Abstract
Upon infection of a mammalian host, Plasmodium parasites first replicate inside hepatocytes, generating thousands of new parasites. Although Plasmodium intra-hepatic development represents a substantial metabolic challenge to the host hepatocyte, how infected cells respond to and integrate this stress remains poorly understood. Here, we present proteomic and transcriptomic analyses, revealing that the endoplasmic reticulum (ER)-resident unfolded protein response (UPR) is activated in host hepatocytes upon Plasmodium berghei infection. The expression of XBP1s—the active form of the UPR mediator XBP1—and the liver-specific UPR mediator CREBH is induced by P. berghei infection in vivo. Furthermore, this UPR induction increases parasite liver burden. Altogether, our data suggest that ER stress is a central feature of P. berghei intra-hepatic development, contributing to the success of infection.
Keywords: CREBH, liver, Plasmodium, UPR, XBP1
See also: A Kaushansky & SHI Kappe (August 2015)
Introduction
Plasmodium spp., the causative agents of malaria, are obligate intracellular parasites with a complex life cycle involving both a mosquito and a mammalian host. In mammals, Plasmodium life cycle is initiated when a motile sporozoite is injected with the bite of an infected Anopheles mosquito. Sporozoites travel, through the bloodstream, to the liver and infect hepatocytes, where each sporozoite develops and multiplies into thousands of merozoites 1. Once released into the bloodstream, each merozoite infects a red blood cell (RBC) where a new replication cycle occurs, culminating in the production of new merozoites. The continuous cycle of invasion, intracellular development and proliferation and release of merozoites from RBCs is central to malaria-associated pathology 2. Plasmodium, as an obligate intracellular parasite, depends on host cell resources to support its development. This feature is particularly important during the liver stage as the Plasmodium replication rate in the liver is exceedingly higher than that in the blood.
Hepatocytes are responsible for a myriad of metabolic processes, including protein synthesis and metabolism of lipids, carbohydrates and other nutrients and micronutrients (such as iron) 3. The key players of these pathways exist, entirely or partially, in the lumen or membrane domains of the endoplasmic reticulum (ER). As a consequence, hepatocytes are unusually rich in both smooth and rough ER.
In hepatocytes, the ability of the ER to respond to metabolic demands is crucial for cell homeostasis. The unfolded protein response (UPR) of the ER is an elaborate stress signalling pathway activated upon conditions that challenge ER function, in particular the accumulation of unfolded proteins in the lumen owing to an increased demand on its synthesis capacity that the cell cannot cope with, termed ER stress 4. In eukaryotic cells, three ER transmembrane proteins mediate the canonical UPR: the two kinases, IRE1 (inositol requiring enzyme 1) and PERK (PKR-like eukaryotic initiation factor 2α kinase), and the transcription factor precursor ATF6 (activating transcription factor 6). IRE1, the most conserved signalling branch of the UPR response, functions by activating the transcription factor XBP1 (X-box binding protein 1). XBP1 mRNA is activated by an IRE1-dependent unconventional splicing and generates a mature (spliced) XBP1 transcription factor with a potent transactivation domain 5. Together, they activate signalling pathways that restore the folding capacity of the ER. Several recent studies have revealed that ER stress and UPR activation regulatory actions are broad-acting and intersect at certain metabolic pathways including lipid and glucose metabolism, particularly in the liver 6. The metabolic role of the UPR in the liver is further enhanced by the existence of a hepatocyte-specific UPR branch, mediated by the ER transcription factor CREBH (cAMP responsive element-binding protein, hepatocyte specific). This UPR branch does not activate protein folding transcriptional programmes but rather regulates liver metabolic pathways 7-9. As such, ER stress and UPR are activated in several metabolic syndromes, including obesity and type II diabetes, as well as in specific liver diseases including fatty liver disease and viral hepatitis 10.
Plasmodium infection leads to alterations in the hepatocyte transcriptome, in particular by activating metabolic processes 11. Plasmodium development thus represents a metabolic challenge to the host cell. How hepatocytes cope with a developing parasite and how intracellular host signalling events shape the infection outcome remain unclear. Here, we hypothesized that Plasmodium development inside hepatocytes impacts ER function and modulates UPR signalling pathways. We now show that Plasmodium infection induces ER stress and that activation of the UPR strongly increases Plasmodium liver infection through both the XBP1 and CREBH pathways.
