Conserved poly(A) tracks in coding regions are attenuators of translation.
Keywords: gene regulation, mRNA, polybasic amino acid runs, poly(A) nucleotide tracks, lysine, ribosome stalling, synonymous mutations
Abstract
Regulation of gene expression involves a wide array of cellular mechanisms that control the abundance of the RNA or protein products of that gene. We describe a gene regulatory mechanism that is based on polyadenylate [poly(A)] tracks that stall the translation apparatus. We show that creating longer or shorter runs of adenosine nucleotides, without changes in the amino acid sequence, alters the protein output and the stability of mRNA. Sometimes, these changes result in the production of an alternative “frameshifted” protein product. These observations are corroborated using reporter constructs and in the context of recombinant gene sequences. About 2% of genes in the human genome may be subject to this uncharacterized yet fundamental form of gene regulation. The potential pool of regulated genes encodes many proteins involved in nucleic acid binding. We hypothesize that the genes we identify are part of a large network whose expression is fine-tuned by poly(A) tracks, and we provide a mechanism through which synonymous mutations may influence gene expression in pathological states.
Gene expression in cells is a multistep process that involves transcription of genetic material from DNA to RNA and ultimately translation of mRNA into protein. These processes are subject to stringent control at all levels. Translational regulation generally controls the amount of protein generated from a given mRNA. Although most translational regulation mechanisms target the recruitment of ribosomes to the initiation codon, the protein synthesis machinery can also modulate translation elongation and termination (1, 2).
Pausing during the translational cycle—so-called ribosome stalling—is one mechanism by which the level of translation elongation can be regulated. Ribosome stalling is recognized by components of mRNA surveillance pathways, no-go decay (NGD), and nonstop decay (NSD), resulting in endonucleolytic cleavage of the stalled mRNA, ribosome rescue, and proteolytic degradation of incomplete protein products (3). NGD and NSD act on aberrant mRNAs that trigger translational arrest, as observed with damaged bases, stable stem-loop structures (4), rare codons (5), or mRNAs lacking stop codons (nonstop mRNAs) (6). However, these mechanisms also act on more specific types of translational pauses, such as runs of codons that encode consecutive basic amino acids (7, 8). It is thought that polybasic runs, as well as translation of the polyadenylate [poly(A)] tail in the case of nonstop mRNAs, cause ribosome stalling through interaction of the positively charged peptide with the negatively charged ribosome exit channel (9). Presumably, the strength of the stall is dependent on the length and composition of the polybasic stretch, and thus, the impact on overall protein expression might vary (3). Given this logic, it seems plausible that such an amino acid motif may act as a gene regulatory element that would define the amount of protein translated and the stability of the mRNA. For example, structural and biophysical differences between lysine and arginine residues, as well as potential mRNA sequence involvement, could act to further modulate this process.
Most studies investigating the effects of polybasic sequences during translation have used reporter sequences in Escherichia coli (10), yeast (8, 11), or in vitro rabbit reticulocyte lysate (9). However, detailed mechanistic information about the nature of the stall in endogenous targets through genome-wide analyses has not yet been conducted. Here, we report on translational regulation induced by poly(A) coding sequences in human cells, demonstrating that these sequences unexpectedly induce ribosome pausing directly, without a role for the encoded basic peptide.
Bioinformatic analysis can be used as an initial approach to determine whether there are evolutionary constraints that limit the abundance of polybasic amino acid residues. Runs of polybasic residues in coding sequences of genes from many eukaryotic organisms are underrepresented when compared to runs of other amino acids (12). Polyarginine runs have a similar abundance to polylysine runs at each segment length across multiple organisms (fig. S1). We developed a series of mCherry reporters to evaluate the effects of polybasic sequences on translation efficiency (output). The reporter construct consists of a double hemagglutinin (HA) tag, a run of control or polybasic sequences, followed by the mCherry reporter sequence (HA-mCherry, Fig. 1A). As a control for DNA transfection and in vivo fluorescence measurements, we also created a construct with green fluorescent protein (GFP). We used our reporters to determine whether the polybasic sequences influence the translation of reporter sequences in neonatal human dermal fibroblasts (HDFs) as well as in Drosophila S2 cells and Chinese hamster ovary (CHO) cells (Fig. 1, B and C, and figs. S2 and S3). We followed the expression of the mCherry reporter using fluorescence at 610 nm in vivo or Western blot analyses of samples collected 48 hours after transfection (Fig. 1, B and C). The stability of reporter mRNAs was determined using standard quantitative reverse transcription polymerase chain reaction (qRT-PCR) (13) assay (Fig. 1D). By careful primer design, this method allows us to estimate the level of endonucleolytic cleavage on mRNAs with stalled ribosome complexes.
Fig. 1. Effects of different lysine codons on mCherry reporter expression and mRNA stability.
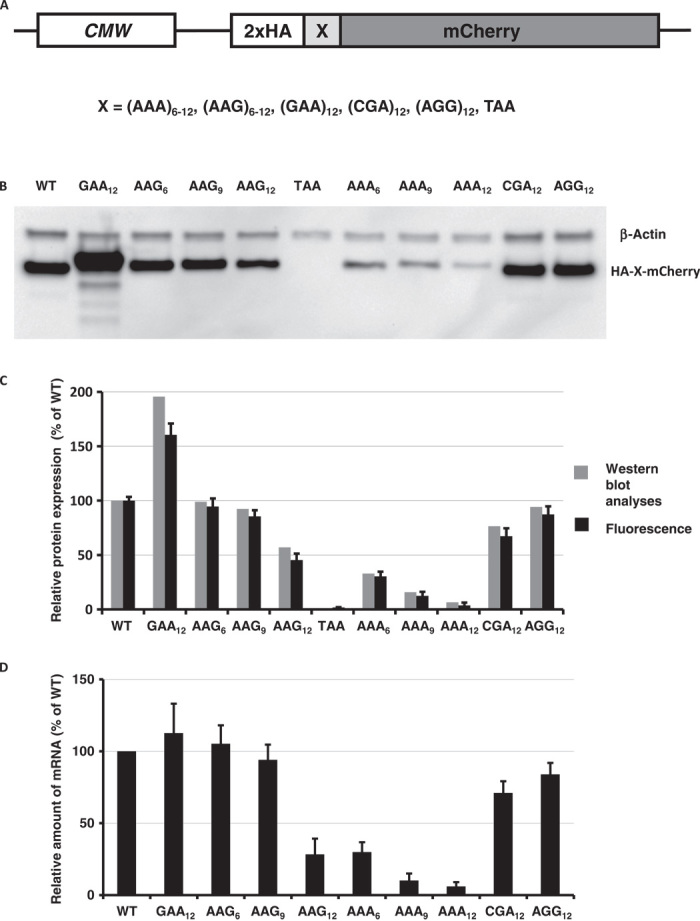
(A) Cartoon of reporter constructs used in electroporation experiments. (B) Western blot analyses of HA-X-mCherry constructs 48 hours after electroporation (HA and β-actin antibodies). (C) Normalized protein expression using LI-COR Western blot analyses or in vivo mCherry fluorescence measurement. β-Actin or fluorescence of coexpressed GFP construct was used for normalization of the data. Each bar represents the percentage of wild-type mCherry (WT) expression/fluorescence. (D) Normalized RNA levels of HA-X-mCherry constructs. Neomycin resistance gene was used for normalization of qRT-PCR data. Each bar represents the percentage of wild-type mCherry (WT) mRNA levels.
