Abstract
Studies during the last two decades have revealed the involvement of epigenetic modifications in the development of human cancer. It is now recognized that the interplay of DNA methylation, post-translational histone modification, and non-coding RNAs can interact with genetic defects to drive tumorigenesis. The early onset, reversibility, and dynamic nature of such epigenetic modifications enable them to be developed as promising cancer biomarkers and preventive/therapeutic targets. In addition to the recent approval of several epigenetic therapies in the treatment of human cancer, emerging studies have indicated that dietary phytochemicals might exert cancer chemopreventive effects by targeting epigenetic mechanisms. In this review, we will present the current understanding of the epigenetic alterations in carcinogenesis and highlight the potential of targeting these mechanisms to treat/prevent cancer. The latest findings, published in the past three years regarding the effects of dietary phytochemicals in modulating epigenetic mechanisms will also be discussed.
Keywords: Epigenetic, chemoprevention, phytochemicals
1. Introduction
Cancer is a disease involving dynamic changes in the genome. The activation of oncogenes and the loss of function of tumor suppressor genes due to genetic mutations have long been considered the driving force of neoplasia [1]. However, the important contribution of epigenetic events to the malignant phenotype has been recognized with the help of significant advancements in the field of cancer epigenetics [2]. The definition of “epigenetic” has evolved over time from the impact of chromatin structure on embryonic development to its implication in a wide variety of biological processes [3]. Currently, the term “epigenetic” refers to the study of heritable alterations in gene expression without changes in the primary DNA sequence [4]. These heritable alterations are primarily established and maintained through cell differentiation and division, enabling the cells with the same genetic information to have distinct identities. The major epigenetic mechanisms for regulating these heritable gene alterations are the methylation of cytosine bases in DNA, covalent modifications of histones, and post-transcriptional gene regulation by microRNAs (miRNAs) [2]. The disruption of these epigenetic modifications is associated with abnormalities of various signaling pathways and can lead to the induction and maintenance of many disease states, including cancer [5].
It is now widely accepted that epigenetic abnormalities and genetic alterations in cancer cells may interact at all stages to initiate and promote cancer [6, 7]. In contrast to genetic mutations, epigenetic modifications are potentially reversible. For example, genes with repressed transcriptional activity by epigenetic silencing can be reactivated through epigenetic interventions because the genes themselves are still intact, whereas genetic mutations are permanent. This fact may explain why increasing attention and effort had been focused on the discovery and development of epigenetic-targeted therapeutics to treat cancer in recent years. To date, several small-molecule epigenetic therapies targeting chromatin-modifying enzymes have been developed and approved for cancer treatment by the U.S. Food and Drug Administration (FDA). These drugs include DNA methyltransferase (DNMT) inhibitors (azacitidine and decitabine) and histone deacetylase (HDAC) inhibitors (vorinostat and romidepsin) [8]. A number of clinical trials are also underway with these agents and many other newly developed epigenetic agents in a variety of cancer types. Moreover, the synergistic effects between epigenetic drugs and conventional antitumor therapies are quite promising [9]. Other than these small-molecule agents, accumulating evidence suggests that the epigenetic landscape is largely influenced by dietary and environmental factors [10]. With their relatively low toxicity, feasible long exposure, and promising effects observed in vitro and in vivo [11, 12], dietary phytochemicals may become potential chemopreventive agents by targeting epigenetic modifications.
In this review, we will discuss the current understanding of the epigenetic mechanisms that occur during carcinogenesis and highlight their potential roles in cancer chemoprevention. Studies published in the past three years regarding the impact of dietary chemopreventive phytochemicals in modulating epigenetic alterations will also be reviewed and discussed.
2. DNA methylation
DNA methylation, the addition of a methyl group by DNMTs to the cytosine bases located 5′ to a guanosine in a CpG dinucleotide, is perhaps the most extensively investigated epigenetic modification in mammals [13]. CpG dinucleotides are not evenly distributed across the entire genome but are clustered in short regions known as CpG islands that are 0.5–4 kb in length [13]. These CpG islands are known to be preferentially located in the proximal promoter end of approximately 60% of genes in the genome and generally remain unmethylated in normal cells [14, 15], allowing access to transcription factors and chromatin-associated proteins for active transcription. In cancer, however, CpG islands in promoter regions become hypermethylated, and this event is believed to cause inappropriate transcriptional silencing of numerous tumor suppressors and other genes with important functions in carcinogenesis (Figure 1) [16]. The recruitment of transcriptional proteins to DNA is reduced by hypermethylated CpG islands, thus resulting in gene silencing [17]. Alternatively, methylated CpG islands provide binding sites for various methyl-binding proteins (MBDs), such as MBD1-MDB4 and methyl CpG binding protein 2 (MeCP2), which can mediate gene repression by interacting with HDACs [18]. Surprisingly, promoter CpG island hypermethylation-mediated gene silencing is at least as common as mutational alterations in the classic tumor-suppressor genes in human cancer [19]. The list of cancer-related genes that are inactivated by CpG hypermethylation is ever-growing with advances in techniques. Examples of these genes include hMLH1 (human mutL homolog 1), MGMT (O6-alkylguanine DNA alkyltransferase) [20, 21], p16INK4a, p15INK4b [22, 23], Bcl-2 (B-cell lymphoma), and DAPK (death-associated protein kinase 1) [24, 25]. The studies conducted in our group demonstrated that Nrf2 [nuclear factor (erythroid-derived 2)-like 2] expression is down-regulated in TRAMP C1 cells and JB6 P+ cells due to promoter hypermethylation, and the expression of these genes can be restored by reducing the promoter methylation status with various phytochemical treatments [26–30]. This effect will be further reviewed in Section 5. Other than the hypermethylation of promoter CpG islands, global DNA hypomethylation in tumor cells compared with normal cells has been reported repeatedly (Figure 1) [31, 32]. Genome-wide hypomethylation is suggested to be associated with enhanced genomic instability and can thereby facilitate tumor progression [33]. Thus, an imbalance of DNA methylation between genome-wide hypomethylation and regional hypermethylation may characterize human neoplasia [34].
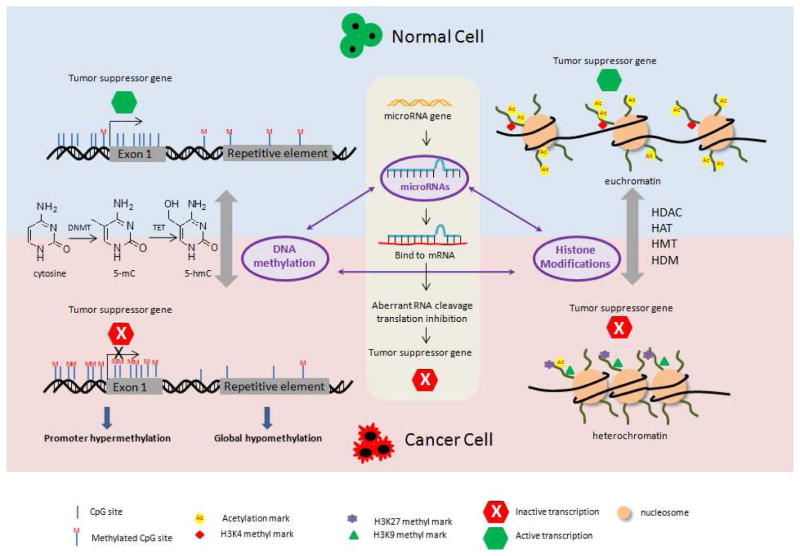
Figure 1.
Schematic representation showing epigenetic modifications of tumor suppressor genes in normal cell and in cancer cell. CpG island of promoter region remains hypomethylated to facilitate active transcription of tumor suppressor genes in normal cells. In cancer cells, however, promoter hypermethylation of tumor suppressor genes is frequently detected. In addition, genome-wide hypomethylation in cancer cells has been reported. The enzymes such as DNMT and TET dynamically regulate the DNA methylation. Acetylation and methylation on the histone tails influence the chromatin structure. For example, lysine acetylation and H3K4 methylation are associated with active transcription in euchromatin in normal cells. In cancer cells, loss of lysine acetylation and methylation at H3K9 and H3K27 leads to the repression of some tumor suppressor genes. The enzymes such as HDAC, HAT, HDM, and HMT catalyze histone acetylation and methylation. miRNAs bind and interfere with mRNAs and specifically target mRNA degradation or translation inhibition of tumor suppressor genes in cancer cells. The interplay of epigenetic pathways is shown in the center of the figure.
