure was essentially identical to the monomers of the E•IMP crystal. All of the other monomers contained both IMP and NADPH. In all cases, IMP was positioned as in the binary E•IMP complex (Supplementary Figure 7), and the surrounding residues also had very similar conformations, with the exception of the side chain of Ser270, which was rotated away from the purine ring in three monomers. The distance between the Cys186 thiol and C2 was compressed relative to the binary complex (2.3 ± 0.1 Å). This unusually short distance indicated that the presence of the cofactor further predisposed the enzyme to attack the purine ring.
Computational confirmation of the Cys186-IMP distance
Density functional theory (DFT) calculations were performed on a model of the active site conformation to ensure that the unusually short distance between the Cys186 thiol and C2 of IMP was not an artifact of refinement. IMP, Cys186, NADP+ and the Met269-Ser270 peptide bond were included in the calculation (Supplementary Figure 8a). When Cys186 was deprotonated, the calculations yielded a distance of 1.91 Å between the thiol and C2 of IMP. No interaction was observed in the presence of NADPH, in the absence of cofactor, or when Cys186 was protonated. These results could be rationalized by considering the alternative resonance form of IMP, where C2 was positive and O6 was negative (Supplementary Figure 8b). This resonance form was stabilized by the presence of NAD+ and the Cys186 thiolate as well as by the hydrogen bond between O6 and the NH of Ser270. The positive C2 interacts with the negative thiol of Cys186 to yield the short distance. Though these effects were exaggerated in the small model used in the DFT calculations, similar forces are clearly at play in the GMPR active site.
The cofactor binding site
Curiously, the cofactor binding site of GMPR was located in a different region of the (β/α)8-barrel than in IMPDH (Figure 4a). While the nicotinamide ribotide portion of NADPH had two different positions, the 2'-phosphoadenosine occupied the same place, and interacted with the same residues, in all seven monomers (Figure 4b, c). The adenine ring interacted with residues from the adjacent subunit, forming hydrogen bonds with Ser314 and Thr317. The adenosine was in the syn conformation, in contrast to most NAD(P) binding enzymes (Supplementary Table 3 contains detailed descriptions of the cofactor conformations)29. Segment 25–28, which was disordered in E•IMP, was ordered in the ternary complex; Ser26 and Arg27 interacted with the 2'-phosphate of the NADPH in the adjacent subunit. Segment 279–286 formed the remainder of the 2'-phosphate binding pocket. This observation suggests that cofactor binding activates the E•GMP complex by moving Arg286 from its position protecting GMP, exposing the purine ring for reaction.
Figure 4.
NADPH has two conformations in the E•IMP complex of hGMPR2. (a) The cofactor binding site is located in different regions in IMPDH and GMPR. The NAD analog tiazofurin adenine dinucleotide (pink) is shown bound to IMPDH (brown; 1LRT 17). NADPH (green) is shown in the "in" conformation of GMPR (blue, subunits A and D). (b) The interactions of the 2'-phosphoadenosine in GMPR. Subunit A is shown in blue, subunit D in slate blue. Residues within 4 Å are shown. Hydrogen bonds are depicted by cyan lines. (c) The "in" conformation of NADPH (subunit C). (d) The "out" conformation of NADPH (subunit E). (e) Interactions of nicotinamide ribotide in the "in" conformation. Residues within 4 Å of NADP are shown (subunit C; the 2'-phosphoadenosine is omitted). (f) Interactions of nicotinamide ribotide in the "out" conformation. Residues within 4 Å of NADP are shown (subunit E; the 2'-phosphoadenosine is omitted). This figure was rendered with Chimera 50.
NADPH was found in two conformations in the seven monomers, an "in" conformation and an "out" conformation (Figure 4c-e). NADPH was unusually compact in both cases, with only ~7–9 Å separating the C2 of the nicotinamide and C6 of the adenine (Figure 4c,d). In contrast, the cofactor typically binds in an extended conformation with 14–17 Å between the nicotinamide and adenine rings in IMPDH and other NAD/NADP-utilizing enzymes (Supplementary Table 3, 29).
Monomers B, C, and F displayed the "in" conformation, where the nicotinamide was stacked against IMP as in IMPDH (Figure 4e). The C2 of IMP and C4 of the nicotinamide were separated by 2.6–2.9 Å and were aligned for transfer of the proS hydride from NADPH. Therefore the "in" conformation displayed the catalytically relevant alignment of substrate and cofactor required for the hydride transfer reaction. If a similar complex can form with NADP+, GMPR should react with IMP to form E-XMP*.
