Abstract
Translocation of E. coli across the gut epithelium can result in fatal sepsis in post-surgical patients. In vitro and in vivo experiments have identified the existence of a novel pathotype of translocating E. coli (TEC) that employs an unknown mechanism for translocating across epithelial cells to the mesenteric lymph nodes and the blood stream in both humans and animal models. In this study the genomes of four TEC strains isolated from the mesenteric lymph nodes of a fatal case of hospitalised patient (HMLN-1), blood of pigs after experimental shock (PC-1) and after non-lethal haemorrhage in rats (KIC-1 and KIC-2) were sequenced in order to identify the genes associated with their adhesion and/or translocation. To facilitate the comparison, the genomes of a non-adhering, non-translocating E. coli (46–4) and adhering but non-translocating E. coli (73–89) were also sequenced and compared. Whole genome comparison revealed that three (HMLN-1, PC-1 and KIC-2) of the four TEC strains carried a genomic island that encodes a Type 6 Secretion System that may contribute to adhesion of the bacteria to gut epithelial cells. The human TEC strain HMLN-1 also carried the invasion ibeA gene, which was absent in the animal TEC strains and is likely to be associated with host-specific translocation. Phylogenetic analysis revealed that the four TEC strains were distributed amongst three distinct E. coli phylogroups, which was supported by the presence of phylogroup specific fimbriae gene clusters. The genomic comparison has identified potential genes that can be targeted with knock-out experiments to further characterise the mechanisms of E. coli translocation.
Introduction
Bacterial translocation (BT) is one of the main causes of sepsis in hospitalised and immunocompromised patients and is defined as the passage of viable bacteria (and their products) through the gut epithelium to mesenteric lymph nodes (MLN) and further to the blood and sterile organs [1]. Although E. coli forms a small population of the gut microbial community, it is one of the most commonly isolated species from surgical patients with septicaemia accounting, in some cases, for up to 37.5% mortality rate [2]. Previous studies have demonstrated that the process of BT occurs independently of the E. coli gut population size, and that adherence and the subsequent translocation is a selective event [3, 4]. In other words, only a few of the many E. coli strains found in the gut are capable of adhering to the gut epithelium, and even fewer strains are capable of translocating to extra-intestinal sites. These results suggest that BT is a process dictated to a great extent by the ability of the bacteria to translocate.
None the less, this selective process has been found to occur in different animal hosts, and has led to isolation of E. coli strains capable of translocating with higher efficiency than other resident E. coli strains in these particular hosts [4–7]. These include a strain isolated from a case of fatal pancreatitis where the bacterium was found in blood, MLN and peritoneal cavity of the deceased person (HMLN-1) [7], an E. coli strain isolated from pigs subjected to experimental shock (PC-1)[8], and two strains of E. coli isolated from rat subjected to starvation with or without haemorrhage (KIC-1 and KIC-2) [5].
Animal studies have shown that the efficiency of translocation is dependent on host specificity with the human and pig translocating E. coli (TEC) strains having a higher rate of translocation in the pig model than the rat TEC strains [6]. The high translocation rate of the human TEC strain in pigs is not surprising since the physiology of the pig intestine and its microbial community is similar to humans [9, 10].
In order to elucidate the pathogenesis of TEC strains, Ramos et al, (2011) investigated the virulence characteristics and the interleukin-8 response to infection by the above TEC strains [11]. They also tested for the presence of 47 virulence genes associated with intestinal and extra-intestinal E. coli and found that, among TEC strains, the virulence gene coding for Group III poly sialic capsule synthesis was carried by HMLN-1 and PC-1 only and the enteroaggregative stable toxin-1 (EAST1) gene was carried by KIC-2 only. However, it was found that TEC strains elicited significantly higher IL-8 response in both gastrointestinal cell lines (i.e. Caco-2 and HT-29 cells) and monocyte THP-1 cells than non-TEC strains [11]. These in vitro experiments also revealed that most TEC strains (except KIC-1) could adhere with greater efficiency to gut epithelial cell lines than non-TEC strains [11]. The KIC-1 strain adhered to these cells at the same level as non-TEC strains, suggesting that the degree of adhesion is not solely associated with translocation. Based on these results, we postulated that TEC strains might harbour certain properties which are probably not seen in other pathotypes of E. coli and that they may have unique virulence genes contributing to their translocation.
