Abstract
Silencing of transposable elements (TEs) in the metazoan germline is critical for genome integrity and is primarily dependent on Piwi proteins and associated RNAs, which exert their function through both transcriptional and posttranscriptional mechanisms. Here, we report that the evolutionarily conserved Pelo (Dom34)-Hbs1 mRNA surveillance complex is required for transposon silencing in the Drosophila germline. In pelo mutant gonads, mRNAs and proteins of some selective TEs are up-regulated. Pelo is not required for piRNA biogenesis, and our studies suggest that Pelo may function at the translational level to silence TEs: This function requires interaction with Hbs1, and overexpression of RpS30a partially reverts TE-silencing defects in pelo mutants. Interestingly, TE silencing and spermatogenesis defects in pelo mutants can also effectively be rescued by expressing the mammalian ortholog of Pelo. We propose that the Pelo-Hbs1 surveillance complex provides another level of defense against the expression of TEs in the germline of Drosophila and possibly all metazoa.
Keywords: Drosophila, germline, Hbs1, Pelota/Dom34, transposon silencing
Introduction
The Drosophila Piwi proteins and associated piRNAs defend against transposable elements (TEs) in the germline at both transcriptional and posttranscriptional levels 1. In the cytoplasm, the transcripts are subjected to endonucleolytic cleavage by RNA-induced silencing complexes (RISCs) containing Aub or Ago3, two Piwi family proteins that generate piRNAs in the ping-pong cycle to amplify the response to the actively transcribed TEs 2,3. In the nucleus, the Piwi-containing RISCs are guided to the chromatin via piRNAs to establish repressive chromatin state at the transposable DNA elements and other targeting loci 4-6. Murine members of Piwi and associated piRNAs are found to be associated with ribosomes and regulate translation during spermatogenesis 7,8, but whether TEs could be regulated at the translational level is unknown.
Piwi, the founding member of the Drosophila piRNA pathway, was initially identified as an essential regulator of germline stem cell maintenance in the Drosophila ovary 9. Later studies demonstrate that Piwi is required in both the somatic niche cells and the germline for the maintenance, proliferation, and differentiation of germline stem cells 10-12. Pelo encodes a eukaryotic release factor (eRF1)-like protein, and similar to piwi mutants, pelo homozygous null females have rudimentary ovaries because of its requirement for germline stem cell maintenance 13. Insights on the molecular function of Pelo came from yeast in which Dom34, the Pelo ortholog, is found to form a complex with a small GTPase Hbs1 to regulate endonucleolytic cleavage of mRNAs whose secondary structure causes ribosome stalling during translational elongation, a process known as no-go decay (NGD) 14. The structure of Dom34-Hbs1 complex is similar to that of eRF1 and eRF3, but Dom34 lacks the motifs for codon recognition and peptide release 15,16. Along with studies from biochemical analyses, it is proposed that the Dom34-Hbs1 complex binds to the ribosomal A site to promote subsequent dissociation of ribosome subunits for ribosomal recycle 17,18. The molecular connection between Dom34-Hbs1-mediated ribosomal recycle and mRNA cleavage, and the identity of the responsible endonuclease remain unclear. In addition to the RNA quality control in the NGD pathway, Dom34-Hbs1 is also important for non-stop decay, decay of non-functional 18S rRNAs and mRNAs with premature stop codon 19-21. Therefore, the Dom34-Hbs1 complex appears to represent a major RNA surveillance pathway in eukaryotes to detect and rescue the ribosomes stalled on mRNAs during translation.
Here, we report that the Drosophila Pelo is required for silencing of germline transposons in the ovary and testis. Pelo is not required for piRNA biogenesis or function, and our analyses indicate that Pelo might function together with Hbs1, possibly via a NGD-like mechanism, to prevent translation of transposon mRNAs.
Results and Discussion
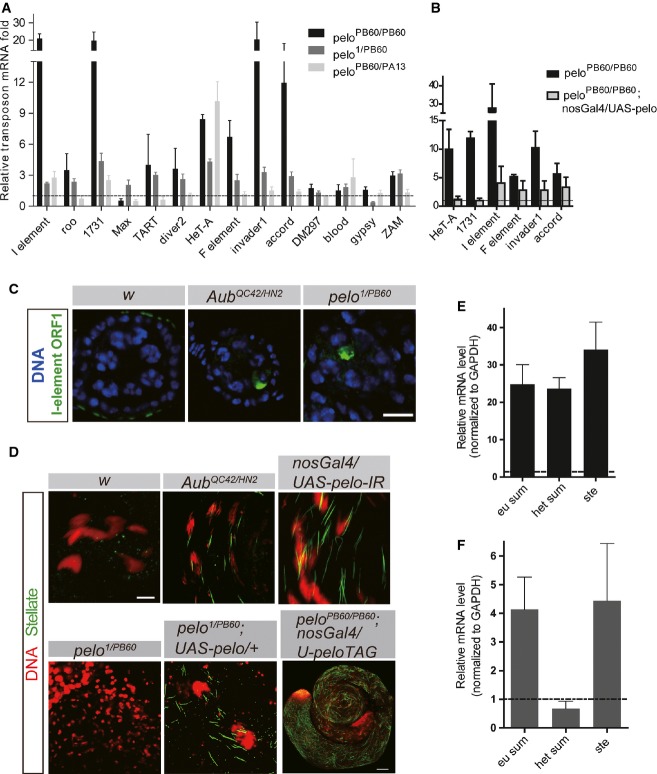
To facilitate the study on the potential role of Pelo in transposon silencing, we generated a few hypomorphic alleles of pelo by imprecise excision of P-elements inserted in the pelo locus (FigEV1). Based on molecular lesion, gene expression abundance, and ovary phenotype, we considered peloPB60 as the strongest loss-of-function allele, peloPA13 the weakest, and pelo1 in-between (see Materials and Methods, and FigEV1). By real-time PCR analysis, we observed moderate but significant up-regulation of selective germline transposons in ovaries of pelo homozygous or transheterozygous mutants (Fig1A). The magnitude of TE up-regulation was correlated with the strength of pelo mutations (Fig1A) and was further enhanced by ambient temperature (Fig EV2). In addition, up-regulation of multiple TE elements, such as HeT-A, 1731, and I-element, found in peloPB60 homozygous ovaries could be significantly suppressed by germline expression of a pelo transgene (Fig1B). Using antibodies to I-element ORF1, we also observed protein accumulation in pelo ovaries in the oocyte of the stage 2 egg chambers in all ovarioles examined (23/23). As controls, accumulation of I-element protein was only observed in the oocyte of developing egg chambers in aub mutant ovaries, but not in wild-type ovaries (Fig1C). Taken together, these observations suggest that pelo is required for transposon silencing in the Drosophila ovary whose mutation causes up-regulation of some germline TEs at both mRNA and protein levels.
