Abstract
The mammalian target of rapamycin complex 1 (mTORC1) is a central regulator of physiological adaptations in response to changes in nutrient supply. Major downstream targets of mTORC1 signalling are the mRNA translation regulators p70 ribosomal protein S6 kinase 1 (S6K1p70) and the 4E-binding proteins (4E-BPs). However, little is known about vertebrate mRNAs that are specifically controlled by mTORC1 signalling and are engaged in regulating mTORC1-associated physiology. Here, we show that translation of the CCAAT/enhancer binding protein beta (C/EBPβ) mRNA into the C/EBPβ-LIP isoform is suppressed in response to mTORC1 inhibition either through pharmacological treatment or through calorie restriction. Our data indicate that the function of 4E-BPs is required for suppression of LIP. Intriguingly, mice lacking the cis-regulatory upstream open reading frame (uORF) in the C/EBPβ-mRNA, which is required for mTORC1-stimulated translation into C/EBPβ-LIP, display an improved metabolic phenotype with features also found under calorie restriction. Thus, our data suggest that translational adjustment of C/EBPβ-isoform expression is one of the key processes that direct metabolic adaptation in response to changes in mTORC1 activity.
Keywords: C/EBPβ, calorie restriction, metabolism, mTORC1, translation
See also: V Albert & MN Hall (August 2015)
Introduction
C/EBPβ is a transcriptional regulator with a broad tissue expression including liver and adipose tissue (http://www.genecards.org). It controls genes related to glucose and fat metabolism as well as other cellular processes 1,2. The Cebpb gene is intronless, and from its mRNA three different protein isoforms are expressed through usage of alternative translation initiation sites (FigEV1A). The isoforms LAP* and LAP (liver activating protein) are transcriptional activators that consist of transactivation domains and a DNA-binding domain. The truncated isoform LIP (liver inhibitory protein) lacks the N-terminal transactivation domains but still possesses the DNA-binding domain. LIP can therefore act as a competitive inhibitor of LAP*/LAP function 3. However, LIP may also have additional and distinct functions. Hence, the ratio between LAP and LIP is crucial for the biological functions elicited by C/EBPβ. Translation from both the LAP* and LAP AUG codons is achieved by regular translation initiation, although translation into LAP* is often weaker since this AUG codon lacks a Kozak consensus sequence required for efficient recognition by the ribosome 4,5. Expression of LIP from a distal initiation codon depends on a cis-regulatory uORF located in the 5′ UTR of the C/EBPβ-mRNA. The limited size of the uORF allows the small ribosomal subunit to remain attached to the mRNA after translation termination and to resume scanning along the mRNA. After reloading of the ribosomal complex with initiator tRNA, translation of LIP from the downstream initiation codon can be re-initiated. Mutation of the uORF consequently results in diminished LIP expression 4,5 (see also schematic representation in FigEV1A).
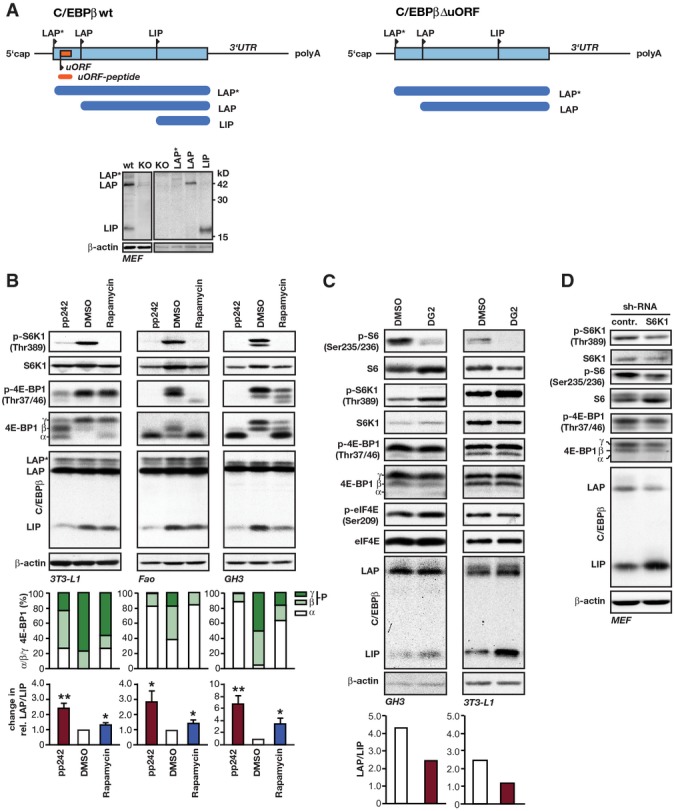
Analyses of LAP/LIP C/EBPβ-isoform expression and the mTORC1 signalling pathway
- Schematic view of C/EBPβ wt (left) and ΔuORF (right) mRNA structure and translated isoforms LAP*, LAP and LIP as indicated. The immunoblots at the left show wt MEFs and KO MEFs derived from C/EBPβ KO mice and at the right KO MEFs ectopically expressing empty vector control, LAP* (weakly expressed in MEFs), LAP or LIP. β-actin was used as a loading control.
- Immunoblots of extracts from 3T3-L1 mouse adipocytes (24-h treatment), Fao rat hepatoma cells (6-h treatment) and GH3 rat pituitary cells (24-h treatment) treated with the pan-mTOR inhibitor pp242 (1 μM) or the allosteric mTORC1 inhibitor rapamycin (1 μM) compared to solvent (DMSO) showing phosphorylation (p-) in relation to total levels of the indicated proteins. β-actin was used as a loading control. Upper bar graphs show quantification of percentages of 4E-BP1 α- (hypophosphorylated), β- and γ-bands (hyperphosphorylated) of the pan-4E-BP1 blot shown. The lower bar graphs show quantification of the relative changes in LAP/LIP-isoform ratio by pp242 or rapamycin compared to solvent (3T3-L1, n = 4; Fao, n = 6; GH3, n = 3). All values are mean ± SEM. P-values were determined with Student’s t-test, *P < 0.05; **P < 0.01.
- Immunoblots from extracts of GH3 and 3T3-L1 cells treated with the S6K1 inhibitor DG2 (20 μM) or solvent for 24 h. Phosphorylation (p-) in relation to total protein levels of indicated proteins is shown. β-actin was used as a loading control. The bar graph shows quantification of the relative change in LAP/LIP-isoform ratio by DG2 compared to solvent (n = 1).
- Immunoblots from extracts of MEFs after retroviral transduction with control-sh or S6K1-sh expression vector showing phosphorylation (p-) in relation to total levels of the indicated proteins. β-actin was used as a loading control (n = 1).
Data information: Quantification of the LAP/LIP C/EBPβ-isoform ratios was done from X-ray films for (B) and by chemiluminescence digital imaging for (C).
Reducing signalling through mTORC1 by pharmacological treatment, mutations, restricted calorie intake or low protein:carbohydrate macronutrient ratio enhances metabolic health and increases life span in many species up to mammals. On the contrary, hyper-activation of mTORC1 is believed to promote metabolic disorders resulting from overfeeding such as diabetes 6-8,9-11,12-14,15. Our earlier studies pointed to an involvement of mTORC1 signalling in the regulation of C/EBPβ-LIP expression, since mTORC1 inhibition by rapamycin reduced LIP expression in a uORF-dependent manner 4,16.
Here, we show that interventions that cause a reduction in mTORC1 signalling also decrease the translation of the C/EBPβ-LIP isoform. Genetic elimination of the mTORC1-sensitive uORF of the C/EBPβ-mRNA in mice similarly results in a reduction of the LIP isoform 5. Our data show that mice with this mutation display an improved metabolic phenotype, including reduced fat accumulation and increased β-oxidation, improved insulin sensitivity and glucose tolerance as well as enhanced activity. Thus, pharmacological targeting of C/EBPβ-isoform expression may provide a promising strategy for the treatment of metabolic diseases such as obesity and type II diabetes, thereby extending health span.
Results
mTORC1 controls C/EBPβ-LIP expression
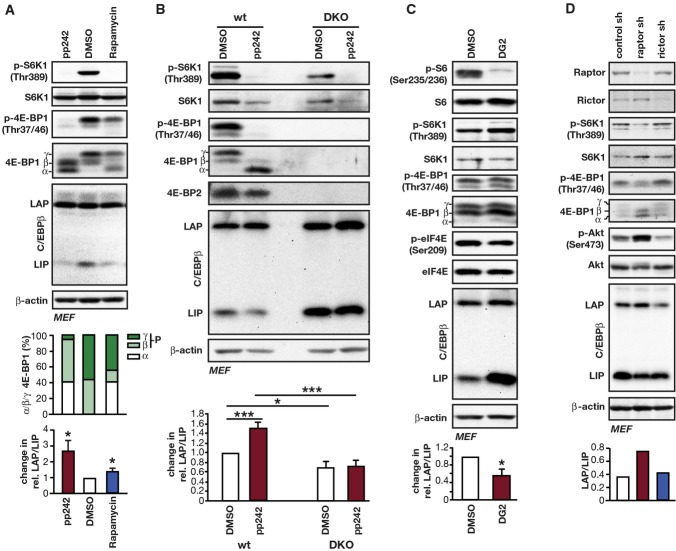
To clarify the regulation of C/EBPβ-isoform expression by mTORC1, we treated different cell lines with the catalytic pan-mTOR inhibitor pp242 or the allosteric mTOR inhibitor rapamycin. Rapamycin primarily acts on mTORC1 but was shown to also affect mTORC2 after prolonged treatment 17. Treatment with pp242 resulted in a strong reduction in LIP levels without affecting LAP expression in all cell lines tested (Figs1A and EV1B). pp242 treatment consistently resulted in strong de-phosphorylation of the mTORC1 targets S6K1 (Thr389) and 4E-BP1 (Thr37/46), which was analysed with phospho-specific antibodies. Reduced phosphorylation of 4E-BP1 is also visible in the pan-4E-BP1 immunoblots as a decrease in γ/β-phosphorylation signals and an increase in α-hypophosphorylation signals. Treatment with rapamycin strongly reduced S6K1 phosphorylation in all cases. However, the effect on 4E-BP1 phosphorylation and LIP levels was weaker and varied between different cell types (Figs1A and EV1B). The correlation between 4E-BP1 phosporylation state and LIP expression levels suggests that decreased phosphorylation of 4E-BP1 and the resulting restrain of eukaryotic initiation factor 4E (eIF4E) are important for reducing LIP expression through mTORC1 inhibition in these cells. To discriminate between effects on C/EBPβ-isoform expression by 4E-BPs or S6K1, we used 4E-BP1/4E-BP2 double knockout (4E-BP DKO) MEFs (mouse embryonic fibroblasts), or MEFs treated with either the S6K1 inhibitor DG2 or an sh-RNA targeting S6K1. It has been shown that 4E-BP DKO MEFs contain more accessible eIF4E 18,19. In 4E-BP DKO cells, expression of LIP was strongly increased compared to control cells. This was concomitant with a lower LAP/LIP ratio, which could not be reversed by pp242 treatment (Fig1B). Treatment with the S6K1 inhibitor DG2 also did not reduce LIP expression in wt MEFs, GH3 or 3T3-L1 cells although S6K1 activity was completely abolished as shown by the lack of S6 (Ser235/236) phosphorylation (Figs1C and EV1C). On the contrary, LIP levels were enhanced by DG2 treatment. Similarly, S6K1 knockdown did not reduce but rather stimulated LIP expression (FigEV1D) by a yet to be identified mechanism. Therefore, our data indicate that mTORC1 inhibition decreases LIP expression rather through 4E-BPs than through S6K1. Since pp242 inhibits both mTORC1 and mTORC2 20, we employed a knockdown of either the mTORC1-specific component raptor or the mTORC2-specific component rictor to clarify their involvement in the regulation of LIP expression. Knockdown of raptor resulted in decreased LIP expression (higher LAP/LIP ratio) and a concomitant decrease in phosphorylation of S6K1 and 4E-BP1, while knockdown of rictor reduced the rictor-mTORC2-sensitive Ser473-Akt phosphorylation but did not change the LAP/LIP ratio, although we observed reduced expression levels of all protein isoforms (Fig1D). These data demonstrate that only mTORC1 specifically regulates the ratio of LAP/LIP expression.