In the "out" conformation, the nicotinamide was too far away from IMP for hydride transfer to occur (C4 of the nicotinamide was 6.6 ± 0.1 Å from C2 of IMP; Figure 4f). The proR hydrogen of NADPH was closest to the C2 of IMP, further demonstrating that this conformation was not competent for the hydride transfer reaction. A density, which we modeled as water, was found 2.9 Å from C2, on the opposite face of the hypoxanthine ring as Cys186. This position is expected to be occupied by the departing ammonia. This putative water molecule formed hydrogen bonds with Oγ of Thr188 and the amide group of the NADPH. These observations suggested that the "out" conformation of NADPH was required for the deamination reaction, and that the amide group of NADPH worked in concert with Thr188/Glu289 to activate the leaving group.
Deamination of GMP to form E-XMP*
We performed a series of mutagenesis and analog experiments with EcGMPR to probe the role of Thr188/Glu289 and the nicotinamide in the deamination reaction (Table 1). The substitution of Thr188 with Ala decreased the value of kcat for the GMP reaction by a factor of 400, but only decreased kcat for the 2-Cl-IMP reaction by a factor of 15. These observations demonstrated that Thr188 played a major role in activation of the leaving group (Table 1). Likewise, substitution of Glu289 with Gln decreased the value of kcat for GMP reaction by a factor of 900, but had more modest effects on the 2-Cl-IMP reaction, indicating that Glu289 was also required for the deamination of GMP. Thus, unlike the IMPDH reaction, the presence of Glu289 was critical for GMPR.
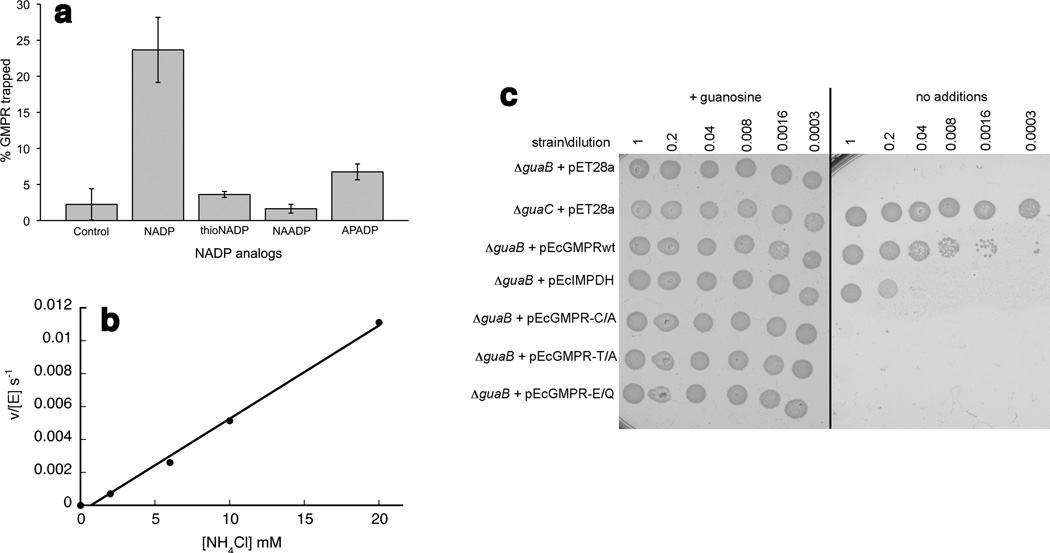
To probe the importance of the cofactor in the deamination reaction, the reaction was performed with acetylpyridine adenine dinucleotide phosphate (APADPH), where the amide group is replaced with a methylketone. No reaction with GMP and APADPH was detected with EcGMPR, in agreement with an investigation of human GMPR (Table 1; 26). Likewise, only a minor amount of E-XMP* was detected when 8-[14C]-GMP was incubated with enzyme and APADP+, and no E-XMP* was formed in the presence of thionicotinamide adenine dinucleotide phosphate or nicotinic acid adenine dinucleotide phosphate (Figure 5a). In contrast, EcGMPR catalyzed the reaction between 2-Cl-IMP and APADPH (Table 1; KM(APADPH) = 11.0 ± 2.0 µM) and kcat of 0.08 ± 0.01 s−1). These observations indicated that the amide group of the cofactor played a crucial role in the deamination of GMP.