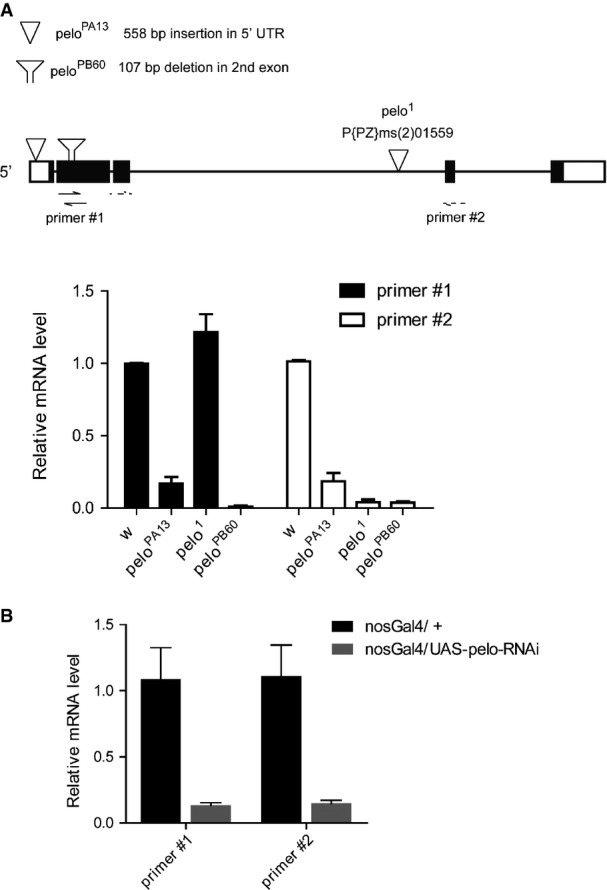
The molecular lesions and transcripts expression analysis of pelo alleles
- Upper diagram: a schematic drawing for the molecular lesions of different pelo alleles and the primer sets used for quantitative RT–PCR analysis. pelo1 is an allele with P-element inserted in the 3rd intron. peloPB60 and peloPA13 were generated by using P-element excision from peloKG06646. peloPB60 has a 107-bp deletion in the second exon, resulting in early stop codon. peloPA13 has 558-bp insertion at 5′ UTR. Lower plot: quantitative RT–PCR analysis of relative pelo expression in ovaries of the indicated genotypes using two different sets of primers. Values are means ± SEM, n = 3.
- Quantitative RT–PCR analysis to measure pelo-RNAi efficiency. Germline knockdown of pelo by nos-Gal4; UAS-pelo-RNAi caused approximately 90% reduction of pelo expression in testes. Values are means ± SEM, n = 4.
Figure 1.
Pelo is required for transposon silencing in the Drosophila germline
- A, B Quantitative RT–PCR analysis of retro-transposon transcripts in ovaries of the indicated genotypes. The expression levels were relative to that in peloPB60/+ ovaries. Germline expression of Pelo transgene suppressed TE up-regulation in peloPB60 mutant ovaries (B). Values are means ± SEM, n = 3–4.
- C Egg chambers immunostained with anti-I-element ORF1. I-element ORF1p was detected in newly formed egg chambers in AubQC42/HN2 and pelo1/PB60 ovaries but is absent in wild-type ovaries (63× magnification); scale bar represents 10 μm.
- D Testes with the indicated genotypes were immunostained with anti-Stellate (Ste, green). The needle-like Ste crystals were found in aub mutant and pelo weak mutant testes. DAPI staining is shown in red. Scale bars represent 20 μm except in the last image, which is 100 μm.
- E, F Quantitative RT–PCR analysis of stellate transcript in AubQC42/HN2 (D) and pelo-RNAi (E) testes. Values are means ± SEM, n = 4.
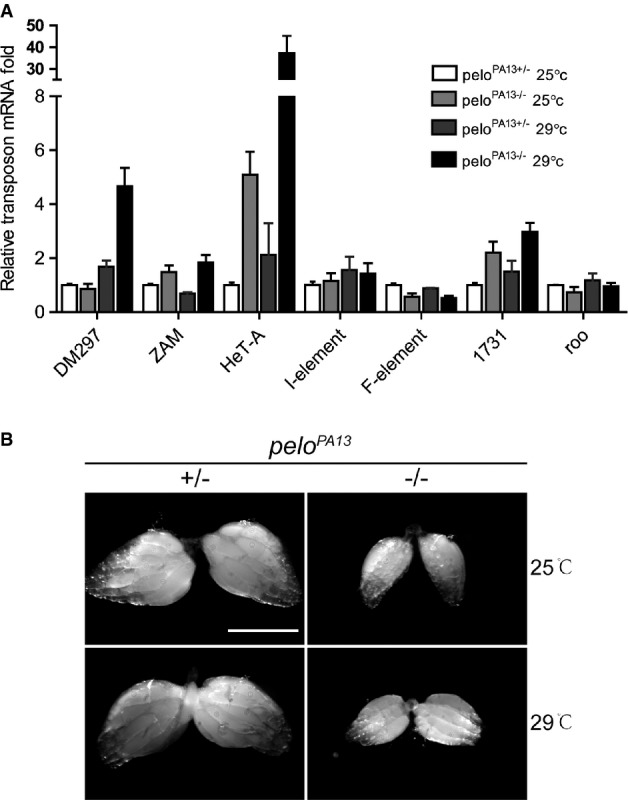
HeT-A mRNA is further up-regulated in peloPA13 ovaries under a high-temperature condition
- Quantitative RT–PCR analysis to detect the relative amount of transposon mRNAs from ovaries of peloPA13+/− and peloPA13−/− under a normal (25°C) condition or a high-temperature (29°C) condition. Values are means ± SEM, n = 3.