Results and Discussion
Plasmodium hepatocyte infection induces ER stress and UPR activation
Our recent transcriptomic analysis of P. berghei sporozoite-infected mouse Hepa 1.6 cells at different time points 11 identified transcriptional upregulation of several key ER stress markers, as early as 6 h after infection. These include Atf4 and Chop, Atf6 and Atf3, Cebpb, Herpud1 and Trib3 (Fig1A), all with a well-established role in ER stress 12. These results were independently validated by analysing the expression of 4 of those transcripts (Atf4, Chop, Atf3 and Herpud1) by quantitative real-time PCR (qRT–PCR) of purified Hepa 1.6 GFP-expressing P. berghei ANKA-infected cells infected with GFP-expressing P. berghei ANKA sporozoites 13 at 6 and 24 h after infection (Fig1B). Uninfected cells (Hepa 1.6 GFP-negative cells), from both time points, were used to control for basal mRNA expression. Additionally, comparative quantitative proteomic analysis of infected and uninfected cells (V. Zuzarte-Luís and M. Mota, unpublished data) identified 59 ER proteins that were differentially expressed (DE) within 12 h of Huh7 human hepatoma cells being infected with GFP-expressing P. berghei ANKA sporozoites (Fig1C). Among these, gene ontology (GO) analysis identified an enrichment of proteins involved in protein folding processes, as demonstrated by the induced expression of calnexin (Canx) and calreticulin (Calr). These factors play important roles in the ER quality control apparatus through retention of incorrectly folded proteins 14,15: the ER resident protein 29 (Erp29) 16 and ERp44 (Erp44) 17, both important to correct protein folding, together with ERp19 (Txndc12) and ERp46 (Txndc5), members of the thioredoxin family of ER proteins that are highly expressed in the liver 18, and protein-modifying enzymes, including dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunits 1 and 2 (Rpn1/Rpn2) and mannosyl-oligosaccharide glucosidase (Mogs) 19 (Fig1C). Proteins involved in the ER-associated protein degradation (ERAD) pathway were also overrepresented. These include endoplasmin (Hsp90b1), both components of the ERLIN1/ERLIN2 complex, and the UBX domain-containing protein 4 (Ubxn4, also known as Ubxd2) 20 (Fig1C). The upregulation of the protein folding capacity together with ERAD activation is consistent with the induction of the unfolded protein response (UPR), in response to ER stress. In fact, ERp44 and UBX domain-containing protein 4 are induced, at the protein level, with ER stress 17,21. Moreover, the major ER stress marker, the 78 kDa glucose-regulated protein also known as BiP (Hspa5) and protein disulphide-isomerase (PDI, P4hb) 22, were clearly upregulated with infection (Fig1C). Notably, transcriptomic data from hepatocytes infected with a different rodent model, P. yoelii, show a similar alteration in ER response 11. Altogether, our data analysis on both the transcriptional and ER protein responses to infection suggests that Plasmodium liver stage parasites induce a clear signature of UPR activation in response to ER stress.
Figure 1.
Plasmodium sporozoite infection induces ER stress and activates the UPR in hepatocytes
- Heatmap of DE transcripts involved in ER stress/UPR pathways in parasitized hepatocytes at 6, 12, 18 and 24 h after infection. Each row of the plot is a gene and was colour-coded according to the log base 2 of the expression fold changes for each transcript, with red meaning upregulation and blue meaning downregulation. Original data from Albuquerque et al 11.
- Quantitative real-time PCR (qRT–PCR) analysis of Atf4, Chop, Atf3 and Herpud1 mRNA in sorted Hepa 1.6 cells infected with P. berghei at 6 and 24 h after infection relative to its GFP-negative control (dashed line), normalized to hypoxanthine-guanine phosphoribosyltransferase (Hprt). *P < 0.05, one-sample t-test. Results are expressed as means ± SEM (n = 3 independent experiments).
- Heatmap of DE proteins identified as ER proteins by gene ontology (GO) (GO_0044432). Each row of the plot is a protein and was colour-coded according to row-normalized log intensity (z-score) with red meaning upregulation and blue meaning downregulation. Each row is identified with gene name and UniProt accession number. Each column represents a replicate. Highlighted in blue are all the proteins mentioned in the text. Data are available via ProteomeXchange with identifier PXD002269.
In vivo UPR activation strongly increases exoerythrocytic forms (EEFs) in the liver
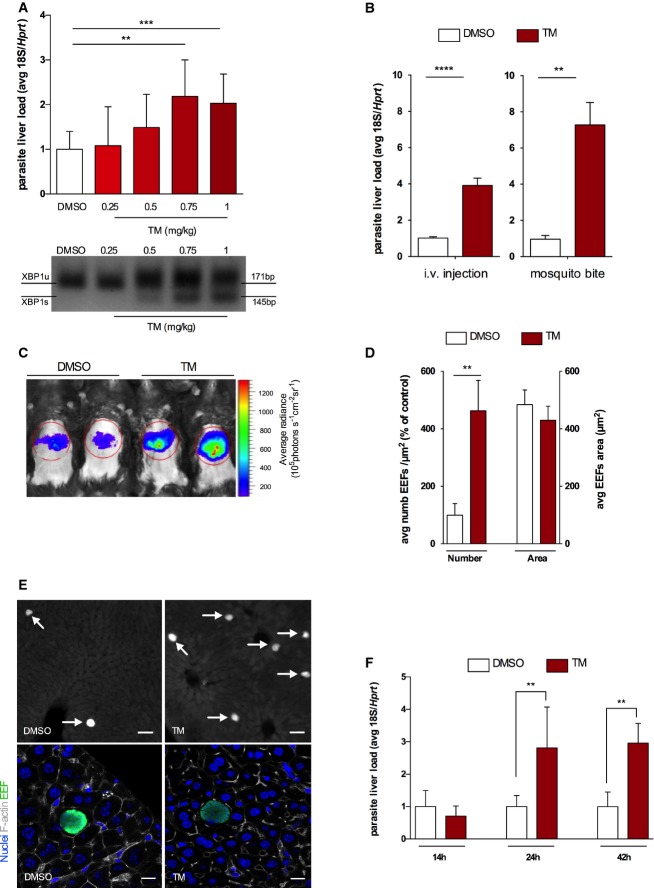
We next sought to determine how UPR activation impacts Plasmodium hepatocyte infection. To that end, we have used a well-described pharmacological ER stress inducer, tunicamycin (TM) 7,12. We performed a dose–response analysis of TM impact on P. berghei ANKA liver stage infection 13. Mice were intraperitoneally treated with different TM concentrations (or DMSO as control) 8 h prior to intravenous (i.v.) injection of 50,000 P. berghei ANKA sporozoites. Liver infection was quantified by qRT–PCR and ER stress induction was confirmed by XBP1 splicing assay. While all tested concentrations induced ER stress response within 8 h after intraperitoneal injection (data not shown), the results show that a sustained ER stress response throughout infection was only observed using 0.75 and 1 mg/kg body weight (bw) of TM. Notably, a clear and significant increase in infection was observed for mice treated with these two concentrations of TM (Fig2A). To exclude any toxic effect of TM concentrations on mouse livers, we performed histopathological analysis on liver sections of TM-treated and DMSO control mice. No histomorphological differences between control and TM-treated mice were found (Appendix Fig S1A). Additionally, none of these treatments had an impact on liver weights (Appendix Fig S1B). As such, 1 mg/kg bw was chosen for the subsequent experiments. Confirmation of TM impact on infection was obtained by initiating infection either through i.v. injection of P. berghei ANKA sporozoites or mosquito bite (the natural route of infection) and analysing parasite liver load by both in vivo light emission quantification of luciferase-expressing P. berghei liver stage parasites 23 and qRT–PCR (Fig2B and C). To exclude a direct effect of TM on parasites, P. berghei ANKA sporozoites were incubated with TM (10 μg/ml) prior to in vitro infection. No differences in sporozoites’ infectivity were found (Appendix Fig S1C).