The precise DNA methylation patterns in the mammalian genome are known to be regulated by DNMTs (Figure 1). DNMT3a and DNMT3b act cooperatively to establish de novo methylation independent of replication, whereas DNMT1 maintains methylation patterns during DNA replication by preferentially methylating hemimethylated DNA [35]. A fourth member, DNMT-3L was first isolated in 2000 and had been shown to facilitate DNA methylation by interacting with DNMT3a and 3b [36, 37]. Given that the DNMT enzymes are modestly overexpressed in many types of tumor cells and that the inhibition of DNMTs has been found to reduce tumor formation in various mouse models [38, 39], the search for and studies of DNMT inhibitors has become extremely popular. Successful examples include FDA-approved anticancer drugs, potent DNMT inhibitors under clinical trials, and numerous dietary chemopreventive phytochemicals that have been identified in pre-clinical models. Although the enzymes that regulate DNA methylation have been well characterized, those that mediate methyl group removal are still elusive. A novel TET (ten-eleven translocation) enzyme family that is capable of modifying 5-methylcytosine to 5-hydroxymethylcytosine through oxidation has been discovered in recent years (Figure 1) [40, 41]. We anticipate many more exciting discoveries regarding the mechanistic roles of TET in the dynamic regulation of DNA methylation to enhance our understanding of DNA methylation in tumorigenesis.
3. Histone modification
The covalent modification of histone proteins also plays a critical role in regulating gene expression, chromatin structure, cellular identity, and ultimately, carcinogenesis. Histone proteins (H3, H4, H2A, H2B and H1) are at the heart of chromatin structure and act as scaffolds to wrap ~146 bp of eukaryotic DNA into repeating nucleosomes, which are further folded into compact chromatin fibers (~30 nm) [42]. The chromatin structure, which is closely involved in gene transcription, replication, and repair, is regulated by the “histone code”, known as the language of histone modification [43]. The two distinct chromatin structures, namely heterochromatin and euchromatin, represent a tightly packed structure with repressed gene transcription or a loosely packed structure with active gene transcription, respectively (Figure 1) [44]. While highly conserved, specific residues such as lysine, arginine, and serine, on the N-terminal tails of histones can undergo extensive post-translational modifications, including methylation, acetylation, phosphorylation, ubiquitination, sumoylation, and ADP ribosylation [45].
Histone modifications can lead to either gene activation or repression, depending on which residues are modified and what types of modifications are involved. Usually, lysine acetylation alters nucleosomal conformation by neutralizing the positive charge, thereby increasing the accessibility of transcriptional factors to chromatin and resulting in transcriptional activation (Figure 1) [46]. Histone acetylation is dynamically catalyzed by enzymes that add (HATs, histone acetyltransferases) and remove (HDACs, histone deacetyltransferases) acetyl groups (Figure 1). To date, 18 HDACs and 25 HATs enzymes have been identified and classified into several families, and these enzymes are capable of controlling various physiological functions [47]. The loss of acetyl groups in H4-lysine 16 and the overexpression of certain HDACs (1, 2, and 6) have been demonstrated in a number of cancers [48]. Notably, two HDAC inhibitors have already been approved by the FDA, and more novel inhibitors are currently undergoing clinical investigations for the treatment of a broad range of cancers [8]. It is exciting to note that some dietary phytochemicals may be involved in chromatin remodeling by targeting HDACs and HATs, highlighting their potential in cancer chemoprevention [49].
Unlike lysine acetylation, methylation at lysine residues appears to activate or repress transcription depending upon which residue is methylated and the degree of the methylation. For example, methylated H3K4, H3K36, and H3K79 are generally associated with active genes in euchromatin, whereas the methylation at H3K9, H3K27, and H4K20 leads to gene repression (Figure 1) [50]. Moreover, histone methylation has been suggested to cooperate with DNA methylation. For example, DNA methylation is associated with H3K9 methylation [51]. Histone methyltransferases (HMTs) and histone demethylases (HDMs) dynamically regulate histone methylation (Figure 1). In contrast to HATs, HMTs specifically target certain lysine residues; for example, EZH2 (enhancer of zeste homolog 2) is primarily responsible for H3K27 methylation [52]. Investigations in recent years have implicated hyperactive EZH2 in the development of prostate and breast cancer via its histone methylation-induced repression of tumor suppressors [53], making this enzyme a promising chemotherapeutic target.
4. microRNAs
MicroRNAs (miRNAs) are small non-coding RNAs approximately 22 nucleotides in length that are increasingly recognized as important players in epigenetic gene regulation in mammals. By specifically targeting mRNA degradation or translation inhibition, miRNAs can bind and interfere with a wide spectrum of transcripts and profoundly influence cancer-related processes, such as proliferation, apoptosis, differentiation, cell cycle, and migration (Figure 1) [54]. Since the deregulation of miRNA in cancer was first documented in 2002 [55], the network of miRNAs identified in the cancer-related processes, their tissue distributions, and their potential targets have rapidly grown, elucidating their extensive roles in carcinogenesis and chemotherapy. For example, miR-155 and miR-21 have been found to be overexpressed in many cancer types [56, 57], and the attenuated expression of miR-let7 was observed in human lung cancers [58]. Interestingly, the expression of miRNAs can be controlled by epigenetic mechanisms, such as DNA methylation and histone modifications. Moreover, miRNAs can target key enzymes, such as DNMTs and EZH2, which mediate epigenetic mechanisms, thereby modulating the epigenetic landscape of cells (Figure 1) [59]. Progress had been made in utilizing miRNAs in cancer prognosis and therapy. Notably, the first miRNA mimic entered the clinic for the treatment of liver cancer patients in 2013 [60]. In addition, the interaction between dietary phytochemicals and miRNAs has been investigated in cancer cells. Hence, miRNAs might be a promising target for chemopreventive dietary phytochemicals.
5. Dietary phytochemicals modulate epigenetic modifications
Environmental and dietary factors can influence the pathological progression of diseases, including cancer. Some naturally occurring phytochemicals that are common secondary metabolites in fruits and vegetables have been demonstrated to be beneficial for human health through various actions, including ameliorating oxidative stress, inducing detoxification enzymes, inhibiting nitrosamine formation, binding/diluting carcinogens in the digestive tract, altering hormone metabolism, and modulating carcinogenic cellular and signaling events [61]. Recently, accumulating research has demonstrated that dietary phytochemicals can alter the epigenome and may help to prevent and treat human cancer. Here we review the most recent studies regarding the epigenetic role of dietary phytochemicals, including polyphenols [quercetin, apigenin, (−)-epigallocatechin-3-gallate (EGCG), genistein, resveratrol, and curcumin], organosulfur compounds [sulforaphane (SFN), phenethyl isothiocyanate (PEITC), diallyl disulfide (DADS)], and indoles [diindolylmethane (DIM)] in cancer chemoprevention and therapy. We also discussed the latest progress in the identification of chemopreventive phytochemicals from Chinese herbal medicine in modulating epigenetic mechanisms. The epigenetic modifications regulated by phytochemicals are summarized in Table 1.
Table 1.