Figure 1.
Regulation of LAP/LIP C/EBPβ-isoform expression through the mTORC1 signalling pathway
- Immunoblots of extracts from MEFs treated with the pan-mTOR inhibitor pp242 (1 μM) or the allosteric mTORC1 inhibitor rapamycin (1 μM) compared to solvent (DMSO) for 12 h showing phosphorylation (p-) in relation to total levels of the indicated proteins. Upper bar graph shows quantification of percentages of 4E-BP1 α- (hypophosphorylated), β- and γ-bands (hyperphosphorylated) of the pan-4E-BP1 blot shown. The lower bar graphs show quantification of the relative changes in LAP/LIP-isoform ratio by pp242 or rapamycin compared to solvent (n = 4 independent experiments).
- Immunoblots of extracts from wt and 4E-BP DKO MEFs treated with pp242 (1 μM) or solvent (DMSO) for 12 h showing phosphorylation (p-) in relation to total protein levels as indicated. The bar graph shows quantification of the relative change in LAP/LIP-isoform ratio by pp242 compared to solvent (n = 4, independent experiments).
- Immunoblots of extracts from MEFs treated with the S6K1 inhibitor DG2 (20 μM) or solvent (DMSO) for 12 h. Phosphorylation (p-) in relation to total protein levels of indicated proteins is shown. The bar graph shows quantification of the relative change in LAP/LIP-isoform ratio by DG2 compared to solvent (n = 4, independent experiments).
- Immunoblots of extracts from MEFs with sh-RNA-mediated knockdown of raptor, rictor or control with detection of the indicated proteins and their phosphorylation (p-). The bar graph shows quantification of the relative changes in LAP/LIP-isoform ratio (n = 1).
Data information: LAP/LIP C/EBPβ-isoform ratios were quantified by chemiluminescence digital imaging or using ImageJ software from film scans. All values are mean ± SEM. P-values were determined with Student’s t-test, *P < 0.05; ***P < 0.005. β-actin was used as a loading control.
Limiting mTORC1 activity through inhibition by rapamycin or caloric restriction inhibits C/EBPβ-LIP expression in vivo
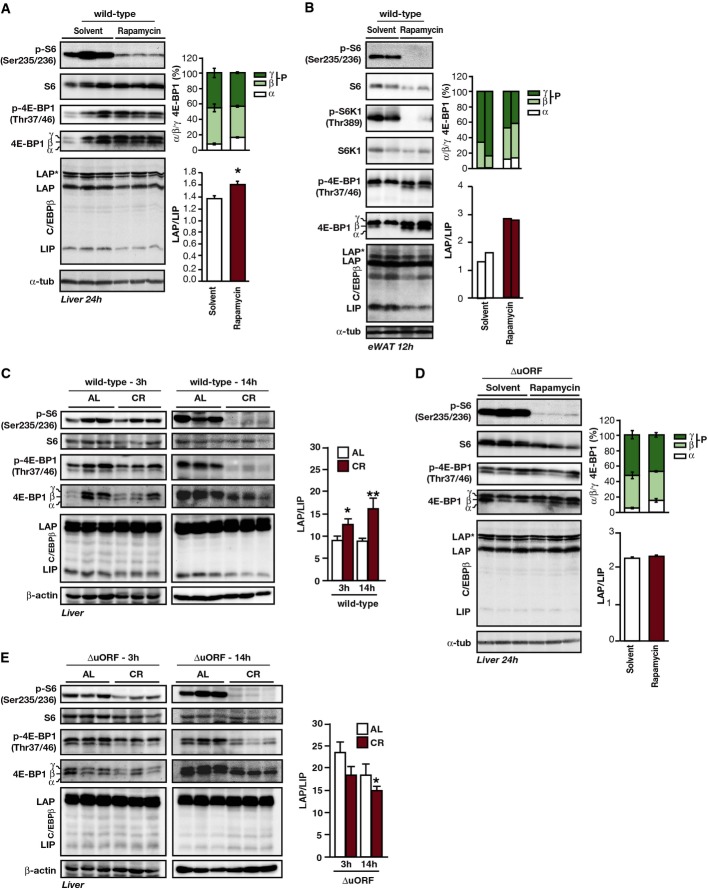
To verify mTORC1-dependent C/EBPβ-isoform expression in vivo, mice were injected intraperitoneally with rapamycin and analysed for expression of C/EBPβ on protein level. In liver (Fig2A) and epididymal white adipose tissue (eWAT) (Fig2B), treatment with rapamycin led to higher LAP/LIP ratios mainly resulting from decreased LIP expression. The efficacy of mTORC1 inhibition through treatment with rapamycin was shown by reduced phosphorylation of S6K1, S6 or 4E-BP1.
Figure 2.
The LAP/LIP C/EBPβ-isoform ratio in vivo is regulated by rapamycin or CR and is uORF dependent
- Immunoblots of extracts from livers of fed wt mice 24 h after i.p. injection of rapamycin (8 μg/g body weight) or solvent. Phosphorylation (p-) in relation to total protein levels of indicated proteins is shown. α-tubulin was used as a loading control. The upper bar graph shows quantification of percentages of 4E-BP1 α- (hypophosphorylated), β- and γ-bands (hyperphosphorylated) of the pan-4E-BP1 blot, and the lower bar graph shows quantification of the LAP/LIP-isoform ratio (n = 3).
- Immunoblots of extracts from epididymal (e)WAT of fed wt mice 12 h after i.p. injection of rapamycin (8 μg/g) or solvent showing phosphorylation (p-) of proteins in relation to total levels. α-tubulin was used as a loading control. The upper bar graph shows quantification of percentages of 4E-BP1 α- (hypophosphorylated), β- and γ-bands (hyperphosphorylated) of the pan-4E-BP1 blot, and the lower bar graph shows quantification of the LAP/LIP-isoform ratio (n = 2).
- Immunoblots of extracts from of wt mice either fed ad libitum (AL) or under caloric restriction (CR) for 4 weeks and sacrificed either 3 or 14 h past-feeding. Phosphorylation (p-) in relation to total protein levels of indicated proteins is shown. β-actin was used as a loading control. The bar graph shows quantification of LAP/LIP-isoform ratio (n = 3).
- Immunoblots of extracts from liver of fed C/EBPβΔuORF/BL6 mice 24 h after rapamycin (8 μg/g) or solvent injection (i.p.). Phosphorylation (p-) in relation to total levels of indicated proteins is shown. α-tubulin was used as a loading control. The upper bar graph shows percentages of 4E-BP1 α- (hypophosphorylated), β- and γ-bands (hyperphosphorylated) of the pan-4E-BP1 blot, and the lower bar graph shows quantification of the LAP/LIP-isoform ratio (n = 3).
- Immunoblots of extracts from of C/EBPβΔuORF/BL6 mice either fed ad libitum (AL) or under caloric restriction (CR) for 4 weeks and sacrificed either 3 or 14 h past-feeding. The phosphorylation (p-) in relation to total protein levels is shown. β-actin was used as a loading control. The bar graph shows quantification of LAP/LIP-isoform ratio (n = 3).
Data information: Quantification of the C/EBPβ LAP/LIP-isoform ratio of the blots was done by chemiluminescence digital imaging for the CR experiment or from X-ray films for rapamycin and is shown as bar graphs. All values are mean ± SEM. P-values were determined with Student’s t-test, *P < 0.05; **P < 0.01.
The mTORC1 signalling pathway is an important regulator of metabolic adaptations in response to nutritional changes. Therefore, we examined mTORC1-mediated C/EBPβ regulation in adult wt mice fed either ad libitum (AL) or caloric restricted (CR) for 4 weeks. As described by 21, mice on a CR regime consume their daily single food allotment immediately followed by a prolonged period of absence of food (mice fed AL spread their food intake over the day). This results in a pronounced change in whole body fuel selection with an initial nutrition phase of high carbohydrate utilisation and a prolonged starvation phase of primarily fat utilisation that lasts until the next feeding 21. We verified the dynamics of fuel selection in AL and CR mice by calculating the respiratory exchange ratio (RER) between the amount of CO2 exhaled and O2 inhaled from mice kept individually in metabolic chambers (RER = VCO2/VO2 = 1.0 for pure carbohydrate usage; RER = 0.7 for pure fat usage) (see FigEV2A and legend for further explanation). This analysis allows covering these accentuated nutritional states for analysing mTORC1 activity and C/EBPβ-isoform expression. We chose 3 and 14 h after feeding as time points of analysis since they represent the maximal usage of carbohydrate or fat in CR fed mice, respectively. At 3 h post-feeding, mTORC1 activity was slightly reduced in livers from CR compared to mice fed AL as reflected by the levels of phosphorylated S6 and 4E-BP1 (Fig2C). This correlated with a moderate reduction in LIP levels and resulted in a slight change in the LAP/LIP ratio in the CR fed mice. However, in the starvation phase at 14 h, both mTORC1 activity and LIP expression were reduced to a higher extent in the CR fed mice compared to mice fed AL (Fig2C). Mice fed AL display more moderate diurnal cycles of fuel selection with a relative high mTORC1 activity and a LAP/LIP ratio that stays constant at 3 and 14 h (Fig2C). These data show that mTORC1 signalling alternates between the activated and suppressed state in liver during the diurnal cycle of fuel selection under CR. Furthermore, these data demonstrate that LIP expression levels follow the changes in mTORC1 activity also under these physiologically induced conditions.
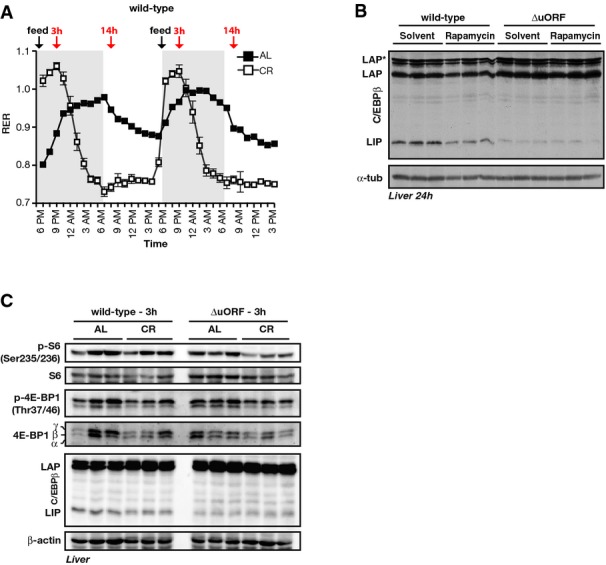
C/EBPβΔuORF/BL6 mice data
- Respiratory exchange ratio (RER) over 43 h of wt mice fed AL (black squares) or CR (open squares) for 4 weeks based on Oxymax measurements (n = 10). The dark phases are marked by grey boxes, and time of feeding is indicated as well as time points (3 and 14 h) used for immunoblotting shown in Fig2C and E. All values are mean ± SEM.