This pilot study aimed to employ a genomic approach to identify genes and/or genetic mechanisms that could potentially contribute to adhesion and translocation of TEC strains. Genomes of the above TEC and non-TEC strains were sequenced and compared and, here, we report all genetic differences identified between these two groups of E. coli and discuss their possible role in the translocation ability of these bacteria.
Materials and Methods
Bacterial strains and growth condition
Four translocating and two non-translocating E. coli strains were included in this study. Table 1 shows details of their sources of isolation and other characteristics of each strains. All bacterial strains were grown in Luria-Bertani (LB) broth, to log phase (3–4 h), at 37°C with agitation (100 strokes per min). The cultures were centrifuged and resuspended in phosphate-buffered saline (PBS). Total genomic DNA of the strains was extracted using DNeasy blood and tissue kit (QIAGEN, Australia) as per manufacturer’s instructions. DNA quality from the strains was visually assessed using gel electrophoresis using a 1% agarose (Amresco, Astral Scientific, Australia) gel in 0.6 x Tris/Borate/EDTA (TBE) buffer, pre-stained with ethidium bromide, run at 130 V for 60 min. Bands were visualized under ultraviolet (UV). DNA was then submitted for sequencing on ice.
Table 1. Summary of Escherichia coli genomes sequenced in this study.
| Strain | Source of isolate (reference no.) | No. of Contigs | Genes | Plasmids | Accession Number |
|---|---|---|---|---|---|
| Translocating strains (TEC) | |||||
| HMLN-1 | Isolated from MLNs, blood and peritoneal fluid of a patient with fatal haemorrhagic pancreatitis [7] | 59 | 4678 | 1 | LFKC00000000 |
| PC-1 | Isolated from MLNs and blood of pigs subjected to ischemia/ reperfusion or peritonitis [8] | 125 | 4725 | 1 | LFKB00000000 |
| KIC-1 | Isolated from MLNs of rats subjected to 24 or 48 h of starvation with and without haemorrhage [5] | 70 | 4590 | 1 | LFKA00000000 |
| KIC-2 | Isolated from MLNs of rats subjected to 24 or 48 h of starvation with and without haemorrhage [5]. | 175 | 4816 | 0 | LFJZ00000000 |
| Non-translocating strains (controls) | |||||
| 73–89 | Isolated from the caecal epithelium of rats subjected to 48 h of starvation with haemorrhage [11] | 124 | 5110 | 1 | LFJY00000000 |
| 46–4 | Isolated from the caecal contents of rats subjected to 48 h starvation with haemorrhage [11] | 66 | 4595 | 1 | LFJX00000000 |
Genome sequencing and phylogenetics
Genomic DNA of all six strains was sequenced at the Australian Genome Research Facility (AGRF) using an Illumina MiSeq to produce paired-end 101-bp reads. Read quality was checked with FASTQC and Spades 3.0.0 was used to assemble the E. coli genomes with k-mer values of 15, 21, 33, 51 and 71 [12] The evolutionary relationship of the E. coli strains sequenced in this study compared to 29 complete E. coli and one E. fergusonnii genome was predicted by phylogenetic analysis using concatenated nucleotide sequences of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA and recA), as previously described [13]. Sequences were aligned in Muscle v3.8.31 with default settings. The Neighbour-Joining method of MEGA5 was used to infer the evolutionary history, with distances computed by the Jukes-Cantor method. The phylogenetic tree was rooted using E. fergusonnii as an out-group.
Genome comparative analysis
The Rapid Annotation using Subsystems Technology (RAST) server provided annotation for the six E. coli genomes. Manual curation was also performed to ensure the accuracy of the annotation with particular attention to regions of difference, prophage regions and genomic islands using Artemis [14]. BLAST ring image generator (BRIG) [15], Easyfig [16] and Artemis Comparison Tool (ACT) [17] were used to visualise the comparison of the six E. coli genomes sequenced in this study and 41 previously completed E. coli genomes (S1 Table). The presence/absence of chaperone-usher (CU) fimbriae gene clusters were determined with BLAST using 38 CU fimbrial operons defined in Wurpel et al [18] and visualized with BRIG.