Epigenetic modifications by phytochemicals
| Category | Phytochemicals | Sources | Structure | Epigenetic modification(s) | Effect(s) | Citation |
|---|---|---|---|---|---|---|
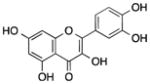
| Polyphenols | Quercetin | Citrus fruits, onion, parsley, berries |
|
Demethylated p16INK4a gene promoter, inhibited p300/HAT activity and HDACs, influenced miRNA expression (let-7, miR-146, miR-26, miR-17, miR-142-3p) | Suppressed the growth of colon cancer cells and pancereatic ductal adenocarcinoma cells, reduced COX-2 expression in breast cancer cells, induced apoptosis in human leukemia HL-60 cells, induced senescence in glioma cells | [63–68] |
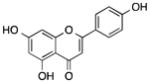
| Apigenin | Parsley, celery, chamomile tea |
|
Demethylated Nrf2 promoter, reduced the expression of DNMTs and HDACs, increased global acetylation of histone H3 and H4, induced the expression of miR-138 | Activated Nrf2 pathway in skin epidermal JB6 P+ cells, induced growth arrest and apoptosis in human prostate cancer cells and malignant neuroblastoma cells | [69–71] | |
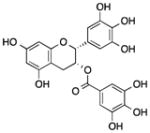
| (−)-Epigallocatech in-3-gallate (EGCG) | Tea leaves |
|
Demethylated WIF-1 promoter, inhibited the expression of HDAC1, MeCP2, and DNMT1, increased acetylation level of H3K9/14, H4K5/12/16, decreased methylation level of H3K9, decreased EZH2 localization and H3K27 trimethylation enrichment, increased histone H3K9/18 acetylation, induced the expression of miR-210, suppressed the expression of p53-targeted miRNAs (miR-25, miR-92, miR-141, miR-200) | Restored the expression of WIF-1 in lung cancer cells, reactivated ERα, PRB, TMS1, cyclin D2, and MGMT gene in MCF-7 cells, reactivated p16INK4a and Cip1/p21 in A431 cells, delayed breast cancer progression and invasion, reduced proliferation rate and anchorage-independent growth of lung cancer cells | [72, 73, 75–77, 74] | |
| Genistein | Soy beans |
|
Suppressed global DNA methylation, DNMT activity, and DNMT1 expression, induced histone modifications (H3K9-me2, H3K9-me3, and H3K27-me3), down-regulated onco-miR-1260b and miR-27a | Increased expression of ATM, APC, PTEN, and SERPINB5 in breast cancer cells, activated sFRP1, Smad4, inhibited proliferation of prostate cancer cells, enhanced apoptosis of pancreatic cancer cells | [78–80] | |
| Resveratrol | Blueberries, cranberries, Grapes |
|
Reduced DNA methylation of RASSF1A, suppressed DNMT3b, increased the expression of miR-129, -204, and -489, inhibited miR-21-mediated pathway, acted as a HDAC inhibitor | Inhibited prostate cancer growth and metastasis, promoted the apoptosis of pancreatic cancer cells | [81–85] | |
| Curcumin | Turmeric |
|
Act as DNMT inhibitor, suppressed DNA methylation in Nrf2 and Neurog 1 promoter, inhibited HDAC and HAT activity, increased global level of H3K18ac and H4K16ac, induced miRNA-9-mediated Akt/FOXO1 pathway, up-regulated miR-181b | Restored the expression of Nrf2 and Neurog 1 in TRAMP C1 and LnCap cells, restored the expression of SOCS1 and SOCS3 in K562 and HEL cells, inhibited cell proliferation of MCF-7 cells, induced apoptosis of SKOV3 cells, suppressed the expression of CXCL1 and CXCL2 in breast cancer cells | [91, 92, 86, 94, 87, 93] | |
| Organosulfur compounds | Sulforaphane (SFN) | Broccoli, cabbage, brussels sprouts |
|
Suppressed DNA methylation in Nrf2 promoter by inhibiting DNMTs and HDACs, epigenetically restored cyclin D2 expression, restored miR-140, up-regulated miR-200c | Restored Nrf2 expression, reduced breast tumor growth, inhibited proliferation of LnCap cells, inhibited EMT process in human bladder cancer T24 cells | [96, 98, 95, 30, 97] |
| Phenethyl isothiocyanate (PEITC) | Cruciferous vegetables |
|
Demethylated GSTP1 promoter, modified the acetylation and methylation of H3, increased the expression of miR-17, decreased the expression of PCAF | Reactivated GSTP1 in LnCap cells, reduced inflammation-related genes in SW480 cells, inhibited prostate cancer cell growth | [101, 100, 99] | |
| Diallyl disulfide (DADS) | Garlic |
|
Inhibited HDAC and increased the acetylation of H4, up-regulated miR-200b and miR-22 | Enhanced apoptosis in human gastric cells and xenograft models. | [103, 102] | |
| Indoles | 3, 3′-Diindolylmeth ane (DIM) | Broccoli, cabbage, brussels sprouts |
|
Altered the DNA methylation of cancer-associated gene promoters such as Nrf2, induced proteasome-mediated degradation of HDACs, influenced miR-21-mediated Cdc25A degradation, up-regulated miR-let-7, down-regulated the expression of EZH2 | Exerted chemopreventive effects in prostate tumorigenesis by up-regulating Nrf2, triggered cell cycle arrest and apoptosis in HT29 cells, inhibited cell proliferation in MCF-7 and MDA-MB-468 cells, attenuated prostate cancer aggressiveness | [107, 106, 105, 104, 29] |
| Chinese herbal medicine | Compound K | Ginseng |
|
Demethylated RUNX3 promoter | Reactivated RUNX3 and inhibited the proliferation of HT29 cells | [108] |
| Ginsenoside Rh2 | Ginseng |
|
Regulated a network of miRNAs such as miR-128 | Inhibited the proliferation of human glioma cells | [110, 109] | |
| Z-Ligustilide (Lig) | Radix angelicae sinensis (Danggui) |
|
Hypomethylated the Nrf2 promoter | Restored Nrf2 expression in TRAMPC1 cells | [28] | |
| Tanshinone IIA | Salvia miltiorrhiza, (Danshen) |

|
Reduced the methylation of Nrf2 promoter, suppressed DNMTs and HDACs, inhibited the over-expressed miR-155 | Blocked TPA-mediated JB6 transformation through restoration of Nrf2 signaling, decreased inflammatory responses in LPS-induced macrophages | [111, 112] | |
| Tanshinone I | Salvia miltiorrhiza, (Danshen) |
|
Reduced H3 acetylation levels in Aurora A promoter | Triggered cell cycle arrest in breast cancer cells | [113] | |
| Tanshindiols | Salvia miltiorrhiza, (Danshen) |
|
Potential EZH2 inhibitor | Inhibited the growth of cancer cell lines | [114] | |
| Other dietary phytochemicals | Boswellic acids | Boswellia serrata |
|
Inhibited DNMT activity, induced genome-wide demethylation, up-regulated tumor-suppressive miRNAs such as let-7 and miR-200 | Restored the expression of SAMD14 and SMPD3 in colon cancer cells, inhibited growth of colon cancer xenografts in nude mice | [116, 115] |
| Ursolic acid | Apples, berries, Thyme, rosemary |
|
Influenced miR-21 pathway | Suppressed proliferation of human glioma cell line | [117] |
5.1 Polyphenols
Quercetin, a flavonol with yellow color, is widely found in fruits, vegetables and grains. Quercetin was shown to inhibit the recombinant prokaryotic SssI DNMT- and human DNMT1-mediated DNA methylation [62]. Quercetin suppresses the growth of the human colon cancer cell line RKO via demethylation of the p16INK4a gene promoter [63]. Quercetin has been found to block the binding of the transactivators CREB2 (cAMP-response element-binding protein 2), C-Jun, C/EBPβ (CCAAT-enhancer-binding protein beta), and NF-κB to the COX-2 promoter. In addition, quercetin suppresses COX-2 expression in breast cancer cells by attenuating p300/HAT-mediated signaling [64]. Moreover, quercetin induces Fas ligand related apoptosis through the activation of the c-jun/AP-1 signaling pathway, the induction of HAT, and the inhibition of HADC in HL-60 cells [65]. Quercetin was also found to induce senescence in glioma cells via the inhibition of HDACs [66]. A quercetin-rich diet has been reported to influence miRNA expression in human lung cancer tissues, including the tumor suppressor let-7 family and carcinogenesis-related miR-146, miR-26, and miR-17 [67]. Quercetin also up-regulates miR-142-3p, a negative regulator of heat shock protein 70, which is related to the inhibition of the cell proliferation of pancreatic ductal adenocarcinoma cells (MIA PaCa-2, Capan-1, and S2-013) [68].
Apigenin is a yellow flavone compound in fruits and vegetables, especially in parsley, celery and chamomile tea. In our recent study, we found that apigenin effectively demethylated the Nrf2 promoter, resulting in an increase in the mRNA and protein expression of Nrf2 and the Nrf2 downstream target gene NQO1 (NAD[P]H:quinine oxidoreductase-1) in skin epidermal JB6 P+ cells. This effect was associated with the reduced expression of epigenetic proteins, including DNMT1, DNMT3a, DNMT3b, and some HDACs [69]. Apigenin also induces growth arrest and apoptosis in human prostate cancer cells through the up-regulation of global histone H3 and H4 acetylation and hyperacetylation of histone H3 on the p21/waf1 promoter in prostate cancer PC-3 and 22Rv1 cells. These effects may be caused by the inhibitive effect of apigenin on HDAC enzyme activity and the expression of HDAC1 and HDAC3 [70]. The tumor suppressor miR-138 is correlated with telomerase activity in many human cancers, and apigenin-induced overexpression of miR-138 has been demonstrated to powerfully induce apoptosis of human malignant neuroblastoma in cell culture and animal models [71].
(−)-Epigallocatechin-3-gallate (EGCG) is one of the most abundant catechins in tea leaves and has been identified as a non-nucleoside DNMT inhibitor. The restoration of WIF-1 (Wnt inhibitory factor 1) expression by EGCG treatment, occurring via the demethylation of the WIF-1 promoter, has been found in lung cancer H460 and A549 cell lines [72]. A recent study reported that EGCG treatment inhibits DNMT transcript levels and the protein expression of DNMT1, HDAC1, and MeCP2, effectively reactivating genes silenced by promoter methylation, such as ERα (estrogen receptor α), PRB (progesterone receptor B), TMS1 (target of methylation induced silencing-1), Cyclin D2 (G1/S-specific cyclin-D2), and MGMT in MCF-7 cells [73]. EGCG treatment was found to reactivate the tumor suppressor gene p16INK4a and Cip1/p21 by reducing DNA methylation and increasing histone acetylation in human epidermoid carcinoma A431 cells [74]. EGCG may delay breast cancer progression and invasion via the induction of matrix metalloproteinase-3 (TIMP-3) expression. The proposed mechanism for this effect is that EGCG decreases EZH2 localization and H3K27 trimethylation enrichment at the TIMP-3 promoter, with a concomitant increase in histone H3K9/18 acetylation, in breast cancer cells [75]. EGCG also induces the expression of miR-210, a major miRNA regulated by hypoxia-induced factor (HIF)-1α, in lung cancer cells, resulting in a reduced cell proliferation rate and anchorage-independent growth [76]. EGCG can suppress the expression of p53-targeting miRNAs, including miR-25, miR-92, miR-141, and miR-200a, which are induced by the environmental carcinogen benzo[a]pyrene (BaP) in multiple myeloma, a common and deadly cancer of blood plasma cells [77].