The results of DNA transfections indicate that strings of lysine codons specifically inhibit translation and decrease the stability of the mCherry reporter mRNA, whereas up to 12 arginine codons (AGG and CGA) have much less, if any, effect on either translation or mRNA stability (Fig. 1, B to D, and figs. S2 and S3). The potency of translational repression by lysine codons is clearly seen with as few as six AAA-coded lysines (AAA6) and increases with the length of the homopolymeric amino acid run. We also note that the levels of expressed mCherry reporters (Fig. 1, B and C) correlate with the stability of their mRNAs (Fig. 1D), consistent with earlier published observations (4, 6, 11). To control for possible transcriptional artifacts due to the effects of homopolymeric sequence on transcription by RNA polymerase, we electroporated mRNAs synthesized in vitro by T7 RNA polymerase directly into HDF cells. Previous studies established that T7 RNA polymerase can transcribe such homopolymeric sequences with high fidelity (10, 14). Results of our mRNA electroporation work reproduced DNA transfection experiments, consistent with models of translational repression triggered by lysine codons (fig. S4). To assess whether the stability of polylysine reporter mRNAs is dependent on translation, we introduced the translation initiation inhibitor harringtonine (15) into HDF cells before mRNA electroporation. In this case, we did not observe any significant change in mRNA stability between wild-type and polylysine-encoding mCherry constructs (fig. S5); these data indicate that accelerated decay of polylysine mCherry mRNAs is dependent on translation. Consistent with this observation, the insertion of 36 A’s (sequence equivalent to 12 lysine AAA codons) after the stop codon, in the 3′ untranslated region, did not affect the protein expression level or mRNA stability of the assayed construct (fig. S6). Insertion of polylysine codons at different positions along the coding sequence drastically reduced reporter expression and mRNA levels independent of the relative position in the construct. Hence, it follows that the observed changes in mRNA stability (Fig. 1D) result from a translation-dependent process.
The most striking observation from these data is that the production of polylysine constructs is codon-dependent; runs of polylysine residues coded by AAA codons have a much larger effect on the protein output from reporter constructs than an equivalent run of lysine AAG codons (Fig. 1, B to D, and figs. S2 to S7). This effect is unlikely to be driven by the intronless nature of our reporter because constructs containing human hemoglobin gene (delta chain; HBD) with two introns showed the same effect on protein output and RNA stability (fig. S7). We also note that this effect is unlikely to be simply due to tRNALys abundance, because the relative protein expression and mRNA stability are comparable in cells from various species that do not share similar transfer RNA (tRNA) abundance profiles (http://gtrnadb.ucsc.edu/; Fig. 1 and figs. S2 to S7). Furthermore, the human genome encodes a comparable number of tRNA genes for AAA and AAG codons (http://gtrnadb.ucsc.edu/Hsapi19/), and general codon usage is similar (0.44 versus 0.56, AAA versus AAG). The generality of codon-dependent polylysine protein production was recently documented in E. coli cells, where a single tRNALys(UUU) decodes both AAA and AAG codons (10).
In light of these experimental observations, we systematically explored codon usage and the distribution of lysine codons in polylysine tracks in various species (fig. S8). Remarkably, we find a strong underrepresentation of poly(A) nucleotide runs in regions coding for iterated lysines (even with as few as three lysines) in human genes (fig. S8). When there are four iterated lysine residues, the difference between expected (from data for all lysine residues) and observed codon usage for four AAA codons in a row is more than one order of magnitude (fig. S9). Notably, similar patterns of codon usage in lysine poly(A) tracks are observed in other vertebrates (fig. S10).
Ribosome profiling data have the potential to reveal features of pausing on polybasic stretches throughout the genome (16). A cumulative analysis of three ribosome profiling data sets from human cells for regions encoding four lysines in a row revealed that the occupancy pattern on four lysines encoded by three AAA and one AAG codon is different from the pattern for two, three, and four AAG codons in four lysine tracks (Fig. 2A). The latter three resemble the occupancy pattern for tracks of arginines (fig. S11), which is similar to the ribosome stalling on runs of basic amino acids observed by other researchers (17). This suggests that the observed effect on protein output and mRNA stability is dependent on nucleotides not simply on the amino acid sequence. The first example (with three AAA and one AAG codon) has a region of increased ribosome occupancy found additionally after the analyzed region (Fig. 2A). Together, these data suggest that attenuation of translation on poly(A) nucleotide tracks occurs via a different mechanism than just the interaction of positively charged residues with the negatively charged ribosomal exit tunnel.
Fig. 2. The effect of codon usage in polylysine tracks on translation and protein levels.
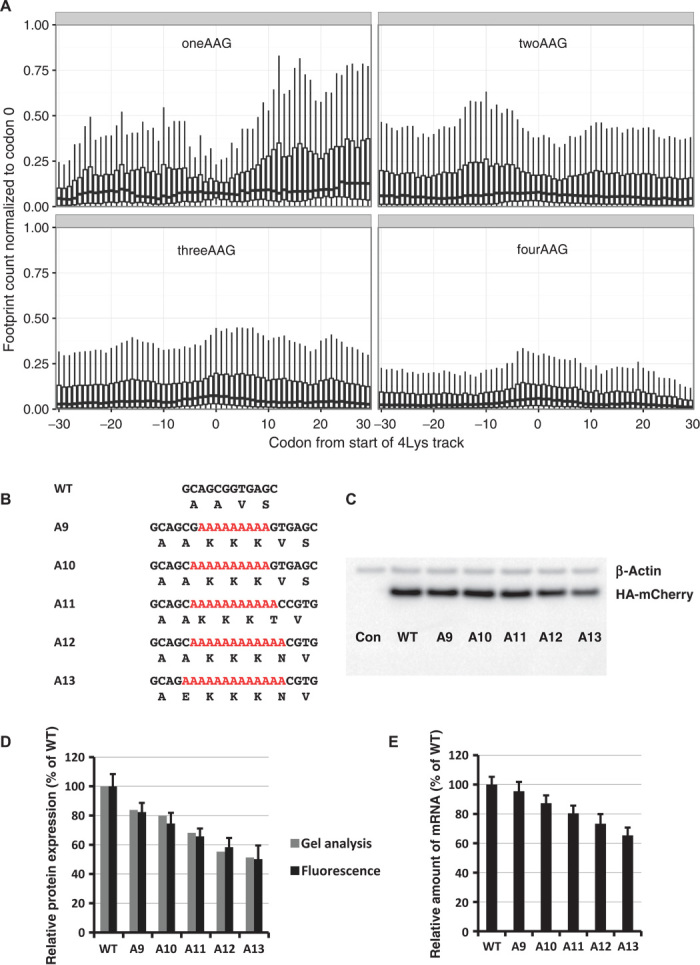
(A) Occupancy of ribosomal footprints for regions around different codon combinations for four lysine tracks. All combinations of one, two, three, and four AAG codons per group are shown. Data for four AAA codons are not shown because only a single gene has such a sequence. The upper and lower “hinges” correspond to the first and third quartiles (the 25th and 75th percentiles). The upper and lower whiskers extend from hinges up or down at a maximum of 1.5*IQR (interquartile range) of the respective hinge. (B) Sequences of HA-(A9–A13)-mCherry constructs used in electroporation experiments. (C) Western blot analyses of HA-(A9–A13)-mCherry constructs 48 hours after electroporation (HA and β-actin antibodies). (D) Normalized protein expression using LI-COR Western blot analyses or in vivo mCherry fluorescence measurement. β-Actin or fluorescence of coexpressed GFP construct was used for normalization of the data. Each bar represents the percentage of wild-type mCherry (WT) expression/fluorescence. (E) Normalized RNA levels of HA-X-mCherry constructs. Neomycin resistance gene was used for normalization of qRT-PCR data. Each bar represents the percentage of wild-type mCherry (WT) mRNA levels.