- Whole immunoblot as shown separately in Fig2A and D of extracts from livers of fed wt and C/EBPβΔuORF/BL6 mice 24 h after i.p. injection of rapamycin (8 &# x03BC;g/g body weight) or solvent. Phosphorylation (p-) in relation to total protein levels of indicated proteins is shown. α-tubulin was used as a loading control (n = 3).
- Whole immunoblot of extracts of livers from wt and C/EBPβΔuORF/BL6 mice (as shown separately in Fig2C und E) either fed ad libitum (AL) or under caloric restriction (CR) for 4 weeks, sacrificed 3 h past-feeding (6 p.m. for CR). Phosphorylation (p-) in relation to total protein levels of indicated proteins is shown. β-actin was used as a loading control, n = 3.
Taken together, our data show that expression of the C/EBPβ-LIP isoform correlates with mTORC1/4E-BP1 signalling in vitro and in vivo and follows mTORC1 activity upon changes in calorie supply or pharmacological inhibition of mTORC1.
Permanent reduction of C/EBPβ-LIP expression in C/EBPβΔuORF/BL6 mice enhances fat metabolism
To investigate whether a permanently altered C/EBPβ-isoform ratio affects metabolic performance, we analysed mice deficient in the cis-regulatory uORF that is required for LIP expression (C/EBPβΔuORF/BL6 mice; based on mice described in 5, but back-crossed in C57BL/6J background, see also FigEV1A). These mice display a diminished LIP expression that was not further reduced by rapamycin treatment (Figs2D and EV2B) or through caloric restriction (Fig2E). Thus, the C/EBPβΔuORF/BL6 mice with their invariably low LIP expression mimic reduced mTORC1 activity at the level of C/EBPβ translation. mTORC1 activity itself does not seem to be influenced by the C/EBPβΔuORF/BL6 mutation or the reduced LIP levels since neither S6 phosphorylation nor 4E-BP1 phosphorylation was noticeably altered in livers from C/EBPβΔuORF/BL6 mice compared to wt mice (3 h post-feeding) (Fig EV2C).
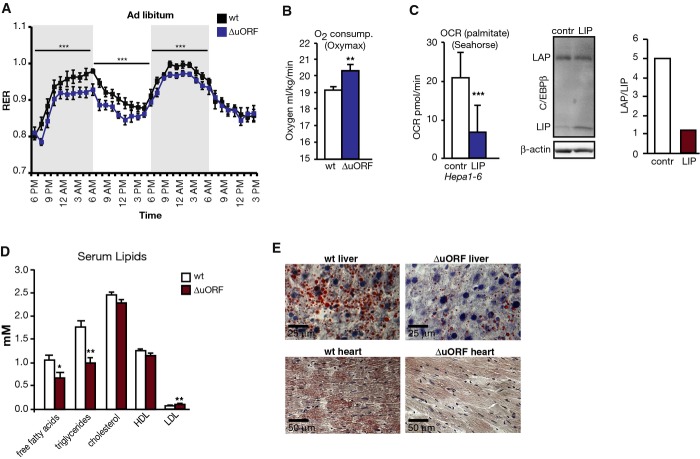
We found that C/EBPβΔuORF/BL6 mice fed AL displayed a reduced body weight compared to wt mice accumulating to a difference of 8% at 26 weeks of age (Fig3A). This was not due to changes in body length, food intake or caloric utilisation (Fig3B–D). C/EBPβΔuORF/BL6 mice even showed a slightly increased food intake. To investigate why the body weight was reduced, we performed an abdominal computed tomography (CT) to analyse body composition. The data demonstrated that both visceral and subcutaneous fat volumes were clearly reduced in C/EBPβΔuORF/BL6 mice compared to age- and sex-matched littermate controls (Fig3E), while there was no significant change in the lean body mass (Fig3F). Histological analyses of epididymal fat pads revealed an average reduction of white adipocyte cell size of 30% in C/EBPβΔuORF/BL6 male mice compared to wt control tissue (Fig3G). C/EBPβ is a known regulator of adipogenesis and induces transcription of the adipogenic transcription factors C/EBPα and PPARγ. mRNA expression levels of C/EBPα and PPARγ were similar in visceral adipose tissue of C/EBPβΔuORF/BL6 mice compared to control littermates. This indicates that the reduced fat accumulation in WAT is not caused by deficiencies of key adipogenic transcription factors (Fig EV3A). Notably, MEFs derived from C/EBPβΔuORF/BL6 mice displayed increased differentiation into adipocytes in cell culture compared to MEFs derived from wt mice, as was revealed by Oil Red O staining of lipid droplets (FigEV3B). Vice versa, experimental induction of LIP in 3T3-L1 adipocytes resulted in less efficient adipogenic differentiation and a reduction in fat accumulation (FigEV3C) reminiscent of what has been described before 4. In accordance with the lower fat content of C/EBPβΔuORF/BL6 adipocytes, adiponectin levels were increased (Fig3H), while leptin levels were unchanged (Fig EV3D) in the blood plasma of C/EBPβΔuORF/BL6 mice compared to wt controls. High levels of the adipocyte-derived hormone adiponectin correlate with increased fatty acid oxidation, reduced lipid accumulation in non-adipose tissues and increased insulin sensitivity 22-24. This prompted us to examine whether whole body fatty acid oxidation is increased in the C/EBPβΔuORF/BL6 mice by determination of the RER using metabolic cages. The RER curves of both C/EBPβΔuORF/BL6 and wt mice fed ad libitum reflected the diurnal rhythm with a higher RER in the active (dark) phase representing mostly carbohydrate usage and a lower RER in the resting (light) phase in which more fatty acids are oxidised. As shown in Fig4A, we measured continuously lower RER values both over the active and over the resting phases for C/EBPβΔuORF/BL6 mice compared to wt mice. Thus, C/EBPβΔuORF/BL6 mice have a moderate but significant daily increase in fatty acid oxidation over carbohydrate oxidation under normal feeding conditions. Furthermore, oxygen consumption of the C/EBPβΔuORF/BL6 mice was increased compared to wt mice, indicating that the C/EBPβΔuORF/BL6 mice have a higher energy expenditure (Fig4B). To examine whether altering the LAP/LIP ratio results in a cell intrinsic shift in β-oxidation, we ectopically expressed LIP in the mouse hepatoma cell line Hepa 1-6 and studied palmitate-substrate fatty acid oxidation (FAO) using the Seahorse FX extracellular flux analyser. Ectopic expression of LIP (low LAP/LIP ratio) resulted in a reduced FAO compared to FAO in the parent cells (high LAP/LIP ratio) (Fig4C). Hence, the LAP-/LIP-associated β-oxidation changes found in cell culture support the phenotype found in C/EBPβΔuORF/BL6 mice.
Figure 3.
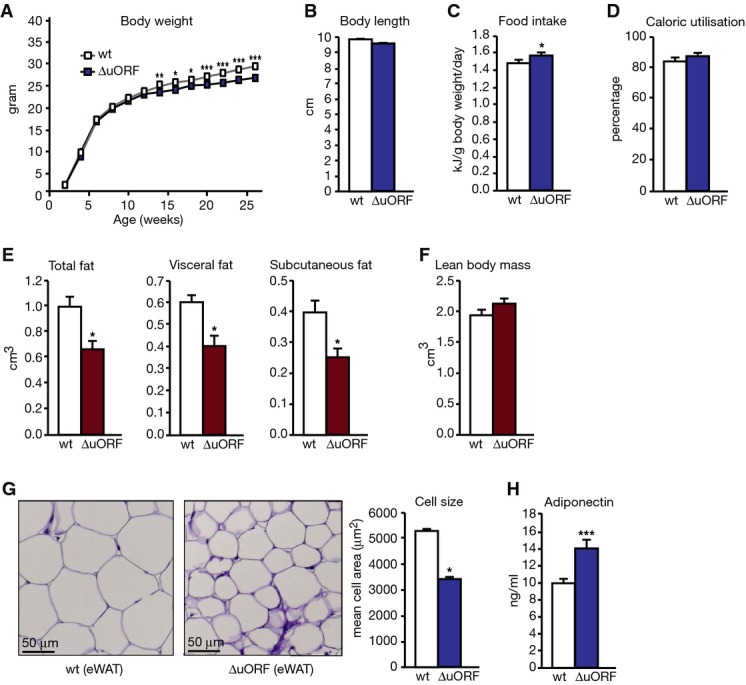
Fat accumulation in WAT is reduced in C/EBPβΔuORF/BL6 mice
- Growth curves of wt and C/EBPβΔuORF/BL6 mice on a normal diet, ad libitum fed (wt, n = 14; C/EBPβΔuORF/BL6, n = 13).
- Body length at week 19 of mice on a normal diet, ad libitum fed (wt, n = 6; C/EBPβΔuORF/BL6, n = 5).
- Daily food intake per mouse, normalised to body weight as determined over 18 weeks (mice on a normal diet, ad libitum fed, n = 6).
- Efficiency of caloric utilisation measured by bomb calorimetry of food and faeces (wt, n = 6; C/EBPβΔuORF/BL6, n = 7).
- Volume of total fat discriminated between visceral and subcutaneous fat as measured by abdominal CT analyses (n = 5).
- Volume of lean body mass measured by abdominal CT analyses (n = 5).
- Histological haematoxylin and eosin (H&E) staining of epididymal WAT (scale bar corresponds to 50 μm). Quantification of the fat cell area is shown at the right (n = 6, 10 adjacent cells are measured per mouse).
- Adiponectin levels in blood plasma (n = 6, measured in 8-month-old mice).
Data information: All values are mean ± SEM. P-values were determined with Student’s t-test, *P < 0.05; **P < 0.01; ***P < 0.005.
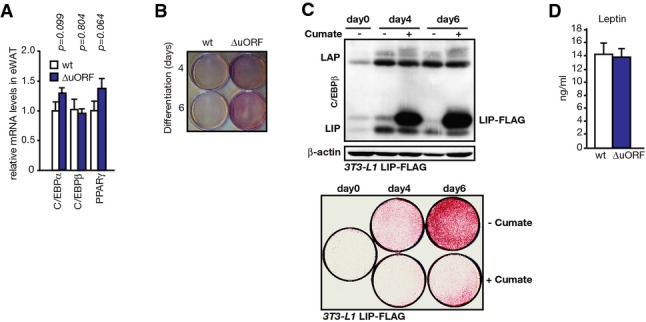
Analyses of adipogenic function
- mRNA expression levels in epididymal WAT from wt and C/EBPβΔuORF/BL6 mice of genes important for adipogenic differentiation analysed by qRT–PCR (wt, n = 5; C/EBPβΔuORF/BL6, n = 6).
- Lipid staining (Oil Red O) of primary MEFs isolated from wt or C/EBPβΔuORF/BL6 mice 4 and 6 days after adipogenic differentiation as indicated at the left side.
- Lipid staining (Oil Red O) of 3T3-L1 cells containing an ectopic cumate-inducible LIP-FLAG cassette either undifferentiated (day 0) or after 4 and 6 days of adipogenic differentiation with (+; LIP-FLAG induced) or without (−; not induced) cumate. Immunoblot shows induction of LIP-FLAG. β-actin was used as a loading control.