Figure 2.
UPR activation increases EEF numbers in the liver
- Upper panel: P. berghei liver load quantification on mice treated with DMSO (control) or different tunicamycin (TM) doses 8 h prior to sporozoite injection. qRT–PCR of Plasmodium berghei 18S ribosomal RNA at 42 h after sporozoite infection normalized to hypoxanthine-guanine phosphoribosyltransferase (Hprt) expression. ***P < 0.001 and **P < 0.01, t-test. Results are expressed as means ± SD (n = 5 mice per group, two independent experiments) and TM expressed as fold change compared to DMSO. Lower panel: PCR analysis of XBP1 maturation in infected mouse livers previously treated with DMSO and TM. XBP1 unspliced (U) and spliced (S) mRNA species were resolved by high-density agarose gel (3%) (image representative of two independent experiments with n = 5 mice per group).
- Plasmodium berghei liver load quantification on mice treated with DMSO or TM 8 h prior to sporozoite delivery by either i.v. injection or mosquito bite. qRT–PCR of parasite 18S ribosomal RNA at 42 h after sporozoite infection normalized to Hprt expression. ****P < 0.0001 and **P < 0.01, t-test. Results are expressed as means ± SD (n = 5 mice per group, six independent experiments, for i.v. injection; n = 5 mice per group, three independent experiments for mosquito bite) and TM expressed as fold change compared to DMSO.
- Luciferase-expressing P. berghei infection load on DMSO and TM-treated mice at 42 h after infection (image representative of 3 independent experiments).
- Fluorescent microscopy quantification of EEF density (number) and size (area) on DMSO and TM liver sections at 42 h after P. berghei sporozoite injection. **P < 0.01, t-test. Results are expressed as means ± SD (n = 4 mice per group, three independent experiments).
- Representative fluorescence images of liver sections of DMSO- and TM-treated mice, 42 h after infection with GFP-expressing P. berghei sporozoites; arrows indicate parasite EEFs, left scale bars 50 μm; parasite in green, DNA stained with Hoechst (blue); and F-actin with phalloidin Alexa 555 (white), right scale bars 11 μm.
- Plasmodium berghei liver load quantification at 14, 24 and 42 h after i.v. sporozoite injection on mice treated with DMSO and TM for 8 h. Infection measured by qRT–PCR of parasite 18S ribosomal RNA normalized to Hprt. **P < 0.01, t-test. Results are expressed as means ± SD (n = 4 mice per group, two independent experiments).
The observed increase in parasite load can result from either a higher number of parasites that manage to reach and establish a successful liver infection or an increase in parasite replication while the number of infected cells remains constant. To determine the cause of this increase, we isolated liver sections from TM- and DMSO-treated mice and quantified GFP-positive parasite numbers and growth by fluorescence microscopy. Microscopic examination revealed that the increase in infection is due to higher numbers of exoerythrocytic forms (EEFs) as opposed to increased EEF size (Fig2D and E). Hence, our data suggest that UPR activation potentiates infection by increasing Plasmodium-infected hepatocyte numbers. Importantly, this effect on parasite numbers could not be ascribed to a defect in initial invasion of hepatocytes, as there was no difference in liver infection at 14 h after infection between TM- and DMSO-treated mice (Fig2F). The increase in infection in TM-treated mice only became apparent 24 h after sporozoite delivery (Fig2F).