Genistein, an isoflavone, is a major phytoestrogen compound in soy beans (Glycine max). Genistein has been demonstrated to suppress global DNA methylation, DNMT activity, and DNMT1 expression. These effects lead to promoter hypomethylation and increased mRNA expression of multiple tumor suppressor genes, including ataxia telangiectasia mutated (ATM), adenomatous polyposis coli (APC), phosphatase and tensin homolog (PTEN), and mammary serpin peptidase inhibitor (SERPINB5), in human breast cancer MCF-7 and MDA-MB-231 cells [78]. Genistein induces the expression of two tumor suppressor genes, sFRP1 (secreted frizzled-related protein 1) and Smad4 (mothers against decapentaplegic homolog 4), via the demethylation of their promoter regions and histone modifications, such as H3K9-me2, H3K9-me3, and H3K27-me3, in prostate cancer cells [79]. Genistein also down-regulates onco-miRNA-1260b in prostate cancer cells, resulting in the up-regulation of sFRP1 and Smad4 and the inhibition of cell proliferation and invasion [79]. miR-27a down-regulation by genistein leads to enhanced apoptosis and reduced cell growth and invasion in pancreatic cancer cells [80].
Resveratrol is a stilbenoid, a type of natural polyphenol, and is found in blueberries, cranberries, and grapes. DNA methylation of the tumor suppressor gene RASSF1A (Ras association domain-containing protein 1) was reported to be reduced by resveratrol intake (twice daily for 12 weeks) in the breasts of women at high breast cancer risk [81]. Resveratrol suppressed the increase in DNMT3b expression in estradiol-induced mammary tumor tissue in female ACI rats, an effect that may increase the expression of miRNA-129, -204, and -489 [82]. The role of resveratrol as a HDAC inhibitor has also been demonstrated in glioma cells and human-derived hepatoblastoma cells [83]. Recent studies suggested that resveratrol inhibits prostate cancer growth and metastasis and promotes the apoptosis of pancreatic cancer cells by inhibiting a miRNA-21-mediated pathway [84, 85].
Curcumin, a curcuminoid, is the primary component in the most popular Indian spice, turmeric (Curcuma longa). Growing evidence shows that curcumin harbors DNA demethylation potential in various cancer cell lines and might be a DNMT inhibitor [86–89]. For example, studies conducted in our laboratory suggested that curcumin restored the expression of Nrf2 and Neurog1 (Neurogenin-1) in murine prostate cancer Tramp C1 cells and human prostate cancer LnCap cells, respectively, by suppressing DNA methylation in the promoter region [86, 87]. The hypomethylation effect of some novel synthetic curcumin analogs, such as EF31 and UBS109, has also been described to activate silenced genes, including p16, SPARC (secreted protein acidic and rich in cysteine), and E-cadherin (epithelial cadherin), in pancreatic cancer MiaPaCa-2 and PANC-1 cells [90]. Curcumin has also been reported to modulate the activities of HDAC and HAT. Curcumin restored the expression of SOCS1 and SOCS3, suppressors of cytokine signaling, via the inhibition of HDAC activity (especially HDAC8), resulting in increased histone acetylation in the SOCS1 and SOCS3 promoter regions of the myeloproliferative neoplasm cell lines K562 and HEL [91]. In breast tumor MCF-7 cells, the inhibitory effects of curcumin in the activities of HAT have also been demonstrated, with increased global levels of acetylated H3K18 and H4K16, potentially leading to the arrest of cell proliferation [92]. Curcumin may also induce apoptosis of ovarian cancer SKOV3 cells through inducing the miRNA-9-mediated Akt/FOXO1 (forkhead box protein O1) pathway [93]. The up-regulation of miRNA-181b by curcumin was found to suppress the expression of the pro-inflammatory cytokines CXCL1 (chemokine [C-X-C motif] ligand 1) and CXCL2, leading to the diminished proliferation and invasion of breast cancer cells [94].
5.2 Organosulfur compounds
Sulforaphane (SFN) is a bioactive isothiocyanate, a group of organosulfur compounds, which are abundant in cruciferous vegetables, such as broccoli, cabbage, and brussels sprouts. According to our recent studies, SFN suppresses DNA methylation of the Nrf2 promoter in mouse skin JB6 and prostate Tramp C1 cells by down-regulating DNMTs and HDACs. These effects may contribute to its preventive potentials against TPA-induced skin transformation and prostate carcinogenesis, respectively [95, 30]. SFN has also been demonstrated to exhibit anti-proliferative effects on LnCaP prostate cancer cells by epigenetically restoring the expression of cyclin D2 [96]. The restoration of miR-140 by SFN, accompanied by the reduced expression of SOX9 and ALDH1 (aldehyde dehydrogenases 1), has been reported to result in decreased breast tumor growth in vivo [97]. SFN also inhibits the epithelial-to-mesenchymal transition (EMT) process in human bladder cancer T24 cells, and the up-regulation of miRNA-200c by SFN may be one of the mechanisms underlying this effect [98].
Phenethyl isothiocyanate (PEITC), another isothiocyanate, exists in some cruciferous vegetables. PEITC has been reported to be able to demethylate and reactivate GSTP1 (pi-class glutathione S-transferase). This protein is a frequently silenced detoxifying enzyme that is highly associated with prostate carcinogenesis through its regulation of the cross-talk between DNA and chromatin in LNCaP cells [99]. PEITC was also observed to modify the acetylation and methylation of histone 3 in human colon cancer SW480 cells, leading to the down-regulation of some inflammation-related genes, such as CCL2 (chemokine ligand 2), CD40, CXCL10 (C-X-C motif chemokine 10), CSF2 (colony stimulating factor 2), IL-8 (interleukin 8), NF-kB, and TNFaip3 (tumor necrosis factor, alpha-induced protein 3) [100]. PEITC treatment significantly increased the expression of miRNA-17 and decreased the expression of PCAF (p300/CBP-associated factor) in dihydrotestosterone-stimulated LNCaP cells, which might contribute to the inhibitory effect of PEITC against AR (androgen receptor) transcriptional activity and cell growth in prostate cancer [101]
Diallyl disulfide (DADS) is one of the principal sulfur compounds in Allium vegetables, such as garlic (Allium sativum). DADS has been found to exhibit an inhibitory effect on HDAC, resulting in hyperacetylation of histone 4 in the breast cancer MCF-7 cell line [102]. In addition, DADS treatment has been demonstrated to impair proliferation and enhance apoptosis in both human gastric cell lines and xenograft models. This effect occurred through the Wnt-1 signaling pathway and was mediated by the up-regulation of miRNA-200b and miRNA-22 [103].
5.3 Indoles
3, 3′-Diindolylmethane (DIM), an indole compound, is derived from glucosinolate indole-3-carbinol (I3C) in cruciferous vegetables, including broccoli, cabbage, cauliflower, and brussels sprouts. In addition to SFN, DIM can alter the DNA methylation status of many cancer-associated gene promoters in normal PrECs as well as in the prostate cancer cell lines LnCap and PC3 [104]. Similarly, DIM exerts its chemopreventive effects in prostate tumorigenesis by epigenetically demethylating the Nrf2 promoter and up-regulating the expression of Nrf2 and its downstream gene NQO1 [29]. The proteasome-mediated degradation of class I HDACs (HDAC1, HDAC2, HDAC3, and HDAC8) induced by DIM triggers cell cycle arrest and apoptosis in human colon cancer HT-29 cells and in tumor xenografts [105]. DIM also inhibits cell proliferation in human breast cancer MCF-7 (estrogen-dependent) and MDA-MB-468 (estrogen receptor-negative, p53 mutant) cells via miRNA-21-mediated Cdc25A (cell division cycle 25 homolog A) degradation [106]. A phase II clinical study in patients prior to radical prostatectomy suggested that formulated DIM intervention could attenuate prostate cancer aggressiveness via the up-regulation of miRNA let-7 and down-regulation of EZH2 expression in tissue specimens [107].