- Leptin levels in blood plasma (wt, n = 5; C/EBPβΔuORF/BL6, n = 6, measured in 8-month-old mice).
Data information: All values are mean ± SEM. P-values were determined with Student’s t-test.
Figure 4.
Fatty acid oxidation is regulated by the C/EBPβ protein isoform ratio
- Respiratory exchange ratio (RER) of C/EBPβΔuORF/BL6 (blue line) and wt (black line) mice was measured by Oxymax during a period of 45 h (wt, n = 9; C/EBPβΔuORF/BL6, n = 11). The dark phases are marked by grey boxes.
- Whole animal oxygen consumption of C/EBPβΔuORF/BL6 and wt mice during the resting phase was measured with the Oxymax system (n = 10).
- Oxygen consumption rate (OCR) upon usage of palmitate as exogeneous energy source (fatty acid oxidation rate) in Hepa 1-6 cells with ectopic expression of LIP or empty vector control, n = 8. Immunoblot shows expression of LAP and LIP, and bar graph shows quantification of the LAP/LIP-isoform ratio (n = 1).
- Analyses of serum lipids in wt or C/EBPβΔuORF/BL6 mice (n = 5).
- Histological sections of liver and of cardiac muscle of wild-type (wt) or C/EBPβΔuORF/BL6 (ΔuORF) mice. Sections were stained with haematoxylin (blue) and Sudan III for lipid detection visible as red colour.
Data information: All values are mean ± SEM. P-values were determined with Student’s t-test, *P < 0.05; **P < 0.01; ***P < 0.005.
Increased fatty acid oxidation is known to improve the health status by lowering the concentration of free fatty acids in the serum and counteracting lipid accumulation in non-adipose tissues 25. In the serum of C/EBPβΔuORF/BL6 mice, the concentration of free fatty acids (FFA) and triglycerides (TG) was reduced compared to wt mice, while the levels of cholesterol/high-density lipoprotein (HDL)/low-density lipoprotein (LDL) were similar (Fig4D). Furthermore, lipid accumulation in liver and heart was strongly reduced in 8-month-old C/EBPβΔuORF/BL6 mice compared to wt littermates as revealed by Sudan III staining (Fig4E).
C/EBPβΔuORF/BL6 mice display a CR-like metabolic gene expression profile
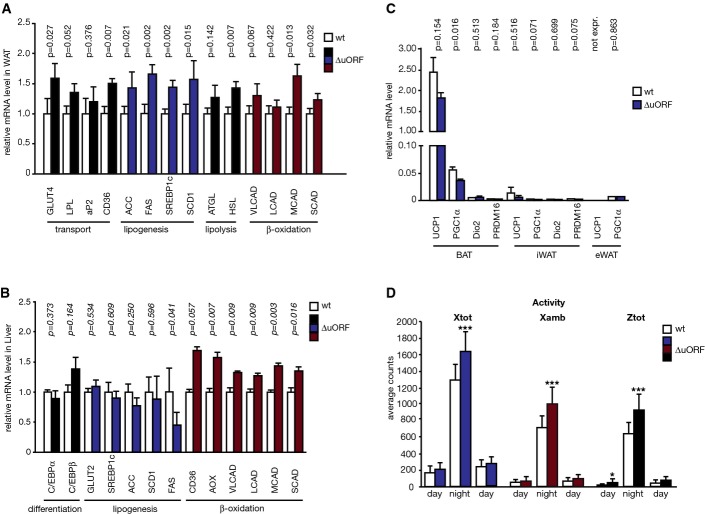
Next, we analysed mRNA expression of genes involved in fat metabolism that could be involved in the metabolic phenotype of C/EBPβΔuORF/BL6 mice. We measured a moderate but consistent shift towards higher transcript levels of genes that are involved in glucose/fat transport, lipogenesis and lipolysis in WAT of C/EBPβΔuORF/BL6 mice compared to littermate controls using quantitative real-time PCR (Fig5A). This points towards an increased fat turnover in adipose tissue. We found elevated transcript levels of the insulin-dependent glucose transporter GLUT4 (1.6-fold, P = 0.027) required for glucose uptake for de novo lipogenesis. Furthermore, we detected increased mRNA levels of fatty acid translocase (FAT)/cluster of differentiation 36 (CD36) (1.5-fold, P = 0.007) that is involved in fatty acid uptake. We observed a small but not significant increase for lipoprotein lipase (LPL) (1.4-fold, P = 0.052) or the intracellular fatty acid binding protein aP2 (1.2-fold, P = 0.376). In addition, transcripts that stimulate lipogenesis were elevated: the sterol regulatory element-binding protein 1c (SREBP1c) (1.5-fold, P = 0.002), acetyl-CoA carboxylase (ACC), which is the flux-determining enzyme of the lipogenic pathway (1.4-fold, P = 0.021), the key enzyme in fatty acid synthesis fatty acid synthase (FAS) (1.7-fold, P = 0.002) and stearoyl-coenzyme A desaturase 1 (SCD1) (1.6-fold, P = 0.015), which is important for the synthesis and regulation of unsaturated fatty acids. Finally, among the transcripts that stimulate lipolysis, the hormone-sensitive lipase (HSL) mRNA was elevated (1.4-fold, P = 0.007), while upregulation of the adipose triglyceride lipase (ATGL) transcript was not significant (1.3-fold, P = 0.142). Immunoblot analyses showed that the moderate increases in mRNA levels correlate with significantly increased protein levels for FAS, ACC and GLUT4. Smaller increases were observed for CD36 or HSL (Fig EV4A). To investigate whether expression of genes involved in β-oxidation is altered in WAT, we examined mRNA expression of the short-chain (SCAD), medium-chain (MCAD), long-chain (LCAD) and very long-chain (VLCAD) acyl-CoA dehydrogenases (Fig5A). Although we observed a general tendency towards enhanced expression, only the MCAD transcript was found to be significantly upregulated (1.7-fold, P = 0.013), which is also reflected on the protein levels (Fig EV4A).
Figure 5.
Gene expression and physical activity changes in C/EBPβΔuORF/BL6 mice
- mRNA levels of depicted genes measured in epididymal WAT of C/EBPβΔuORF/BL6 mice relative to wt mice (wt, n = 5; C/EBPβΔuORF/BL6, n = 6).
- mRNA levels of depicted genes measured in livers of C/EBPβΔuORF/BL6 mice relative to livers of wt mice (wt, n = 5; C/EBPβΔuORF/BL6, n = 6).
- Relative mRNA levels calculated from qRT–PCR data of depicted genes measured in BAT, inguinal iWAT and epididymal eWAT of C/EBPβΔuORF/BL6 mice compared to wt mice (wt, n = 6; C/EBPβΔuORF/BL6, n = 6).
- Activity during day and night phases for total x- (blue) and z-axis (red) movement and ambulatory x-axis (black) movements of C/EBPβΔuORF/BL6 and wt mice (n = 4). Measurements were taken with the Oxymax system.
Data information: All values are mean ± SEM. mRNA levels were determined by qRT-PCR, and corresponding P-values are depicted as determined with Student’s t-test, *P < 0.05; ***P < 0.005.
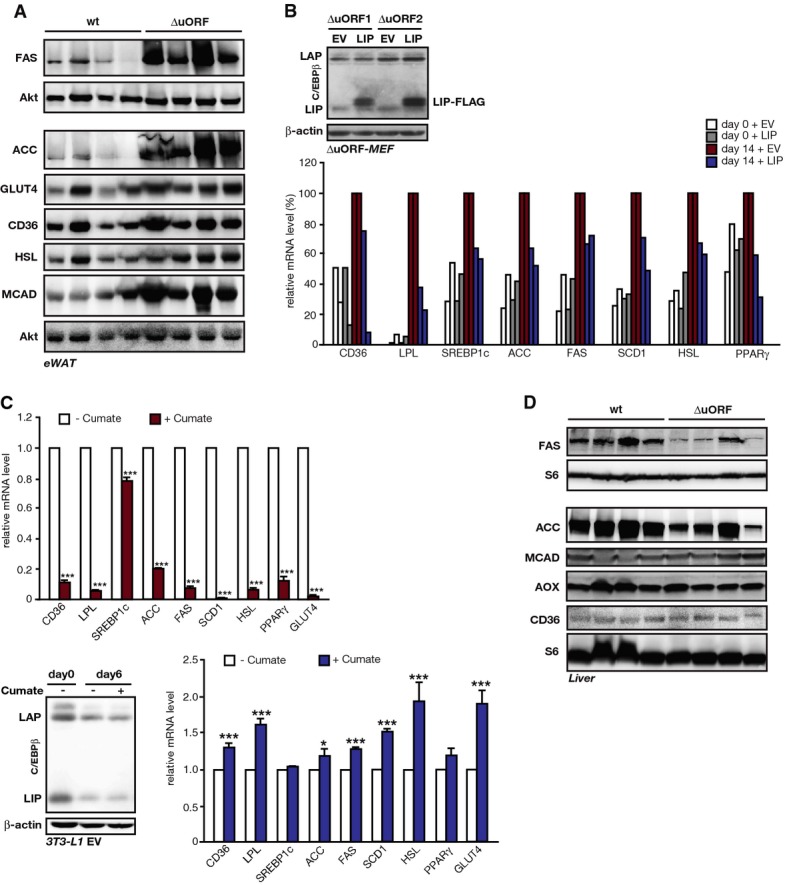
Analyses of gene expression
- Immunoblots of extracts from epididymal WAT of fed wt and C/EBPβΔuORF/BL6 mice with detection of the indicated proteins and Akt as a loading control (n = 4).
- Relative mRNA levels calculated from qRT–PCR data of indicated genes in C/EBPβΔuORF/BL6 MEFs 14 days after adipogenic differentiation compared to undifferentiated cells (day 0). Cells ectopically overexpressing LIP-FLAG (day 0/14 + LIP) were compared to empty vector control cells (day 0/14 + EV). mRNA levels are shown as percentage of day 14 controls. Immunoblot shows induction of LIP-FLAG, n = 2.
- Upper part: Relative mRNA levels calculated from qRT–PCR data of indicated genes in 3T3-L1 cells 6 days after adipogenic differentiation with cumate-inducible LIP-FLAG cultured without cumate (−; non-induced) or with cumate (+; LIP-FLAG induced). mRNA levels are shown relative to non-induced controls. Immunoblot for C/EBPβ expression in LIP-FLAG inducible cells upon differentiation without cumate (−) or with cumate (+) and β-actin as a loading control is shown in FigEV3C. Lower part: Relative mRNA levels calculated from qRT–PCR data of indicated genes in 3T3-L1 cells 6 days after adipogenic differentiation with cumate-inducible empty vector (EV) cultured without cumate (−) or with cumate (+). mRNA levels of + cumate are shown relative to − cumate. Values are depicted as mean ± SD of three technical replicates, and corresponding P-values of the Student’s t-test are depicted *P < 0.05; ***P < 0.005. Immunoblot shows C/EBPβ expression in empty vector (EV)-transfected cells upon differentiation without cumate (−) or with cumate (+). β-actin was used as a loading control.
- Immunoblots of extracts from livers of fed wt and C/EBPβΔuORF/BL6 mice with detection of the indicated proteins and S6 as a loading control (n = 4).