Both XBP1 and CREBH pathways contribute to Plasmodium infection
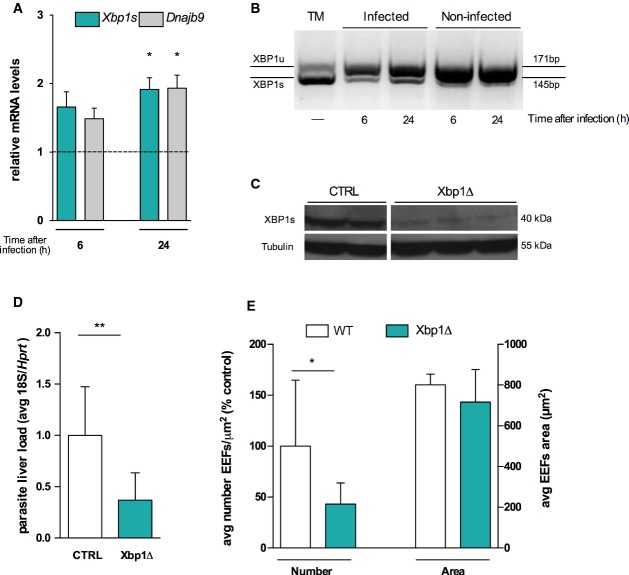
Our results so far showed that infection led to an UPR activation in the liver and that exogenous induction of ER stress led to an increase in the number of productively infected cells. We thus hypothesized that UPR activation during infection may therefore play a role in the maintenance of a successful infection. Owing to the complexity of UPR signalling, the study of its individual pathways is crucial to understand how liver UPR pathways and Plasmodium EEFs crosstalk to determine in vivo liver infection. In the liver, the XBP1 24-26 and CREBH 7-9 pathways have been shown to regulate aspects of hepatic metabolism. We first sought to determine whether Plasmodium infection activated splicing of Xbp1 mRNA. We detected maturation of Xbp1 to its active form, Xbp1s (Fig3A and B), together with an increased expression of its target gene Dnajb9, in P. berghei-infected cells (Fig3A). We next investigated the relevance of our findings to Plasmodium liver infection. We infected mice with an inducible, conditional disruption of the Xbp1 gene in the liver (Xbp1Δ), as previously described 24 (Fig3C). We observed a significant decrease in parasite liver load in Xbp1Δ mice compared to their wild-type (WT) littermates (Fig3D). In agreement with our finding of UPR activation by TM, we observed that the effect in P. berghei liver load is due to an alteration in numbers of EEFs rather than in their development. Our results show that Xbp1Δ livers exhibit a reduction in the number of infected cells (Fig3E), suggesting a direct effect of XBP1 regulated pathways on P. berghei liver stage infection. During this stage, a bulk of lipids is required to support the generation of thousands of merozoites. Plasmodium possesses a type II fatty acid biosynthetic pathway 27; however, several studies have shown that in the liver the parasite relies on its own synthesis pathway (in addition to scavenging host fatty acids) only at the transitional stage from the liver to the blood 28-30. Therefore, during the majority of the liver stage, we hypothesize that the host must fulfil the bulk of the parasite’s lipid needs. The ER has a central role in the regulation of lipid metabolism, and in the liver, XBP1s was shown to regulate the expression of genes involved in fatty acid synthesis 24. Thus, it is tempting to hypothesize a functional link between XBP1-mediated regulation of de novo hepatic fatty acid synthesis and infection. On the other hand, XBP1s overexpression was shown to induce the synthesis of phospholipids, mainly phosphatidylcholine (PC), the primary phospholipid of eukaryotic membranes in general, and the ER membrane in particular 31. Importantly, we have now shown that Plasmodium relies on the abundance of PC within hepatocytes to support infection 32. Whether XBP1 role in infection is mediated by its impact in PC levels remains to be explored.
Figure 3.
Liver Xbp1 deletion inhibits Plasmodium liver infection
- qRT–PCR analysis of Xbp1 maturated form, Xbp1s, and its target Dnajb9 mRNA in sorted Hepa 1.6 cells infected with P. berghei at 6 and 24 h after infection relative to its GFP-negative control (dashed line), normalized to hypoxanthine-guanine phosphoribosyltransferase (Hprt) and glyceraldehyde 3-phosphate dehydrogenase (Gapdh) expression. *P < 0.05, one-sample t-test. Results are expressed as means ± SEM (n = 3 independent experiments).
- Representative PCR analysis of XBP1 maturation in 8 h TM-treated Hepa 1.6 cells (positive control) and in sorted Hepa 1.6 sporozoite-infected (GFP+) and uninfected (GFP−) cells at 6 and 24 h after infection. XBP1 unspliced (U) and spliced mRNA species were resolved by high-density agarose gel (3%).
- Protein expression analysis of XBP1s protein in Xbp1Δ mice and CTRL mice with tubulin protein expression as a loading control.
- Plasmodium berghei liver load quantification on Xbp1Δ and CTRL mice by qRT–PCR of parasite 18S ribosomal RNA at 42 h after sporozoite infection, normalized to Hprt. **P < 0.01, t-test. Results are expressed as means ± SD (n ≥ 4 per group, three independent experiments).
- EEF density (numbers) and size (EEF area) quantified on Xbp1Δ mice and wild-type (WT) control mice by fluorescent microscopy of GFP-expressing P. berghei parasites at 42 h after infection, *P < 0.05, t-test (n ≥ 3 per group, three independent experiments). Results are expressed as means ± SD.