- Morphology of ovaries from indicated genotypes under 25°C (upper panel) or 29°C (lower panel) conditions. Scale bar represents 500 μm.
In Drosophila testis, the repeated Stellate (Ste) element is normally silenced by the piRNA pathway mediated by Su(Ste) piRNAs 22. As expected, in aub mutant testis, where the piRNA pathway is compromised, Ste proteins accumulated and formed needle-like crystals in the spermatocyte (Fig1D). Germline knockdown of pelo by nos-Gal4/UAS-pelo-RNAi also led to Ste crystal formation in the spermatocytes with 100% penetrance (49/49) though the severity varied in different testis (Fig1D). Strong loss-of-function mutants of pelo (pelo1/PB60) did not display the crystal phenotype in spermatocytes (Fig1D), possibly because the mutant spermatocytes have not yet developed to a later stage necessary for Ste expression, as Pelo is required for the progression through meiosis I during spermatogenesis 23. Consistent with this notion, leaky expression of a UAS-pelo transgene (without a GAL4 driver) in pelo1/PB60 mutants also caused Ste crystals in the spermatocytes (Fig1D). We generated another pelo transgene with a GFP tag to its C-terminal and found that this transgene is only partially functional because the transgene allowed the formation of spermatids, but failed to rescue the sterile phenotype of pelo mutant males (data not shown). Germline expression of the pelo-GFP transgene in pelo null testis also led to the formation of Ste crystals (Fig1D). These data suggest that Pelo is required for Ste silencing in the developing spermatocytes.
The Ste elements are distributed in two clusters in the genome, the 12D euchromatic region and the distal X-heterochromatic region. Similar to previous observations, Ste transcripts derived from either locus was significantly up-regulated (approximately 25-fold) in AubQC42/HN2 mutant testes 24 (Fig1E). By contrast, only transcripts derived from the euchromatic locus showed significant up-regulation in pelo-RNAi testis, and the magnitude of mRNA up-regulation was also comparatively less dramatic (approximately fourfold) (Fig1F).
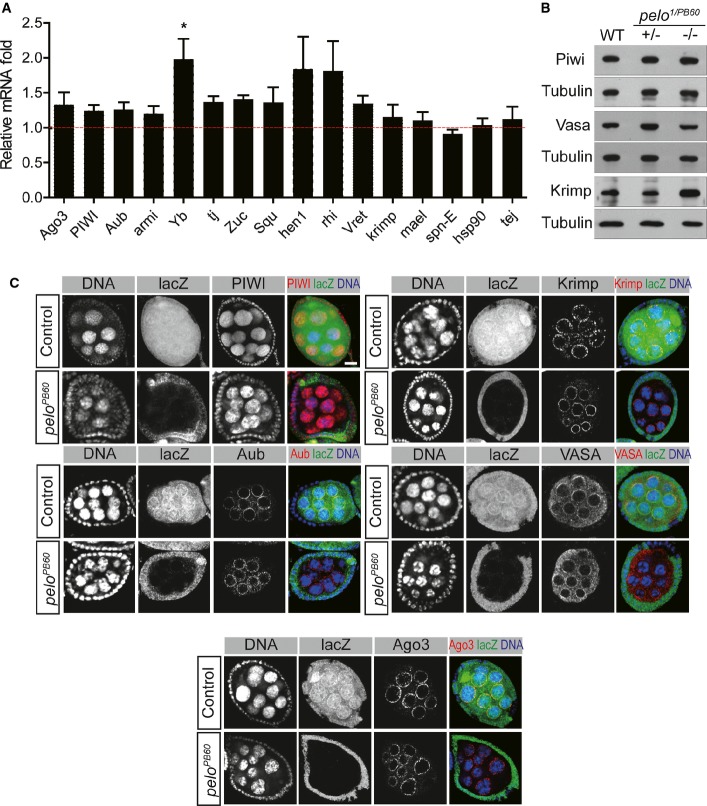
Pelo could regulate TE silencing through the piRNA pathway or other mechanisms. We first determined whether Pelo regulates the expression or function of genes in the piRNA pathway. The mRNA levels of virtually all major known genes involved in the piRNA remained unchanged in pelo mutant ovaries, except for Yb, whose expression was up-regulated (Fig2A). The protein levels for several major piRNA pathway components, such as Piwi, Vasa, and Krimp, remained unaltered as well (Fig2B). In addition, the subcellular distribution patterns of all piRNA proteins examined, including Piwi, Aub, Ago3, Vasa, and Krimp, remained unaltered in pelo mutant germline cysts (Fig2C). The above data suggest that Pelo does not regulate the expression or subcellular localization of major protein components involved in the piRNA pathway.
Figure 2.
Pelo does not affect the expression, stability or localization of core protein components in the piRNA pathway
- Quantitative RT–PCR analysis to detect changes in the expression of indicated genes in pelo1/PB60 ovaries compared to the heterozygous controls. Values are means ± SEM, n = 3. *P < 0.05, t-test.
- Western blot analysis of indicated proteins in wild-type, pelo1/+, and pelo1/PB60 ovaries. Tubulin was used as a loading control.
- Immunostaining to detect subcellular localization of the indicated proteins (red) in peloPB60 and control egg chambers. pelo mutant clones are visualized by the absence of LacZ expression (green). Scale bar represents 10 μm.