To probe the potential impact of the observed disparities in codon distribution for runs of three and four consecutive lysine codons, we inserted runs of three lysine residues with various numbers of consecutive A’s (A9 to A13) into our mCherry reporter construct (Fig. 2B). As in the previous experiments (Fig. 1, B and C), we followed the expression of the mCherry reporter and the stability of the mRNA (Fig. 2, C to E). We find that the insertion of sequences with 12 or more consecutive A’s reduces mCherry reporter expression by more than 50% with comparable effects on mRNA stability. In each construct, no more than three lysines are encoded, so the increasing effect on protein output must result from consecutive A’s, not K’s.
Next, we determined whether polylysine sequences from naturally occurring genes have the same general effect on expression of reporter protein. To take an unbiased approach, we selected different lengths of homopolymeric lysine runs and various distributions of AAA and AAG codons (Fig. 3A). Reporter constructs with lysine runs were electroporated into HDF cells, and relative amounts of reporter expression and mRNA stability were evaluated (Fig. 3, B and C). As with the designed sequences in Fig. 2B, the observed decreases in reporter protein expression and mRNA stability correlated with the number of consecutive A nucleotides and not with the total number of lysine codons in the chosen sequences. Our reporter experiments together (Figs. 1, B to D, 2, B to E, and 3, A to C, and figs. S2 to S7) argue that the repressive effects of polylysine sequence are caused by iterated poly(A) tracks rather than by runs of encoded lysine residues. Similar effects were recently documented in in vivo and in vitro experiments with E. coli cells or a purified translational system, respectively (10). The differences that we observe in expression of reporter sequences with poly(A) nucleotide tracks from human genes favor the possibility that such regions in natural genes play a “translational attenuator” role that can modulate overall protein expression.
Fig. 3. Native poly(A) tracks control reporter mRNA and protein levels.
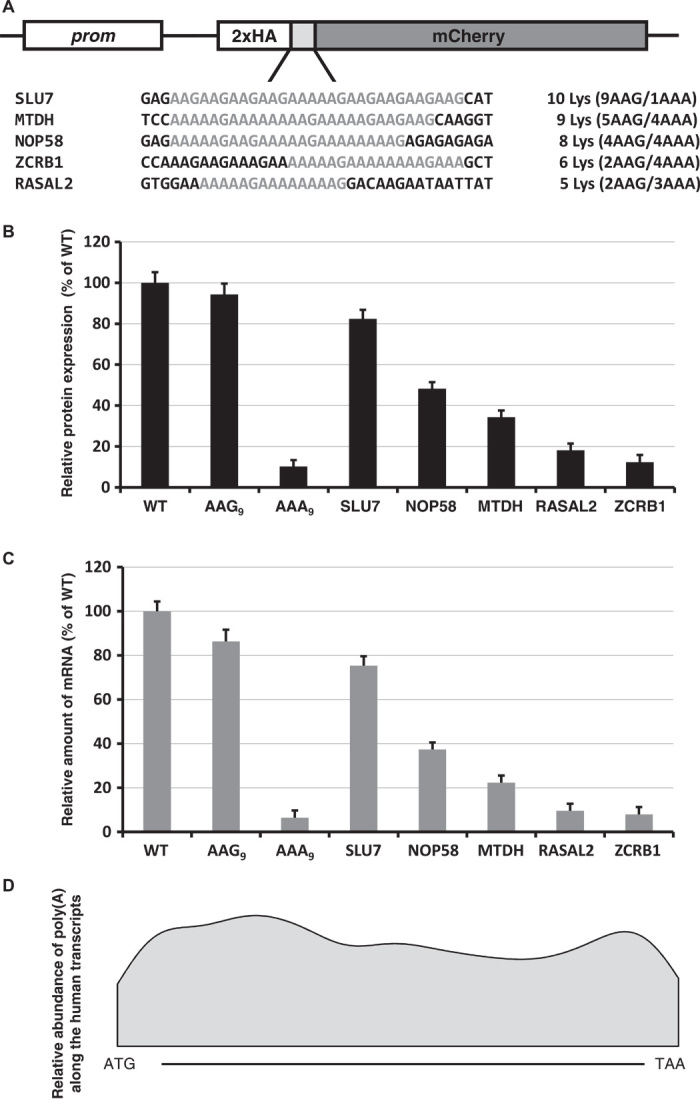
(A) Sequences of polylysine runs from human genes incorporated into HA-X-mCherry constructs. Continuous runs of lysine residues are labeled. The number of lysine residues and the ratio of AAG and AAA codons for each construct are indicated. (B) Normalized protein expression using in vivo mCherry reporter fluorescence. Fluorescence of cotransfected GFP was used to normalize the data. Each bar represents the percentage of wild-type mCherry (WT) expression/fluorescence. (C) Normalized RNA levels of HA-X-mCherry constructs. Neomycin resistance gene was used for normalization of qRT-PCR data. Each bar represents the percentage of wild-type mCherry (WT) mRNA levels. (D) Smoothed Gaussian kernel density estimate of positions of poly(A) tracks along the gene. Position of poly(A) segment is expressed as a ratio between the number of the first residue of the poly(A) track and the length of the gene.
On the basis of our results with insertion of 12 consecutive A nucleotides (Fig. 2C) and endogenous A-rich sequences (Fig. 3B), we propose that a run of 11 A’s in a stretch of 12 nucleotides (12A-1 pattern) will typically yield a measurable effect on protein expression. Because we did not require the A string to begin in any particular codon frame, the sequence may not necessarily encode four consecutive lysines. Hence, we have used the 12A-1 pattern to search the complementary DNA (cDNA) sequence database for multiple organisms [National Center for Biotechnology Information (NCBI) RefSeq resource (18)]. This query revealed more than 1800 mRNA sequences from more than 450 human genes; the proportion was similar in other vertebrates (table S1). Gene ontology analyses revealed an overrepresentation of nucleic acid binding proteins, especially RNA binding and poly(A) RNA binding proteins (table S2). The positions of poly(A) tracks are distributed uniformly along these identified sequences with no significant enrichment toward either end of the coding region (Fig. 3D). The proteins encoded by these mRNAs are often conserved among eukaryotes; of the 7636 protein isoforms coded by mRNA with poly(A) tracks from human, mouse, rat, cow, frog, zebrafish, and fruit fly, 3877 are classified as orthologous between at least two organisms. These orthologous proteins share very similar codon usage in the polylysine track, as seen in the example of the RASAL2 tumor suppressor protein (19) (fig. S12). These observations are consistent with the idea that poly(A) tracks may regulate specific sets of genes in these different organisms. Additional analyses of the ribosome profiling data for mRNAs from selected pools of genes (12A-1 pattern genes) showed an increased number of ribosome footprints in sequences following the poly(A) tracks (fig. S11). The observed pattern was similar to, albeit more pronounced than, the pattern observed for four lysine tracks encoded by three AAA codons and one AAG (Fig. 2A), despite the fact that in many cases, the selected pattern did not encode four lysines.