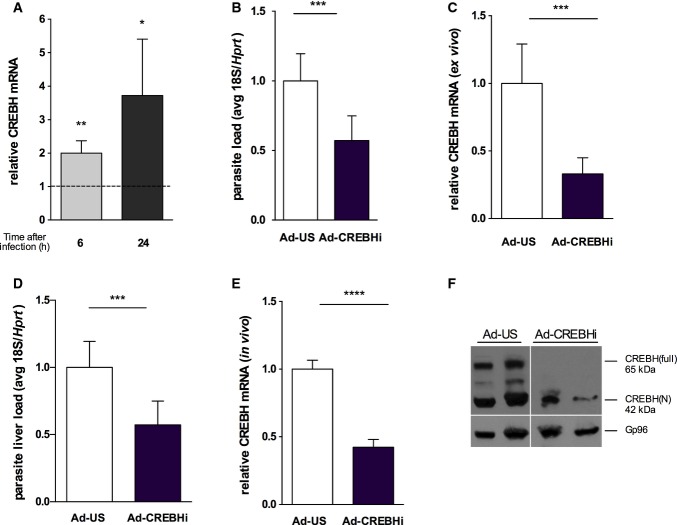
CREBH is an ER-bound transcription factor that is specifically and highly expressed in the liver and small intestine 9,33 and is activated upon ER stress 33. We tested whether Crebh mRNA is induced during Plasmodium infection. Results showed that Crebh mRNA expression was highly up-regulated at 6 and 24 h after infection in infected Hepa 1.6 cells (Fig4A). To understand the functional significance of Crebh upregulation, we infected mouse primary hepatocytes with adenovirus expressing a small hairpin RNA for Crebh (Ad-CREBHi) prior to sporozoite infection. A significant decrease in infection was observed upon Crebh knock-down (Fig4B and C), without affecting cell viability (Appendix Fig S1D). Additionally, administration of Ad-CREBHi in vivo, which led to a significant decrease in expression of both mRNA and protein, resulted in a marked decrease in liver infection (Fig4D–F). Crebh has been shown to regulate the expression of the major regulator of mammalian iron homeostasis, hepcidin (encoded by Hamp) 7, which we have shown to influence liver stage infection 34. Iron is an essential nutrient that has been shown to be required for Plasmodium liver infection 34-36 and infected hepatoma cells significantly increase the iron importer divalent metal transporter-1 (DMT1), while ferroportin expression is highly reduced 11. It is thus tempting to postulate that CREBH role in infection might be mediated by modulation of iron levels and availability. Future research will establish whether that is the case or not.
Figure 4.
CREBH knock-down, ex vivo and in vivo, restricts Plasmodium liver infection
- qRT–PCR analysis of hepatic Crebh mRNA in sorted Hepa 1.6 cells infected with P. berghei sporozoites and analysed at 6 and 24 h after infection relative to its GFP-negative control (dashed line) normalized to hypoxanthine-guanine phosphoribosyltransferase (Hprt). *P < 0.05 and **P < 0.01, t-test. Results are expressed as means ± SEM (n = 5 independent experiments).
- Plasmodium berghei infection quantification on mouse primary hepatocytes transduced ex vivo with adenovirus expressing a short hairpin RNA for Crebh (Ad-CREBH RNAi) and control adenovirus (Ad-US) by qRT–PCR for parasite 18S ribosomal RNA at 44 h after sporozoite delivery normalized to Hprt expression. Results are expressed as means ± SD, ***P < 0.001, t-test (n = 3 independent experiments).
- qRT–PCR analysis of Crebh mRNA in primary hepatocytes after Ad-US and Ad-CREBHi transduction normalized to Hprt expression. Results are expressed as means ± SD, ***P < 0.001, t-test (n = 3 independent experiments).
- Plasmodium berghei liver load quantification at 44 h after sporozoite delivery in mice transduced with Ad-US and Ad-CREBHi 48 h prior to infection by qRT–PCR for parasite 18S ribosomal RNA normalized to Hprt, ***P < 0.001, t-test. Results are expressed as means ± SD (n ≥ 4 mice per group, two independent experiments).
- qRT–PCR analysis of Crebh mRNA livers of mice transduced with Ad-US and Ad-CREBHi transduction normalized to Hprt expression. Results are expressed as means ± SD, ****P < 0.0001, t-test (n ≥ 4 mice per group, two independent experiments).
- Western blot analysis of CREBH precursor (CREBH-full) and the processed nuclear form (CREBH-N) of whole-liver lysates showing the complete absence of the precursor and a significant decrease in the processed form upon adenovirus-mediated CREBH knock-down (Ad-CREBHi). Gp96 served as a loading control.
Altogether, our data show that Plasmodium infection induces ER stress in hepatocytes via both XBP1 and CREBH pathways being activated and contributing significantly to infection. This raises the question of what is the nature of disrupted ER homeostasis mechanisms in the context of malaria liver infection? The liver, a highly metabolically active organ, has a protein synthesis rate of ∼13 million secretory proteins per minute 37. Thus, in hepatocytes, even subtle perturbations to protein synthesis can lead to ER stress. Moreover, alterations to ER fatty acid/lipid composition can also induce ER stress and activate the UPR 6,38. We have recently reported a global analysis of the total lipid repertoire of Plasmodium-infected hepatocytes, which revealed an enrichment of neutral lipids as well as the major membrane phospholipid, PC 32. Whether these alterations are the underlying cause of the ER stress during Plasmodium liver stage infection remains to be elucidated. Many pathogens induce ER stress and use diverse strategies to activate or modulate it. Viral replication, for example, co-opts ER functions to produce viral glycoproteins, which leads to induction of the UPR 39. On the other hand, several bacteria seem to induce this response via the secretion of virulence factors into the host cell 40-42,43. Interestingly, an active Plasmodium export machinery may be operating in infected hepatocytes and at least one parasite protein—the circumsporozoite (CS) protein—has been reported to be exported to the hepatocyte cytoplasm where it can impact host inflammatory responses 44. Whether other parasite factors are exported to interfere with host pathways in hepatocytes, such as the UPR, represents an exciting and yet to be explored field. Our work sheds light on a crucial and unknown aspect of the cell biology of Plasmodium–liver interactions, with the identification of the host ER as an important new determinant, and paves the way for future studies trying to better understand the implications of the newly revealed ER–Plasmodium interaction. Although several pathogens (including viruses, bacteria and parasites) can induce the UPR during infection, it is not clear in each case whether this response benefits the host or the pathogen. Modulation of ER function by these pathogens may promote infection by providing a replicative niche, but at the same time, it has been shown that the resulting disruption of the secretory pathway can aid the innate immune system in recognizing intracellular infection and in mounting an appropriate defence (reviewed in 45). Indeed, it has been claimed that some forms of ER stress may use innate immune pathogen-sensing pathways to augment IRF3-regulated type I IFN response 46, which we have recently shown to play an important role during Plasmodium liver stage infection 47. Furthermore, the UPR is a complex pathway mediated, in the liver, by four different ER sensors. Therefore, the role of PERK-eIF2α and ATF6 pathways in malaria liver infection is still to be determined. Finally, the ER is increasingly being considered an attractive potential therapeutic target, under the premise that maintaining ER function and reducing ER stress may be able to prevent metabolic diseases. In the light of our results, we postulate that resolving ER stress in the liver can mitigate malaria liver infection.