To determine whether Pelo affects piRNA biogenesis, we profiled all small RNAs extracted from pelo1/+ and pelo1/PB60 ovaries by deep sequencing. After filtering out tRNA, rRNA, and degraded snoRNAs, we had about 10M reads for each sample, 60% of which mapped to the repetitive DNA regions in the genome (FigEV3). The RNA profile in general showed that the production of endogenous siRNA (esiRNA), piRNAs, and microRNAs (miRNAs) in pelo1/PB60 ovaries was largely similar to that in the control ovaries. We specifically analyzed small repetitive RNAs uniquely mapped to the two major piRNA clusters, the 240-kb 42AB cluster that exclusively processes germline piRNAs, and the 179-kb flamenco cluster that predominantly produces somatic piRNAs. Both the abundance and distribution of small RNAs derived from these two clusters were virtually unchanged in pelo1/PB60 ovaries (Fig3A). We further examined the small RNAs mapped to specific transposable DNA elements. HeT-A is a germline-specific TE, and ZAM is soma-specific. The abundance of small RNAs derived from both TEs also remained unaltered in pelo1/PB60 ovaries (Fig3B). In parallel, we examined the production levels of several piRNAs, including roo, HeT-A, and AT-chX-1, by polyacrylamide gel electrophoresis and Northern blotting analysis. As a positive control, piRNAs were significantly reduced in AubQC42/HN2 ovaries (Fig3C). We found that the production of these piRNAs remained unchanged in pelo1/PB60 ovaries, and the production of miR-8 was unaltered in either AubQC42/HN2 or pelo1/PB60 ovaries (Fig3D). Finally, Northern blot analysis revealed that Su(Ste)-4 piRNA production in testis was significantly compromised in Aub mutants but remained at comparable level to the control in pelo-RNAi mutants (Fig3E). Taken together, these data agree with the global RNA profiling analysis and suggest that Pelo does not regulate the biogenesis of small RNAs, including piRNAs and miRNAs. In addition, Pelo does not seem to generally regulate the levels of mRNAs. As shown previously, the mRNA expression of many germline-specific genes was largely unaltered in pelo mutants (Fig2A). We also examined a set of housekeeping genes (polymerase genes), and their mRNA expression was virtually unaltered or within twofold of up-regulation in pelo mutants (Fig EV3).
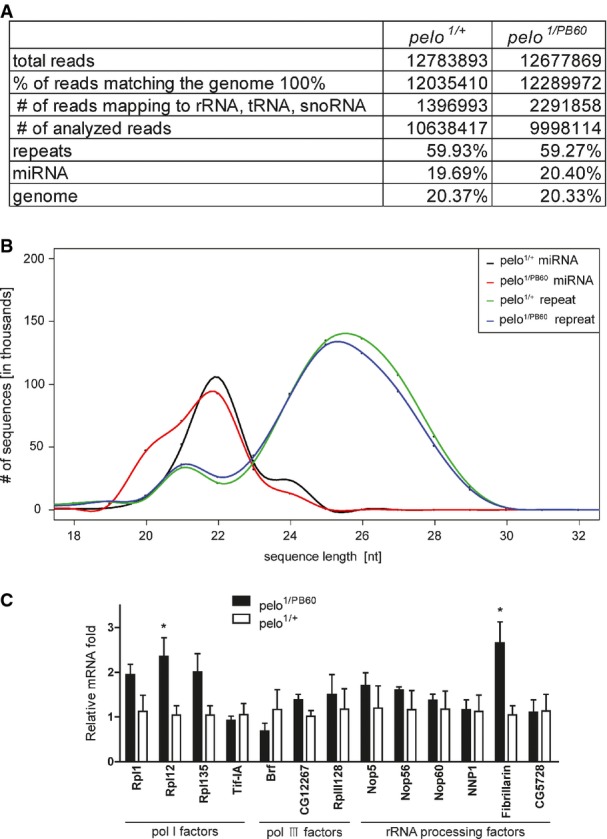
RNA profiles in wild-type and pelo mutant ovaries
- Reads analysis of total small RNAs from pelo1/PB60 and pelo1/+ ovaries.
- Small RNA size profiles correspond to genome-matching reads after excluding rRNA, tRNA, and snoRNA.
- Quantitative RT–PCR analysis of the expression of a set of housekeeping genes in pelo mutant ovaries. Values are means ± SEM, n = 3. *P < 0.05, t-test.
Figure 3.
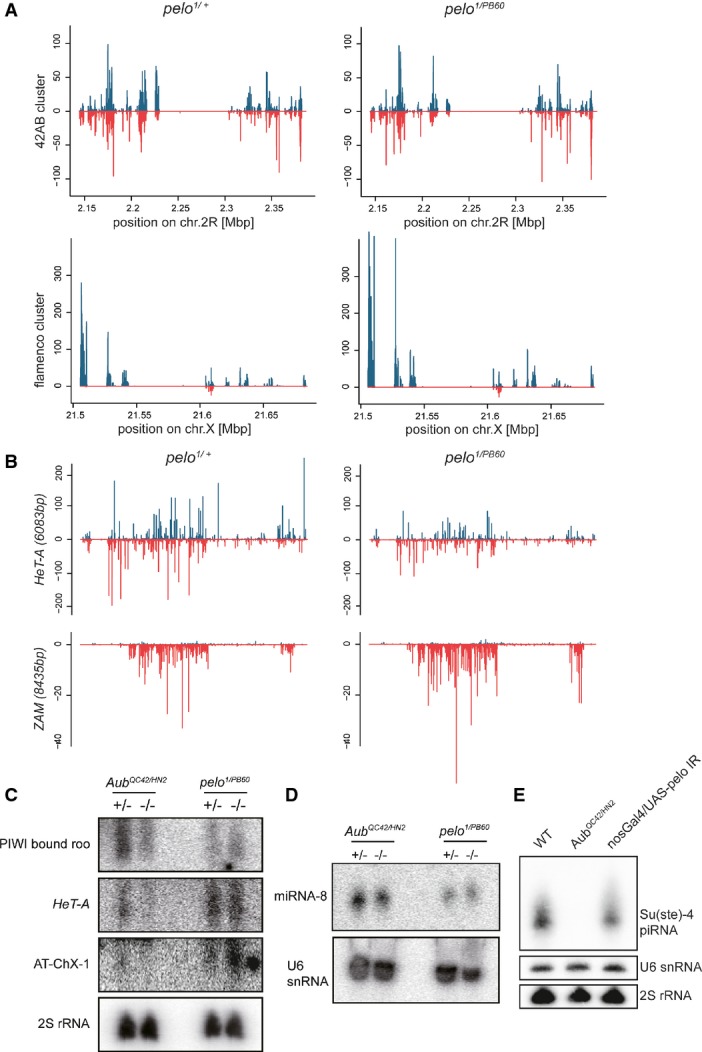
Small RNA biogenesis is not disrupted in pelo mutant ovaries
- A Normalized and calibrated piRNA density profiles mapped to 42AB and flamenco cluster in pelo1/PB60 and pelo1/+ ovaries. Plus and minus strands are shown in blue and red, respectively.