Figure 5.
Reactions of EcGMPR. (a) Formation of E-XMP* from [14C]-GMP and EcGMPR in presence of NADP analogs. Results are the average and standard deviation of two trials. Thio-NADP, thionicotinamide adenine dinucleotide phosphate; NAADP, nicotinic acid adenine dinucleotide phosphate; APAD, acetylpyridine adenine dinucleotide phosphate. (b) EcGMPR catalyzes the reaction of IMP, NADP+ and ammonia to GMP as measured by production of NADPH. (c) Over-expression of EcGMPR can replace IMPDH/GMPS. pET28a, empty vector; pEcIMPDH, expresses wild-type EcIMPDH; pEcGMPRwt, expresses wild-type EcGMPR; pEcGMPRC/A, expresses the Cys186Ala variant; pEcGMPRT/A, expresses the Thr188Ala variant; pEcGMPRE/Q, expresses the Glu289Gln variant. BL21 ΔguaC, which contains a deletion in the gene that encodes GMPR, grows under all conditions as expected. BL21 ΔguaB contains a deletion in the gene that encodes IMPDH. This deletion also attenuates expression of the neighboring gene that encodes GMPS (Supplementary Figure 11a).
Formation of GMP from IMP and ammonia
The unusually short distance between the Cys186 thiol and IMP described above suggested that E-XMP*•NADPH might form even though EcGMPR did not catalyze the oxidation of IMP to XMP. When EcGMPR was incubated with 8-[14C]-IMP and NADP+, radioactivity co-precipitated with protein, suggesting that E-XMP* was formed on 15–20% of the active sites (Supplementary Figure 9a). This finding was corroborated by monitoring the formation of NADPH by stopped-flow spectroscopy (Supplementary Figure 9b); this experiment indicated that ~10% of the active sites formed E-XMP*•NADPH, but the reaction did not proceed further. However, more NADPH was produced when NH4Cl was added to the IMP, NADP and GMPR mixture, demonstrating that EcGMPR catalyzed the formation of GMP (Figure 5b; adjusting for the pKa of ammonia, Km(NH3) ≫ 2 mM). The production of GMP was confirmed by HPLC (Supplementary Figure 10). No GMP was observed when NH4Cl was omitted. These observations demonstrate that EcGMPR selectively reacted with ammonia over water with a preference of at least 105.
GMPR can replace IMPDH/GMPS
The above observations suggested that if sufficient ammonia was available, GMPR can catalyze the synthesis of GMP directly from IMP abrogating the requirement for IMPDH and GMPS. E. coli is typically cultured in 18 mM ammonia, which is similar to the concentration of ammonia in mammalian feces 30. In E. coli, the genes that encode IMPDH and GMPS are part of the same operon (guaBA, guaB encodes IMPDH and guaA encodes GMPS) while GMPR expression is regulated separately 31. We previously constructed a ΔguaB strain of BL21 for expression of recombinant IMPDHs 32. The expression of guaA was also attenuated in this strain as demonstrated by poor growth in the presence of xanthine (Supplementary Figure 11a). The presence of a plasmid that over-expressed IMPDH only partially restored the ability to grow on minimal medium, further demonstrating that guaA was attenuated (Supplementary Figure 11a; SDS-PAGE confirms that IMPDH was expressed, Supplementary Figure 11b). These observations indicated that native GMPR expression was not sufficient to replace IMPDH and GMPS. In contrast, over-expression of GMPR restored the ability of BL21 ΔguaB to grow on minimal media (Figure 5c; SDS-PAGE confirms that GMPR was expressed, Supplementary Figure 11b). GMPR variants containing mutations in the catalytic residues failed to support growth. These experiments demonstrated that GMPR converted IMP directly to GMP with sufficient efficiency to support life.