Materials and Methods
Ethics statement
All in vivo protocols were approved by the Animal Care Committee of the Instituto de Medicina Molecular (AEC_2010_024_MM_RDT_General_IMM) and were performed according to the regulations of the European guidelines 86/609/EEG.
Parasite strains and mice
Sporozoites from GFP-expressing P. berghei (parasite line 259cl2) 13 or luciferase-expressing P. berghei (parasite line 676m1cl1) 23 were dissected from infected female Anopheles stephensi salivary glands 20–24 days after the infectious blood meal. Sporozoite numbers were determined using a Neubauer chamber. Male C57BL/6 mice were obtained from Charles River (Spain) and all experiments performed with mice aged 6–8 weeks. Experimental groups were set up with mice from the same age range, gender and supplier, to exclude variation within groups.
Mosquito bite infection
Mice were intraperitoneally injected with 200 μl of anaesthesia mixture (ketamine, xylazine) diluted in PBS. Each mouse was exposed to A. stephensi mosquitoes infected with GFP-expressing P. berghei for 30 min.
Tunicamycin treatment
For in vivo treatment, tunicamycin (TM) diluted in 150 mM dextrose at 50 μg/ml was intraperitoneally injected at different doses (0.25, 0.5, 0.75 and 1 mg/kg body weight). For sporozoite treatment, TM was added to freshly dissected sporozoites at a concentration of 10μg/ml and incubated for 30 min at room temperature. After incubation, sporozoites were centrifuged for 5 min at 10,000 g, 4°C, the medium with TM removed, and sporozoites added to cells previously seeded.
Quantification of parasite liver load
Livers were collected at the mentioned time point of infection and homogenized in denaturing solution (4 M guanidine thiocyanate; 25 mM sodium citrate pH 7, 0.5% N-lauroylsarcosine and 0.7% β-mercaptoethanol in DEPC-treated water). Total RNA was extracted using RNeasy Mini kit (Qiagen). One microgram of total RNA was reverse transcribed into cDNA using the Transcriptor First Strand cDNA Synthesis kit (Roche) with the following cycle: 25°C for 10 min, 55°C for 30 min and 85°C for 5 min. Parasite burdens in the liver were determined by qRT–PCR using Pb 18S rRNA primers and normalized against the Hprt housekeeping gene. qRT–PCR was performed in a 7500 Fast Applied Bioscience (AB) machine and ViiA™ 7 Real-Time PCR System (both Applied Biosystems) using Power SYBR Green PCR Master Mix (Applied Biosystems) as follows: 50°C for 2 min, 95°C for 10 min, 40 cycles at 95°C for 15 s and 60°C for 1 min; melting stage was done at 95°C for 15 s, 60°C for 1 min and 95°C for 30 s. Primer sequences are listed in Appendix Table S1. Real-time in vivo luminescence measurement of P. berghei liver infection was performed as previously described 23.
Quantification of sporozoite infectivity by luminescence
Sporozoite infectivity was determined 24 h after infection by measuring the luminescence intensity in Hepa 1.6 cells infected with a firefly luciferase-expressing P. berghei line, as previously described 23.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5 software with unpaired Student’s t-tests. Significance is indicated by * and its value is identified in every graph.
Acknowledgments
We thank Fernanda Baptista and Inês Albuquerque for laboratory support, Iset Vera for in vivo adenovirus injections and Ana Parreira for mosquito production and infection. Additionally, we are grateful to Seung-Hoi Koo (Department of Molecular Cell Biology, Sungkyunkwan University School of Medicine, Korea) for providing CREBH adenovirus and to Ann-Hwee Lee (Weill Cornell Medical College) for providing mouse anti-CREBH antibody. We also gratefully thank Laurie Glimcher (Weill Cornell Medical College, New York) for providing us with the Xbp1flox mice. This work was supported by Fundação para a Ciência e Tecnologia (FCT, Portugal) grants EXCL/IMI-MIC/0056/2012 (to M.M.M.) and PTDC/SAU-MIC/113697/2009 (to V.Z.-L.). The work also received funding from the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreement No. 242095 (EVIMalaR) and also the European Research Council’s grant agreement no. 311502 (M.M.M.). Partial funding for this work was also provided by the NIH (R01 AI085584; Principal Investigator D.A.F.). P.I. was supported by the Fundação para a Ciência e a Tecnologia (FCT), Lisboa, Portugal (SFRH/BD/33221/2007), and Bolsas C&T – I&D Concurso 2012 from Fundação Luso-Americana. V.Z.-L. is supported by a post-doctoral fellowship from FCT, Portugal (SFRH/BPD/81953/2011), and was supported in the past by an EMBO Fellowship (ALTF-357-2009).