Given the strong sequence conservation and possible role in modulation of protein expression, we further explored the effects of mutations in poly(A) tracks. We used our reporter constructs containing poly(A) nucleotide tracks from endogenous genes (ZCRB1, MTDH, and RASAL2) to evaluate the effects of synonymous lysine mutations in these poly(A) tracks on protein expression (Fig. 4, A to C, and figs. S13 and S14). In each construct, we made mutations that changed selected AAG codons to AAA, increasing the length of consecutive A’s. Alternatively, we introduced AAA to AAG changes to create interruptions in poly(A) tracks. Reporter constructs with single AAG-to-AAA changes demonstrate consistent decreases in protein expression and mRNA stability. Conversely, AAA-to-AAG changes result in increases in protein expression and mRNA stability (Fig. 4, B and C, and figs. S13 and S14).
Fig. 4. The effect of synonymous mutations in poly(A) tracks of human genes.
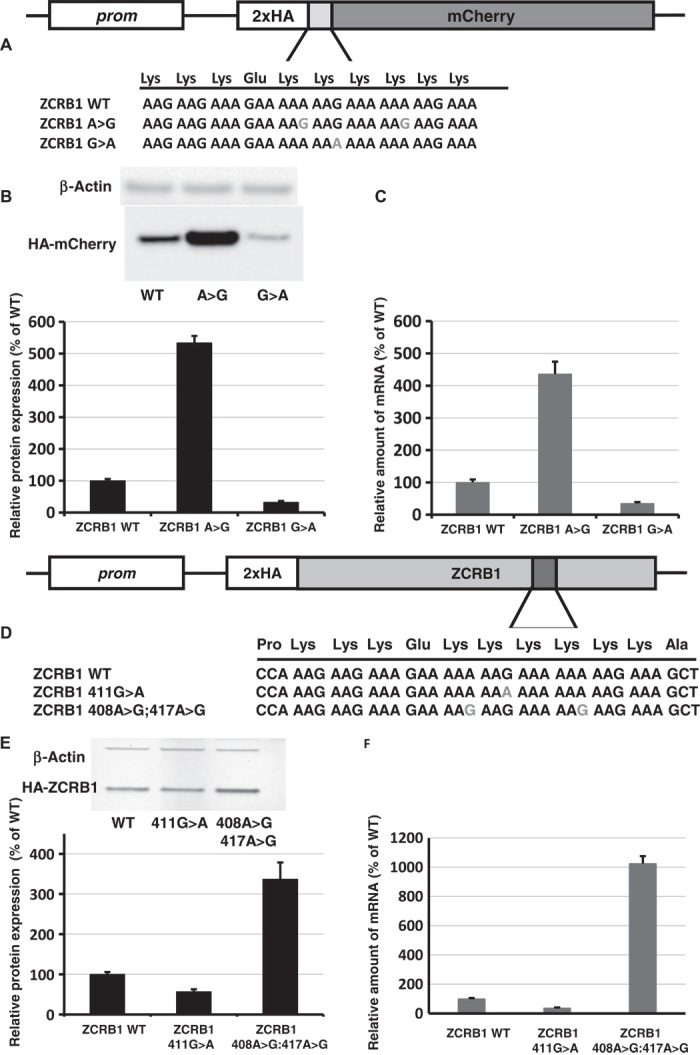
(A) Scheme of constructs with ZCRB1 gene poly(A) tracks used for analyses of synonymous mutations. (B) Western blot analyses and normalized protein expression of ZCRB1 reporter constructs with synonymous mutations (HA and β-actin antibodies). Each bar represents the percentage of wild-type ZCRB1-mCherry (WT) expression. (C) Normalized RNA levels of ZCRB1 reporter constructs with synonymous mutations. Neomycin resistance gene was used for normalization of qRT-PCR data. Each bar represents the percentage of wild-type ZCRB1-mCherry construct (WT) mRNA levels. (D) Scheme of full-length HA-tagged ZCRB gene constructs. Position and mutations in poly(A) tracks are indicated. (E) Western blot analysis and normalized protein expression of ZCRB1 gene constructs with synonymous mutations. Each bar represents the percentage of wild-type HA-ZCRB1 (WT) expression. (F) Normalized RNA levels of ZCRB1 gene constructs. Neomycin resistance gene was used for normalization of qRT-PCR data.
We next determined whether the same synonymous mutations have similar effects when cloned in the full-length coding sequence of the ZCRB1 gene (Fig. 4, D to F, and fig. S15). Indeed, the effects on protein and mRNA levels that we observed with the mCherry reporter sequences are reproduced within the context of the complete coding sequence of the ZCRB1 gene (and mutated variant). Mutation of single AAG-to-AAA codons in the poly(A) track of the ZCRB1 gene (K137K; 411G>A) resulted in a significant decrease in both protein expression and mRNA stability (Fig. 4, E and F, and fig. S15); substitution of two AAA codons with synonymous AAG codons (K136K:408A>G; K139K:417A>G) resulted in increases in both recombinant ZCRB1 protein output and mRNA stability. Generally, mutations resulting in longer poly(A) tracks reduced protein expression and mRNA stability, whereas synonymous substitutions that result in shorter poly(A) nucleotide tracks increased both protein expression and mRNA stability. From these observations, we suggest that synonymous mutations in poly(A) tracks could modulate protein production from these genes.
Poly(A) tracks resemble ribosome “slippery” sequences that have been associated with translational frameshifts (20, 21). Recent studies suggest that poly(A) tracks can induce “sliding” of E. coli ribosomes resulting in frameshifting (10, 22). Therefore, we looked for potential frameshifted products of overexpressed ZCRB1 variants by immunoprecipitation using an engineered N-terminally located HA tag. We observed the presence of a protein product of the expected size that results from possible frameshifting in our construct with increased length A tracts [ZCRB K137K (411G>A) mutant] (Fig. 5A). The presence of potential frameshifted protein products was not observed in wild-type or control double synonymous mutations K136K(408A>G):K139K(417A>G). We note that the K137K synonymous change represents a recurrent cancer mutation found in the COSMIC (Catalogue of Somatic Mutations in Cancer) database (http://cancer.sanger.ac.uk) (23) for the ZCRB1 gene (http://cancer.sanger.ac.uk/cosmic/mutation/overview?id=109189). Similar results were obtained when we compared immunoprecipitations of overexpressed and HA-tagged wild-type MTDH gene and a K451K (1353G>A) variant, yet another cancer-associated mutation (http://cancer.sanger.ac.uk/cosmic/mutation/overview?id=150510; fig. S16).