- B piRNA density profiles mapped to HeT-A and ZAM transposon loci. Plus and minus strands are shown in blue and red, respectively.
- C–E Small RNAs extracted from ovaries (C, D) or testes (E) of the indicated genotypes were hybridized with probes complimentary to roo, HeT-A, AT-chX-1 piRNAs (C), miRNA-8 (D), and Su(Ste)-4 piRNA (E). 2S rRNA and U6 snoRNA were used as loading controls.
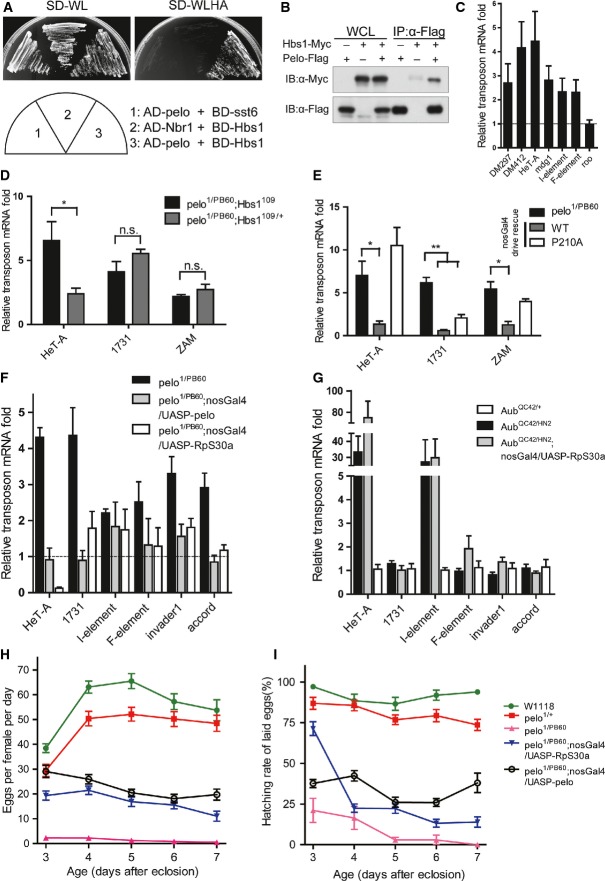
In yeast, Dom34/Pelo complexes with Hbs1 in the NGD process to regulate endonucleolytic cleavage of mRNAs in stalled ribosomes during translational elongation 14. We found that in Drosophila, Pelo displayed abilities to interact with the Drosophila Hbs1 (CG1898) in the yeast two-hybrid assay (Fig4A). Furthermore, Pelo co-immunoprecipitated with Hbs1 in ovarian extracts (Fig4B). To determine whether Pelo functions in a complex with Hbs1 in regulating TE silencing in the Drosophila germline, we generated a Hbs1 null Drosophila strain. Ovaries from Hbs1 mutants showed a moderate but significant increase in TE expression levels compared to the wild-type controls (Fig4C). In addition, mutation of Hbs1 significantly enhanced the up-regulation of HeT-A element expression caused by pelo mutation, although it did not further increase 1731 or ZAM expression (Fig4D). In yeast NGD pathway, mutational analysis suggests that Hbs1 has a much minor role in the process compared to the requirement for Dom34, possibly due to the redundant functions by other eRF3-like proteins 14. Structural and biochemical data have shown that the conserved PGF motif in the central domain of Pelo/Dom34 interacts with a conserved RDF motif in the GTPase domain of Hbs1. The PGF motif is also required for efficient NGD activity as a P to A mutation in this motif, which does not completely abolish the interaction 16, reduces NGD activity 25,26. We therefore generated a transgene for Pelo carrying P210 to A (P210A) mutation in the PGF motif and examined the ability of the mutant Pelo in rescuing the observed defects in pelo1/PB60 ovaries. Indeed, the P210A mutant Pelo had significantly reduced ability to repress TE levels (Fig4E). These observations suggest that Pelo might form a complex with Hbs1 (and possibly other eRF3-like proteins) in the Drosophila germline to regulate transposon silencing.
Figure 4.
Pelo possibly functions at the level of translation in transposon silencing
- A Pelo and Hbs1 interaction was detected by the yeast two-hybrid assay.
- B Co-immunoprecipitation of Pelo with Hbs1 in ovarian extracts.
- C–G Quantitative RT–PCR to detect the relative amount of transposon mRNAs from ovaries of the indicated genotypes. Fold changes were compared to heterozygous controls. (C) Transposon mRNAs were mildly increased in Hbs1−/− ovaries at 29°C. (D) Hbs1 mutation further enhanced HeT-A up-regulation in pelo mutant ovaries. (E) Transgene expression of Pelo (P210A) could not prevent HeT-A and ZAM up-regulation in pelo mutant ovaries. (F, G) Transgene expression of RpS30a reduced TE levels in pelo1/PB60 but not AubQC42/HN2 (G) ovaries. Values are means ± SEM, n = 3–5. n.s., not significant; *P < 0.05, **P < 0.01, t-test.
- H, I Germline expression of RpS30a partially restored egg production (H) and hatching rate (I) of pelo1/PB60 females, similar to the effect caused by germline expression of pelo. Values are means ± SEM, n = 16–40 females.
In yeast, RpS30a, a ribosomal protein, has been identified as a high-copy suppressor of the growth defect caused by Dom34 ablation 27. Interestingly, overexpression of RpS30a is able to complement the NGD defects and allow some cleavage of NGD substrates in a Dom34 strain 25. The underlying mechanism is unclear, but it has been speculated that extra RpS30a might extend the elongation pausing allowed for mRNA cleavage 25. We generated a transgenic fly expressing the Drosophila RpS30a and found that RpS30a overexpression in the germline significantly reduced the TE levels in pelo−/− ovaries, an effect that is largely similar to overexpression of Pelo (Fig4F). By contrast, overexpression of RpS30a failed to reduce the TE levels in AubQC42/HN2 ovaries (Fig4G). Germline overexpression of RpS30a also improved the egg production (Fig4H) and the hatching rate of pelo females, similar to the germline expression of Pelo (Fig4I). Taken together, these data indicate that Pelo might regulate transposon silencing at the translational level, possibly through the NGD pathway to cleave TE transcripts that have been loaded with ribosomes, thereby preventing translation.