5.4 Phytochemicals from Traditional Chinese Herbal Medicine
During the last few decades, great progress had been made in the identification of chemopreventive agents and anticancer drugs in traditional Chinese herbal medicine. Recently, the potential of the components from Chinese herbs to influence epigenetic mechanisms in cancer prevention have been recognized. Ginseng is one of the most commonly used herbs in East Asia. Compound K (20-O-β-(D-glucopyranosyl)-20(S)-protopanaxadiol), the main metabolite of ginseng saponin, was found to inhibit the proliferation of human HT29 human colon cancer cells by demethylating and reactivating RUNX 3 (runt-related transcription factor 3), which is associated with reduced DNMT1 activity [108]. Ginsenoside Rh2 is another biologically active triterpene saponin extracted from ginseng. The chemopreventive effect of Rh2 in inhibiting the proliferation of human glioma cells had been demonstrated to involve epigenetic modifications, such as the regulation of miRNAs. Specifically, the up-regulation of miR-128 by the treatment with Rh2 had been shown to trigger apoptosis-related signaling [109]. Similarly, using miRNA microarray analysis, An et al. identified a network of miRNAs regulated by treatment with Rh2 in non-small cell lung cancer A549 cells, which may contribute to the anti-proliferative effect of Rh2 [110]. A research study from our group demonstrated that the Chinese herb Radix Angelicae Sinensis (RAS; Danggui) and its bioactive component Z-Ligustilide (Lig) are able to hypomethylate the Nrf2 promoter, resulting in the restoration of Nrf2 and downstream targets such as NQO1, HO-1 (heme oxygenase 1), and UGT1A1 (UDP-glucuronosyltransferase 1 family, polypeptide A1) in murine prostate cancer TRAMP C1 cells [28]. Another Chinese herb with great promise in altering epigenetic mechanisms is Salvia miltiorrhiza, also known as Danshen. We found that tanshinone IIA, one of the main active components from Danshen, blocks TPA (12-O-tetradecanoylphorbol-13-acetate)-mediated JB6 transformation through epigenetic regulation of the Nrf2 signaling pathway [111]. Treatment with tanshinone IIA reduced the methylation of the Nrf2 promoter, elevated the expression of Nrf2 and downstream targets, suppressed the protein levels of DNMT1, DNMT3a, DNMT3b, and HDAC3, and inhibited HDAC activity [111]. Another study showed that tanshinone IIA decreases inflammatory responses in LPS-induced macrophages and inhibits the proliferation of inflammation-stimulated colon cancer cells by inhibiting the over-expressed miR-155 in macrophages [112]. Tanshinone I, another main component derived from Danshen, has been shown to trigger cell cycle arrest in several breast cancer cells by down-regulating Aurora A gene expression via the reduction of H3 acetylation levels in the Aurora A promoter [113]. In addition to tanshinones, the primary components, minor components, including tanshindiols, are currently under investigation for their potential antitumor ability by targeting epigenetic modifications. Using molecular docking and an enzyme kinetics approach, Woo et al. proposed that tanshindiols B and C are potential EZH2 inhibitors, resulting in the inhibition of the growth of several cancer cell lines [114].
5.5 Other dietary phytochemicals
In addition to above-mentioned dietary phytochemicals, various other natural compounds are currently under investigation regarding their cancer chemopreventive potential through epigenetic modifications. Boswellic acids, a pentacyclic terpenoid derived from the plant Boswellia serrata, have long been used as anti-inflammatory and cancer chemopreventive agents. Recently, Shen et al. demonstrated that boswellic acids inhibit DNMT activity and induce genome-wide demethylation, permitting the restoration of tumor suppressor genes, such as SAMD14 (sterile α motif domain containing 14) and SMPD3 (sphingomyelin phosphodiesterase 3) in colorectal cancer cells [115]. In addition to modulating DNA methylation, boswellic acids were found to significantly up-regulate tumor-suppressive miRNAs, such as let-7 and miR-200, and to modulate the expression of downstream targets in several colon cancer cells and tumor xenografts in nude mice [116]. Experimental evidence demonstrated that ursolic acid, another naturally occurring pentacyclic triterpene, suppresses proliferation and induces apoptosis in the human glioma cell line U251 by mediating the miR-21 pathway [117]. A recent study proposed that the antitumor activity of rosemary extracts with high contents of phenolic diterpene carnosic acid and carnosol might involve the up-regulation of GCNT3 (glycosyltransferase 3) and down-regulation of miR-15b in colon and pancreatic cancer cells [118].
6 Conclusions and perspectives
Great accomplishments have been made in recent years in advancing our understanding of epigenetic alterations in the development of cancer. These epigenetic abnormalities are now believed to exist in all cancer types and drive tumor progression along with genetic defects. The reversible and dynamic nature of epigenetic modifications strongly encouraged clinicians and pharmaceutical industries to develop epigenetic biomarkers and therapeutic targets in cancer diagnosis and treatment. However, the complexity of epigenetic pathways, including the interplay of the different epigenetic mechanisms in regulating gene transcription and the genetic mutations in epigenetic regulators, need to be addressed before we can fully apply our current understanding to the clinical field. For example, histone modification enzymes such as HDACs might be abnormally regulated by genetic or DNA methylation changes in cancer cells. Thus, further systematic studies may facilitate the development of epigenetic research in preventing and treating cancer.
The approval of several DNMT and HDAC inhibitors for clinical use has opened up a new avenue in cancer therapy. However, it could be reasonably argued that epigenetic interventions may be more effective in hematopoietic malignancies than solid malignancies. Factors such as the microenvironment, epigenetic landscape, drug exposure, and drug metabolism appear to be largely different in solid tumors than in hematopoietic malignances. However, more intensive studies regarding these cellular or epigenetic differences are urgently needed to successfully apply the concept of epigenetic therapy across a broader spectrum. Furthermore, adverse effects and a lack of selectivity have hindered the road towards effective epigenetic therapies. Investigations should be conducted regarding whether a selective subset or large numbers of genes will be influenced by the drugs or phytochemicals that target epigenetic modifications. Additionally, based on the crosstalk between genetic and epigenetic mechanisms, combining conventional antitumor drugs with epigenetic therapies or dietary phytochemicals that target epigenetic mechanisms might be a promising strategy for reducing toxicity and resistance.
Accumulating evidence indicates that some dietary phytochemicals can modulate epigenetic mechanisms. Here, we summarized and discussed the latest findings in the past three years. Together with numerous reports published more than three years ago, it is now clear that these natural compounds hold great promise in cancer prevention via acting on a variety of epigenetic targets. However, we should also notice that the success of epigenetic interventions elicited by phytochemicals was mostly limited in pre-clinical models. Thus, future studies should be carefully designed on the translation of these natural agents’ effects to prevent human malignancies in clinical settings. Moreover, most phytochemicals have been reported to influence a wide range of epigenetic regulators. Therefore, understanding the global patterns of epigenetic modifications that are induced by phytochemicals will help to optimize strategies to prevent and treat cancer.
In summary, aberrant epigenetic modifications, such as DNA methylation, histone modifications, and miRNA, add another layer of complexity to the development of human cancer. The identification of dietary phytochemicals that modulate epigenetic modifications offers promising benefits in the management of human cancer.
Acknowledgments
This work was supported in part by institutional funds and by R01-CA118947, R01-CA152826, from the National Cancer Institute (NCI), R01AT007065 from the National Center for Complementary and Alternative Medicines (NCCAM) and the Office of Dietary Supplements (ODS). We thank all the members in the laboratory for the discussion and preparation of this review.