Author contributions
PI performed the majority of the experimental work. PI and VZ-L performed the collection of sorted infected cells for proteomics screen. VZ-L, NN and MM performed sample processing and proteomics screen. MR and VZ-L performed the in vivo TM dose dependency and sporozoite TM pre-incubation experiments. PI and BF performed the experiments on Xbp1Δ mice. KR performed the analysis of ER proteins from proteomics screen. GM and DF contributed with reagents. PI and MMM conceived the study and designed the experimental procedures. Mmo supervised the study. PI and MMM wrote the manuscript. GM and DF provided insightful comments. All authors read and approved the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Appendix
Review Process File
References
- Prudencio M, Rodriguez A, Mota MM. The silent path to thousands of merozoites: the Plasmodium liver stage. Nat Rev Microbiol. 2006;4:849–856. doi: 10.1038/nrmicro1529. [DOI] [PubMed] [Google Scholar]
- Zuzarte-Luis V, Mota MM, Vigario AM. Malaria infections: what and how can mice teach us. J Immunol Methods. 2014;410:113–122. doi: 10.1016/j.jim.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Protzer U, Maini MK, Knolle PA. Living in the liver: hepatic infections. Nat Rev Immunol. 2012;12:201–213. doi: 10.1038/nri3169. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Fu S, Watkins SM, Hotamisligil GS. The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab. 2012;15:623–634. doi: 10.1016/j.cmet.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Vecchi C, Montosi G, Zhang K, Lamberti I, Duncan SA, Kaufman RJ, Pietrangelo A. ER stress controls iron metabolism through induction of hepcidin. Science. 2009;325:877–880. doi: 10.1126/science.1176639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MW, Chanda D, Yang J, Oh H, Kim SS, Yoon YS, Hong S, Park KG, Lee IK, Choi CS, et al. Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell Metab. 2010;11:331–339. doi: 10.1016/j.cmet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- Lee JH, Giannikopoulos P, Duncan SA, Wang J, Johansen CT, Brown JD, Plutzky J, Hegele RA, Glimcher LH, Lee AH. The transcription factor cyclic AMP-responsive element-binding protein H regulates triglyceride metabolism. Nat Med. 2011;17:812–815. doi: 10.1038/nm.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi H, Kaufman RJ. Endoplasmic reticulum stress in liver disease. J Hepatol. 2011;54:795–809. doi: 10.1016/j.jhep.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque SS, Carret C, Grosso AR, Tarun AS, Peng X, Kappe SH, Prudencio M, Mota MM. Host cell transcriptional profiling during malaria liver stage infection reveals a coordinated and sequential set of biological events. BMC Genom. 2009;10:270. doi: 10.1186/1471-2164-10-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samali A, Fitzgerald U, Deegan S, Gupta S. Methods for monitoring endoplasmic reticulum stress and the unfolded protein response. Int J Cell Biol. 2010;2010:830307. doi: 10.1155/2010/830307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke-Fayard B, Trueman H, Ramesar J, Mendoza J, van der Keur M, van der Linden R, Sinden RE, Waters AP, Janse CJ. A Plasmodium berghei reference line that constitutively expresses GFP at a high level throughout the complete life cycle. Mol Biochem Parasitol. 2004;137:23–33. doi: 10.1016/j.molbiopara.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Schrag JD, Bergeron JJ, Li Y, Borisova S, Hahn M, Thomas DY, Cygler M. The structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol Cell. 2001;8:633–644. doi: 10.1016/s1097-2765(01)00318-5. [DOI] [PubMed] [Google Scholar]
- Williams DB. Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J Cell Sci. 2006;119:615–623. doi: 10.1242/jcs.02856. [DOI] [PubMed] [Google Scholar]
- Sargsyan E, Baryshev M, Szekely L, Sharipo A, Mkrtchian S. Identification of ERp29, an endoplasmic reticulum lumenal protein, as a new member of the thyroglobulin folding complex. J Biol Chem. 2002;277:17009–17015. doi: 10.1074/jbc.M200539200. [DOI] [PubMed] [Google Scholar]
- Anelli T, Alessio M, Mezghrani A, Simmen T, Talamo F, Bachi A, Sitia R. ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J. 2002;21:835–844. doi: 10.1093/emboj/21.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblach B, Keller BO, Groenendyk J, Aldred S, Zheng J, Lemire BD, Li L, Michalak M. ERp19 and ERp46, new members of the thioredoxin family of endoplasmic reticulum proteins. Mol Cell Proteomics. 2003;2:1104–1119. doi: 10.1074/mcp.M300053-MCP200. [DOI] [PubMed] [Google Scholar]
- Rini J, Esko J, Varki A. Glycosyltransferases and glycan-processing enzymes. In: Varki A, Cummings RD, Esko JD, et al., editors. Essentials of glycobiology. New York: Cold Spring Harbor Laboratory Press; 2009. 2nd edn, Chapter 5. [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Yin C, Doong H, Fang S, Peterhoff C, Nixon RA, Monteiro MJ. Characterization of erasin (UBXD2): a new ER protein that promotes ER-associated protein degradation. J Cell Sci. 2006;119:4011–4024. doi: 10.1242/jcs.03163. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ, Scheuner D, Schroder M, Shen X, Lee K, Liu CY, Arnold SM. The unfolded protein response in nutrient sensing and differentiation. Nat Rev Mol Cell Biol. 2002;3:411–421. doi: 10.1038/nrm829. [DOI] [PubMed] [Google Scholar]