Studies with artificially expressed mRNAs have shown that the Dom34-Hbs1 complex can detect and dissociate stalled ribosomes with no-stop mRNAs 14. But the in vivo targets are largely unknown. A recent study also suggests that Dom34 does not generally dissociate ribosomes on coding sequence but on 3′ UTRs 28. Because TE transcripts loaded on the ribosomes could still be potentially recognized by complementary piRNAs, one possibility is that the binding of piRNAs to the transcript may produce no-go mRNAs, followed by endonucleolytic cleavage via NGD. If this is true, Pelo-mediated TE silencing must rely on proper piRNA biogenesis. In Ago3- and Aub-depleted ovaries, where the piRNA biogenesis is reduced, we found that mutation in pelo was able to further enhance Het-A and 1731 up-regulation (FigEV4). In addition, depletion of Pelo could still further enhance TE up-regulation by additionally depleting Dcr2 (FigEV4), which generates esiRNAs that are implicated in TE silencing in somatic cells 29,30. Because disrupting the ping-pong cycle may reduce, but not eliminate the piRNA production, it remains undetermined whether Pelo-mediated TE silencing is dependent or independent of piRNAs.
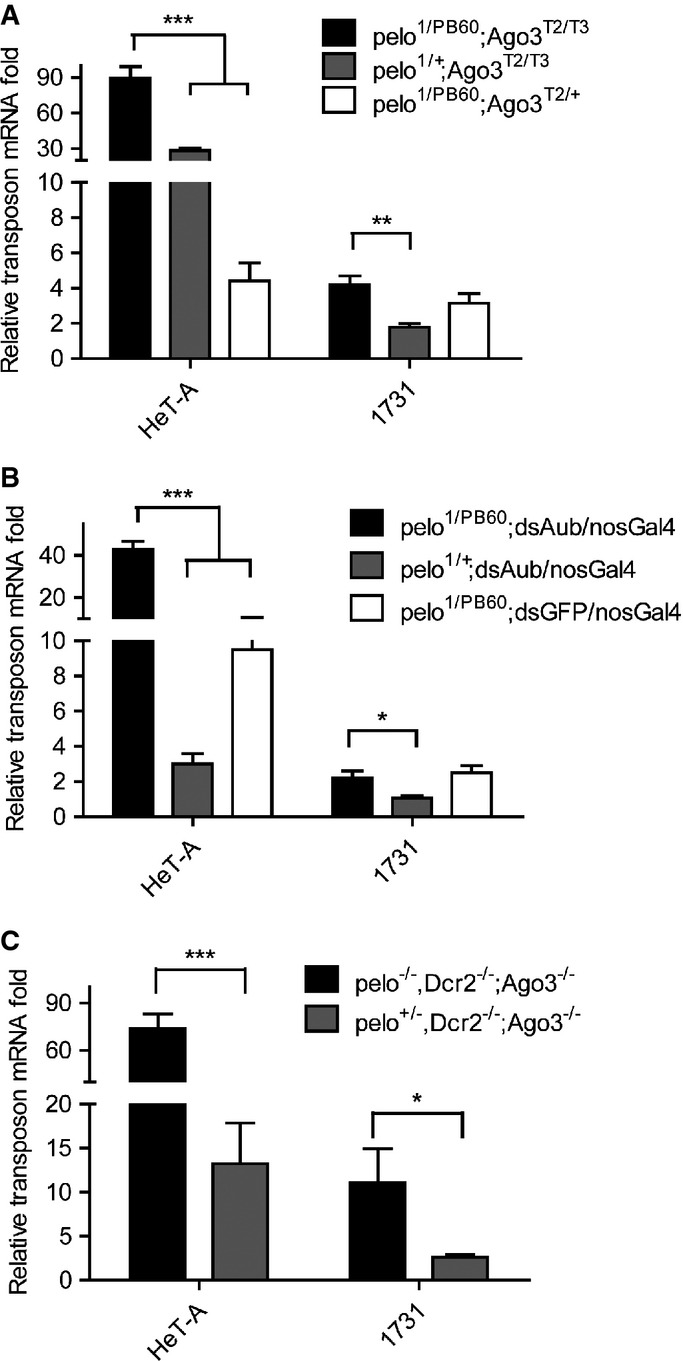
Pelo-mediated TE silencing in Ago3 or Aub mutant ovaries
- A–C Quantitative RT–PCR to detect the relative amount of transposon mRNAs from ovaries of the indicated genotypes. All relative fold changes were compared to the heterozygous controls. Pelo depletion further enhances TE up-regulation in Ago3 mutant ovaries (A), Aub RNAi ovaries (B) and Ago3 and Dcr2 double-mutant ovaries (C). Values are means ± SEM, n = 4. *P < 0.05, **P < 0.001, ***P < 0.0001, t-test.
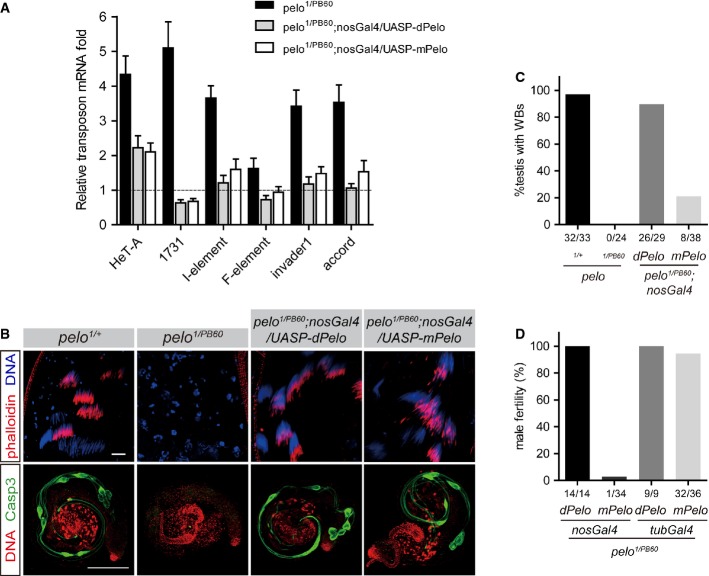
RNA surveillance mediated by Dom34-Hbs1 appears to be an evolutionarily conserved mechanism from yeast to human 14,18. We found that expression of the mouse Pelo (mPelo) gene in Drosophila was able to rescue the observed defects found in pelo1/PB60 mutants, including TE depression (Fig5A) and defects in spermatogenesis (Fig5B and C). Ubiquitous expression but not germline-restricted expression of mPelo effectively rescued the sterility of pelo1/PB60 mutants (Fig5D), suggesting that the function of Pelo in transposon silencing could be conserved in mammals. On another note, insufficient rescue by the germline-restricted expression of Pelo indicates a role for Pelo in somatic cells as well.