Abbreviation
- ALDH1
aldehyde dehydrogenases 1
- APC
adenomatous polyposis coli
- AR
androgen receptor
- ATM
ataxia telangiectasia mutated
- BaP
benzo[a]pyrene
- Bcl-2
B-cell lymphoma 2
- C/EBPβ
CCAAT-enhancer-binding protein beta
- CCL2
chemokine ligand 2
- Cdc25A
cell division cycle 25 homolog A
- CREB2
cAMP-response element-binding protein 2
- CSF2
colony stimulating factor 2
- CXCL1
chemokine (C-X-C motif) ligand 1
- CXCL10
C-X-C motif chemokine 10
- Cyclin D2
G1/S-specific cyclin-D2
- DADS
diallyl disulfide
- DAPK
death-associated protein kinase 1
- DIM
diindolylmethane
- DNMT
DNA methyltransferase
- E-cadherin
epithelial cadherin
- EGCG
(−)-epigallocatechin-3-gallate
- EMT
epithelial-to-mesenchymal transition
- ERα
estrogen receptor α
- EZH2
enhancer of zeste homolog 2
- FOXO1
forkhead box protein O1
- GCNT3
glycosyltransferase 3
- GSTP1
pi-class glutathione S-transferase
- HATs
histone acetyltransferases
- HDAC
histone deacetylase
- HDMs
histone demethylases
- HIF-1α
hypoxia-induced factor
- hMLH1
human mutL homolog 1
- HMTs
histone methyltransferases
- HO-1
heme oxygenase 1
- IL-8
interleukin 8
- MBDs
methyl-binding proteins
- MeCP2
methyl CpG binding protein 2
- MGMT
O6-alkylguanine DNA alkyltransferase
- miRNAs
microRNAs
- Neurog1
Neurogenin-1
- NQO1, NAD[P]H
quinine oxidoreductase-1
- Nrf2
nuclear factor (erythroid-derived 2)-like 2
- PCAF
p300/CBP-associated factor
- PEITC
phenethyl isothiocyanate
- PRB
progesterone receptor B
- PTEN
phosphatase and tensin homolog
- RASSF1A
Ras association domain-containing protein 1
- RUNX 3
runt-related transcription factor 3
- SAMD14
sterile α motif domain containing 14
- SERPINB5
serpin peptidase inhibitor
- SFN
sulforaphane
- sFRP1
secreted frizzled-related protein 1
- Smad4
mothers against decapentaplegic homolog 4
- SMPD3
sphingomyelin phosphodiesterase 3
- SPARC
secreted protein acidic and rich in cysteine
- TET
ten-eleven translocation
- TIMP-3
matrix metalloproteinase-3
- TMS1
target of methylation induced silencing-1
- TNFaip3
tumor necrosis factor, alpha-induced protein 3
- TPA
12-O-tetradecanoylphorbol-13-acetate
- UGT1A1
UDP-glucuronosyltransferase 1 family, polypeptide A1
- WIF-1
Wnt inhibitory factor 1
Footnotes
Conflict of Interest
Yue Guo, Zheng-Yuan Su,and Ah-Ng Tony Kong declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
* Of importance
** Of major importance
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waddington CH. The epigenotype. 1942. International journal of epidemiology. 2012;41(1):10–3. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- 4.Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286(5439):481–6. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 5.Feinberg AP, Tycko B. The history of cancer epigenetics. Nature reviews Cancer. 2004;4(2):143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 6.Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Current opinion in genetics & development. 2012;22(1):50–5. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 7**.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer cell. 2012;22(1):9–20. doi: 10.1016/j.ccr.2012.06.008. An important overview of the crosstalk between epigenetics and genetics in cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8*.Dhanak D, Jackson P. Development and classes of epigenetic drugs for cancer. Biochemical and biophysical research communications. 2014 doi: 10.1016/j.bbrc.2014.07.006. A updated summary of the recent progress in the discovery and development of epigenetic therapeutics in the treatment of cancer. [DOI] [PubMed] [Google Scholar]
- 9.Kantarjian HM, Giles FJ, Greenberg PL, Paquette RL, Wang ES, Gabrilove JL, et al. Phase 2 study of romiplostim in patients with low- or intermediate-risk myelodysplastic syndrome receiving azacitidine therapy. Blood. 2010;116(17):3163–70. doi: 10.1182/blood-2010-03-274753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reproductive toxicology. 2007;23(3):297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Lee KW, Bode AM, Dong Z. Molecular targets of phytochemicals for cancer prevention. Nature reviews Cancer. 2011;11(3):211–8. doi: 10.1038/nrc3017. [DOI] [PubMed] [Google Scholar]
- 12.Surh YJ. Cancer chemoprevention with dietary phytochemicals. Nature reviews Cancer. 2003;3(10):768–80. doi: 10.1038/nrc1189. [DOI] [PubMed] [Google Scholar]
- 13.Bird A. DNA methylation patterns and epigenetic memory. Genes & development. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Leung FC. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics. 2004;20(7):1170–7. doi: 10.1093/bioinformatics/bth059. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature reviews Genetics. 2008;9(6):465–76. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 16.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends in genetics : TIG. 2000;16(4):168–74. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 17.Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251(4990):186–9. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- 18.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393(6683):386–9. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 19.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature reviews Genetics. 2002;3(6):415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 20.Esteller M, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Watkins DN, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer research. 2000;60(9):2368–71. [PubMed] [Google Scholar]
- 21.Simpkins SB, Bocker T, Swisher EM, Mutch DG, Gersell DJ, Kovatich AJ, et al. MLH1 promoter methylation and gene silencing is the primary cause of microsatellite instability in sporadic endometrial cancers. Human molecular genetics. 1999;8(4):661–6. doi: 10.1093/hmg/8.4.661. [DOI] [PubMed] [Google Scholar]
- 22.Hall GL, Shaw RJ, Field EA, Rogers SN, Sutton DN, Woolgar JA, et al. p16 Promoter methylation is a potential predictor of malignant transformation in oral epithelial dysplasia. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17(8):2174–9. doi: 10.1158/1055-9965.EPI-07-2867. [DOI] [PubMed] [Google Scholar]
- 23.Wemmert S, Bettscheider M, Alt S, Ketter R, Kammers K, Feiden W, et al. p15 promoter methylation - a novel prognostic marker in glioblastoma patients. International journal of oncology. 2009;34(6):1743–8. doi: 10.3892/ijo_00000305. [DOI] [PubMed] [Google Scholar]
- 24.Carvalho JR, Filipe L, Costa VL, Ribeiro FR, Martins AT, Teixeira MR, et al. Detailed analysis of expression and promoter methylation status of apoptosis-related genes in prostate cancer. Apoptosis : an international journal on programmed cell death. 2010;15(8):956–65. doi: 10.1007/s10495-010-0508-6. [DOI] [PubMed] [Google Scholar]
- 25.Narayan G, Arias-Pulido H, Koul S, Vargas H, Zhang FF, Villella J, et al. Frequent promoter methylation of CDH1, DAPK, RARB, and HIC1 genes in carcinoma of cervix uteri: its relationship to clinical outcome. Molecular cancer. 2003;2:24. doi: 10.1186/1476-4598-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu S, Khor TO, Cheung KL, Li W, Wu TY, Huang Y, et al. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PloS one. 2010;5(1):e8579. doi: 10.1371/journal.pone.0008579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y, Khor TO, Shu L, Saw CL, Wu TY, Suh N, et al. A gamma-tocopherol-rich mixture of tocopherols maintains Nrf2 expression in prostate tumors of TRAMP mice via epigenetic inhibition of CpG methylation. The Journal of nutrition. 2012;142(5):818–23. doi: 10.3945/jn.111.153114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su ZY, Khor TO, Shu L, Lee JH, Saw CL, Wu TY, et al. Epigenetic reactivation of Nrf2 in murine prostate cancer TRAMP C1 cells by natural phytochemicals Z-ligustilide and Radix angelica sinensis via promoter CpG demethylation. Chemical research in toxicology. 2013;26(3):477–85. doi: 10.1021/tx300524p. [DOI] [PubMed] [Google Scholar]
- 29.Wu TY, Khor TO, Su ZY, Saw CL, Shu L, Cheung KL, et al. Epigenetic modifications of Nrf2 by 3,3′-diindolylmethane in vitro in TRAMP C1 cell line and in vivo TRAMP prostate tumors. The AAPS journal. 2013;15(3):864–74. doi: 10.1208/s12248-013-9493-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30*.Zhang C, Su ZY, Khor TO, Shu L, Kong AN. Sulforaphane enhances Nrf2 expression in prostate cancer TRAMP C1 cells through epigenetic regulation. Biochemical pharmacology. 2013;85(9):1398–404. doi: 10.1016/j.bcp.2013.02.010. This research article proveded Experimental evidence that sulforaphane act as an epigenetic modifier in the regulation of Nrf2 expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 32.Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer research. 1988;48(5):1159–61. [PubMed] [Google Scholar]
- 33.Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer research. 2006;66(17):8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 34.Baylin SB, Makos M, Wu JJ, Yen RW, de Bustros A, Vertino P, et al. Abnormal patterns of DNA methylation in human neoplasia: potential consequences for tumor progression. Cancer cells. 1991;3(10):383–90. [PubMed] [Google Scholar]