DISCUSSION
The IMPDH/GMPR family provides an excellent system to explore how subtle differences in protein structure change reaction outcomes. The fundamental challenge of both reactions is how to catalyze two chemical transformations at a single active site. The two enzymes use contrasting strategies to solve this problem. In IMPDH, the dehydrogenase reaction produces E-XMP*, NADH releases and a protein conformational change repurposes the cofactor binding site for the hydrolysis reaction. GMPR uses the same catalytic machinery to produce E-XMP*, but now the cofactor changes conformation in each chemical transformation (Figure 6). This cofactor mobility comes with a cost: unlike the IMPDH reaction, the dehydrogenase step is rate-limiting in the GMPR reaction. The cofactor also changes conformation during the catalytic cycle of aldehyde dehydrogenases, moving out of the way to allow hydrolysis of an acylenzyme intermediate 33. This movement allows a protein residue to activate water, much as occurs in IMPDH. However, the cofactor is an integral part of the deamination machinery in GMPR. To the best of our knowledge, there are no other examples of NAD(P) serving in a similar role in other enzymes. Perhaps the unusually compact conformation of NADPH is required for this function. More importantly, the "out" conformation is not readily accessible given the configuration of the cofactor in IMPDH. This observation suggests that the position of the ADP portion of the cofactor is a critical determinant of reaction specificity.
Figure 6.
The mechanism of GMPR.
The >105 preference for ammonia over water is the defining feature distinguishing GMPR from IMPDH. Ammonia is ~104 times more nucleophillic than water, which can account for much of this selectivity 34. Nonetheless, the concentration of water is much greater than ammonia in our experiments, so there must also be an ammonia-specific binding site. Although water and ammonia have similar sizes and dipole moments, ammonia has one less hydrogen bond acceptor and one more hydrogen bond donor than water, and is also significantly more lipophillic. These properties can be exploited to build a selective binding site, as demonstrated by ammonia-specific channels such as AmtB and RhAG 35.
Our results also redefine the physiological role of GMPR. Schemes of purine biosynthesis typically show that GMPR provides a route for recycling guanine nucleotides into the adenine nucleotide pool (Figure 1). Such a role is generally consistent with the availability (and toxicity) of ammonia in mammalian cells. As a consequence, the possibility that GMPR may function to produce GMP has been largely ignored. Contrary to previous reports, we find that GMPR can catalyze the formation of GMP with respectable efficiency, suggesting that GMP biosynthesis may be the physiological role of GMPR in ammonia-rich environments. Ammonia concentrations as high as 130 mM have been observed in the gut of insects 36. Ammonia is believed to be membrane permeable, which suggests that the intracellular concentrations will be correspondingly high, so that the synthesis of GMP via GMPR would be facile in this environment. Intriguingly, GMP biosynthesis appears to occur via GMPR in the aphid symbiont Buchnera, a close relative of E. coli. All five sequenced genomes of Buchnera contain GMPR, but are missing both IMPDH and GMPS 37. Buchnera uses waste ammonia to synthesize amino acids 38, so it is reasonable to propose that ammonia is also used to synthesize guanine nucleotides. Lastly, ~20 bacteria/archaea contain enzymes annotated as IMPDH, but appear to be missing GMPS and GMPRs (The SEED, http://theseed.uchicago.edu/FIG/index.cgi accessed June 11, 2011). Therefore we suggest that the metabolic role of GMPR should be revised to reflect its ability to synthesize GMP.
If life emerged in an ammonia-rich reductive environment as is proposed 39, then the IMPDH/GMPR ancestor may well have been a GMPR operating to produce GMP. Some evidence suggests that GMPS arose after IMPDH/GMPR, which would be consistent with this view 40. The importance of Glu289 in the GMPR reaction takes on added significance in this scenario. The analogous substitution has negligible effects on the IMPDH reaction, and Gln is found in this position in the eukaryotic branch of IMPDHs. This substitution may not represent the loss of an obsolete pathway to activate water as previously proposed 41. Instead, the Gln replacement may be a sign of IMPDH specialization and adaptation to a low ammonia environment.
METHODS
Materials
Radiolabeled IMP and GMP were purchased from Moravek Biochemicals (Brea, CA). Synthesis of 2-Cl-IMP, HPLC detection of GMP and methods for expression and purification of the proteins are detailed in the Supplementary Methods.