Next, we studied cell intrinsic effects of LAP/LIP ratio on adipogenic gene expression in primary MEFs or the adipoblast cell line 3T3-L1 in cell culture. Briefly, we transduced confluent C/EBPβΔuORF/BL6 MEFs with a lentivirus expressing either C/EBPβ-LIP or an empty control vector, and induced the adipogenic programme. mRNA expression of all examined adipogenic genes was generally downregulated by ectopic expression of LIP (low LAP/LIP ratio) compared to control C/EBPβΔuORF/BL6 MEFs (high LAP/LIP ratio) (FigEV4B). In 3T3-L1 cells containing an inducible LIP expression cassette (cumate-inducible system), adipogenesis was induced simultaneously with LIP induction (+ cumate) or without ectopic LIP induction (− cumate, solvent treatment) as control. Induction of LIP resulted in reduced expression of adipogenic transcripts measured at day 6 of differentiation (FigEV4C). Cumate treatment of the empty vector control cells had no or a rather stimulatory effect on the expression of adipogenic genes, ruling out that cumate itself acts anti-adipogenic. Therefore, LAP-/LIP-associated regulation of adipogenic transcripts found in cell culture supports the observations we made in mice.
In the liver of C/EBPβΔuORF/BL6 mice, the expression of the following transcripts involved in fatty acid β-oxidation was upregulated compared to wt mice: the peroxisome acyl coenzyme A oxidase (AOX) (1.6-fold, P = 0.007), SCAD (1.3-fold, P = 0.016), MCAD (1.4-fold, P = 0.003), LCAD (1.3-fold, P = 0.009) and VLCAD (1.3-fold, P = 0.009). Only expression of the FAT/CD36 mRNA for fatty acid uptake was not significantly increased (1.7-fold, P = 0.057) (Fig5B). On the contrary, transcript levels of factors fostering lipogenesis were unchanged (GLUT2, SREBP1c, ACC and SCD1) or even decreased (FAS, P = 0.041). Furthermore, immunoblot analyses showed that expression of FAS and ACC proteins (lipogenesis) is decreased, while expression of the MCAD, AOX and CD36 (β-oxidation) was not significantly different (Fig EV4D). To further support the direct regulatory role of C/EBPβ in the examined gene regulation, we analysed the ENCODE database (http://genome.ucsc.edu/ENCODE/) for promoter occupation by chromatin immunoprecipitation sequencing (ChIP-Seq). This analysis revealed that all genes analysed are associated with C/EBPβ at regions that are in most cases associated with the histone H3 lysine 27 acetylation (H3K27Ac) mark characterising active enhancers (Table EV1).
C/EBPβΔuORF/BL6 mice are physically more active
An intriguing aspect of the C/EBPβΔuORF/BL6 phenotype is the lower body weight in spite of similar food intake and caloric utilisation. This points to an increased energy expenditure in C/EBPβΔuORF/BL6 mice, which is supported by the observed increase in oxygen consumption. Possible causes for higher energy use are respiratory uncoupling in BAT, browning of WAT or an increase in physical activity. To examine the potential involvement of altered BAT or WAT function, we analysed mRNA expression levels of genes involved in uncoupling and thermogenesis (UCP1, PGC-1α, Dio2, PRDM16) in BAT, inguinal (i) WAT or epididymal (e) WAT (Fig5C). The expression of those genes was very low in both iWAT and eWAT and not increased in C/EBPβΔuORF/BL6 mice excluding a critical involvement of uncoupling or browning of WAT as a cause of the higher energy expenditure.
To examine if an increase in physical activity could be the cause for this phenotype, we determined physical activity of C/EBPβΔuORF/BL6 mice compared to age- and sex-matched littermate controls using the Oxymax/CLAMS animal motion detection system. As shown in Fig5D, the total activity (Xtot; total counts of fine movements (e.g. grooming) and ambulatory activity), as well as the ambulatory activity (Xamb; “distance”) and vertical activity (Ztot; “rearing”) of the C/EBPβΔuORF/BL6 mice, were higher compared to wt mice during their active period at night. Thus, the reduced body weight of the C/EBPβΔuORF/BL6 mice might be caused by higher energy expenditure due to their increased physical activity.
C/EBPβΔuORF/BL6 mice have improved glucose clearance and insulin sensitivity
Fat metabolism is highly interconnected with glucose metabolism and particularly with insulin responsiveness. Therefore, we analysed glucose metabolism in C/EBPβΔuORF/BL6 mice. The intraperitoneal (i.p.) glucose tolerance test (IPGTT) revealed an enhanced glucose clearance in C/EBPβΔuORF/BL6 mice, which is reflected by a smaller area under the curve (AUC) (Fig6A). The i.p. insulin sensitivity test (IPIST) demonstrated that C/EBPβΔuORF/BL6 mice also display an increase in insulin sensitivity compared to wt control mice (Fig6B). Accordingly, tissue insulin sensitivity was higher in C/EBPβΔuORF/BL6 mice, as revealed by enhanced induction of Akt-Ser473 phosphorylation in muscle and liver after intravenous (i.v.) insulin injection (Figs6C and EV5A). Improved insulin sensitivity is usually accompanied by reduced levels of circulating glucose and insulin, which is also found under caloric restriction 26. In C/EBPβΔuORF/BL6 mice, fasting insulin and fed glucose levels were lower compared to wt mice, while fasting glucose and fed insulin levels showed no significant reduction (Fig6D and E). The significantly lower HOMA-IR (homeostatic model assessment of insulin resistance) supports the conclusion that C/EBPβΔuORF/BL6 mice show improvements in insulin sensitivity (Fig6F). To exclude that the lower insulin levels were due to aberrantly reduced pancreatic β-cell mass, insulin production in the pancreas was examined by quantitative immunofluorescence analysis. We observed no difference in β-cell mass but a decreased insulin production in C/EBPβΔuORF/BL6 mice compared to wt mice (Fig EV5B). This indicates that the improved tissue insulin sensitivity in C/EBPβΔuORF/BL6 mice requires less insulin production for efficient function.
Figure 6.
Enhanced glucose clearing and insulin sensitivity in C/EBPβΔuORF/BL6 mice
- Glucose tolerance test (IPGTT) with the calculated area under the curve (AUC) of C/EBPβΔuORF/BL6 mice and wt mice injected i.p. with glucose (2 g/kg) after a 16-h fast (n = 6).
- Insulin sensitivity test (IPIST) with the calculated area under the curve (AUC) of fed C/EBPβΔuORF/BL6 and wt mice injected i.p. with insulin (0.5 IU/kg) (wt, n = 6; C/EBPβΔuORF/BL6, n = 5).
- Immunoblot showing Akt phosphorylation (p-Akt) (Thr308), Akt and β-actin protein levels in muscle 10 min after i.v. administration of insulin (0.75 IU/kg) in 6 h-fasted C/EBPβΔuORF/BL6 and wt mice (n = 3).
- Concentration of blood glucose measured in the morning of fed or overnight-fasted C/EBPβΔuORF/BL6 and wt mice (n = 4).
- Concentration of blood plasma insulin measured in the morning of fed or overnight-fasted C/EBPβΔuORF/BL6 and wt mice (n = 6).
- HOMA2-IR calculation of fasting glucose and insulin levels (n = 6).
Data information: All values are mean ± SEM. P-values were determined with Student’s t-test, *P < 0.05.
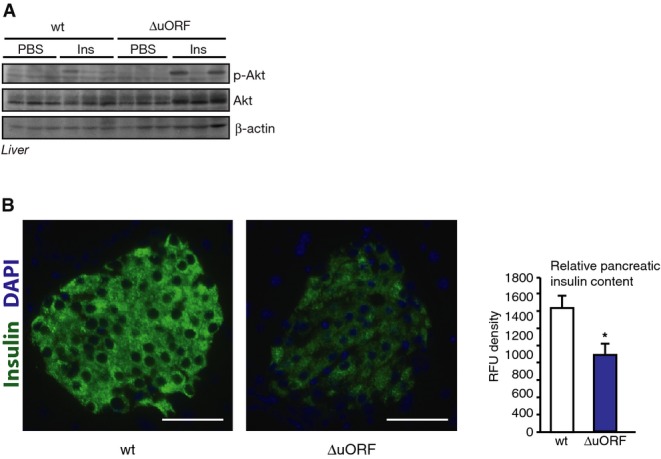
Analysis of insulin signalling and pancreatic β-cells
- Immunoblot showing Akt phosphorylation (p-Akt) (Thr308), Akt and β-actin protein levels in liver 10 min after i.v. administration of insulin (0.75 IU/kg) in 6 h-fasted wt and C/EBPβΔuORF/BL6 mice (n = 3).
- Fluorescent immunohistochemical analysis of the pancreas of wt or C/EBPβΔuORF/BL6 mice with insulin staining in green (anti-insulin antibody and Alexa Fluor 488-conjugated secondary antibody) and DAPI DNA staining in blue. Representative pancreatic islets are shown (scale bar corresponds to 50 μm). Relative quantification of insulin-specific fluorescence is shown at the right side (n = 6 mice, three islets each). Values are mean ± SEM, *P < 0.05.
Discussion
Reduced mTORC1 signalling is thought to be responsible for many of the metabolic improvements under caloric restriction (CR) 7 and is believed to attenuate the development of the metabolic syndrome 15. Here, we show that the C/EBPβΔuORF/BL6 mice display a range of metabolic improvements that are remarkably similar to what has been described for CR, however, without reducing calorie (food) intake. Loss of the C/EBPβ uORF leads to diminished expression of the C/EBPβ-LIP isoform in vitro and in vivo 4,5. This mimics reduced mTORC1 activity at the level of C/EBPβ translation and is sufficient to improve a whole set of metabolic health parameters.
We observed a gradual difference in body weight for the C/EBPβΔuORF/BL6 mice compared to wt mice, which becomes apparent after maturity (week 12) and accumulates to a reduction of 8% in adult C/EBPβΔuORF/BL6 mice (week 26) (Fig3A). The reduced body weight of the C/EBPβΔuORF/BL6 mice is largely due to reduced fat accumulation in WAT. This is accompanied by increased levels of the adipocyte-specific hormone adiponectin and a metabolic shift in whole body energy utilisation towards more fatty acid oxidation (lower RER) (Figs3E and H, 4A). In addition, we found increased expression of lipogenesis and lipolysis genes in WAT and increased expression of β-oxidation genes in liver (Fig5A and B). Together with the enhanced adipocyte differentiation of C/EBPβΔuORF/BL6 MEFs (Fig EV3B) in cell culture, the increased expression of lipogenesis genes in WAT may seem to be inconsistent with the lower fat accumulation and leanness in the C/EBPβΔuORF/BL6 mice. However, increased expression of lipogenesis and lipolysis genes in WAT and increased expression of β-oxidation genes in liver are also observed in mice on CR 21-28,29,30. Intriguingly, in calorie-restricted mice, fatty acid oxidation exceeds fat intake 21. Thus, additionally required amounts of fat are generated from ingested carbohydrates by de novo lipogenesis in WAT followed by lipolysis to meet the increased requirements of fatty acids for energy production. The C/EBPβΔuORF/BL6 mutation may induce a similar metabolic roundabout of enhanced WAT function and fat turnover coupled to increased fatty acid oxidation.
The number of fat cells in WAT of the C/EBPβΔuORF/BL6 mice, as can be calculated from the fat volume (CT analysis) and cell size (histology), does not seem to be altered, indicating that the reduction of fat mass observed in C/EBPβΔuORF/BL6 mice results from less fat storage. This suggests that the enhanced adipogenic differentiation potential observed in MEFs derived from C/EBPβΔuORF/BL6 mice in vitro does not lead to increased number of adipocytes in vivo.