- 35.Kim GD, Ni J, Kelesoglu N, Roberts RJ, Pradhan S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. The EMBO journal. 2002;21(15):4183–95. doi: 10.1093/emboj/cdf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. The Journal of biological chemistry. 2004;279(26):27816–23. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 37.Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65(3):293–8. doi: 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- 38.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, et al. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic acids research. 1999;27(11):2291–8. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: a novel target for prevention and therapy. Frontiers in oncology. 2014;4:80. doi: 10.3389/fonc.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–9. doi: 10.1038/nature12750. A discussion of the new discoveries and implications of DNA demethylation by TET enzymes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98(3):285–94. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 43.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 44.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 45.Fullgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene. 2011;30(31):3391–403. doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- 46.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes & development. 1998;12(5):599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 47**.Thakur VS, Deb G, Babcook MA, Gupta S. Plant phytochemicals as epigenetic modulators: role in cancer chemoprevention. The AAPS journal. 2014;16(1):151–63. doi: 10.1208/s12248-013-9548-5. A overview of the role of plant pytochemicals in targeting epigenetic alterations in carcinogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature genetics. 2005;37(4):391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 49*.Hardy TM, Tollefsbol TO. Epigenetic diet: impact on the epigenome and cancer. Epigenomics. 2011;3(4):503–18. doi: 10.2217/epi.11.71. A review of the dietary factors that could influence the epigenome in carcinogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Molecular and cellular biology. 2006;26(24):9185–95. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416(6880):556–60. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 52.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 53.Kim W, Bird GH, Neff T, Guo G, Kerenyi MA, Walensky LD, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nature chemical biology. 2013;9(10):643–50. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jansson MD, Lund AH. MicroRNA and cancer. Molecular oncology. 2012;6(6):590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(24):15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochimica et biophysica acta. 2009;1792(6):497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 57.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer research. 2004;64(11):3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 59.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(40):15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bouchie A. First microRNA mimic enters clinic. Nature biotechnology. 2013;31(7):577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- 61.Shukla S, Gupta S. Dietary agents in the chemoprevention of prostate cancer. Nutrition and cancer. 2005;53(1):18–32. doi: 10.1207/s15327914nc5301_3. [DOI] [PubMed] [Google Scholar]
- 62.Lee WJ, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Molecular pharmacology. 2005;68(4):1018–30. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- 63.Tan S, Wang C, Lu C, Zhao B, Cui Y, Shi X, et al. Quercetin is able to demethylate the p16INK4a gene promoter. Chemotherapy. 2009;55(1):6–10. doi: 10.1159/000166383. [DOI] [PubMed] [Google Scholar]
- 64.Xiao X, Shi D, Liu L, Wang J, Xie X, Kang T, et al. Quercetin suppresses cyclooxygenase-2 expression and angiogenesis through inactivation of P300 signaling. PloS one. 2011;6(8):e22934. doi: 10.1371/journal.pone.0022934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee WJ, Chen YR, Tseng TH. Quercetin induces FasL-related apoptosis, in part, through promotion of histone H3 acetylation in human leukemia HL-60 cells. Oncology reports. 2011;25(2):583–91. doi: 10.3892/or.2010.1097. [DOI] [PubMed] [Google Scholar]
- 66.Vargas JE, Filippi-Chiela EC, Suhre T, Kipper FC, Bonatto D, Lenz G. Inhibition of HDAC increases the senescence induced by natural polyphenols in glioma cells. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2014;92(4):297–304. doi: 10.1139/bcb-2014-0022. [DOI] [PubMed] [Google Scholar]
- 67.Lam TK, Shao S, Zhao Y, Marincola F, Pesatori A, Bertazzi PA, et al. Influence of quercetin-rich food intake on microRNA expression in lung cancer tissues. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21(12):2176–84. doi: 10.1158/1055-9965.EPI-12-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.MacKenzie TN, Mujumdar N, Banerjee S, Sangwan V, Sarver A, Vickers S, et al. Triptolide induces the expression of miR-142-3p: a negative regulator of heat shock protein 70 and pancreatic cancer cell proliferation. Molecular cancer therapeutics. 2013;12(7):1266–75. doi: 10.1158/1535-7163.MCT-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paredes-Gonzalez X, Fuentes F, Su ZY, Kong AN. Apigenin reactivates Nrf2 anti-oxidative stress signaling in mouse skin epidermal JB6 P + cells through epigenetics modifications. The AAPS journal. 2014;16(4):727–35. doi: 10.1208/s12248-014-9613-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pandey M, Kaur P, Shukla S, Abbas A, Fu P, Gupta S. Plant flavone apigenin inhibits HDAC and remodels chromatin to induce growth arrest and apoptosis in human prostate cancer cells: in vitro and in vivo study. Molecular carcinogenesis. 2012;51(12):952–62. doi: 10.1002/mc.20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chakrabarti M, Banik NL, Ray SK. miR-138 overexpression is more powerful than hTERT knockdown to potentiate apigenin for apoptosis in neuroblastoma in vitro and in vivo. Experimental cell research. 2013;319(10):1575–85. doi: 10.1016/j.yexcr.2013.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao Z, Xu Z, Hung MS, Lin YC, Wang T, Gong M, et al. Promoter demethylation of WIF-1 by epigallocatechin-3-gallate in lung cancer cells. Anticancer research. 2009;29(6):2025–30. [PubMed] [Google Scholar]
- 73.Mirza S, Sharma G, Parshad R, Gupta SD, Pandya P, Ralhan R. Expression of DNA methyltransferases in breast cancer patients and to analyze the effect of natural compounds on DNA methyltransferases and associated proteins. Journal of breast cancer. 2013;16(1):23–31. doi: 10.4048/jbc.2013.16.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nandakumar V, Vaid M, Katiyar SK. (−)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32(4):537–44. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deb G, Thakur VS, Limaye AM, Gupta S. Epigenetic induction of tissue inhibitor of matrix metalloproteinase-3 by green tea polyphenols in breast cancer cells. Molecular carcinogenesis. 2014 doi: 10.1002/mc.22121. [DOI] [PubMed] [Google Scholar]
- 76.Wang H, Bian S, Yang CS. Green tea polyphenol EGCG suppresses lung cancer cell growth through upregulating miR-210 expression caused by stabilizing HIF-1alpha. Carcinogenesis. 2011;32(12):1881–9. doi: 10.1093/carcin/bgr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gordon MW, Yan F, Zhong X, Mazumder PB, Xu-Monette ZY, Zou D, et al. Regulation of p53-targeting microRNAs by polycyclic aromatic hydrocarbons: Implications in the etiology of multiple myeloma. Molecular carcinogenesis. 2014 doi: 10.1002/mc.22175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xie Q, Bai Q, Zou LY, Zhang QY, Zhou Y, Chang H, et al. Genistein inhibits DNA methylation and increases expression of tumor suppressor genes in human breast cancer cells. Genes, chromosomes & cancer. 2014;53(5):422–31. doi: 10.1002/gcc.22154. [DOI] [PubMed] [Google Scholar]
- 79.Hirata H, Hinoda Y, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, et al. Genistein downregulates onco-miR-1260b and upregulates sFRP1 and Smad4 via demethylation and histone modification in prostate cancer cells. British journal of cancer. 2014;110(6):1645–54. doi: 10.1038/bjc.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xia J, Cheng L, Mei C, Ma J, Shi Y, Zeng F, et al. Genistein inhibits cell growth and invasion through regulation of miR-27a in pancreatic cancer cells. Current pharmaceutical design. 2014;20(33):5348–53. doi: 10.2174/1381612820666140128215756. [DOI] [PubMed] [Google Scholar]
- 81.Zhu W, Qin W, Zhang K, Rottinghaus GE, Chen YC, Kliethermes B, et al. Trans-resveratrol alters mammary promoter hypermethylation in women at increased risk for breast cancer. Nutrition and cancer. 2012;64(3):393–400. doi: 10.1080/01635581.2012.654926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qin W, Zhang K, Clarke K, Weiland T, Sauter ER. Methylation and miRNA effects of resveratrol on mammary tumors vs. normal tissue. Nutrition and cancer. 2014;66(2):270–7. doi: 10.1080/01635581.2014.868910. [DOI] [PubMed] [Google Scholar]
- 83.Venturelli S, Berger A, Bocker A, Busch C, Weiland T, Noor S, et al. Resveratrol as a pan-HDAC inhibitor alters the acetylation status of histone [corrected] proteins in human-derived hepatoblastoma cells. PloS one. 2013;8(8):e73097. doi: 10.1371/journal.pone.0073097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu P, Liang H, Xia Q, Li P, Kong H, Lei P, et al. Resveratrol induces apoptosis of pancreatic cancers cells by inhibiting miR-21 regulation of BCL-2 expression. Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2013;15(9):741–6. doi: 10.1007/s12094-012-0999-4. [DOI] [PubMed] [Google Scholar]
- 85.Sheth S, Jajoo S, Kaur T, Mukherjea D, Sheehan K, Rybak LP, et al. Resveratrol reduces prostate cancer growth and metastasis by inhibiting the Akt/MicroRNA-21 pathway. PloS one. 2012;7(12):e51655. doi: 10.1371/journal.pone.0051655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khor TO, Huang Y, Wu TY, Shu L, Lee J, Kong AN. Pharmacodynamics of curcumin as DNA hypomethylation agent in restoring the expression of Nrf2 via promoter CpGs demethylation. Biochemical pharmacology. 2011;82(9):1073–8. doi: 10.1016/j.bcp.2011.07.065. [DOI] [PubMed] [Google Scholar]