Fig. 5. Putative mechanisms through which poly(A) tracks exert their function.
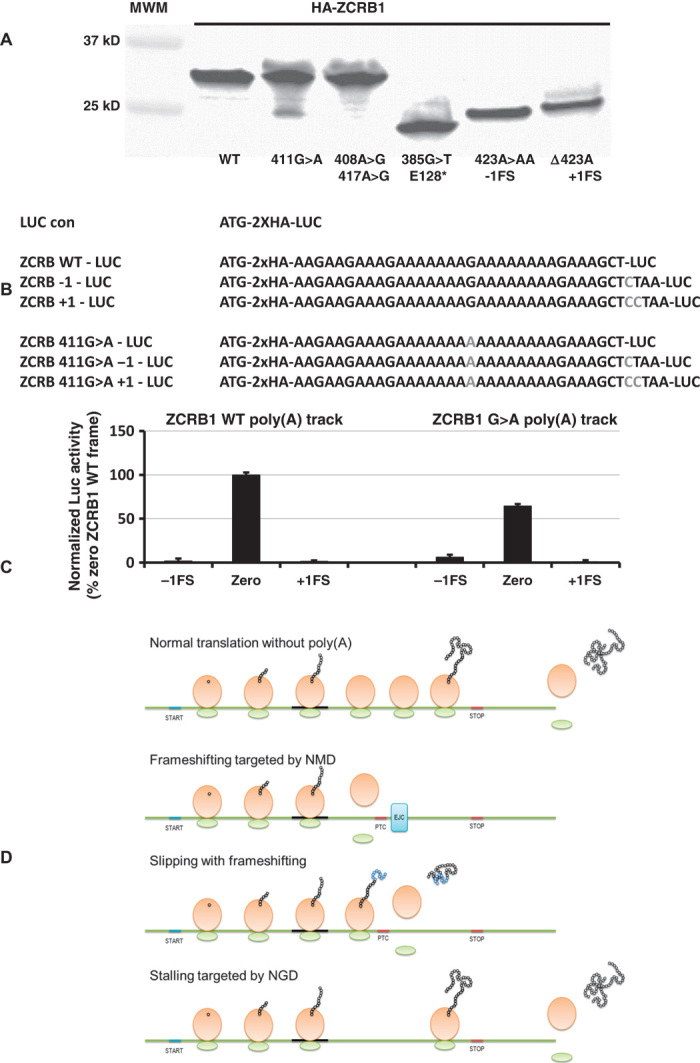
(A) Immunoprecipitation of HA-ZCRB gene constructs using anti-HA magnetic beads. ZCRB1 WT, synonymous (single 411G>A or double 408A>G; 417A>G), nonsense [385G>T, insertion of stop codon before poly(A) track], deletion (423ΔA, equivalent to +1 frameshift), and insertion (423A>AA, equivalent to −1 frameshift) mutant constructs are labeled. (B) Scheme of luciferase constructs used to estimate frameshifting potential for ZCRB1 WT and 411G>A mutant poly(A) tracks. (C) Luciferase levels (activity) from −1, “zero,” and +1 frame constructs of wild-type and G>A mutant ZCRB1 poly(A) tracks are compared. Bars represent the normalized ratio of ZCRB1 G>A and ZCRB1 WT poly(A) tracks, elucidating changes in the levels of luciferase expression in all three frames. (D) Model for function of poly(A) tracks in human genes. Poly(A) tracks lead to three possible scenarios: frameshifting consolidated with NMD, which results in reduced output of wild-type protein; frameshifting with synthesis of both out-of-frame and wild-type protein; and nonresolved stalling consolidated by endonucleolytic cleavage of mRNA and reduction in wild-type protein levels, as in the NGD pathway. Scheme for translation of mRNAs without poly(A) tracks is shown for comparison.
To further document the extent and direction of frameshifting in the ZCRB1 transcript, we introduced poly(A) tracks from wild-type ZCRB1 and a K137K ZCRB1 mutant into a Renilla luciferase reporter gene. We introduced single or double nucleotide(s) downstream in the reporter sequence following the A track, thus creating +1 and −1 frameshift (FS) constructs, respectively (Fig. 5B). When compared to wild-type ZCRB1 poly(A) track, the G>A mutant shows decreases in full-length luciferase protein expression (about 40% reduction in zero frame); additionally, the G>A mutant exhibits an increase in expression of −1FS frame construct [which is not observed in the wild-type ZCRB1 poly(A) track −1FS construct] (Fig. 5C). The total amount of luciferase protein activity from the −1FS ZCRB1 G>A mutant construct is about 10% of that expressed from the zero frame mutant construct (Fig. 5C and fig. S17). No significant change in luciferase expression was detected in samples electroporated with +1FS constructs, where expression from these constructs resulted in background levels of luciferase activity (fig. S17).
Frameshifting and recognition of out-of-frame premature stop codons can lead to nonsense-mediated mRNA decay (NMD) that results in targeted mRNA decay (24, 25). Our recent data suggest that NMD may play a role in determining the stability of poly(A) track–containing mRNAs. Deletion of NMD factor Upf1p in yeast cells partially rescues mRNA levels from constructs with simple poly(A) tracks (10). We have analyzed the complete set of human poly(A) track–containing genes to see whether they would be likely targets for NMD as a result of frameshifting on the poly(A) track [based on the usual rules for NMD (26–29)]. On the basis of the position of the poly(A) tracks, and their position relative to possible premature termination codons (PTCs) in the −1 and +1 frame, and the location of downstream exon-intron boundaries, we find that a part of our genes of interest would likely be targeted by NMD as a result of frameshifting during poly(A)-mediated stalling (these transcripts and position of PTCs are listed in table S3). The considerable number of human poly(A) track genes may not elicit NMD response because PTCs in both −1 and +1 frame following poly(A) tracks are less than 50 nucleotides away from established exon-intron boundaries. Although most frameshift events seem to lead to proteins that would be truncated immediately after poly(A) tracks, in a few cases, a novel peptide chain of substantial length may be produced (table S4). Hence, the outcome of poly(A) track stalling and slipping may include a scenario in which a frameshifted protein product is synthesized in addition to the full-length gene product (scheme shown in Fig. 5D). The possible role and presence of such fragments from poly(A) track genes and their variants is still to be elucidated.
In conclusion, we present evidence that lysine coding poly(A) nucleotide tracks in human genes may act as translational attenuators. We show that the effect is dependent on nucleotide, not amino acid, sequence, and the attenuation occurs in a distinct manner from previously described polybasic amino acid runs. These “poly(A) translational attenuators” are highly conserved across vertebrates, implying that they might play an important role in balancing gene dosage. The presence of such a regulatory function is further supported by negative selection against single-nucleotide variants in human poly(A) segments in both dbSNP and COSMIC databases (Supplementary data D1, table S5, and fig. S18). However, it is not yet clear what the effects stemming from synonymous mutation in poly(A) tracks are. Our results point to either alterations in protein levels (altered gene dosage) or the production of frameshifted products in the cell. Hence, these translational attenuation mechanisms may supplement the already large number of mechanisms through which synonymous mutations can exert biological effects [reviewed in (30)].
