Abstract
CD4 T-cell responses are functionally complex and regulate many aspects of innate and adaptive immunity. Follicular helper (Tfh) cells are CD4 T cells specialized to support B-cell production of isotype-switched, high-affinity antibody. So far, studies of Tfh cells in humans have focused on their differentiation requirements, with little research devoted to their antigen specificity. Here, after separating circulating human memory CD4 T cells based on expression of CXCR5, a signature marker of Tfh, we have quantified and assayed the influenza protein antigen specificity of blood Tfh cells and CD4 T cells lacking this marker. Through the use of peptide pools derived from nucleoprotein (NP) or haemagglutinin (HA) and a panel of human donors, we have discovered that circulating Tfh cells preferentially recognize peptide epitopes from HA while cells lacking CXCR5 are enriched for specificity toward NP. These studies suggest that reactive CD4 T cells specific for distinct viral antigens may have generalized differences in their functional potential due to their previous stimulation history.
Keywords: CD4 T cells, influenza virus, vaccines, B cells
Introduction
CD4 T cells possess a broad range of functions that contribute to protective immunity against influenza. Some of these functions segregate with CD4 T-cell lineage; for example, T helper type 1 and type 17 cells induce pulmonary inflammation, regulatory T cells limit tissue damage, cytotoxic CD4 T cells kill infected cells and T follicular helper (Tfh) cells help support antibody production, (reviewed in refs 1–3). One particular lineage of CD4 T cells, Tfh cells, is particularly critical for generating protection from influenza infection. Tfh cells function in an antigen-specific manner to support B-cell affinity maturation and class switching; both events essential for the generation of durable neutralizing antibody responses.2,4–7 For influenza, the generation of neutralizing antibodies to the virion surface protein haemagglutinin (HA) is most closely associated with vaccine efficacy.
Our recent work has shown that antigen-specific CD4 T-cell help can be a limiting factor in the antibody response to influenza infection and importantly, the delivery of CD4 help is linked to the protein specificity of the B cells, where only HA-specific CD4 T cells help the B-cell response to HA.8 This linked determination between CD4 T-cell and B-cell epitopes, a cornerstone of conjugate-vaccine design, highlights the importance of CD4 T-cell specificity in directing the antibody response to influenza. However, study of Tfh cells in humans has been limited because of the difficulty in sampling lymphoid tissues. Recent studies have shown CD4+ CXCR5+ cells in human peripheral blood to have B-cell helper functions analogous to conventional Tfh cells.9–15 The ability to easily access lymphoid cells from the blood makes the evaluation of CD4+ CXCR5+ cells practical for both acute and longitudinal studies of the immune response.
In this report, we sought to quantify influenza-reactivity in Tfh and non-Tfh cells and to examine the relationship between influenza antigen specificity and potential for helper function. Because of the correlative link between influenza vaccine efficacy and neutralizing anti-HA antibodies, we focused on cells reactive to HA and the internal virion protein, nucleoprotein (NP). Our studies suggest that cells that do or do not have B-cell helper function differ in the frequency of reactivity to alternate influenza proteins.
Materials and methods
Isolation of memory CD4 T-cell subsets and naive B cells
Peripheral blood mononuclear cells were collected from 15 healthy donors (ages 18–50 years). The University of Rochester Research Subjects Review Board approved this study protocol, and human experimentation guidelines of the US Department of Health and Human Services and the University of Rochester were followed. Study procedures were in accordance with the ethical standards of the Declaration of Helsinki and all subjects provided written informed consent. CD4+ CD45RA− T cells were purified using a memory CD4 T-cell isolation kit per the manufacturer’s directions (Miltenyi Biotec, Auburn, CA). Single, CD4+ CD45RA− cells were separated into two populations, based on expression of CXCR5, by flow cytometric sorting. CD19+ cells were enriched from peripheral blood mononuclear cells using a CD19+ multisort kit (Miltenyi Biotec) and further enriched for naive B cells by first releasing cells from the anti-CD19 microbeads, and then enriching for naive cells by depletion with CD27 microbeads (Miltenyi Biotec).
B-cell helper assay
Fifty thousand naive B cells (CD19+ CD27−) were cultured with 50 000 CD4+ CD45RA− CXCR5+ or CD4+ CD45RA− CXCR5− T cells in the presence or absence of 500 ng/ml staphylococcal enterotoxin B (SEB). Culture supernatants, harvested at day 10, were assayed for IgG and IgM by ELISA. Purified anti-IgG (MT145), anti-IgG-alkaline phosphatase (MT78), and human IgM ELISA Kit (alkaline phosphatase) reagents were obtained from MabTech (Cincinnati, OH).