- 87.Shu L, Khor TO, Lee JH, Boyanapalli SS, Huang Y, Wu TY, et al. Epigenetic CpG demethylation of the promoter and reactivation of the expression of Neurog1 by curcumin in prostate LNCaP cells. The AAPS journal. 2011;13(4):606–14. doi: 10.1208/s12248-011-9300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Du L, Xie Z, Wu LC, Chiu M, Lin J, Chan KK, et al. Reactivation of RASSF1A in breast cancer cells by curcumin. Nutrition and cancer. 2012;64(8):1228–35. doi: 10.1080/01635581.2012.717682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Link A, Balaguer F, Shen Y, Lozano JJ, Leung HC, Boland CR, et al. Curcumin modulates DNA methylation in colorectal cancer cells. PloS one. 2013;8(2):e57709. doi: 10.1371/journal.pone.0057709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nagaraju GP, Zhu S, Wen J, Farris AB, Adsay VN, Diaz R, et al. Novel synthetic curcumin analogues EF31 and UBS109 are potent DNA hypomethylating agents in pancreatic cancer. Cancer letters. 2013;341(2):195–203. doi: 10.1016/j.canlet.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 91.Chen CQ, Yu K, Yan QX, Xing CY, Chen Y, Yan Z, et al. Pure curcumin increases the expression of SOCS1 and SOCS3 in myeloproliferative neoplasms through suppressing class I histone deacetylases. Carcinogenesis. 2013;34(7):1442–9. doi: 10.1093/carcin/bgt070. [DOI] [PubMed] [Google Scholar]
- 92.Collins HM, Abdelghany MK, Messmer M, Yue B, Deeves SE, Kindle KB, et al. Differential effects of garcinol and curcumin on histone and p53 modifications in tumour cells. BMC cancer. 2013;13:37. doi: 10.1186/1471-2407-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao SF, Zhang X, Zhang XJ, Shi XQ, Yu ZJ, Kan QC. Induction of microRNA-9 mediates cytotoxicity of curcumin against SKOV3 ovarian cancer cells. Asian Pacific journal of cancer prevention : APJCP. 2014;15(8):3363–8. doi: 10.7314/apjcp.2014.15.8.3363. [DOI] [PubMed] [Google Scholar]
- 94.Kronski E, Fiori ME, Barbieri O, Astigiano S, Mirisola V, Killian PH, et al. miR181b is induced by the chemopreventive polyphenol curcumin and inhibits breast cancer metastasis via down-regulation of the inflammatory cytokines CXCL1 and -2. Molecular oncology. 2014;8(3):581–95. doi: 10.1016/j.molonc.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95*.Su ZY, Zhang C, Lee JH, Shu L, Wu TY, Khor TO, et al. Requirement and epigenetics reprogramming of Nrf2 in suppression of tumor promoter TPA-induced mouse skin cell transformation by sulforaphane. Cancer prevention research. 2014;7(3):319–29. doi: 10.1158/1940-6207.CAPR-13-0313-T. This research article demonstrated that sulforaphane suppressed JB6 cell transforamtion through epigenetic induction of Nrf2. [DOI] [PubMed] [Google Scholar]
- 96.Hsu A, Wong CP, Yu Z, Williams DE, Dashwood RH, Ho E. Promoter de-methylation of cyclin D2 by sulforaphane in prostate cancer cells. Clinical epigenetics. 2011;3:3. doi: 10.1186/1868-7083-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li Q, Yao Y, Eades G, Liu Z, Zhang Y, Zhou Q. Downregulation of miR-140 promotes cancer stem cell formation in basal-like early stage breast cancer. Oncogene. 2014;33(20):2589–600. doi: 10.1038/onc.2013.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shan Y, Zhang L, Bao Y, Li B, He C, Gao M, et al. Epithelial-mesenchymal transition, a novel target of sulforaphane via COX-2/MMP2, 9/Snail, ZEB1 and miR-200c/ZEB1 pathways in human bladder cancer cells. The Journal of nutritional biochemistry. 2013;24(6):1062–9. doi: 10.1016/j.jnutbio.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 99.Wang LG, Beklemisheva A, Liu XM, Ferrari AC, Feng J, Chiao JW. Dual action on promoter demethylation and chromatin by an isothiocyanate restored GSTP1 silenced in prostate cancer. Molecular carcinogenesis. 2007;46(1):24–31. doi: 10.1002/mc.20258. [DOI] [PubMed] [Google Scholar]
- 100.Liu Y, Chakravarty S, Dey M. Phenethylisothiocyanate alters site- and promoter-specific histone tail modifications in cancer cells. PloS one. 2013;8(5):e64535. doi: 10.1371/journal.pone.0064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yu C, Gong AY, Chen D, Solelo Leon D, Young CY, Chen XM. Phenethyl isothiocyanate inhibits androgen receptor-regulated transcriptional activity in prostate cancer cells through suppressing PCAF. Molecular nutrition & food research. 2013;57(10):1825–33. doi: 10.1002/mnfr.201200810. [DOI] [PubMed] [Google Scholar]
- 102.Altonsy MO, Habib TN, Andrews SC. Diallyl disulfide-induced apoptosis in a breast-cancer cell line (MCF-7) may be caused by inhibition of histone deacetylation. Nutrition and cancer. 2012;64(8):1251–60. doi: 10.1080/01635581.2012.721156. [DOI] [PubMed] [Google Scholar]
- 103.Tang H, Kong Y, Guo J, Tang Y, Xie X, Yang L, et al. Diallyl disulfide suppresses proliferation and induces apoptosis in human gastric cancer through Wnt-1 signaling pathway by up-regulation of miR-200b and miR-22. Cancer letters. 2013;340(1):72–81. doi: 10.1016/j.canlet.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 104.Wong CP, Hsu A, Buchanan A, Palomera-Sanchez Z, Beaver LM, Houseman EA, et al. Effects of sulforaphane and 3,3′-diindolylmethane on genome-wide promoter methylation in normal prostate epithelial cells and prostate cancer cells. PloS one. 2014;9(1):e86787. doi: 10.1371/journal.pone.0086787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li Y, Li X, Guo B. Chemopreventive agent 3,3′-diindolylmethane selectively induces proteasomal degradation of class I histone deacetylases. Cancer research. 2010;70(2):646–54. doi: 10.1158/0008-5472.CAN-09-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jin Y. 3,3′-Diindolylmethane inhibits breast cancer cell growth via miR-21-mediated Cdc25A degradation. Molecular and cellular biochemistry. 2011;358(1–2):345–54. doi: 10.1007/s11010-011-0985-0. [DOI] [PubMed] [Google Scholar]
- 107.Kong D, Heath E, Chen W, Cher ML, Powell I, Heilbrun L, et al. Loss of let-7 up-regulates EZH2 in prostate cancer consistent with the acquisition of cancer stem cell signatures that are attenuated by BR-DIM. PloS one. 2012;7(3):e33729. doi: 10.1371/journal.pone.0033729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kang KA, Kim HS, Kim DH, Hyun JW. The role of a ginseng saponin metabolite as a DNA methyltransferase inhibitor in colorectal cancer cells. International journal of oncology. 2013;43(1):228–36. doi: 10.3892/ijo.2013.1931. [DOI] [PubMed] [Google Scholar]
- 109.Wu N, Wu GC, Hu R, Li M, Feng H. Ginsenoside Rh2 inhibits glioma cell proliferation by targeting microRNA-128. Acta pharmacologica Sinica. 2011;32(3):345–53. doi: 10.1038/aps.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.An IS, An S, Kwon KJ, Kim YJ, Bae S. Ginsenoside Rh2 mediates changes in the microRNA expression profile of human non-small cell lung cancer A549 cells. Oncology reports. 2013;29(2):523–8. doi: 10.3892/or.2012.2136. [DOI] [PubMed] [Google Scholar]
- 111.Wang L, Zhang C, Guo Y, Su ZY, Yang Y, Shu L, et al. Blocking of JB6 Cell Transformation by Tanshinone IIA: Epigenetic Reactivation of Nrf2 Antioxidative Stress Pathway. The AAPS journal. 2014;16(6):1214–25. doi: 10.1208/s12248-014-9666-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tu J, Xing Y, Guo Y, Tang F, Guo L, Xi T. TanshinoneIIA ameliorates inflammatory microenvironment of colon cancer cells via repression of microRNA-155. International immunopharmacology. 2012;14(4):353–61. doi: 10.1016/j.intimp.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 113.Gong Y, Li Y, Abdolmaleky HM, Li L, Zhou JR. Tanshinones inhibit the growth of breast cancer cells through epigenetic modification of Aurora A expression and function. PloS one. 2012;7(4):e33656. doi: 10.1371/journal.pone.0033656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Woo J, Kim HY, Byun BJ, Chae CH, Lee JY, Ryu SY, et al. Biological evaluation of tanshindiols as EZH2 histone methyltransferase inhibitors. Bioorganic & medicinal chemistry letters. 2014;24(11):2486–92. doi: 10.1016/j.bmcl.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 115.Shen Y, Takahashi M, Byun HM, Link A, Sharma N, Balaguer F, et al. Boswellic acid induces epigenetic alterations by modulating DNA methylation in colorectal cancer cells. Cancer biology & therapy. 2012;13(7):542–52. doi: 10.4161/cbt.19604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Takahashi M, Sung B, Shen Y, Hur K, Link A, Boland CR, et al. Boswellic acid exerts antitumor effects in colorectal cancer cells by modulating expression of the let-7 and miR-200 microRNA family. Carcinogenesis. 2012;33(12):2441–9. doi: 10.1093/carcin/bgs286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang J, Li Y, Wang X, Jiang C. Ursolic acid inhibits proliferation and induces apoptosis in human glioblastoma cell lines U251 by suppressing TGF-beta1/miR-21/PDCD4 pathway. Basic & clinical pharmacology & toxicology. 2012;111(2):106–12. doi: 10.1111/j.1742-7843.2012.00870.x. [DOI] [PubMed] [Google Scholar]
- 118.Gonzalez-Vallinas M, Molina S, Vicente G, Zarza V, Martin-Hernandez R, Garcia-Risco MR, et al. Expression of microRNA-15b and the glycosyltransferase GCNT3 correlates with antitumor efficacy of Rosemary diterpenes in colon and pancreatic cancer. PloS one. 2014;9(6):e98556. doi: 10.1371/journal.pone.0098556. [DOI] [PMC free article] [PubMed] [Google Scholar]