- Ploemen IH, Prudêncio M, Douradinha BG, Ramesar J, Fonager J, van Gemert GJ, Luty AJ, Hermsen CC, Sauerwein RW, Baptista FG, et al. Visualisation and quantitative analysis of the rodent malaria liver stage by real time imaging. PLoS ONE. 2009;4:e7881. doi: 10.1371/journal.pone.0007881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, et al. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- Lee J, Sun C, Zhou Y, Gokalp D, Herrema H, Park SW, Davis RJ, Ozcan U. p38 MAPK-mediated regulation of Xbp1s is crucial for glucose homeostasis. Nat Med. 2011;17:1251–1260. doi: 10.1038/nm.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph SA, van Dooren GG, Waller RF, Crawford MJ, Fraunholz MJ, Foth BJ, Tonkin CJ, Roos DS, McFadden GI. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol. 2004;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- Pei Y, Tarun AS, Vaughan AM, Herman RW, Soliman JM, Erickson-Wayman A, Kappe SH. Plasmodium pyruvate dehydrogenase activity is only essential for the parasite’s progression from liver infection to blood infection. Mol Microbiol. 2010;75:957–971. doi: 10.1111/j.1365-2958.2009.07034.x. [DOI] [PubMed] [Google Scholar]
- Yu M, Kumar TR, Nkrumah LJ, Coppi A, Retzlaff S, Li CD, Kelly BJ, Moura PA, Lakshmanan V, Freundlich JS, et al. The fatty acid biosynthesis enzyme FabI plays a key role in the development of liver-stage malarial parasites. Cell Host Microbe. 2008;4:567–578. doi: 10.1016/j.chom.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan AM, O’Neill MT, Tarun AS, Camargo N, Phuong TM, Aly AS, Cowman AF, Kappe SH. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell Microbiol. 2009;11:506–520. doi: 10.1111/j.1462-5822.2008.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoe MA, Sampaio JL, Cabal GG, Real E, Zuzarte-Luis V, March S, Bhatia SN, Frischknecht F, Thiele C, Shevchenko A, et al. Host cell phosphatidylcholine is a key mediator of malaria parasite survival during liver stage infection. Cell Host Microbe. 2014;16:778–786. doi: 10.1016/j.chom.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- Portugal S, Carret C, Recker M, Armitage AE, Gonçalves LA, Epiphanio S, Sullivan D, Roy C, Newbold CI, Drakesmith H, et al. Host-mediated regulation of superinfection in malaria. Nat Med. 2011;17:732–737. doi: 10.1038/nm.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goma J, Renia L, Miltgen F, Mazier D. Iron overload increases hepatic development of Plasmodium yoelii in mice. Parasitology. 1996;112:165–168. doi: 10.1017/s0031182000084729. [DOI] [PubMed] [Google Scholar]
- Stahel E, Mazier D, Guillouzo A, Miltgen F, Landau I, Mellouk S, Beaudoin RL, Langlois P, Gentilini M. Iron chelators: in vitro inhibitory effect on the liver stage of rodent and human malaria. Am J Trop Med Hyg. 1988;39:236–240. doi: 10.4269/ajtmh.1988.39.236. [DOI] [PubMed] [Google Scholar]
- Mullins C. The Biogenesis of Cellular Organelles. Georgetown, TX: New York, NY: Kluwer Academic/Plenum; 2005. Landes Bioscience/Eurekah.com; [Google Scholar]
- Fu S, Yang L, Li P, Hofmann O, Dicker L, Hide W, Lin X, Watkins SM, Ivanov AR, Hotamisligil GS. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watowich SS, Morimoto RI, Lamb RA. Flux of the paramyxovirus hemagglutinin-neuraminidase glycoprotein through the endoplasmic reticulum activates transcription of the GRP78-BiP gene. J Virol. 1991;65:3590–3597. doi: 10.1128/jvi.65.7.3590-3597.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Lee MS, Cherla RP, Tesh VL. Shiga toxin 1 induces apoptosis through the endoplasmic reticulum stress response in human monocytic cells. Cell Microbiol. 2008;10:770–780. doi: 10.1111/j.1462-5822.2007.01083.x. [DOI] [PubMed] [Google Scholar]
- He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006;13:393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- Pillich H, Loose M, Zimmer KP, Chakraborty T. Activation of the unfolded protein response by Listeria monocytogenes. Cell Microbiol. 2012;14:949–964. doi: 10.1111/j.1462-5822.2012.01769.x. [DOI] [PubMed] [Google Scholar]
- Wolfson JJ, May KL, Thorpe CM, Jandhyala DM, Paton JC, Paton AW. Subtilase cytotoxin activates PERK, IRE1 and ATF6 endoplasmic reticulum stress-signalling pathways. Cell Microbiol. 2008;10:1775–1786. doi: 10.1111/j.1462-5822.2008.01164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP, Buscaglia CA, Wang Q, Levay A, Nussenzweig DR, Walker JR, Winzeler EA, Fujii H, Fontoura BM, Nussenzweig V. Plasmodium circumsporozoite protein promotes the development of the liver stages of the parasite. Cell. 2007;131:492–504. doi: 10.1016/j.cell.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Celli J, Tsolis RM. Bacteria, the endoplasmic reticulum and the unfolded protein response: friends or foes? Nat Rev Microbiol. 2015;13:71–82. doi: 10.1038/nrmicro3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YP, Zeng L, Tian A, Bomkamp A, Rivera D, Gutman D, Barber GN, Olson JK, Smith JA. Endoplasmic reticulum stress regulates the innate immunity critical transcription factor IRF3. J Immunol. 2012;189:4630–4639. doi: 10.4049/jimmunol.1102737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liehl P, Zuzarte-Luís V, Chan J, Zillinger T, Baptista F, Carapau D, Konert M, Hanson KK, Carret C, Lassnig C, et al. Host-cell sensors for Plasmodium activate innate immunity against liver-stage infection. Nat Med. 2014;20:47–53. doi: 10.1038/nm.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Review Process File