MATERIALS AND METHODS
Experimental protocols
Cell culture. HDF cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) and supplemented with 10% fetal bovine serum, 5% minimum essential medium nonessential amino acids (100×, Gibco), 5% penicillin and streptomycin (Gibco), and l-glutamine (Gibco). T-Rex-CHO cells were grown in Ham’s F12K medium (American Type Culture Collection) with the same supplements. Drosophila S2 cells were cultured in Express Five SFM Medium (Invitrogen) supplemented with penicillin (100 U/ml), streptomycin (100 U/ml) (Gibco), and 45 ml of 200 mM l-glutamine (Gibco) per 500 ml of medium.
Plasmids and mRNA were introduced to the cells by the Neon Transfection System (Invitrogen) with 100-μl tips according to cell-specific protocols (www.lifetechnologies.com/us/en/home/life-science/cell-culture/transfection/transfection---selection-misc/neon-transfection-system/neon-protocols-cell-line-data.html). Cells electroporated with DNA plasmids were harvested after 48 hours if not indicated differently. Cells electroporated with mRNA were harvested after 4 hours, if not indicated differently. All transfections in S2 cells were performed using Effectene reagent (Qiagen).
DNA constructs. mCherry reporter constructs were generated by PCR amplification of an mCherry template with forward primers containing the test sequence at the 5′ end and homology to mCherry at the 3′ end. The test sequence for each construct is listed in the following table. The PCR product was purified by NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel) and integrated into the pcDNA-DEST40, pcDNA-DEST53, or pMT-DEST49 expression vector by the Gateway cloning system (Invitrogen). Luciferase constructs were generated by the same method.
Whole gene constructs were generated by PCR amplification from gene library database constructs from Thermo (MTDH clone ID: 5298467) or Life Technologies GeneArt Strings DNA Fragments (ZCRB1) and cloned in pcDNA-DEST40 vector for expression. Synonymous mutations in the natural gene homopolymeric lysine runs were made by site-directed mutagenesis. Human β-globin gene (delta chain; HBD) was amplified from genomic DNA isolated from HDF cells. Insertions of poly(A) track, AAG codons, or premature stop codon in HBD constructs were made by site-directed mutagenesis. The sequences of inserts are given in table S6.
In vitro mRNA synthesis. Capped and polyadenylated mRNA was synthesized in vitro using a mMESSAGE mMACHINE T7 Transcription Kit (Life Technologies) following the manufacturer’s procedures. The quality of mRNA was checked by electrophoresis and sequencing of RT-PCR products.
RNA extraction and qRT-PCR. Total RNA was extracted from cells using the RiboZol RNA extraction reagent (Amresco) according to the manufacturer’s instructions. RiboZol reagent (400 μl) was used in each well of 6- or 12-well plates for RNA extraction. Precipitated nucleic acids were treated with Turbo deoxyribonuclease (Ambion), and total RNA was dissolved in ribonuclease-free water and stored at −20°C. RNA concentration was measured by NanoDrop (OD260/280). iScript Reverse Transcription Supermix (Bio-Rad) was used with 1 μg of total RNA following the manufacturer’s protocol. iQ SYBR Green Supermix (Bio-Rad) protocol was used for qRT-PCR on the CFX96 Real-Time system with Bio-Rad CFX Manager 3.0 software. Cycle threshold (Ct) values were normalized to the neomycin resistance gene expressed from the same plasmid.
Western blot analysis. Total cell lysates were prepared with passive lysis buffer (Promega). Blots were blocked with 5% milk in 1× tris-buffered saline–0.1% Tween 20 (TBST) for 1 hour. Horseradish peroxidase–conjugated or primary antibodies were diluted according to the manufacturer’s recommendations and incubated overnight with membranes. The membranes were washed four times for 5 min in TBST and prepared for imaging, or secondary antibody was added for additional 1 hour of incubation. Images were generated by Bio-Rad Molecular Imager ChemiDoc XRS System with Image Lab software by chemiluminescence detection or by the LI-COR Odyssey Infrared Imaging System. Blots imaged by the LI-COR system were first incubated for 1 hour with Pierce DyLight secondary antibodies.
Immunoprecipitation. Total cell lysates were prepared with passive lysis buffer (Promega) and incubated with Pierce anti-HA magnetic beads overnight at 4°C. Proteins were eluted by boiling the beads with 1× SDS sample buffer for 7 min. Loading of protein samples was normalized to total protein amounts.
Cell imaging. HDF cells were electroporated with the same amount of DNA plasmids and plated in six-well plates with optically clear bottom. Before imaging, cells were washed with fresh DMEM without phenol red and incubated for 20 min with DMEM containing 0.025% Hoechst 33342 dye for DNA staining. Cells were washed with DMEM and imaged in phenol red–free medium with an EVOS FL microscope using a 40× objective. Images were analyzed using EVOS FL software.
Bioinformatics analysis
Sequence data and variation databases. Sequence data were derived from a NCBI RefSeq resource (18) on February 2014. Two variations of databases were used: dbSNP (31), build 139 and COSMIC, build v70 (23).
mRNA mapping. Because we observed some inconsistencies between transcripts and proteins in some of the sequence databases, before starting the analyses, we mapped protein sequences to mRNA sequences using the exonerate tool (32), using protein2genome model and requiring a single best match. In case of multiple best matches (when several transcripts had given identical results), the first one was chosen because the choice of corresponding isoform (this was the most common reason for multiple matches) did not influence downstream analyses.
Ribosome profiling data. Three independent studies of ribosome profiling data from human cells were analyzed: (i) GSE51424 prepared by Gonzalez and co-workers (33), from which samples SRR1562539, SRR1562540, and SRR1562541 were used; (ii) GSE48933 prepared by Rooijers and co-workers (34), from which samples SRR935448, SRR935449, SRR935452, SRR935453, SRR935454, and SRR935455 were used; and (iii) GSE42509 prepared by Loayza-Puch and co-workers (35), from which samples SRR627620 to SRR627627 were used. The data were analyzed similarly to the original protocol created by Ingolia and co-workers (36), with modifications reflecting the fact that reads were mapped to RNA data instead of genome.
Raw data were downloaded and adapters specific for each experiments were trimmed. Then, the reads were mapped to human noncoding RNAs with bowtie 1.0.1 (37) (bowtie -p 12 -t --un), and unaligned reads were mapped to human RNAs (bowtie -p 12 -v 0 -a -m 25 --best --strata --suppress 1,6,7,8). The analysis of occupancy was originally done in a similar way to Charneski and Hurst (17); however, given that genes with poly(A) were not highly expressed and the data were sparse (several positions with no occupancy), instead of mean of 30 codons before poly(A) position, we decided to normalize only against occupancy of codon at the position 0 multiplied by the average occupancy along the gene. Occupancy data were visualized with R and ggplot2 library using geom_boxplot aesthetics. On all occupancy graphs, the upper and lower hinges correspond to the first and third quartiles (the 25th and 75th percentiles). The upper and lower whiskers extend from hinges at 1.5*IQR of the respective hinge.