Crystallization
hGMPR2-IMP complex was crystallized using the hanging drop vapor diffusion method with 1µl protein solution (7.5 mg/mL or 5 mg/mL) + 1 µl reservoir solution. 7.5 mM GMP was added to the protein prior to setting the drops. The drops were equilibrated against a reservoir solution containing 11% PEG3350, 0.1 M sodium citrate pH 5.8. Crystals formed after several days. Crystals was transferred into a cryo solution containing well solution with the addition of 20% glycerol and then flash frozen in liquid nitrogen. In order to obtain the ternary complex, crystals grown in 9% PEG3350, 0.1 M sodium citrate pH 5.5 were soaked in 6 mM NADPH for 1 hour. Crystals were then transferred into a cryo solution containing well solution with the addition of 20% glycerol and 6 mM NADPH and then flash frozen in liquid nitrogen.
Data collection and processing
Synchrotron diffraction data were collected at BESSY beamline BL14.1. Wavelength for data collection, IMP complex 0.954 Å; IMP + NADPH complex 0.9537 Å. Temperature for data collections were 100 K. The data was processed and scaled using MOSFLM and SCALA (IMP complex) 42 or XDS and XSCALE (IMP+NADPH complex) 43. Crystal data and refinement statistics are shown in Supplementary Table 2.
Structure determination and refinement
The structure of the IMP complex was solved by molecular replacement using MOLREP 44 with the GMPR1 structure (PDB ID 2BLE) as template. The asymmetric unit for both complexes contained two tetramers. Iterative rounds of model building and refinement was carried out using Refmac 5 44 and Coot 45. MolProbity was used to analyze Ramachandran distributions46. Ramachandran zone distribution, favored/allowed (%); IMP complex 98/100; IMP + NADPH complex 98.5/100. Coordinates and structure factors for the IMP complex and the IMP+NADPH complex were deposited in the PDB under the accession code 2BZN and 2C6Q respectively.
Density functional theory (DFT) calculations
Geometries were optimized using the B3LYP functional in conjunction with the 6-31G(d,p) basis set, as implemented in the Gaussian03 program package 47.
Enzyme Kinetics
Standard GMPR assays were conducted at 25 °C in 75 mM Tris, pH 7.8, 100 mM KCl, 1 mM EDTA, and 1 mM DTT (GMPR assay buffer). Enzyme activity was monitored spectrophotometrically at 340 nm. Initial velocity data was fit to the Michaelis-Menten equation (eq 1) where v is the initial velocity, Vm is the maximal velocity, Km is the Michaelis constant and S is the substrate concentration.
| (1) |
Primary deuterium isotope effects were determined by varying the concentration of 4S-[2H]-NADPH at saturating GMP to determine the values of Vm and V/Km for NADPH.
Stopped-flow kinetics
A stopped-flow spectrophotometer (Applied Photophysics SX.17MV) was utilized to conduct pre-steady-state experiments. The consumption of NADPH was monitored by absorbance or fluorescence at 340 nm at 25 °C, with a 420 nm cut off filter. GMPR was incubated with saturating concentrations of NADPH and mixed with an equal volume of saturating GMP. To observe hydride transfer for the reverse reaction, 39 µM GMPR was incubated with 2 mM NADP+ and mixed with an equal volume 2 mM IMP.
Determination of the stereospecificity of hydride transfer
Purified 4R and 4S-[2H]-NADPH (see Supporting Methods) were incubated with GMP in 10 mM K2HPO4, pH 7.8. EcGMPR was added to the mixture and the reaction was stirred at room temperature. Upon completion, the reaction was lyophilized and exchanged in D2O twice. The reaction mixture was dissolved in 500 µL of D2O with 0.05% TSP and 1H NMR spectra was acquired on a 400-MHz Varian Unity Inova NMR spectrometer.
Ammonia release
Ammonia release was determined with ninhydrin as described previously 48. Standard curves of NH4Cl were linear up to 150 nmol NH4/aliquot.
Labeling GMPR with [14C]-GMP or [14C]-IMP
EcGMPR (4–8 µM) was incubated with 210 µM 8-[14C]-GMP and 1 mM NADPH in GMPR assay buffer at 25 °C, . Alternatively, 8 µM EcGMPR was incubated 400 µM 8-[14C]-IMP and 1 mM NADP+. The reactions were allowed to proceed for various times (15 s to 5 min) and quenched with addition of 33% TCA to give a final TCA concentration of 10%. Assays were incubated on ice for 30 min and then passed over 0.45 um nitrocellulose membranes to collect precipitated enzyme. Membranes were prewashed with 30 mL of 10% TCA and washed with 45 mL 10% TCA after enzyme addition. The amount of radiolabel incorporated into the protein was determined by scintillation counting on the nitrocellulose paper in 5 mL of counting cocktail. NADP+, thioNADP+, NAADP+, or APADP+ (all 1 mM) were substituted for NADPH in trapping 8-[14C]-GMP as necessary. These cofactors were incubated with enzyme for 5 min prior to the acid quench. Control reactions where 8-[14C]-GMP, NADPH, or NADP+ were omitted from the assay mixture were also analyzed.