The prolonged period of increased fatty acid oxidation is thought to contribute to the healthy phenotype induced by CR 21. This is also supported by a study in flies, which shows that the reduction in fatty acid oxidation limits CR-induced life span extension 31. Therefore, we hypothesise that C/EBPβ is an important factor in regulating the CR type of fat metabolism. Moreover, we postulate that the increased LAP/LIP ratio as a result of the ΔuORF mutation retains this metabolic state in C/EBPβΔuORF/BL6 mice without reduction in food intake. A high rate of fatty acid oxidation prevents the accumulation of lipids in the liver and in other non-adipose organs 25. Reduction in liver fat has been shown to increase insulin sensitivity 32. We found decreased levels of free fatty acids in the serum and strongly reduced lipid accumulation in liver and heart of C/EBPβΔuORF/BL6 mice compared to wt littermates. Moreover, we found lower glucose and insulin levels in the serum with concomitantly increased glucose tolerance and insulin sensitivity 29. Together with the increased fatty acid oxidation, these findings underscore the healthy metabolic condition of the mice.
The C/EBPβΔuORF/BL6 mice have a similar food intake compared to wt mice but are leaner most probably due to higher energy expenditure as indicated by their increased oxygen consumption. An important question deals with the underlying cause of the higher energy expenditure. Our data indicate that neither increased respiratory uncoupling in BAT nor browning of WAT are causative factors for the higher energy expenditure. However, the C/EBPβΔuORF/BL6 mice display an increased physical activity, suggesting that the associated higher energy expenditure contributes to the lower body weight (Fig5D). Elevated physical activity is also associated with rapamycin treatment 33,34 or CR 35-37. This suggests that the C/EBPβΔuORF/BL6 mutation might mimic effects of mTORC1 inhibition on physical activity. Intriguingly, a study using flies demonstrated that increased activity is not only associated with CR but is also at least partially required to induce the beneficial effects of CR 31.
The transcriptional effects we observed in the C/EBPβΔuORF/BL6 mice are moderate (Fig5A and B), although they translate into changes on protein levels in most cases. The more subtle changes in gene expression may support the improved overall metabolic phenotype: a small and consistent shift in gene regulation resulting in a continuous shift in metabolism as suggested by the continuously lower RER (Fig4A) that is still in a physiological range. Stronger transcriptional effects may result in a fully unbalanced metabolism with detrimental metabolic effects. In addition to its direct involvement in gene regulation, the C/EBPβΔuORF/BL6 mutation most likely results in systemic (hormonal) alterations such as increased serum concentration of adiponectin that contributes to the improved metabolic parameters.
The phenotypes observed in C/EBPβΔuORF/BL6 mice are similar to those described for C/EBPβ−/− complete knockout mice with diet- or genetically induced obesity. C/EBPβ−/− mice on a high fat diet display a reduced accumulation of body fat, are resistant to steatosis and have enhanced fatty acid oxidation compared to wt mice that develop an obese phenotype 38. Leprdb/db mice are homozygous for a loss of function mutation in the Leptin receptor and become obese at ∼4 weeks of age. Complete C/EBPβ deficiency in Leprdb/db mice results in a general healthier metabolic phenotype with reduced total body fat and weight gain, less steatosis, enhanced fatty acid oxidation and better glucose homeostasis 39. The fact that the metabolic phenotype of C/EBPβΔuORF/BL6 mice (normal expression levels of LAP) resembles the one of C/EBPβ−/− knockout mice suggests that the lack of the LIP isoform and nor the lack of LAP is decisive for the phenotype. Therefore, the complete C/EBPβ knockout may display the beneficial metabolic phenotypes because of its deficiency for the metabolic “harmful” LIP isoform that is under control of mTORC1.
Many open questions remain how CR and the associated metabolic changes influence health span. However, it is intriguing that mutation of a single mRNA-translation cis-regulatory element in a single gene results in a phenotype that (at least partially) resembles the phenotype induced by CR. This implies that pharmacological alteration of the C/EBPβ-isoform ratio may provide a promising therapeutic strategy to intervene with metabolism-related disorders, thereby increasing health span.
Materials and Methods
Cell culture
HEK293T, Fao, 3T3-L1 (all obtained from ATCC), Hepa 1-6 cells, p53−/− MEFs, 4E-BP wt and DKO MEFs 19, immortalised C/EBPβΔuORF/BL6 and C/EBPβ KO MEFs and freshly isolated C/EBPβΔuORF/BL6 and C/EBPβ KO MEFs and the corresponding wt MEFs were maintained in DMEM supplemented with 10% FCS, 1% HEPES and penicillin/streptomycin. GH3 cells were maintained in F12K medium supplemented with 15% horse serum, 2.5% FCS, 1% HEPES and penicillin/streptomycin. For mTOR repression, cells were incubated with pp242 (1 μM) or rapamycin (1 μM or 200 nM for GH3 cells) for different time periods (6 h for Fao, 12 h for MEFs, 24 h for 3T3-L1 and GH3). For S6K1 inhibition, cells were incubated with DG2 (20 μM) for different time periods (12 h for MEFs, 24 h for 3T3-L1 and GH3). For inhibitor treatment, 3T3-L1 cells had been differentiated for 4 days (as described for primary MEFs but in the absence of troglitazone).
DNA constructs
The mouse S6 kinase 1 shRNA expression vector was generated by annealing the oligonucleotides sh-a 5′-CCG GAC ATT GTT ACA CAG CCA GTA TCT CGA GAT ACT GGC TGT GTA ACA ATG TTT TTT-3′ and sh-b 5′AAT TAA AAA ACA TTG TTA CAC AGC CAG TAT CTC GAG ATA CTG GCT GTG TAA CAA TGT-3′ and ligating them into the Tet-pLKO-puro vector (Addgene plasmid 21915, described in 40). For generating the C/EBPβ-LAP or C/EBPβ-LIP expression vectors, the rat C/EBPβ mutants ΔD and C, respectively, that are described in 4 were cloned into pCDNA3 or pZeoSV2 vector (both from Invitrogen). A FLAG-tagged version of rat C/EBPβ-LIP 16 was cloned into the lentiviral pLVX-IRES-neo expression vector (Clontech), and for the generation of the cumate-inducible C/EBPβ-LIP-FLAG lentiviral construct, it was cloned into the pCDH-CuO-MCS-IRES-GFP-EF1-CymR-T2A-Puro-All-in-one vector (System Biosciences).
Isolation and differentiation of primary MEFs
Mouse embryonic fibroblasts (MEFs) were isolated from embryos at embryonic day 14.5 following standard protocols. Cells from passage 3 were seeded into 10-cm dishes for differentiation assays. Adipogenic differentiation was induced 2 days after cells reached confluency by replacing the medium with differentiation medium (DMEM containing 1 μM dexamethasone, 0.5 mM methylisobuthylxanthine, 10 μg/ml insulin, 10 μM troglitazone and 10% FCS). After 2 days of incubation, the medium was replaced by DMEM supplemented with 10 μg/ml insulin and 10% FCS and then replaced every second day. At different days of the differentiation protocol, cells were fixed with 4% PFA and stained with Oil Red O.
Immortalisation of primary MEFs
Primary MEFs of passage 2 were retrovirally infected with a pSUPER-retro-based shRNA construct targeting p19ARF using the PhoenixE producer cell line as described in 4 and selected with puromycin (1.5 μg/ml).
Transfection
For lentivirus production, HEK293T cells were seeded to a density of 3 × 106 cells in 10-cm culture dishes. Twenty-four hours later, transfection was carried out using the calcium phosphate method. For stable C/EBPβ-LIP expression, a pCDNA3-based LIP expression vector was transfected into Hepa 1-6 cells using the Fugene HD transfection reagent (Promega) according to the protocol of the manufacturer. Transfected cells were selected with 0.9 mg/ml G418. For stable expression of C/EBPβ-LAP*, C/EBPβ-LAP and C/EBPβ-LIP immortalised C/EBPβ KO MEFs were transfected with pZeoSV2-based expression vectors using the Fugene transfection reagent and selected with zeocin (0.1 mg/ml).
Lentiviral transduction
p53−/− MEFs were infected following a standard protocol with pLKO.1 lentiviral constructs containing shRNAs against mouse raptor: sh 5′-CCG GCC TCA TCG TCA AGT CCT TCA ACT CGA GTT GAA GGA CTT GAC GAT GAG GTT TTT G-3′ (Addgene plasmid 21339); mouse rictor: 5′- CCG GGC CAG TAA GAT GGG AAT CAT TCT CGA GAA TGA TTC CCA TCT TAC TGG CTT TTT G-3′ (Addgene plasmid 21341), both described in 41, mouse S6 kinase 1: 5′-CCG GAC ATT GTT ACA CAG CCA GTA TCT CGA GAT ACT GGC TGT GTA ACA ATG TTT TTT-3′ or non-target shRNA control (Sigma-Aldrich). Two days after infection, puromycin was added (final concentration 1.5 μg/ml). Cells were harvested for analysis 4 days after infection. Due to leakiness of the inducible S6 kinase-shRNA Tet-pLKO-puro construct, cells treated with doxycycline (100 ng/ml, 48 h) were compared with doxycycline-treated control shRNA-expressing cells. One day before induction of adipogenic differentiation, confluent plates of primary MEFs were infected with a pLVX-IRES-neo-based C/EBPβ-LIP-FLAG construct. Forty-eight hours after infection, G418 was added (0.4 mg/ml), which was replenished upon every medium change. 3T3L1 cells were infected with a cumate-inducible C/EBPβ-LIP-FLAG construct or an empty vector construct and selected with puromycin (1.5 μg/ml). Cells from an individual clone that showed good inducibility were differentiated as described for primary MEFs but in the absence of troglitazone. During the whole differentiation period, puromycin was added in a concentration of 1 μg/ml and cells were treated with cumate (12 μg/ml) or solvent (ethanol) starting at day 0 of the differentiation protocol.
Fatty acid oxidation assay
Fatty acid oxidation was determined using a Seahorse XF96 Extracellular Flux analyser (Seahorse Bioscience). 2 × 104 Hepa 1-6 cells per well were seeded into a 96-well XF cell culture microplate 24 h prior to the assay. Sixteen hours before the assay, cells were washed and the medium was replaced with DMEM containing 0.5 mM glucose, 1 mM glutamine, 0.5 mM carnitine and 1% FCS to deplete the cells from oxidation substrates. One hour before the assay, the cells were washed twice with FAO assay buffer (111 mM NaCl, 4.7 mM KCl, 1.25 mM CaCl2, 2.2 mM MgSO4, 1.2 mM NaH2PO4, 2.5 mM glucose, 0.5 mM carnitine, 5 mM HEPES, pH 7.4), and 15 min before the assay, the oxidation substrate palmitate-BSA or a BSA control (Seahorse Bioscience) was added and the oxygen consumption rate (OCR) with or without palmitate-BSA was measured.