Figure 5.
Rescue of pelo mutant phenotypes by the mouse Pelo (mPelo) transgene
- Quantitative RT–PCR results of transposon mRNAs from ovaries of the indicated genotypes. Expression of mouse Pelo (mPelo) significantly reduced TE up-regulation in pelo1/PB60 ovaries. Values are means ± SEM, n = 5.
- Rescuing spermatogenesis by transgene expression of mPelo in pelo1/PB60 testes. Immunostaining of individualization complex (IC) in testes with indicated genotypes (upper panels). IC was visualized by phalloidin staining (red). Immunostaining of anti-caspase-3 (green) in testes with indicated genotypes (lower panels). Normally, individual IC travels along the axonemes (visualized by anti-caspase-3) to remove cytoplasmic content, which accumulates in the waste bags (WB, green) at the apical end of testes.
- Quantitative data of WB formation in testes of the indicated genotypes.
- Fertility test of male flies with the indicated genotypes.
Our studies here suggest an interesting hypothesis that in addition to the transcriptional and posttranscriptional levels of TE silencing by Piwi proteins and piRNAs, the mRNA surveillance complex Pelo-Hbs1 may provide another level of defense against TE transcripts that have escaped from the piRNA pathway-mediated endonucleolytic cleavage. One plausible mechanism would be that these escaped transcripts are loaded with ribosomes for translation, but the NGD machinery is able to recognize them, followed by degradation. How the loaded TE mRNAs are recognized by the NGD machinery is unclear, but the involvement of piRNAs remains to be a possibility worthy of future investigation. Given that TEs are particularly active in the germline, the involvement of multiple levels of defense against TEs in the germline ensures the integrity of genetic information to be passed to the next generation. We propose that the Pelo-Hbs1 RNA surveillance complex as another gatekeeper against the expression of transposons could be a common feature in the metazoan germline.
Materials and Methods
Additional methods are described in the Appendix file.
Drosophila strains
Flies were cultured on standard food media with yeast paste added to the food surface. The culture temperature was 25°C unless otherwise noted. Strains used in this study were as follows: Ago3T2, Ago3T3, AubQC42, and AubHN2 (gifts from Phillip Zamore); pelo1 is a P-element insertional allele 23, and RT–PCR analysis suggests that this allele produces a C-terminal truncated product. peloPB60 and peloPA13 are insertion and deletion alleles, respectively, generated by imprecise excision of P-element insertions: peloPB60 has a 107-base-pair deletion in the 2nd exon which creates an early stop codon and can be considered as a genetic null allele; peloPA13 has a 558-base-pair insertion in the 5′ UTR, and RT–PCR analysis suggests that the mutation causes approximately 80% reduction of gene product (see FigEV1); Hbs1109 (a null allele, details will be described elsewhere); nos-Gal4VP16 31; UAS-pelo-RNAi (VDRC #34770, #34771); and tub-Gal4 (BDSC #5138).
Generation of transgenic flies
To make a UASp-RpS30a construct, the RpS30a cDNA was cloned from a w1118 ovarian cDNA library and then subcloned into a pUASP vector with KpnI and XbaI. Mouse pelo cDNA from C57BL/6 mice (The Jackson Laboratory) was cloned into a pUASp vector with KpnI and XbaI. The Gateway cloning technology (Invitrogen) was used to generate UASp-pelo-GFP, UASp-pelo-Flag, and UASp-Myc-Hbs1 constructs. The P210A point mutant was generated by QuickChange Site-Directed Mutagenesis Kit (Stratagene). All the plasmids were verified by DNA sequencing. The plasmid DNA was introduced to w1118 embryo by a standard procedure to generate transgenic flies.
Immunostaining and microscopy
Drosophila ovaries were dissected and immunostained as described previously 32. The following primary antibodies were used: anti-Piwi, anti-Ago3, anti-Aub (1:1,000, gifts from Gregory J. Hannon), anti-Krimp (1:10,000, a gift from Toshie Kai), anti-Vasa (DSHB, 1:100), anti-Stellate (1:1,000, a gift from William E. Theurkauf), anti-I-element ORF1p (1:50, a gift from David Finnegan), anti-β-gal (DSHB, 1:50), and anti-cleaved caspase-3 (Cell Signaling, 1:300). For IC staining, fixed testes were incubated with rhodamine-conjugated phalloidin (Molecular Probes, 1:10) at 37°C for 1 h. Secondary antibodies, including goat anti-rabbit and goat anti-mouse IgGs, conjugated to Alexa (488 or 568) (Molecular Probes) were used at a dilution of 1:300 (DAPI (4′,6-diamidino-2-phenylindole), Sigma; 0.1 mg/ml, 5 min incubation). Images were collected by either a Zeiss Meta 510 confocal microscope system or a Zeiss Imager Z1 equipped with an ApoTome. All acquired images were processed in Adobe Photoshop and Illustrator.
RNA isolation and qPCR assays
Total RNA from 10-20 ovaries was extracted by TRIzol reagent (Invitrogen). After DNase treatment, complementary DNA (cDNA) was synthesized using an oligo dT primer and High-Fidelity cDNA Synthesis Kit (Roche). RT–qPCR was performed in three duplicates using SYBR Premix Ex Taq RT-PCR Kit (Takara) on an ABI PRISM 7500 fast Real-time PCR System (Applied Biosystems). Endogenous Actin5c mRNA levels were measured for normalization. Fold changes for mRNA were calculated using ΔΔCt method 33. The primers used are listed in Table EV1.