Peptides
Seventeen-mer peptides overlapping by 11 amino acids spanning the entire Influenza H1 [NR-2602: HA, A/New Caledonia/20/99 (H1N1)] and NP [NR-2611: NP, A/New York/348/03 (H1N1)] sequences were used, provided by the NIH Biodefense and Emerging Infections Research Resources Repository (Bethesda, MD). Peptide sequence data can be obtained at http://www.beiresources.org referencing the NR numbers. All peptides from the HA library were pooled together and used for stimulation ‘HA’ with each peptide at a final concentration of 2 μm. The same was done for NP.
T-cell EliSpot assays
EliSpot assays were performed as previously described:16,17 150 000 CD4+ CD45RA− CXCR5+ or CD4+ CD45RA− CXCR5− cells were cultured with 100 000 autologous antigen-presenting cells (APC; peripheral blood mononculear cells depleted of CD4+ and CD8+ cells) and pools of peptides containing either the entire HA or NP sequence (2 μm of each peptide) for 36 hr at 37°. Plates were washed and processed as previously described for the detection of cytokine-producing cells.16,17 Quantification of cytokine-secretion spot counts was performed with an Immunospot reader series 2A, using Immunospot software, version 5.0.9.19.
CD137/CD69 assay
Sorted CD4+ CD45RA− T cells were cultured with autologous APC at a 1 : 3 ratio in the presence of peptides for 26–30 hr at 37°, washed and stained with Live/Dead Fixable Aqua Dead Cell Stain Kit (Life Technologies, Carlsbad, CA), washed again and resuspended in Fc Blocking Reagent (Miltenyi Biotec). The staining cocktail was as follows anti-CD8-phycoerythrin (PE)-Cy7 (RPA-T8), anti-HLA-DR-PE-Cy7 (G46-6), anti-CD19-PE-Cy7 (SJ25C1), anti-CD14-PE-Cy7 (M5E2), anti-CD4-FITC (RPA-T4), and anti-CD45RA-allophycocyanin (HI100) antibodies obtained from BD Biosciences (San Jose, CA); anti-CXCR5-PE (MU5UBEE) antibody obtained from eBioscience (San Diego, CA); anti-CD69-allophycocyanin-Cy7 (FN50) and anti-CD137-BV421 (4B4-1) antibodies obtained from BioLegend (San Diego, CA). Data were collected with a FACSCanto flow cytometer (BD Biosciences), and analysed with FlowJo 8.6 software (Tree Star Inc., Ashland, OR).
Statistical analysis
GraphPad PRISM V5 software (GraphPad Software, La Jolla, CA) was used for all statistical tests. Statistical significance was evaluated using Wilcoxon signed-rank test with a 95% confidence interval.
Results and discussion
Purification strategy and B-cell helper function of peripheral blood CD4+ CD45RA− CXCR5+ and CXCR5− cells
Recent investigations have shown that most CD4 T-cell help for B cells in human peripheral blood is contained within the CD4+ CXCR5+ population.9–15 We sought to measure the influenza antigen-specificity of these circulating CD4 T cells and compare it with the specificity of circulating CD4 T cells that lacked B-cell helper function. Accordingly, we separated populations of highly enriched, antigen-experienced CD4 cells from the blood of healthy human donors into two populations: CD4+ CD45RA− CXCR5+ (CXCR5+) and CD4+ CD45RA− CXCR5− (CXCR5−) T cells with purities generally > 98% (Fig.1a).
Figure 1.