ESI Mass Spectrometry of GMPR
Thrombin treated EcGMPR (22 µM) was incubated at room temperature for 15 minutes in 100 µL reaction with 2 mM NADP+ and 2.5 mM 2-Cl-IMP (see Supporting Methods) in GMPR assay buffer to force the formation E-XMP*. The samples were placed into 10000 MWCO Slide-A-lyzer Mini Dialysis Units (Thermo Scientific) and dialyzed with 4 changes of 500 mL of 10 mM NH4HCO3 in 22 hours. The protein was lyophilized and sent to the Mass Spectrometry Laboratory at the University of Illinois at Urbana-Champaign for analysis by ESI MS.
Assays of GMPR mutant enzymes
EcGMPR mutant enzymes (4–5 µM) were assayed in the presence of 1 mM GMP and 1 mM NADPH in quartz cuvets with a 0.2 mm path length. The maximum decrease in absorbance at these substrate concentrations were used to extract a kcat value for each mutant. Alternatively, 0.8 mM 2-Cl-IMP was used in place of GMP in these assays. Controls in which these assays lacked enzyme or GMP or 2-Cl-IMP were also performed.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Simon Andrews at the University of Sheffield for supplying the pUC plasmid carrying the E. coli K12 GMPR gene. We also are grateful for construction of pET28a GMPR by Iain S. MacPherson. The authors also thank the staff at BESSY (Berlin, Germany) for support at beamline BL14.1. This work was supported by the National Institutes of Health NIH GM54403 (L.H.) and by grants from the Swedish Research Council (PN and PS), the Swedish Cancer Society (PN), the Wenner-Gren Foundations (PS) and the Swedish Foundation for Strategic Research (P.S). The Q-Tof Ultima mass spectrometer at the University of Illinois was purchased in part with a grant from the National Science Foundation, Division of Biological Infrastructure (DBI-0100085). Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR001081). The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research and the Wellcome Trust.
Footnotes
AUTHOR CONTRIBUTIONS
GCP, DRG, CTS and LH conceived and designed the mechanism experiments. GCP, DRG and CTS performed and analyzed these experiments. PS, HS and PN conceived and designed the structural experiments. PS, HS, SF and PK performed these experiments and PS, HS, PK, PN and HE analyzed the data. RS and FH conceived, performed and analyzed DFT computations. LH, GCP, HE, PS and PN wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors have no competing financial interests.
REFERENCES
- 1.Glasner ME, Gerlt JA, Babbitt PC. Evolution of enzyme superfamilies. Curr Opin Chem Biol. 2006;10:492–497. doi: 10.1016/j.cbpa.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Soskine M, Tawfik DS. Mutational effects and the evolution of new protein functions. Nat Rev Genet. 2010;11:572–582. doi: 10.1038/nrg2808. [DOI] [PubMed] [Google Scholar]
- 3.Zalatan JG, Herschlag D. The far reaches of enzymology. Nat Chem Biol. 2009;5:516–520. doi: 10.1038/nchembio0809-516. [DOI] [PubMed] [Google Scholar]
- 4.Nagano N, Orengo CA, Thornton JM. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J Mol Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 5.Gerlt JA, Raushel FM. Evolution of function in (beta/alpha)8-barrel enzymes. Curr Opin Chem Biol. 2003;7:252–264. doi: 10.1016/s1367-5931(03)00019-x. [DOI] [PubMed] [Google Scholar]
- 6.Wise EL, Rayment I. Understanding the importance of protein structure to nature's routes for divergent evolution in TIM barrel enzymes. Acc Chem Res. 2004;37:149–158. doi: 10.1021/ar030250v. [DOI] [PubMed] [Google Scholar]
- 7.Lo Conte L, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res. 2002;30:264–267. doi: 10.1093/nar/30.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orengo CA, et al. CATH--a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 9.Anantharaman V, Aravind L, Koonin EV. Emergence of diverse biochemical activities in evolutionarily conserved structural scaffolds of proteins. Curr Opin Chem Biol. 2003;7:12–20. doi: 10.1016/s1367-5931(02)00018-2. [DOI] [PubMed] [Google Scholar]