Results
Draft genomes of TEC
The draft genomes of four previously described efficiently translocating E. coli strains isolated from humans (HMLN-1), pigs (PC-1) and rats (KIC-1 and KIC-2) were sequenced. The four TEC strains were selected based on their ability to adhere and translocate across human gut epithelial Caco-2 and HT-29 cells [11]. The draft genomes were assembled into 59 contigs for strain HMLN-1, 125 contigs for PC-1, 70 contigs for KIC-1 and finally 175 contigs for the strain KIC-2 (Table 1). In addition, draft genomes were sequenced from two non-translocating E. coli strains (46–4 and 73–89) isolated from rats. The non-TEC strains were used as controls for testing TEC strain phenotypes [11]. The draft genomes of E. coli strains 46–4 and 73–89 assembled into 66 contigs and 124 contigs, respectively. Three of the TEC strains (HMLN-1, PC-1 and KIC-1) and both non-TEC strains genomes carried a single plasmid ranging from 53 kbp to 101 kbp that encodes a conjunctive transfer system and several hypothetical proteins. No plasmids were found in the genome of the strain KIC-2.
Relationship between TEC and other E. coli pathotypes
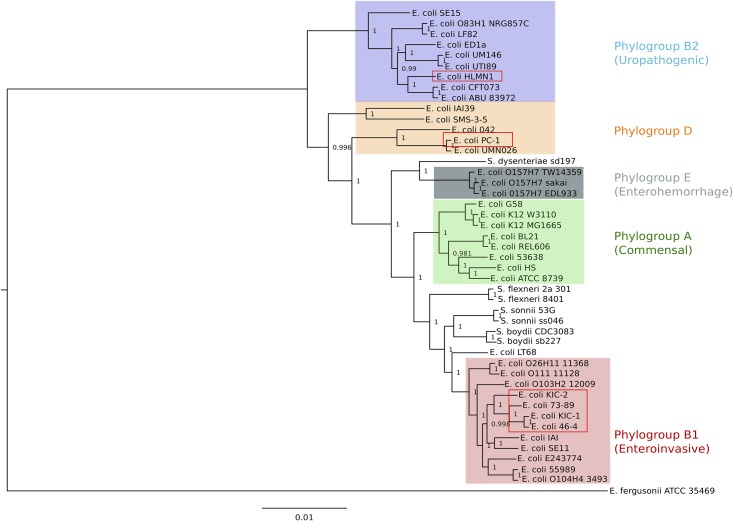
To determine the relationship between the TEC strains sequenced in this study and other E. coli strains, a neighbour-joining phylogenetic tree was constructed using the concatenated nucleotide sequence of seven housekeeping genes with a total length of 9016 bp. The four TEC strains were separated into three distinct phylogenetic groups (Fig 1). The E. coli population is often divided into five phylogenetic groups (A, B1, B2, D and E) based on differences in their phylogenetic relationships [19]. The human and the pig strains belong to the phylogenetic groups B2 and D, respectively. These two phylogroups usually contain extraintestinal pathogenic E. coli (ExPEC) strains [20, 21]. On the other hand, the rat TEC strains KIC-1 and KIC-2 and the two non-TEC strains 46–4 and 73–89 belonged to the B1 phylogroup, which together with phylogenetic group A, are considered as commensals [22, 23] or intestinal pathogenic strains.
Fig 1. Evolutionary context of translocating E. coli.
The phylogeny of E. coli strains inferred using the Neighbour Joining method on the concatenated nucleotide sequence of seven housekeeping genes (~9 kb) with E. fergusonii ATCC 35469 as an out-group. Coloured boxes show the phylogroups (A, B1, B2, D and E). E. coli strains sequenced in this study are marked with red boxes.