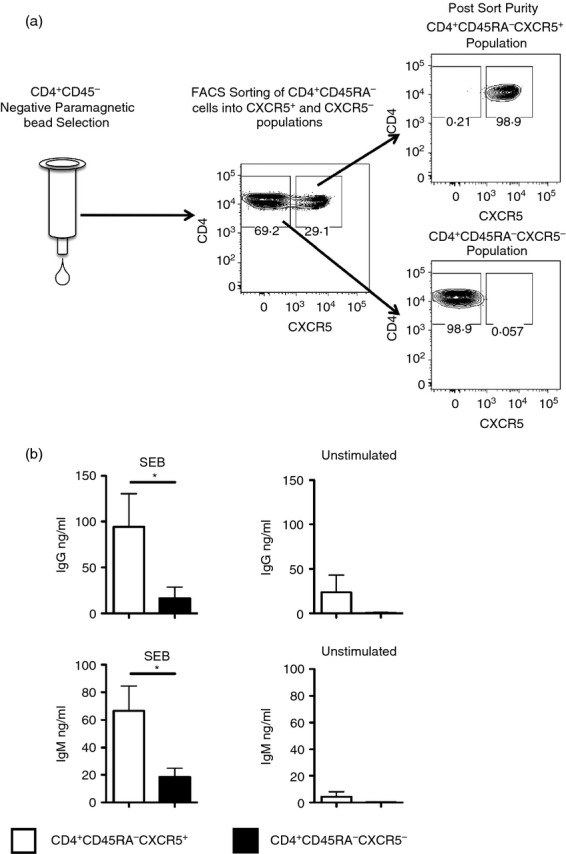
(a) Purification of CXCR5+ and CXCR5– memory CD4 T cells from human peripheral blood mononuclear cells (PBMC). PBMC from healthy human donors were enriched for CD4+ CD45RA– cells with negative paramagnetic bead selection. Pools of enriched cells were then stained for CD4, CD45RA and CXCR5. Stained cells were separated into CD4+ CD45RA− CXCR5+ and CD4+ CD45RA− CXCR5− cells. After sorting, the separated cells were typically > 98% pure. (b) B-cell help is concentrated within the CXCR5+ population of CD4+ CD45RA− human blood cells. Fifty thousand isolated CXCR5+ (open bars) or CXCR5– CD4+ CD45RA− T cells (filled bars) were cultured with 50 000 autologous naive B cells in the presence or absence of 500 ng/ml of staphylococcal enterotoxin B (SEB). Day 10 culture supernatants were tested for total IgG and IgM by ELISA. The average levels of IgG (a) and IgM (b) in supernatant are shown from SEB-stimulated or unstimulated cultures, with the error bars representing the SEM. Data were analysed using the Wilcoxon signed-rank test and * indicates a P-value < 0·05.
To confirm that these methods separated functionally distinct populations of memory CD4 T cells, the ability of the sorted CXCR5+ and CXCR5− cells to support antibody production and class switching by naive B cells was assayed, using SEB to facilitate cognate interactions between CD4 T cells and B cells as described.11,15,18 After a 10-day co-culture, IgG (Fig.1b top panel) and IgM (Fig.1b bottom panel) in the supernatants was quantified by ELISA. These results indicate that the CXCR5+ population was selectively competent to support production of antibody production and class switching of naive B cells.
Distribution of influenza antigen-specificity between circulating memory CD4 T-cell subsets
After enriching for blood Tfh and non-Tfh cells, single cell assays were used to measure influenza antigen reactivity and specificity. Both populations were cultured with overlapping pools of peptides corresponding to the full coding sequence of the HA and NP in the presence of autologous APC. Reactive cells were quantified by cytokine EliSpots. Use of peptide pools allows each potential CD4 T-cell epitope to be accessible to bind to host MHC and stimulate epitope-specific CD4 T cells. The HA and NP responses were selected for this study because our previous studies have shown that most human subjects have readily detectable CD4 T-cell responses to epitopes contained in these influenza proteins.16,17,19 Additionally, HA-reactive Tfh cells are particularly important to monitor, as these are likely to be the most crucial for neutralizing antibody production.17,20
Figure2 shows the results of these experiments, where the summed frequency of influenza NP-specific and HA-specific CD4 T cells in blood Tfh and non-Tfh cells are presented in Fig.2(a), with each donor indicated by a unique symbol. Strikingly, there were some individuals noted with low frequencies of antigen-reactive Tfh cells, despite abundant influenza-specific CXCR5− cells (e.g. Fig.2b, subjects 2, 3, 4). The individuals with few influenza-specific Tfh cells had similar numbers of total circulating Tfh cells, identified by staining, as those subjects showing high influenza reactivity [indicated in Fig.2(b) below the subject number]. This result indicates that some humans are selectively deficient in influenza-specific blood circulating Tfh cells.
Figure 2.