Variation analysis. To assess the differences in single-nucleotide polymorphisms (SNPs) in poly(A) regions versus random regions of the same length in other genes, we needed to use the same distribution of lengths in both cases. The distribution of lengths for poly(A) regions identified as mentioned above (12 A’s allowing for one mismatch) up to length 19 (longer are rare) is presented in fig. S19. Using the same distribution of lengths, we selected one random region of length drawn from the distribution randomly placed along each gene from all human protein coding RNAs. The distributions of the number of SNPs per segment for all poly(A) segments and for one random segment for each mRNA were compared using Welch’s two-sample t test, Wilcoxon rank sum test with continuity correction, and two-sample permutation test with 100,000 permutations.
Abundance of polytracks in protein sequences. Abundance was expressed by the following equation:
where NP is the number of proteins with K+ polytrack (at least 2, at least 3, etc.) and NR is the total number of occurrences of a particular amino acid. This is to normalize against variable amino acid presence in different organisms. All isoforms of proteins were taken into account.
Other analyses. The list of human essential genes was obtained from the work of Georgi and co-workers (38). Gene Ontology analyses were done using Term Enrichment Service at http://amigo.geneontology.org/rte. Most of the graphs were prepared using R and ggplot2 library. For Fig. 3A, the values of the y axis were computed by one-dimensional Gaussian kernel density estimates implemented in the R software. Custom Perl scripts were used to analyze and merge the data.
Acknowledgments
We thank D. Owyoung, J. T. Mendell, J. Coller, and T. Schedl for helpful comments. Funding: This work was supported by the NIH (grant T32 GM: 007067 to L.L.A. and grant F32 GM100608 to K.S.K.) and the American Cancer Society (grant IRG-58-010-58-2 to S.D.). Author contributions: L.L.A. and S.P.D.: designed and conducted experiments, analyzed and interpreted biochemical data, drafted and revised the article. K.S.K., R.G., and S.D.: conception and design, analyzed and interpreted data, drafted and revised the article. P.S.: conception and design, analyzed and interpreted bioinformatics data, drafted and revised the article. Competing interests: The authors declare that they have no competing interests.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/6/e1500154/DC1
Fig. S1. Distribution of polyarginine (A) and polylysine (B) runs of different length in several organisms.
Fig. S2. Expression of HA-X-mCherry reporters in CHO cells.
Fig. S3. Expression of HA-X-mCherry reporters in Drosophila S2 cells.
Fig. S4. Expression of HA-X-mCherry reporters from T7-RNA polymerase in vitro transcribed mCherry mRNAs in HDFs.
Fig. S5. Differential stability of electroporated mRNAs from HA-X-mCherry reporters is translation-dependent.
Fig. S6. Insertion of polylysine mCherry constructs in the coding sequence results in the same protein reduction and decreased mRNA stability.
Fig. S7. Expression of HA-tagged hemoglobin (delta chain; HBD) constructs with natural introns in HDF cells.
Fig. S8. Comparison of usage of AAA in single, double, and triple lysine runs across several organisms.
Fig. S9. Observed codon usage in all isoforms of human proteins versus expected (based on the proportions 0.44 to 0.56, AAA to AAG for all lysines) in the tracks of four consecutive lysines.
Fig. S10. Codon distribution in four-lysine tracks in different organisms.
Fig. S11. Occupancy of ribosomal footprints from three different data sets: (A) region around poly(A) tracks; (B) region around four arginine tracks, all codon combinations together.
Fig. S12. Sequence conservation of RAS activating-like protein 2 gene (RASAL2) at DNA and protein sequences.
Fig. S13. Synonymous mutations in mCherry reporter with metadherin [MTDH, Lyric(Lyr)] poly(A) track.
Fig. S14. Synonymous mutations in mCherry reporter with RASAL2 poly(A) track.
Fig. S15. Expression analysis of N-terminally HA-tagged and C-terminally GFP-tagged ZCRB1 gene and its synonymous mutants in HDF cells using EVOS FL microscope.
Fig. S16. Introduction of COSMIC database reported synonymous mutation K447K (1341G>A) in full-length recombinant MTDH gene.
Fig. S17. Frameshifting efficiency of poly(A) tracks from ZCRB1 wild type (A) and ZCRB G>A mutant (B) measured by luciferase activity.
Fig. S18. Proportion of mutation types in poly(A) segments versus all mutation types.
Fig. S19. The normalized distribution of lengths for poly(A) regions identified as 12 A’s allowing for one mismatch up to length 19 in human transcripts.
Table S1. Statistics of occurrences of transcripts containing poly(A) tracks in different organisms.
Table S2. Overrepresentation of Gene Ontology terms for 456 genes containing poly(A) tracks in their coding regions up to P value of 0.05.
Table S3. Table of mRNAs that have intron-exon boundary closer than 50 nucleotides downstream from a stop codon arising from frameshifting over poly(A) tracks.
Table S4. Peptides arising from possible frameshifting on poly(A) tracks.
Table S5. Table of genes with mutations within poly(A) region reported in COSMIC database.
Table S6. Sequences of mCherry inserts.
Data D1. Analysis of dbSNP database.