Genomic features of translocating E. coli strains
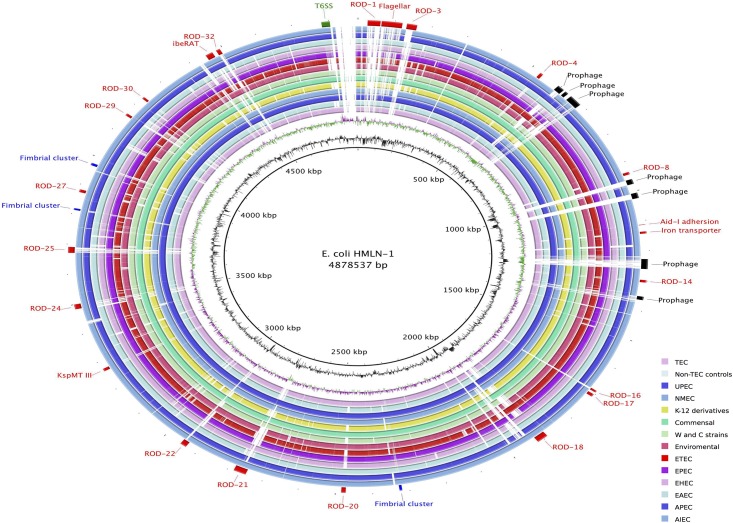
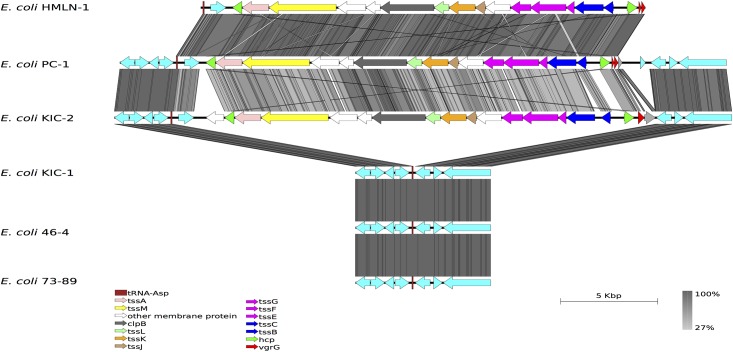
The genomes of the four TEC strains were compared to the two non-TEC genomes to determine the genetic differences that may be linked to adhesion and translocation across epithelial cells. The comparisons were also extended to include 41 publically available E. coli genomes representing all the major pathotypes. The comparison revealed that the genomic backbone of E. coli HMLN-1 was most similar to the genomes of Adherent-Invasive E. coli (AIEC) strains LF82, NRG857C and UM146. In E. coli HMLN-1, we have defined 29 regions of differences (RODs) and five putative prophage regions that are sporadically distributed amongst the other 46 E. coli genomes (Table 2 and Fig 2). The five putative prophage regions, of which three are unique to HMLN-1, could be distinguished from the E. coli genomic backbone by the presence of phage structural/replication genes and skews in the average GC-content. Amongst the RODs, the most significant was the presence of the genomic island GI-argU, which encodes a Type VI Secretion System (T6SS). GI-argU is a 22.4 kbp region inserted next to the tRNAargU and was present in three of the TEC strains; HMLN-1, PC-2 and KIC-2 but absent in KIC-1 and both of the non-TEC control strains, E. coli 46–4 and 73–89 (Fig 3) Genes encoding for a T6SS are commonly found in most other E. coli pathotypes but they are absent in the K-12 derivatives, environmental and enterotoxigenic E. coli strains (Fig 2). The 18 genes located within the GI-argU region in E. coli HMLN-1, PC-1 and KIC-2 encode all the structural proteins of the T6SS and the two T6SS-associated secreted proteins; hemolysin-coregulated protein (Hcp) and valine-glycine repeat G (VgrG) [24].
Table 2. Major regions of difference in E. coli HMLN-1.