Mice
C/EBPβΔuORF mice 5 were back-crossed for 6 generations into the C57BL/6 genetic background (C/EBPβΔuORF/BL6). Male mice that were kept at a standard 12-h light/dark cycle at 22°C in a pathogen-free animal facility were used for all experiments. Numbers of mice used in the separate experiments can be retrieved from the figure legends. Body weight and food consumption (standard chow) were measured weekly for 26 weeks. Body length was determined from nose to anus with an Ultra-Cal IV Electronic Digital Calliper (Ted Pella). C/EBPβ KO mice were obtained from The Jackson Laboratory (STOCK Cebpbtm1Vpo/J, Jackson Laboratory stock no: 006873) and only used for generating C/EBPβ KO MEFs. All animal experiments were performed in compliance with protocols approved by the Institutional Animal Care and Use Committee.
Feeding experiments
Male mice were caged individually, and daily food consumption was measured for 7 days. In the subsequent 4 weeks, the mice received every day 70% of their normal food consumption at 6 p.m. (caloric restriction).
Rapamycin treatment
Mice were injected i.p. with 8 μg rapamycin per gram body weight (1.2 mg/ml rapamycin, 0.25% (w/v) PEG, 0.25% (v/v) Tween-20 in H2O) or solvent. Twelve or twenty-four hours after injection, mice were sacrificed and tissue samples were taken for immunoblot analyses.
Body composition
The abdominal region from lumbar vertebrae 5 to 6 of anaesthetised mice was analysed with an Aloka LaTheta Laboratory Computed Tomograph LCT-100A (Zinsser Analytic). Scans were performed with a resolution of 1.00 mm with high X-ray voltage and fast speed settings. Visceral and subcutaneous fat was discriminated with the supplied software (for visceral fat measurement).
Caloric utilisation
Faeces and food samples were collected and dried in a speed vacuum drier for 5 h at 60°C and then grinded and pressed into tablets. Energy content of food and faeces was determined by bomb calorimetry (IKA-Calorimeter C 5000). Energy efficiency was calculated by subtracting energy loss in faeces from consumed energy.
Energy expenditure
O2 consumption and CO2 output were measured simultaneously through indirect calorimetry with an Oxymax Comprehensive Lab Animal Monitoring System CLAMS open circuit system (Columbus Instruments). Mice were placed in individual metabolic cages with free access to water and food. Measurements were performed every 18 min for 48 h starting at 6 p.m. For graphical presentation, the average of four measurements was taken.
Animal activity measurement
Activity was monitored with the Oxymax CLAMS open circuit system (Columbus Instruments) over 18 h starting at 10 a.m. Movements were recorded for 18-min intervals for the x-axis and z-axis and total movements and ambulatory movements counted. For graphic presentation and analyses, average values for day and night (6 p.m.–6 a.m.) phases were used.
Blood tests
Mice were anaesthetised with isoflurane, and whole blood was collected from the suborbital node with heparinised capillaries into heparin blood collection tubes. Plasma was separated from cells by centrifugation at 5,000 × g for 10 min at 4°C. Levels of insulin, adiponectin and leptin were measured by enzyme-linked immuno-sorbent assay (ELISA) according to the instructions of the manufacturer (Biocat). Lipids in serum were determined by standard clinical laboratory techniques. Free fatty acids (FFA) were determined by enzymatic conversion to H2O2 followed by a colorimetric peroxidase assay. Cholesterol, LDL, HDL and triglycerides were analysed with the Architect System from Abbott. For measuring LDL and HDL, all non-LDL and non-HDL were removed prior to analysis, respectively. The procedure for the determination of triglycerides is described in 42,43; however, 4-chlorophenol was used instead of 2-hydroxy-3,5-dichlorobenzene sulphonate.
Glucose tolerance and insulin sensitivity
For the i.p. glucose tolerance test, mice were fasted overnight (16 h) and injected i.p. with 10 μl of a 20% (w/v) glucose solution per gram body weight. Blood glucose concentration was measured with a glucometer (Accu Chek Aviva, Roche). For the i.p. insulin sensitivity test, mice that had free access to food before, but not during the experiment, were i.p. injected with 0.5 IU/kg insulin (0.05 IU/ml insulin in 1× PBS with 0.08% BSA fatty acid-free) and blood glucose concentration was measured as described before. For determination of insulin sensitivity in muscle, mice were fasted for 6 h and injected i.v. with 0.75 IU/kg insulin (0.21 IU/ml insulin in 1× PBS) or solvent. Ten minutes after injection, mice were sacrificed and tissue samples were collected and analysed in an immunoblot using Akt- and phospho-Akt-specific antibodies (see below). HOMA2-IR was calculated using the HOMA2 Calculator v2.2.3 from the Diabetes Trials Unit, University of Oxford (http://www.dtu.ox.ac.uk/homa).
Immunoblot analyses
For protein extraction, mouse tissue was homogenised in 500 μl tissue lysis buffer (60 mM Tris pH 6.8; 1% SDS, supplemented with protease and phosphatase inhibitors) with a glass douncer on ice. Protein extracts were sonicated and centrifuged for 5 min at 10,000 × g at 4°C, and the fatty layer and cell debris were removed. For protein extraction from cells, these were washed twice with ice-cold 1× PBS and lysed in 50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, supplemented with protease and phosphatase inhibitors followed by sonication. Equal amounts of total protein were separated by SDS–PAGE, transferred to a PVDF or nitrocellulose membrane and analysed by using the following antibodies: C/EBPβ (C-19), 4E-BP1 (C-19) and α-tubulin (TU-02) from Santa Cruz; phospho-p70S6K (Thr389) (108D2 or 1A5) and p70S6K (#9202) phospho-S6 ribosomal protein (Ser235/236) (2F9), S6 ribosomal protein (54D2), phospho-Akt (Ser473) (D9E) and (Thr308) (L32A4), Akt (#9272), phospho-4E-BP1 (Thr37/46) (#9459), phospho-eIF4E (Ser209) (#9741), eIF4E (#9742), 4E-BP2 (#2845), raptor (24C12), rictor (53A2), acetyl-CoA carboxylase (ACC, #3676), fatty acid synthase (FAS, #3180) and HSL (34107) from Cell Signaling, β-actin (AC-40) A3853 from Sigma, Glut4 (#NBP1-49533) and CD36 (#NB400-144) from Novusbio, AOX1 (#10957) from Proteintech and MCAD (#ab129420) from abcam and the following secondary antibodies: rabbit IgG, HRP-linked antibody from donkey and mouse IgG HRP-linked antibody from sheep from GE Healthcare. For detection, the Western lightning Plus-ECL reagent (PerkinElmer) was used. For re-probing, membranes were incubated in Restore Western Blot Stripping buffer (Perbio). Quantification of the protein bands was performed using the FluorChemE Imager (Proteinsimple/Biozym) or the ImageQuant LAS 4000 Mini Imager (GR Healthcare) and the supplied software or the ImageJ software 44 in case of films.
Histology and immunofluorescence
Pieces of tissue were fixed with 4% paraformaldehyde for 24 h and embedded in paraffin. 5-μm-thick sections were stained with haematoxylin and eosin (H&E) using the Autostainer XL (Leica). For Sudan III staining, 10-μm cryosections fixed with 4% paraformaldehyde were stained for 30 min with Sudan III staining solution (3% (w/v) Sudan III in 10% ethanol and 90% acetic acid). For immuno-histofluorescence, paraffin sections were treated with 10 mM citrate buffer pH 6.0 for 10 min at sub-boiling point for antigen retrieval. Sections were cooled down to RT and washed three times with 1× PBS. After blocking for 1 h with blocking buffer (5% (v/v) goat serum, 1% (w/v) BSA, 0.4% (v/v) Triton X-100 in 1× PBS), sections were incubated with the primary antibody (rabbit IgG insulin (H-86) antibody, Santa Cruz) at 4°C over night in a humidified chamber. After washing with 1× PBS, slides were incubated with Alexa Fluor 488-conjugated secondary antibody (Invitrogen) for 2 h at RT. After washing with PBS, sections were sealed with cover slips using DAPI containing mounting medium. All microscopic analyses were done with the Imager ApoTome Axiovert 200 and Axiovision (Zeiss) software.
qRT–PCR analyses
Tissue pieces were homogenised with the Precellys 24 system (Peqlab) in the presence of 1 ml QIAzol reagent (QIAGEN), and RNA was isolated from tissue samples using the RNeasy® Lipid Tissue Mini Kit (QIAGEN). After incubation for 30 min at 37°C with RQ1 RNase-Free DNase (Promega), the RNA was further purified with the RNeasy® Plus Mini Kit (QIAGEN) according to the manufacture’s protocol starting at step 4. For cDNA synthesis, 1 μg RNA was reverse transcribed with the Transcriptor First Strand cDNA Synthesis Kit (Roche) using Oligo(d)T primers. qRT–PCR was performed using the LightCycler® 480 SYBR Green I Master Mix (Roche). The following primer pairs were used:
β-actin: 5′-AGA GGG AAA TCG TGC GTG AC-3′ and 5′-CAA TAG TGA TGA CCT GGC CGT-3′, GH: 5′-CTT CTC GCT GCT GCT CAT C-3′ and 5′-ATC TTC CAG CTC CTG CAT CA-3′, UCP1: 5′-CTG GGC TTA ACG GGT CCT C-3′ and 5′-CTG GGC TAG GTA GTG CCA GTG-3′, C/EBPα: 5′-CAA GAA CAG CAA CGA GTA CCG-3′ and 5′-GTC ACT GGT CAA CTC CAG CAC-3′, C/EBPβ: 5′-CTG CGG GGT TGT TGA TGT-3′ and 5′-ATG CTC GAA ACG GAA AAG GT-3′, PPARγ: 5′-GCC CTT TGG TGA CTT TAT GG-3′ and 5′-CAG CAG GTT GTC TTG GAT GT-3′, GLUT4: 5′-CTG TCG CTG GTT TCT CCA AC-3′ and 5′-CAG GAG GAC GGC AAA TAG AA-3′, CD36: 5′-TGG CCT TAC TTG GGA TTG G-3′ and 5′-CCA GTG TAT ATG TAG GCT CAT CCA-3′, LPL: 5′-GGG CTC TGC CTG AGT TGT AG-3′ and 5′-TGG AAC ACT TTG TAG GGC ATC-3′, SREBP1c: 5′-AAC GTC ACT TCC AGC TAG AC-3′ and 5′-CCA CTA AGG TGC CTA CAG AGC-3′, FAS: 5′-ACA CAG CAA GGT GCT GGA G-3′ and 5′-GTC CAG GCT GTG GTG ACT CT-3′, HSL: 5′-CTC CAC ATG CCC CTC TAC AC-3′ and 5′-CAG AGC GCA AGC CAC AAG-3′, ATGL: 5′-GGA ACC AAA GGA CCT GAT GA-3′ and 5′-ACT CCA ACA AGC GGA TGG T-3′, AOX: 5′-AAG AGT TCA TTC TCA ACA GCC C-3′ and 5′-CTT GGA CAG ACT CTG AGC TGC-3′, ACC (liver): 5′- GGG ACT TCA TGA ATT TGC TGA TTC TCA GTT-3′ and 5′-GTC ATT ACC ATC TTC ATT ACC TCA ATC TC-3′, ACC (WAT): 5′-TCC ACG AAA AGA GCT GAC CT-3′ and 5′-ACT AAG GAT GCT CCC CAC CT -3′, SCD1: 5′- CCG GAG ACC CTT AGA TCG A-3′ and 5′-TAG CCT GTA AAA GAT TTC TGC AAA CC-3′, aP2: 5′-GGA TGG AAA GTC GAC CAC AA-3′ and 5′-GCT CAT GCC CTT TCA TAA ACT C-3′, GLUT2: 5′-GCA ACT GGG TCT GCA ATT TT-3′ and 5′-CCA GCG AAG AGG AAG AAC AC-3′, VLCAD: 5′-TTG TCA ACG AGC AGT TCC TG-3′ and 5′-AGC CTC AAT GCA CCA GCT AT-3′, LCAD: 5′-GCT GCC CTC CTC CCG ATG TT-3′ and 5′-ATG TTT CTC TGC GAT GTT GAT G-3′, MCAD: 5′-GGT TTG GCT TTT GGA CAA TG-3′ and 5′-TGA CGT GTC CAA TCT ACC ACA-3′, SCAD: 5′-CCT GCA ACC GAG AAG AAA TC-3′ and 5′-CCT GTC CTG TCC CTT GTG TT-3′, PGC1a: 5′-GTA AAT CTG CGG GAT GAT GG-3′ and 5′-GGT GGA AGC AGG GTC AAA A-3′, PRDM16: 5′-GAC ATT CCA ATC CCA CCA GA-3′ and 5′-CAC CTC TGT ATC CGT CAG CA-3′, Dio2: 5′-CAG TGT GGT GCA CTG CTC CAA TC-3′ and 5′-TGA ACC AAA GTT GAC CAC CAG-3′.