REFERENCES AND NOTES
- 1.Dinman J. D., Berry M. J., 22 Regulation of Termination and Recoding. Cold Spring Harb. Monogr. Arch. 48, 625–654 (2007); https://cshmonographs.org/index.php/monographs/article/view/3291. [Google Scholar]
- 2.Hershey J. W., Sonenberg N., Mathews M. B., Principles of translational control: An overview. Cold Spring Harb. Perspect. Biol. 4, a011528 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shoemaker C. J., Green R., Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 19, 594–601 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doma M. K., Parker R., Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–564 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Letzring D. P., Dean K. M., Grayhack E. J., Control of translation efficiency in yeast by codon–anticodon interactions. RNA 16, 2516–2528 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimitrova L. N., Kuroha K., Tatematsu T., Inada T., Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 284, 10343–10352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuroha K., Akamatsu M., Dimitrova L., Ito T., Kato Y., Shirahige K., Inada T., Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep. 11, 956–961 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C. C., Li G. W., Zhou S., King D., Shen P. S., Weibezahn J., Dunn J. G., Rouskin S., Inada T., Frost A., Weissman J. S., A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J., Deutsch C., Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 384, 73–86 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koutmou K. S., Schuller A. P., Brunelle J. L., Radhakrishnan A., Djuranovic S., Green R., Ribosomes slide on lysine-encoding homopolymeric A stretches. eLife 4, e05534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuboi T., Kuroha K., Kudo K., Makino S., Inoue E., Kashima I., Inada T., Dom34:Hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol. Cell 46, 518–529 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Karlin S., Brocchieri L., Bergman A., Mrazek J., Gentles A. J., Amino acid runs in eukaryotic proteomes and disease associations. Proc. Natl. Acad. Sci. U.S.A. 99, 333–338 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Djuranovic S., Nahvi A., Green R., miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336, 237–240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barr J. N., Wertz G. W., Polymerase slippage at vesicular stomatitis virus gene junctions to generate poly(A) is regulated by the upstream 3′-AUAC-5′ tetranucleotide: Implications for the mechanism of transcription termination. J. Virol. 75, 6901–6913 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fresno M., Jiménez A., Vázquez D., Inhibition of translation in eukaryotic systems by harringtonine. Eur. J. Biochem. 72, 323–330 (1977). [DOI] [PubMed] [Google Scholar]
- 16.Ingolia N. T., Ribosome profiling: New views of translation, from single codons to genome scale. Nat. Rev. Genet. 15, 205–213 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Charneski C. A., Hurst L. D., Positively charged residues are the major determinants of ribosomal velocity. PLOS Biol. 11, e1001508 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruitt K. D., Brown G. R., Hiatt S. M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C. M., Hart J., Landrum M. J., McGarvey K. M., Murphy M. R., O’Leary N. A., Pujar S., Rajput B., Rangwala S. H., Riddick L. D., Shkeda A., Sun H., Tamez P., Tully R. E., Wallin C., Webb D., Weber J., Wu W., DiCuccio M., Kitts P., Maglott D. R., Murphy T. D., Ostell J. M., RefSeq: An update on mammalian reference sequences. Nucleic Acids Res. 42, D756–D763 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLaughlin S. K., Olsen S. N., Dake B., De Raedt T., Lim E., Bronson R. T., Beroukhim R., Polyak K., Brown M., Kuperwasser C., Cichowski K., The RasGAP gene, RASAL2, is a tumor and metastasis suppressor. Cancer Cell 24, 365–378 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belfield E. J., Hughes R. K., Tsesmetzis N., Naldrett M. J., Casey R., The gateway pDEST17 expression vector encodes a −1 ribosomal frameshifting sequence. Nucleic Acids Res. 35, 1322–1332 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J., Petrov A., Johansson M., Tsai A., O’Leary S. E., Puglisi J. D., Dynamic pathways of −1 translational frameshifting. Nature 512, 328–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan S., Wen J. D., Bustamante C., Tinoco I. Jr, Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell 160, 870–881 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forbes S. A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S., Kok C. Y., Jia M., De T., Teague J. W., Stratton M. R., McDermott U., Campbell P. J., COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belew A. T., Advani V. M., Dinman J. D., Endogenous ribosomal frameshift signals operate as mRNA destabilizing elements through at least two molecular pathways in yeast. Nucleic Acids Res. 39, 2799–2808 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belew A. T., Meskauskas A., Musalgaonkar S., Advani V. M., Sulima S. O., Kasprzak W. K., Shapiro B. A., Dinman J. D., Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway. Nature 512, 265–269 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lykke-Andersen J., Shu M. D., Steitz J. A., Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103, 1121–1131 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Le Hir H., Gatfield D., Izaurralde E., Moore M. J., The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20, 4987–4997 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang Y.-F., Imam J. S., Wilkinson M. F., The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76, 51–74 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Popp M. W.-L., Maquat L. E., Organizing principles of mammalian nonsense-mediated mRNA decay. Annu. Rev. Genet. 47, 139–165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunt R. C., Simhadri V. L., Iandoli M., Sauna Z. E., Kimchi-Sarfaty C., Exposing synonymous mutations. Trends Genet. 30, 308–321 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Sherry S. T., Ward M. H., Kholodov M., Baker J., Phan L., Smigielski E. M., Sirotkin K., dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slater G. S., Birney E., Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6, 31 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez C., Sims J. S., Hornstein N., Mela A., Garcia F., Lei L., Gass D. A., Amendolara B., Bruce J. N., Canoll P., Sims P. A., Ribosome profiling reveals a cell-type-specific translational landscape in brain tumors. J. Neurosci. 34, 10924–10936 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rooijers K., Loayza-Puch F., Nijtmans L. G., Agami R., Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nat. Commun. 4, 2886 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loayza-Puch F., Drost J., Rooijers K., Lopes R., Elkon R., Agami R., p53 induces transcriptional and translational programs to suppress cell proliferation and growth. Genome Biol. 14, R32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingolia N. T., Brar G. A., Rouskin S., McGeachy A. M., Weissman J. S., The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Georgi B., Voight B. F., Bućan M., From mouse to human: Evolutionary genomics analysis of human orthologs of essential genes. PLOS Genet. 9, e1003484 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/6/e1500154/DC1
Fig. S1. Distribution of polyarginine (A) and polylysine (B) runs of different length in several organisms.
Fig. S2. Expression of HA-X-mCherry reporters in CHO cells.
Fig. S3. Expression of HA-X-mCherry reporters in Drosophila S2 cells.
Fig. S4. Expression of HA-X-mCherry reporters from T7-RNA polymerase in vitro transcribed mCherry mRNAs in HDFs.
Fig. S5. Differential stability of electroporated mRNAs from HA-X-mCherry reporters is translation-dependent.
Fig. S6. Insertion of polylysine mCherry constructs in the coding sequence results in the same protein reduction and decreased mRNA stability.
Fig. S7. Expression of HA-tagged hemoglobin (delta chain; HBD) constructs with natural introns in HDF cells.
Fig. S8. Comparison of usage of AAA in single, double, and triple lysine runs across several organisms.
Fig. S9. Observed codon usage in all isoforms of human proteins versus expected (based on the proportions 0.44 to 0.56, AAA to AAG for all lysines) in the tracks of four consecutive lysines.
Fig. S10. Codon distribution in four-lysine tracks in different organisms.
Fig. S11. Occupancy of ribosomal footprints from three different data sets: (A) region around poly(A) tracks; (B) region around four arginine tracks, all codon combinations together.
Fig. S12. Sequence conservation of RAS activating-like protein 2 gene (RASAL2) at DNA and protein sequences.
Fig. S13. Synonymous mutations in mCherry reporter with metadherin [MTDH, Lyric(Lyr)] poly(A) track.
Fig. S14. Synonymous mutations in mCherry reporter with RASAL2 poly(A) track.
Fig. S15. Expression analysis of N-terminally HA-tagged and C-terminally GFP-tagged ZCRB1 gene and its synonymous mutants in HDF cells using EVOS FL microscope.
Fig. S16. Introduction of COSMIC database reported synonymous mutation K447K (1341G>A) in full-length recombinant MTDH gene.
Fig. S17. Frameshifting efficiency of poly(A) tracks from ZCRB1 wild type (A) and ZCRB G>A mutant (B) measured by luciferase activity.
Fig. S18. Proportion of mutation types in poly(A) segments versus all mutation types.
Fig. S19. The normalized distribution of lengths for poly(A) regions identified as 12 A’s allowing for one mismatch up to length 19 in human transcripts.
Table S1. Statistics of occurrences of transcripts containing poly(A) tracks in different organisms.
Table S2. Overrepresentation of Gene Ontology terms for 456 genes containing poly(A) tracks in their coding regions up to P value of 0.05.
Table S3. Table of mRNAs that have intron-exon boundary closer than 50 nucleotides downstream from a stop codon arising from frameshifting over poly(A) tracks.
Table S4. Peptides arising from possible frameshifting on poly(A) tracks.
Table S5. Table of genes with mutations within poly(A) region reported in COSMIC database.
Table S6. Sequences of mCherry inserts.
Data D1. Analysis of dbSNP database.