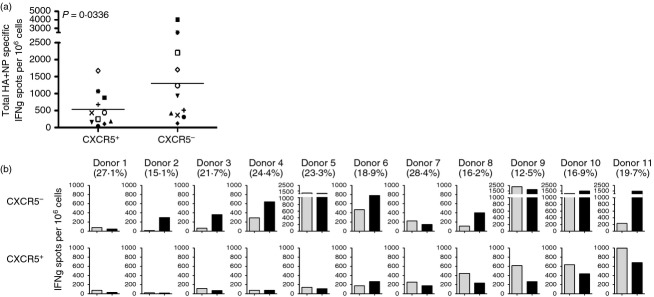
(a) Quantification of influenza-reactive circulating follicular helper T (Tfh) and non-Tfh cells. CXCR5+ and CXCR5– CD4+ CD45RA− T cells were cultured with 2 μm pools of peptide for haemagglutinin (HA) or nucleoprotein (NP) influenza proteins in the presence of autologous antigen-presenting cells (APC). Antigen-specific CD4 T cells were measured by interferon-γ (IFN-γ) EliSpot at 36–40 hr of stimulation and the abundance was summed. (b) Influenza protein-specific reactivity in circulating Tfh and non-Tfh. Frequency of HA-specific (grey bars) or NP-specific (black bars) IFN-γ secreting cells per 1 000 000 sorted CXCR5– (top row) or CXCR5+ (bottom row) for individual donors. The percentage of CXCR5+ circulating Tfh found in each subject is indicated below the subject number at the top of each panel.
When we analysed the distribution of influenza-reactive circulating memory Tfh and non-Tfh cells toward HA and NP (Fig.2b), an intriguing pattern emerged. NP reactivity was highly enriched within the memory non-Tfh population in most of the subjects (Fig.2b, top panel). In the circulating Tfh population (Fig.2b bottom panel) the opposite trend was observed, with Tfh from most subjects displaying enhanced reactivity to HA. To compare the distribution of HA and NP-specific cells within the CXCR5+ and CXCR5− populations, the ratio of NP : HA-specific cells was determined (Fig.3a). This representation, shown as a ratio of NP : HA for each donor, allows for normalization of reactivity among individuals who differ in their abundance of influenza-reactive CD4 T cells. This representation shows that the relative reactivity to NP is enriched in the non-Tfh whereas most Tfh have equal or greater reactivity to HA.
Figure 3.
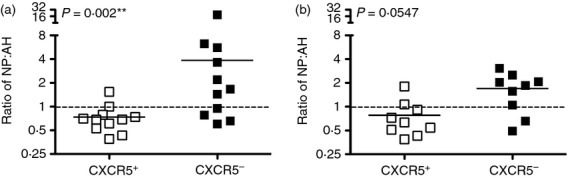
Relative reactivity of follicular helper T (Tfh) and non-Tfh cells for haemagglutinin (HA) and nucleoprotein (NP) in individual healthy human donors. The distribution of HA-specific and NP-specific, interferon-γ (IFN-γ) secreting cells as determined by IFN-γ cytokine assay (left) or the surface expression of CD137 and CD69 after 30 hr of simulation with peptide pools (right) from each donor was calculated as a ratio of total NP-specific/HA-specific cells (NP/HA). Data were analysed using the Wilcoxon signed-rank test.
Because there might be T-cell lineage-biased production of cytokines, we also used a cytokine-independent method of measuring influenza specificity. Additional subjects were recruited and a flow cytometry panel was developed to quantify the surface expression of CD137 and CD69, markers known to be rapidly upregulated upon T-cell receptor stimulation.21–25 The results using the cytokine-independent assay were in agreement with the cytokine EliSpot data (Fig.3b). Both assays indicated that the CXCR5− population contains a higher frequency of NP-specific to HA-specific cells than did the CXCR5+ population. Most individuals (> 65%) displayed this pattern and some subjects showed dramatic disparities in these ratios. Collectively, these findings highlight a pattern for the Tfh cells in circulating influenza specific memory CD4 T cells among many healthy human subjects to be most enriched for HA-specific cells, while NP-specific cells were generally more abundant within the CXCR5− population.
We find it intriguing that among our individual donors, each of which is presumed to have distinct infection and exposure history, many displayed the pattern of high NP reactivity in non-Tfh cells and preferential reactivity toward HA in Tfh populations. This finding was unexpected because most adults have multiple but distinct exposures to influenza though both clinical vaccination and overt and subclinical infections that occur but are not formally documented. We can put forward at least four mechanisms to account for these patterns of specificity. The first is that the glycosylated HA that can bind to cell surface sialic acid could allow a unique pathway of uptake by APC or allow HA to bind to other soluble scavengers after vaccination/infection. Second, as an RNA-binding protein, NP may intrinsically activate signalling through RNA-sensing receptors. Also, the impact of circulating antibody during vaccination or infection may differ between these proteins. Pre-existing circulating NP-specific antibody to highly conserved NP may block B-cell receptor uptake whereas novel B-cell receptor epitopes within HA created by antigenic drift may allow uptake of HA by HA-specific B cells. This would allow enhanced presentation of HA epitopes on B cells, driving more HA-specific CD4 T cells into the Tfh lineage. Finally, intramuscular vaccines are enriched for HA proteins26,27 and CD4 T cells specific for HA may more frequently be boosted by vaccination, perhaps enriching for Tfh cells. Further analyses of CD4 T-cell reactivity to other membrane-associated influenza proteins such M1 and H3, as well as the other internal genetically conserved proteins, should help to distinguish among these possibilities.