- 10.Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- 11.Bergthorsson U, Andersson DI, Roth JR. Ohno's dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci U S A. 2007;104:17004–17009. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 13.Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 14.Khersonsky O, Tawfik DS. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 15.Hedstrom L. IMP Dehydrogenase: structure, mechanism and inhibition. Chem. Rev. 2009;109:2903–2928. doi: 10.1021/cr900021w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber G, Nakamura H, Natsumeda Y, Szekeres T, Nagai M. Regulation of GTP biosynthesis. Adv. Enzyme Regul. 1992;32:57–69. doi: 10.1016/0065-2571(92)90008-n. [DOI] [PubMed] [Google Scholar]
- 17.Gan L, Petsko GA, Hedstrom L. Crystal structure of a ternary complex of Tritrichomonas foetus inosine 5'-monophosphate dehydrogenase: NAD+ orients the active site loop for catalysis. Biochemistry. 2002;41:13309–13317. doi: 10.1021/bi0203785. [DOI] [PubMed] [Google Scholar]
- 18.Prosise GL, Luecke H. Crystal Structures of Tritrichomonas foetus Inosine Monophosphate Dehydrogenase in Complex with Substrate, Cofactor and Analogs: A Structural Basis for the Random-in Ordered-out Kinetic Mechanism. J Mol Biol. 2003;326:517–527. doi: 10.1016/s0022-2836(02)01383-9. [DOI] [PubMed] [Google Scholar]
- 19.Colby TD, Vanderveen K, Strickler MD, Markham GD, Goldstein BM. Crystal structure of human type II inosine monophosphate dehydrogenase: implications for ligand binding and drug design. Proc. Natl. Acad. Sci. USA. 1999;96:3531–3536. doi: 10.1073/pnas.96.7.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riera TV, Wang W, Josephine HR, Hedstrom L. A kinetic alignment of orthologous inosine-5'-monophosphate dehydrogenases. Biochemistry. 2008;47:8689–8696. doi: 10.1021/bi800674a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gan L, et al. The immunosuppressive agent mizoribine monophosphate forms a transition state analog complex with IMP dehydrogenase. Biochemistry. 2003;42:857–863. doi: 10.1021/bi0271401. [DOI] [PubMed] [Google Scholar]
- 22.Guillén Schlippe YV, Riera TV, Seyedsayamdost MR, Hedstrom L. Substitution of the Conserved Arg-Tyr Dyad Selectively Disrupts the Hydrolysis Phase of the IMP Dehydrogenase Reaction. Biochemistry. 2004;43:4511–4521. doi: 10.1021/bi035823q. [DOI] [PubMed] [Google Scholar]
- 23.Guillén Schlippe YV, Hedstrom L. Is Arg418 the Catalytic Base Required for the Hydrolysis Step of the IMP Dehydrogenase Reaction? Biochemistry. 2005;44:11700–11707. doi: 10.1021/bi048342v. [DOI] [PubMed] [Google Scholar]
- 24.Min D, et al. An enzymatic atavist revealed in dual pathways for water activation. PLoS Biol. 2008;6:e206. doi: 10.1371/journal.pbio.0060206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, et al. Crystal structure of human guanosine monophosphate reductase 2 (GMPR2) in complex with GMP. J Mol Biol. 2006;355:980–988. doi: 10.1016/j.jmb.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 26.Spector T, Jones TE, Miller RL. Reaction mechanism and specificity of human GMP reductase. Substrates, inhibitors, activators, and inactivators. J Biol Chem. 1979;254:2308–2315. [PubMed] [Google Scholar]
- 27.Deng Y, et al. NADPH-dependent GMP reductase isoenzyme of human (GMPR2). Expression, purification, and kinetic properties. Int J Biochem Cell Biol. 2002;34:1035–1050. doi: 10.1016/s1357-2725(02)00024-9. [DOI] [PubMed] [Google Scholar]
- 28.Martinelli LK, et al. Recombinant Escherichia coli GMP reductase: kinetic, catalytic and chemical mechanisms, and thermodynamics of enzyme-ligand binary complex formation. Molecular bioSystems. 2011;7:1289–1305. doi: 10.1039/c0mb00245c. [DOI] [PubMed] [Google Scholar]