| Identifier | Contig | Start | End | Size (bp) | Insertion site | No. of genes | GC Content (%) | Description | Notable virulence factor |
|---|---|---|---|---|---|---|---|---|---|
| ROD-1 | 3 | 2182 | 36168 | 33986 | - | 40 | 42 | Hypothetical proteins | |
| ROD-2 | 3 | 40411 | 54202 | 13791 | - | 14 | 45 | Flagellar | |
| ROD-3 | 4 | 54244 | 81785 | 27541 | tRNA-Thr-CGT | 26 | 39 | Sugar transportor | |
| ROD-4 | 7 | 8799 | 17778 | 8979 | - | 9 | 52 | Glycoside degradation | |
| ROD-5 | 8 | 1 | 20003 | 20002 | tRNA-Lys-TTT | 22 | 46 | Prophage | |
| ROD-6 | 8 | 26182 | 36669 | 10487 | - | 10 | 45 | Prophage | |
| ROD-7 | 9 | 1 | 35407 | 35406 | - | 51 | 46 | Prophage | |
| ROD-8 | 9 | 292659 | 300796 | 8137 | - | 6 | 43 | Curli biogenesis | |
| ROD-9 | 9 | 317516 | 333578 | 16062 | - | 16 | 36 | Prophage | |
| ROD-10 | 10 | 1 | 17530 | 17529 | tRNA-Lys-TTT | 20 | 37 | Prophage | |
| ROD-11 | 10 | 106173 | 107692 | 1519 | - | 3 | 42 | Aid-I adhersion | |
| ROD-12 | 10 | 129437 | 135863 | 6426 | - | 6 | 52 | Iron transporter | |
| ROD-13 | 11 | 6885 | 39719 | 32834 | - | 35 | 54 | Prophage | |
| ROD-14 | 11 | 76907 | 84399 | 7492 | - | 5 | 51 | RND multidrug effux pump | |
| ROD-15 | 11 | 131644 | 140713 | 9069 | - | 12 | 46 | Prophage | |
| ROD-16 | 17 | 240004 | 245998 | 5994 | - | 3 | 42 | sel1 protein | |
| ROD-17 | 17 | 253605 | 262317 | 8712 | - | 8 | 46 | thiamine biosynthesis | |
| ROD-18 | 18 | 1 | 30512 | 30511 | tRNA-Asn-GTT | 12 | 58 | ABC transporter | |
| ROD-19 | 18 | 433292 | 440290 | 6998 | - | 7 | 51 | Fimbrial cluster | |
| ROD-20 | 19 | 90511 | 102325 | 11814 | - | 4 | 50 | Virulence related | RatA, SinI |
| ROD-21 | 22 | 1 | 34254 | 34253 | tRNA-Met-CAT | 25 | 52 | uncharacterized Type VI-like secretion system | |
| ROD-22 | 22 | 184315 | 202088 | 17773 | tRNA-Phe-GAA | 12 | 42 | Capsular polysaccharide export system | KspMT III |
| ROD-23 | 22 | 516756 | 525252 | 8496 | - | 8 | 46 | Ribose ABC transporter system | |
| ROD-24 | 26 | 25349 | 41193 | 15844 | - | 16 | 48 | metabolic enzymes | |
| ROD-25 | 28 | 99422 | 118783 | 19361 | tRNA-SeC(p)-TCA | 22 | 43 | Hypothetical proteins | |
| ROD-26 | 28 | 241596 | 247361 | 5765 | - | 5 | 45 | fimbrial cluster | |
| ROD-27 | 29 | 11045 | 19997 | 8952 | - | 9 | 50 | Transporters | |
| ROD-28 | 29 | 104873 | 112534 | 7661 | - | 7 | 39 | fimbrial cluster | |
| ROD-29 | 31 | 50454 | 57616 | 7162 | - | 5 | 51 | Hypothetical proteins | |
| ROD-30 | 31 | 121132 | 127253 | 6121 | - | 6 | 49 | D-allose ABC transporter | |
| ROD-31 | 31 | 342824 | 363000 | 20176 | YjiD | 14 | 46 | phosphotransferase and ibeRAT operon | IbeA |
| ROD-32 | 31 | 374777 | 386028 | 11251 | - | 7 | 46 | Type I restriction modification system | |
| ROD-33 | 32 | 1 | 22898 | 22897 | - | 19 | 53 | Type VI secretion system |
Fig 2. Unfiltered BLASTn comparison of the E. coli HMLN-1 compared against selected E. coli genomes coloured according to pathotype or strain group (collapsed into individual rings).