It is interesting to consider the implications of these studies. First, variability between donors demonstrates that substantial heterogeneity exists in both the magnitude and pattern of influenza-specific CD4 T cells within the circulating memory compartment. In some subjects, this memory contained fewer than 200 influenza-specific cells per million Tfh cells, but most subjects possessed thousands of HA-reactive and NP-reactive cells per million non-Tfh cells. We do not know the infection or vaccination history of the healthy individuals surveyed here, but all our healthy donors tested display evidence of previous encounter with influenza, based on serum reactivity to H1, H3 and NP (data not shown). It is interesting to speculate that those individuals with scarce influenza-specific Tfh cells, particularly those specific for HA, may exhibit selective deficiencies in their neutralizing antibody responses to vaccination. In contrast to memory circulating Tfh cells, the non-Tfh cells may supply distinct but also crucial effector functions, such as the recruitment of innate and adaptive effector cells to infected or compromised tissues, the support of expansion of CD8 T cells, and the mediation of direct killing of antigen-bearing cells during influenza infection (reviewed in ref. 1,3). Understanding how and why antigen-specificity partitions the different functional subsets of CD4 T cells will be paramount to developing vaccine approaches that enhance the characteristics of immunity in the host that provide the needed protection.
Acknowledgments
We would like to thank the following: the URMC clinical core for donor recruitment and sample processing, the URMC Flow Core for operating the flow sorter, and Francisco A. Chaves for peptide preparation. We would also like to thank Dr Jennifer L. Nayak and Zackary Knowlden for thoughtful editorial suggestions and discussions during the preparation of this manuscript. This work was supported by the following grants HHSN27220201200005C, HHSN266200700008C, HHSN272201300005C and 5T32AI007285 from the National Institutes of Health.
Disclosures
All the authors declare no conflict of interest.
References
- McKinstry KK, Strutt TM, Swain SL. Hallmarks of CD4 T cell immunity against influenza. J Intern Med. 2011;269:507–18. doi: 10.1111/j.1365-2796.2011.02367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale JS, Ahmed R. Memory T follicular helper CD4 T cells. Front Immunol. 2015;6:16. doi: 10.3389/fimmu.2015.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zens KD, Farber DL. Memory CD4 T cells in influenza. Curr Top Microbiol Immunol. 2015;386:399–421. doi: 10.1007/82_2014_401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LM, Yu D. Follicular helper T-cell memory: establishing new frontiers during antibody response. Immunol Cell Biol. 2014;92:57–63. doi: 10.1038/icb.2013.68. [DOI] [PubMed] [Google Scholar]
- Crotty S. Follicular helper CD4 T cells (TFH. Annu Rev Immunol. 2011;29:621–63. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- Ma CS, Deenick EK, Batten M, Tangye SG. The origins, function, and regulation of T follicular helper cells. J Exp Med. 2012;209:1241–53. doi: 10.1084/jem.20120994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa CG, Cook MC. Blood relatives of follicular helper T cells. Immunity. 2011;34:10–2. doi: 10.1016/j.immuni.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Alam S, Knowlden ZA, Sangster MY, Sant AJ. CD4 T cell help is limiting and selective during the primary B cell response to influenza infection. J Virol. 2014;88:314–24. doi: 10.1128/JVI.02077-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D, Preite S, Reboldi A, Ronchi F, Ansel KM, Lanzavecchia A, Sallusto F. Persistent antigen and germinal center B cells sustain T follicular helper cell responses and phenotype. Immunity. 2013;38:596–605. doi: 10.1016/j.immuni.2012.11.020. [DOI] [PubMed] [Google Scholar]
- Chevalier N, Jarrossay D, Ho E, Avery DT, Ma CS, Yu D, Sallusto F, Tangye SG, Mackay CR. CXCR5 expressing human central memory CD4 T cells and their relevance for humoral immune responses. J Immunol. 2011;186:5556–68. doi: 10.4049/jimmunol.1002828. [DOI] [PubMed] [Google Scholar]