Data analyses
Sample size was chosen based on our previous experiments and published studies in which the same experimental procedures were used. Animals were randomly assigned for measurements or treatments. Experimental groups were created concerning similarity in age and body weight. Animals that became sick or died during the experiment and those with which the experimental performance was not successful were excluded from analyses. For all data, normal distribution was assumed and the unpaired, two-tailed Student’s t-test was used to calculate statistical significance of results. All graphs show average ± standard error of the mean (s.e.m.). *P < 0.05; **P < 0.01; ***P < 0.005. No blinding of investigators was done.
Acknowledgments
We thank Susanne Klaus and Susanne Keipert (DIfE, Potsdam) for help with bomb calorimetry; Kay Stötzer (Diagnostic Laboratory of the University Hospital Jena) for the plasma and urine analysis; David Sabatini (Whitehead Institute, Cambridge) for providing the raptor and rictor shRNA plasmids; and James Kirkland and Tamar Tchkonia (Mayo Clinic, Rochester) for advise on fat metabolism and ageing. At the FLI, we thank Jan Tuckermann for the CRE deleter (pCX-CRE) mouse line and advice; the staff of the animal house facility for embryo transfer and advice on mouse experiments; Andreas Krämer for help on quantitative microscope analysis; Maik Baldauf for help with histology; and Heike Heuer for providing the GH3 cell line. We also thank Janine Kruit (UMCG, Groningen) for providing the Hepa 1-6 cell line. L.M.Z. was supported by the Deutsche Forschungsgemeinschaft (DFG) through a grant (CA 283/1-1) to C.F.C.
Author contributions
LZ, TA, GH, SE, GK, MK and CM carried out experiments; LZ, MK, AL, NS, ZQW, JM, CM and CC were engaged in designing and/or advising on the experiments and they were involved in the interpretation of the results; LZ, CM and CC wrote the manuscript with contributions from JM; and CM and CC supervised the project.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Expanded View Figures PDF
Table EV1
Review Process File
References
- Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- Roesler WJ. The role of C/EBP in nutrient and hormonal regulation of gene expression. Annu Rev Nutr. 2001;21:141–165. doi: 10.1146/annurev.nutr.21.1.141. [DOI] [PubMed] [Google Scholar]
- Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- Calkhoven CF, Muller C, Leutz A. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 2000;14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- Wethmar K, Begay V, Smink JJ, Zaragoza K, Wiesenthal V, Dorken B, Calkhoven CF, Leutz A. C/EBPbetaDeltauORF mice–a genetic model for uORF-mediated translational control in mammals. Genes Dev. 2010;24:15–20. doi: 10.1101/gad.557910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493:338–345. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, III, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon-Biet SM, McMahon AC, Ballard JW, Ruohonen K, Wu LE, Cogger VC, Warren A, Huang X, Pichaud N, Melvin RG, et al. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 2014;19:418–430. doi: 10.1016/j.cmet.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- Wu JJ, Liu J, Chen EB, Wang JJ, Cao L, Narayan N, Fergusson MM, Rovira II, Allen M, Springer DA, et al. Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell Rep. 2013;4:913–920. doi: 10.1016/j.celrep.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jundt F, Raetzel N, Muller C, Calkhoven CF, Kley K, Mathas S, Lietz A, Leutz A, Dorken B. A rapamycin derivative (everolimus) controls proliferation through down-regulation of truncated CCAAT enhancer binding protein {beta} and NF-{kappa}B activity in Hodgkin and anaplastic large cell lymphomas. Blood. 2005;106:1801–1807. doi: 10.1182/blood-2004-11-4513. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr, Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- Le Bacquer O, Petroulakis E, Paglialunga S, Poulin F, Richard D, Cianflone K, Sonenberg N. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest. 2007;117:387–396. doi: 10.1172/JCI29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruss MD, Khambatta CF, Ruby MA, Aggarwal I, Hellerstein MK. Calorie restriction increases fatty acid synthesis and whole body fat oxidation rates. Am J Physiol Endocrinol Metab. 2010;298:E108–E116. doi: 10.1152/ajpendo.00524.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernstedt Asterholm I, Scherer PE. Enhanced metabolic flexibility associated with elevated adiponectin levels. Am J Pathol. 2010;176:1364–1376. doi: 10.2353/ajpath.2010.090647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Kamon J, Waki H, Imai Y, Shimozawa N, Hioki K, Uchida S, Ito Y, Takakuwa K, Matsui J, et al. Globular adiponectin protected ob/ob mice from diabetes and ApoE-deficient mice from atherosclerosis. J Biol Chem. 2003;278:2461–2468. doi: 10.1074/jbc.M209033200. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Kamon J, Waki H, Terauchi Y, Kubota N, Hara K, Mori Y, Ide T, Murakami K, Tsuboyama-Kasaoka N, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- Stefanovic-Racic M, Perdomo G, Mantell BS, Sipula IJ, Brown NF, O’Doherty RM. A moderate increase in carnitine palmitoyltransferase 1a activity is sufficient to substantially reduce hepatic triglyceride levels. Am J Physiol Endocrinol Metab. 2008;294:E969–E977. doi: 10.1152/ajpendo.00497.2007. [DOI] [PubMed] [Google Scholar]
- Mulligan JD, Stewart AM, Saupe KW. Downregulation of plasma insulin levels and hepatic PPARgamma expression during the fir st week of caloric restriction in mice. Exp Gerontol. 2008;43:146–153. doi: 10.1016/j.exger.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higami Y, Pugh TD, Page GP, Allison DB, Prolla TA, Weindruch R. Adipose tissue energy metabolism: altered gene expression profile of mice subjected to long-term caloric restriction. FASEB J. 2004;18:415–417. doi: 10.1096/fj.03-0678fje. [DOI] [PubMed] [Google Scholar]
- Selman C, Kerrison ND, Cooray A, Piper MD, Lingard SJ, Barton RH, Schuster EF, Blanc E, Gems D, Nicholson JK, et al. Coordinated multitissue transcriptional and plasma metabonomic profiles following acute caloric restriction in mice. Physiol Genomics. 2006;27:187–200. doi: 10.1152/physiolgenomics.00084.2006. [DOI] [PubMed] [Google Scholar]
- Huffman DM, Moellering DR, Grizzle WE, Stockard CR, Johnson MS, Nagy TR. Effect of exercise and calorie restriction on biomarkers of aging in mice. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1618–R1627. doi: 10.1152/ajpregu.00890.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Miura J, Lu LX, Bernier M, DeCabo R, Lane MA, Roth GS, Ingram DK. Circulating adiponectin levels increase in rats on caloric restriction: the potential for insulin sensitization. Exp Gerontol. 2004;39:1049–1059. doi: 10.1016/j.exger.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Katewa SD, Demontis F, Kolipinski M, Hubbard A, Gill MS, Perrimon N, Melov S, Kapahi P. Intramyocellular fatty-acid metabolism plays a critical role in mediating responses to dietary restriction in Drosophila melanogaster. Cell Metab. 2012;16:97–103. doi: 10.1016/j.cmet.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppala-Lindroos A, Vehkavaara S, Hakkinen AM, Goto T, Westerbacka J, Sovijarvi A, Halavaara J, Yki-Jarvinen H. Fat accumulation in the liver is associated with defects in insulin suppression of glucose production and serum free fatty acids independent of obesity in normal men. J Clin Endocrinol Metab. 2002;87:3023–3028. doi: 10.1210/jcem.87.7.8638. [DOI] [PubMed] [Google Scholar]
- Neff F, Flores-Dominguez D, Ryan DP, Horsch M, Schröder S, Adler T, Afonso LC, Aguilar-Pimentel JA, Becker L, Garrett L, et al. Rapamycin extends murine lifespan but has limited effects on aging. J Clin Invest. 2013;123:3272–3291. doi: 10.1172/JCI67674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JE, Burmeister L, Brooks SV, Chan CC, Friedline S, Harrison DE, Hejtmancik JF, Nadon N, Strong R, Wood LK, et al. Rapamycin slows aging in mice. Aging Cell. 2012;11:675–682. doi: 10.1111/j.1474-9726.2012.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bross TG, Rogina B, Helfand SL. Behavioral, physical, and demographic changes in Drosophila populations through dietary restriction. Aging Cell. 2005;4:309–317. doi: 10.1111/j.1474-9726.2005.00181.x. [DOI] [PubMed] [Google Scholar]
- McCarter RJ, Shimokawa I, Ikeno Y, Higami Y, Hubbard GB, Yu BP, McMahan CA. Physical activity as a factor in the action of dietary restriction on aging: effects in Fischer 344 rats. Aging. 1997;9:73–79. doi: 10.1007/BF03340130. [DOI] [PubMed] [Google Scholar]
- Weed JL, Lane MA, Roth GS, Speer DL, Ingram DK. Activity measures in rhesus monkeys on long-term calorie restriction. Physiol Behav. 1997;62:97–103. doi: 10.1016/s0031-9384(97)00147-9. [DOI] [PubMed] [Google Scholar]
- Millward CA, Heaney JD, Sinasac DS, Chu EC, Bederman IR, Gilge DA, Previs SF, Croniger CM. Mice with a deletion in the gene for CCAAT/enhancer-binding protein beta are protected against diet-induced obesity. Diabetes. 2007;56:161–167. doi: 10.2337/db06-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder-Gloeckler JM, Rahman SM, Janssen RC, Qiao L, Shao J, Roper M, Fischer SJ, Lowe E, Orlicky DJ, McManaman JL, et al. CCAAT/enhancer-binding protein beta deletion reduces adiposity, hepatic steatosis, and diabetes in Lepr(db/db) mice. J Biol Chem. 2007;282:15717–15729. doi: 10.1074/jbc.M701329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederschain D, Wee S, Chen L, Loo A, Yang G, Huang A, Chen Y, Caponigro G, Yao YM, Lengauer C, et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8:498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossati P, Prencipe L. Serum triglycerides determined colorimetrically with an enzyme that produces hydrogen peroxide. Clin Chem. 1982;28:2077–2080. [PubMed] [Google Scholar]
- McGowan MW, Artiss JD, Strandbergh DR, Zak B. A peroxidase-coupled method for the colorimetric determination of serum triglycerides. Clin Chem. 1983;29:538–542. [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Review Process File