The innermost rings represent the GC content (black) and GC skew (purple/green) of the E. coli HLMN-1 genome. The subsequent coloured rings show the results of BLAST searches of E. coli genomes against HLMN-1. Inner to outer coloured circles are as follows: TEC strains PC-1, KIC1 and KIC2; Non-TEC strains 73–89 and 46–4; UPEC strains UTI89, 536, ABU83972, CFT073, IAI39 and UMN026; NMEC strains IHE3034 and S88; K-12 derivative strains W3110, MG1655 and DH10B; commensal strains BL21, REL606, SE11, IAI1, HS, ED1a and SE15; clone W and ATCC8739 (clone C) strains; Environmental SMS-3-5 strain; Enterotoxigenic E. coli (ETEC); Enteropathogenic E. coli (EPEC); Enterohemorrhagic E. coli (EHEC); Enteroaggregative E. coli (EAEC); Avian pathogenic E. coli (APEC) and adherent-invasive E. coli (AIEC). Colour intensity is proportional to BLASTn identity with bold regions having high nucleotide identity and light regions having low nucleotide identity. E. coli HLMN-1 genomic features are annotated around the outmost ring: prophages regions in black, fimbria clusters in blue, Type VI Secretion System in green and other features and region of differences (RODs) in red.
Fig 3. Visual comparison of the TEC Type VI Secretion System.
tBLASTx comparison of the Type VI Secretion System (T6SS) genomic island and flanking regions in E. coli HLMN-1, E. coli PC-1, E. coli KIC-1, E. coli KIC-2, E. coli 46–4 and E. coli 73–89. Grey vertical blocks indicated regions of conserved nucleotide sequence determined using tBLASTx and the arrows represent genes and are coloured.
A significant region in E. coli HMLN-1 was ROD-31, which was 20.3 kbp in length and contained the virulence ibeRAT operon. ROD-31 was present in E. coli HMLN-1 but absent in the other TEC strains and both of the non-TEC strains and it can also be found in extraintestinal pathogenic E. coli (ExPEC). The ibeRAT is a three-gene operon encoding a regulatory protein (IbeR) and two invasion proteins (IbeA and IbeT). The role for IbeA in pathogenesis has been reported for E. coli such as avian pathogenic E. coli and adherent-invasive E. coli and is linked with invasion of the intestinal epithelium and survival in macrophages [25]. The IbeT protein has been shown to affect adhesion and invasion of brain endothelial cells [26]. The ibeR gene encodes an RpoS-like regulator that is predicted to regulate the ibeA and ibeT genes. Another important feature of E. coli HMLN-1 genome was the presence of a 17 kbp genomic island (ROD-22), which was inserted next to tRNAPhe and contained the components necessary for the expression of Group III capsular polysaccharide. ROD-22 includes the genes, kpsM and kpsT, whose products are critical for the export of the polysaccharide capsule [27].
The E. coli PC-1 genome contained 12 RODs that were not found in the non-TEC strains and three of which encode proteins that are homologous with proteins that have adhesion and invasion functions (S2 Table). In the genome of the rat TEC strain E. coli KIC-2 there were 14 RODs, which included six prophages (S3 Table) while the E. coli KIC-1 had only three RODs (S4 Table). The extra gene content of KIC-1 and KIC-2 has no suggested roles in translocation. However, the KIC-2 genome did carry additional mechanisms potentially associated with adhesion (T6SS and Aid-I like adhesion protein) while KIC-1 did not, which is supported by the higher adhesion rate seen in in vitro experiments with KIC-2 [11].