Small RNA isolation and Northern blot
Small RNA was enriched by PEG8000 precipitation from total ovaries RNA, and Northern blot was performed according to Qi et al 34. 32P-end-labeled oligonucleotides complimentary to small RNA sequence were used as probes which are listed in Table EV1.
Fertility test
To test female fertility, two virgins were collected and mated with two 5- to 7-day-old w1118 males in a small cage with 35-mm apple juice agar plate with yeast paste as described 35. After 2 days, the plate was replaced by a new one every day. The number of eggs per female per day was counted every 24 h, and the hatching rate was scored 48 h after the plate was changed. At least eight plates were scored in total for each genotype at each time point. To test male fertility, each 2- to 5-day-old male was collected and mated with three 3- to 7-day-old w1118 virgins. Fertility was determined by examining the appearance of larvae 5 days after mating.
Small RNA cloning and sequencing
Ten micrograms enriched small RNA were separated on a 15% denaturing polyacrylamide gel.
18- to 30-nt RNAs were purified according to RNA oligo markers and ligated to adapters to generate the libraries for subsequent sequencing on the Illumina GA II instrument. The small RNA-seq library was prepared by following the manufacturer’s instructions.
The GEO accession code for the RNA-seq data is GSE69468.
Statistical analysis
Data are presented as mean ± SEM. P-values were calculated using one-way ANOVA or unpaired two-tailed Student’s t-test by GraphPad Prism 5 (GraphPad Software Inc.).
Acknowledgments
We thank Phillip Zamore, Greg Hannon, David Finnegan, Toshi Kai, William Theurkauf, the Bloomington Drosophila Stock Center, and Vienna Drosophila Resource Center for providing fly stocks and reagents; Xijuan Li and Anying Kang for technical assistance; and members of the Xi Laboratory for helpful comments and discussion. This work was supported by National Basic Science 973 grants (2011CB812700 and 2014CB849700) from the Chinese Ministry of Science and Technology.
Author contributions
FY, RZ and RX conceived and designed the experiments, FY and RX analyzed the data and wrote the manuscript. FY, RZ, XF, HH, YX, YM, HC, TC, and YQ performed the experiments and analysis.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Appendix
Expanded View Figures PDF
Table EV1
Review Process File
References
- Guzzardo PM, Muerdter F, Hannon GJ. The piRNA pathway in flies: highlights and future directions. Curr Opin Genet Dev. 2013;23:44–52. doi: 10.1016/j.gde.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Hammell M, Hannon GJ. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013;27:400–412. doi: 10.1101/gad.209767.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H. A major epigenetic programming mechanism guided by piRNAs. Dev Cell. 2013;24:502–516. doi: 10.1016/j.devcel.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci USA. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J Biol Chem. 2009;284:6507–6519. doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Jin Z, Flynt AS, Lai EC. Drosophila piwi mutants exhibit germline stem cell tumors that are sustained by elevated Dpp signaling. Curr Biol. 2013;23:1442–1448. doi: 10.1016/j.cub.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Wang S, Do T, Song X, Inaba M, Nishimoto Y, Liu LP, Gao Y, Mao Y, Li H, et al. Piwi is required in multiple cell types to control germline stem cell lineage development in the Drosophila ovary. PLoS ONE. 2014;9:e90267. doi: 10.1371/journal.pone.0090267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi R, Doan C, Liu D, Xie T. Pelota controls self-renewal of germline stem cells by repressing a Bam-independent differentiation pathway. Development. 2005;132:5365–5374. doi: 10.1242/dev.02151. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Armache JP, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. Structure of the no-go mRNA decay comple x Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- Chen L, Muhlrad D, Hauryliuk V, Cheng Z, Lim MK, Shyp V, Parker R, Song H. Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nat Struct Mol Biol. 2010;17:1233–1240. doi: 10.1038/nsmb.1922. [DOI] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell. 2009;34:440–450. doi: 10.1016/j.molcel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, Inada T. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol Cell. 2012;46:518–529. doi: 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- Saito S, Hosoda N, Hoshino S. The Hbs1-Dom34 protein complex functions in non-stop mRNA decay in mammalian cells. J Biol Chem. 2013;288:17832–17843. doi: 10.1074/jbc.M112.448977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Eberhart CG, Wasserman SA. The pelota locus encodes a protein required for meiotic cell division: an analysis of G2/M arrest in Drosophila spermatogenesis. Development. 1995;121:3477–3486. doi: 10.1242/dev.121.10.3477. [DOI] [PubMed] [Google Scholar]
- Kotelnikov RN, Klenov MS, Rozovsky YM, Olenina LV, Kibanov MV, Gvozdev VA. Peculiarities of piRNA-mediated post-transcriptional silencing of Stellate repeats in testes of Drosophila melanogaster. Nucleic Acids Res. 2009;37:3254–3263. doi: 10.1093/nar/gkp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos DO, Doma MK, Shoemaker CJ, Muhlrad D, Green R, Weissman J, Hollien J, Parker R. Analysis of Dom34 and its function in no-go decay. Mol Biol Cell. 2009;20:3025–3032. doi: 10.1091/mbc.E09-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Elzen AM, Henri J, Lazar N, Gas ME, Durand D, Lacroute F, Nicaise M, van Tilbeurgh H, Seraphin B, Graille M. Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways. Nat Struct Mol Biol. 2010;17:1446–1452. doi: 10.1038/nsmb.1963. [DOI] [PubMed] [Google Scholar]
- Davis L, Engebrecht J. Yeast dom34 mutants are defective in multiple developmental pathways and exhibit decreased levels of polyribosomes. Genetics. 1998;149:45–56. doi: 10.1093/genetics/149.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, Green R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- Li X, Han Y, Xi R. Polycomb group genes Psc and Su(z)2 restrict follicle stem cell self-renewal and extrusion by controlling canonical and noncanonical Wnt signaling. Genes Dev. 2010;24:933–946. doi: 10.1101/gad.1901510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Zhao R, Xuan Y, Li X, Xi R. Age-related changes of germline stem cell activity, niche signaling activity and egg production in Drosophila. Aging Cell. 2008;7:344–354. doi: 10.1111/j.1474-9726.2008.00379.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Review Process File