- Morita R, Schmitt N, Bentebibel SE, et al. Human blood CXCR5+ CD4+ T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–21. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerli P, Willimann K, Lang AB, Lipp M, Loetscher P, Moser B. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J Exp Med. 2000;192:1553–62. doi: 10.1084/jem.192.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N, Ueno H. Blood Tfh cells come with colors. Immunity. 2013;39:629–30. doi: 10.1016/j.immuni.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Tsai LM, Leong YA, et al. Circulating precursor CCR7lo PD-1hi CXCR5+ CD4+ T cells indicate Tfh cell activity and promote antibody responses upon antigen reexposure. Immunity. 2013;39:770–81. doi: 10.1016/j.immuni.2013.09.007. [DOI] [PubMed] [Google Scholar]
- Locci M, Havenar-Daughton C, Landais E, et al. Human circulating PD-1+ CXCR3– CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–69. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KA, Topham D, Chaves FA, Sant AJ. Cutting edge: CD4 T cells generated from encounter with seasonal influenza viruses and vaccines have broad protein specificity and can directly recognize naturally generated epitopes derived from the live pandemic H1N1 virus. J Immunol. 2010;185:4998–5002. doi: 10.4049/jimmunol.1001395. [DOI] [PubMed] [Google Scholar]
- Nayak JL, Fitzgerald TF, Richards KA, Yang H, Treanor JJ, Sant AJ. CD4+ T-cell expansion predicts neutralizing antibody responses to monovalent, inactivated 2009 pandemic influenza A(H1N1) virus subtype H1N1 vaccine. J Infect Dis. 2013;207:297–305. doi: 10.1093/infdis/jis684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J Exp Med. 2001;193:1373–81. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KA, Nayak J, Chaves FA, DiPiazza A, Knowlden ZA, Alam S, Treanor JJ, Sant AJ. Seasonal influenza can poise hosts for CD4 T-cell immunity to H7N9 avian influenza. J Infect Dis. 2015;212:86–94. doi: 10.1093/infdis/jiu662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S, Knowlden ZA, Sangster MY, Sant AJ. CD4 T cell help is limiting and selective during the primary B cell response to influenza virus infection. J Virol. 2014;88:314–24. doi: 10.1128/JVI.02077-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehler TC, Nonn M, Brandt B, et al. Targeting the activation-induced antigen CD137 can selectively deplete alloreactive T cells from antileukemic and antitumor donor T-cell lines. Blood. 2007;109:365–73. doi: 10.1182/blood-2006-04-014100. [DOI] [PubMed] [Google Scholar]
- Wolfl M, Kuball J, Ho WY, Nguyen H, Manley TJ, Bleakley M, Greenberg PD. Activation-induced expression of CD137 permits detection, isolation, and expansion of the full repertoire of CD8+ T cells responding to antigen without requiring knowledge of epitope specificities. Blood. 2007;110:201–10. doi: 10.1182/blood-2006-11-056168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms PE, Ellis TM. Utility of flow cytometric detection of CD69 expression as a rapid method for determining poly- and oligoclonal lymphocyte activation. Clin Diagn Lab Immunol. 1996;3:301–4. doi: 10.1128/cdli.3.3.301-304.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso A, Licenziati S, Corulli M, et al. Flow cytometric analysis of activation markers on stimulated T cells and their correlation with cell proliferation. Cytometry. 1997;27:71–6. doi: 10.1002/(sici)1097-0320(19970101)27:1<71::aid-cyto9>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Biselli R, Matricardi PM, D’Amelio R, Fattorossi A. Multiparametric flow cytometric analysis of the kinetics of surface molecule expression after polyclonal activation of human peripheral blood T lymphocytes. Scand J Immunol. 1992;35:439–47. doi: 10.1111/j.1365-3083.1992.tb02879.x. [DOI] [PubMed] [Google Scholar]
- Gerdil C. The annual production cycle for influenza vaccine. Vaccine. 2003;21:1776–9. doi: 10.1016/s0264-410x(03)00071-9. [DOI] [PubMed] [Google Scholar]
- Milian E, Kamen AA. Current and emerging cell culture manufacturing technologies for influenza vaccines. Biomed Res Int. 2015;2015:504831. doi: 10.1155/2015/504831. [DOI] [PMC free article] [PubMed] [Google Scholar]