Fimbria clusters of TEC
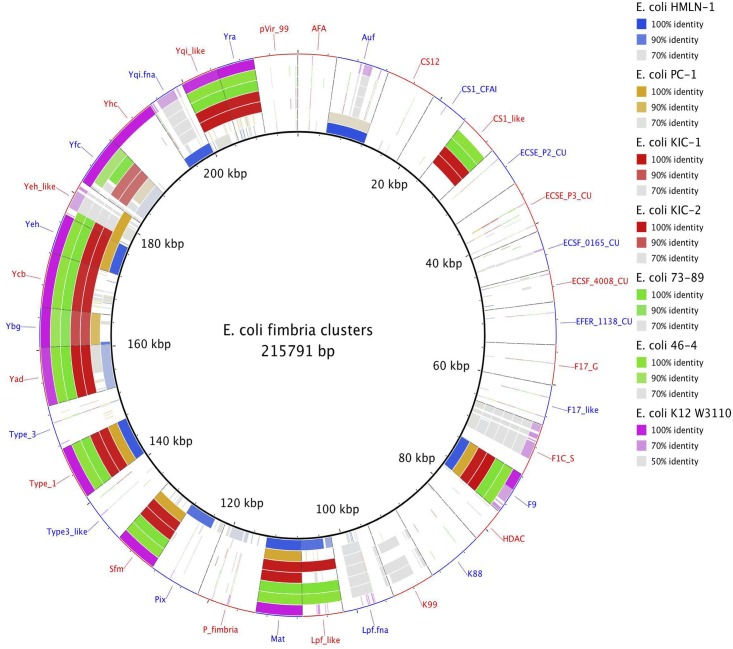
The presence and distribution of chaperone-usher (CU) fimbrial gene clusters were investigated to see if there was a link between fimbriae and the observed phenotypes of the TEC strains. Fig 4 shows the distribution of 38 CU fimbrial operons in the four TEC strains and the two non-TEC strains as well as a reference strains, E. coli K-12 W3110. The distribution of CU fimbrial operons in the TEC strains was consistent with the phylogroups that each of the strains belonged too. All the TEC and non-TEC strains possessed the core-associated E. coli CU fimbrial operons including the Type 1, Yad, Yeh, Yfc, Mat, F9 and Ybg fimbriae. The sfm fimbriae was present in all TEC and non-TEC genomes sequenced in this study expected for E. coli HMLN-1, however, the sfm fimbriae is usually absent in E. coli strains from the B2 phylogroup [18]. In addition to the core CU fimbrial operons, the TEC strains KIC-1 and KIC-2 as well as the non-TEC strains 46–4 and 73–89 also carried clade-specific CU fimbriae commonly found in strains that are part of the B1 phylogroup such as CS1-like, Yra, Yqi-like and Ycb [18]. E. coli HMLN-1 carried three B2-clade specific CU fimbrial gene clusters (Pix, Yqi and P-fimbriae) that were not present in the other genome sequences. The intact Auf fimbrial operon that is commonly found in both B2 and D phylogroup strains was present in both the HMLN-1 and PC-1 genomes.
Fig 4. Distribution of chaperone-usher (CU) fimbrial gene clusters in translocating E. coli, control E. coli strains sequenced in this study and E. coli K-12 W3110.
The inner ring represents the concatenated nucleotide sequences of 38 fimbrial operons. Subsequent rings indicate the presence of the CU fimbrial gene clusters in each genome. Each segment is labelled according to the name of the CU fimbrial families.
Discussion
Translocating E. coli is a unique pathogroup that was identified by their ability to translocate across gut epithelial cells in animals and humans [6]. The human TEC strain tested in this study was associated with a case of fatal haemorrhagic pancreatitis suggesting that TEC strains are a public health concern [7]. However, it is currently unknown what genes are involved in translocation since the most common E. coli virulence genes are absent in TEC strains [11]. Therefore, this study employed whole genome sequencing to compare TEC and non-TEC strains to identify the genes involved in the unique phenotypes exhibited by this pathogroup. The E. coli strains selected for sequencing were TEC strains that were previously tested for adhesion and translocation in animal studies [4] and in in vitro experiments [11]. In addition, non-TEC strains that were used as a control in the in vitro experiments were sequenced so that the genomes can be compared to identify regions of differences that may be associated with the translocation phenotype.
The phylogenetic relationship of the TEC strains have been previously explored using an unweighted pair group method with arithmetic averages (UPGMA) approach based on the similarity between the biochemical phenotypes of these strains [11]. However, using the genome sequences a more accurate phylogenetic relationship was determined for the TEC strains. The neighbour joining tree based on seven housekeeping genes employed in this study, showed that the TEC strains were distributed amongst three distinct E. coli phylogroups. The dissemination of the TEC strains in the E. coli phylogenetic tree are independent of disease phenotype, similar to what has been observed with AIEC strains [28]. Most of the TEC strains were part of the same phylogroup that was predicted with the UPGMA approach. However, in the neighbour joining tree, E. coli HMLN-1 was part of phylogroup B2 instead of phylogroup D as originally predicted [11]. E. coli strains belonging to this phylogenetic group have been associated with pyelonephritis and meningitis [19, 29]. The closely related TEC strain to HMLN-1 was the PC-1 strain, which was originally isolated from the blood of pigs subjected to pancreatitis and ischemic reperfusion. These two strains have previously been shown to have identical biochemical phenotypes and serotypes [11] however, the PC-1 strains belonged to phylogenetic group D which is also found among UPEC strain. The KIC-1 and KIC-2 strains together with the non-TEC strains isolated from rats belonged to phylogroup B1, which are previously regarded as commensal gut strains. Ramos et al, [11] showed that these rat-origin TEC strains were not as efficient at translocating in a cell line representing human gut epithelium as HMLN-1 and PC-1, although they translocated efficiently in rats [4].