- 29.Bellamacina CR. The nicotinamide dinucleotide binding motif: a comparison of nucleotide binding proteins. FASEB J. 1996;10:1257–1269. doi: 10.1096/fasebj.10.11.8836039. [DOI] [PubMed] [Google Scholar]
- 30.Hughes R, Magee EA, Bingham S. Protein degradation in the large intestine: relevance to colorectal cancer. Curr Issues Intest Microbiol. 2000;1:51–58. [PubMed] [Google Scholar]
- 31.Roberts RE, Lienhard CI, Gaines CG, Smith JM, Guest JR. Genetic and molecular characterization of the guaC-nadC-aroP region of Escherichia coli K-12. J Bacteriol. 1988;170:463–467. doi: 10.1128/jb.170.1.463-467.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macpherson IS, et al. The structural basis of Cryptosporidium -specific IMP dehydrogenase inhibitor selectivity. J Am Chem Soc. 2010;132:1230–1231. doi: 10.1021/ja909947a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talfournier F, Pailot A, Stines-Chaumeil C, Branlant G. Stabilization and conformational isomerization of the cofactor during the catalysis in hydrolytic ALDHs. Chem Biol Interact. 2009;178:79–83. doi: 10.1016/j.cbi.2008.10.045. [DOI] [PubMed] [Google Scholar]
- 34.Minegishi S, Mayr H. How constant are Ritchie's "constant selectivity relationships"? A general reactivity scale for n-, pi-, and sigma-nucleophiles. Journal of the American Chemical Society. 2003;125:286–295. doi: 10.1021/ja021010y. [DOI] [PubMed] [Google Scholar]
- 35.Musa-Aziz R, Chen LM, Pelletier MF, Boron WF. Relative CO2/NH3 selectivities of AQP1, AQP4, AQP5, AmtB, and RhAG. Proc Natl Acad Sci U S A. 2009;106:5406–5411. doi: 10.1073/pnas.0813231106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji R, Brune A. Nitrogen mineralization, ammonia accumulation, and emission of gaseous NH3 by soil-feeding termites. Biogeochemistry. 2006;78:267–283. [Google Scholar]
- 37.van Ham RC, et al. Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci U S A. 2003;100:581–586. doi: 10.1073/pnas.0235981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen AK, Moran NA. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci U S A. 2011;108:2849–2854. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zahnle K, Schaefer L, Fegley B. Earth's earliest atmospheres. Cold Spring Harb Perspect Biol. 2010;2:a004895. doi: 10.1101/cshperspect.a004895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim HS, Mittenthal JE, Caetano-Anolles G. MANET: tracing evolution of protein architecture in metabolic networks. BMC Bioinformatics. 2006;7:351. doi: 10.1186/1471-2105-7-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang R, et al. Differential signatures of bacterial and mammalian IMP dehydrogenase enzymes. Curr. Med. Chem. 1999;6:537–543. [PubMed] [Google Scholar]
- 42.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Jt. CCP4 + ESF-EAMCB Newsl. Protein Crystallogr. 1992;26 [Google Scholar]
- 43.Kabsch W. Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J. Appl. Crystallogr. 1988;21:916–924. [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica. 2010;D66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frisch MJ, et al. In: Gaussian 03, Revision C.02. Gaussian I, editor. Wallingford CT: 2004. [Google Scholar]
- 48.Brady K, Abeles RH. Inhibition of chymotrypsin by peptidyl trifluoromethyl ketones: determinants of slow-binding kinetics. Biochemistry. 1990;29:7608–7617. doi: 10.1021/bi00485a010. [DOI] [PubMed] [Google Scholar]
- 49.Zhang R-G, et al. Characteristics and crystal structure of bacterial inosine-5'-monophosphate dehydrogenase. Biochemistry. 1999;38:4691–4700. doi: 10.1021/bi982858v. [DOI] [PubMed] [Google Scholar]
- 50.Pettersen EF, et al. UCSF Chimera- a visualization system for exploratory research and analysis. J. Comp. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.