A genomic island encoding a T6SS was found in the three TEC strains (HMLN-1, PC-1 and KIC-2) that can efficiently adhere to epithelial cells [11]. The T6SS however, was absent in E. coli KIC-1 and the two non-TEC strains, which adhered to epithelial cells to a lesser degree than the other strains. The T6SS is commonly found in pathogenic E. coli strains isolated from patients with inflammatory bowel disease [30]. The T6SS has also been identified in commensal E. coli, including E. coli strain W [31]. The presence of similar T6SS in both pathogenic and commensal E. coli suggests that it has a role in pathogenesis and/or intra-bacterial competiveness [32]. The T6SS consists of 14 genes that encode for the core structural apparatuses. Studies of other bacterial pathogens have revealed that the T6SS is involved in adhesion to host cells [33, 34]. Therefore, the presence of the T6SS in TEC strains with a high adhesion rate and its absent in the TEC strain KIC-1 suggests that the T6SS plays a role in adhesion for TEC strains rather than translocation. Another unique finding of this comparative study was the presence of the ibeRAT operon in E. coli HMLN-1 that was missing from other TEC strains. The ibeA and ibeT genes encodes for invasive proteins [26] while the ibeR gene encodes a regulator [35]. Experimental work on ibeA in E. coli has shown that the encoded protein contributes to invasion by improving bacterial survival in macrophages by functioning as an oxidoreductase, which provides resistance to reactive oxygen species [25, 36]. The presence of the ibeA gene likely contributes to the pathogenic potential of E. coli HMLN-1 and may assist with translocation. However, the absence of the ibeA gene in the other TEC strains suggest that there are other mechanisms associated with translocation, which may include other unknown invasion genes located on the chromosome or plasmids. It is also possible that ibeA gene is involved in a host-specific adhesion mechanism, a view which is supported by the study of Katouli et al [4] who showed that translocation of these four TEC strains was host-specific.
Fimbriae are long proteinaceous organelles that extend from the surface of many bacteria and mediate diverse functions, including adherence and biofilm formation [37]. In Gram-negative bacteria, one of the ways those fimbriae are assembled and transported from the cytoplasm to the cell surface is via the chaperone-usher pathway [38]. The fimbriae structure genes are clustered together with the chaperone and usher genes. A previous study has identified 38 distinct CU fimbrial operons in E. coli based on an usher protein phylogeny [18]. The presence of clade specific CU fimbriae operons in TEC strains was consistent with the phylogroups that each strain belonged too. In addition, the presence of the Ygi and Pix CU fimbrial operons in E. coli HMLN-1 further support that this strain belongs to the B2 phylogroup, since the Ygi and Pix fimbriae are only found in strains from the B2 phylogroup [18]. These observations therefore suggest that the variation in the CU operons is not linked to TEC strain adhesion and translocation phenotypes.
In conclusion, the genomic comparison strongly suggests that the T6SS is involved in adhesion of the TEC strains to epithelial cells rather than translocation. The invasion gene ibeA in E. coli HMLN-1 may also contributed to translocation and disease outcome of this particular TEC strain. It is likely that translocation is mediated by an assortment of different invasion proteins that may vary between strains from different hosts. A future direction for the investigation of TEC will be to “knock-out” the T6SS ATPase gene and the ibeA gene in the E. coli HMLN-1 to determine the effect they may have on adhesion and translocation.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All genome files are available from the National Center for Biotechnology Information (accession numbers PRJNA284559-PRJNA284564).
Funding Statement
The authors have no support or funding to report.
References
- 1. Berg RD, Garlington AW. Translocation of certain indigenous bacteria from the gastrointestinal tract to the mesenteric lymph nodes and other organs in a gnotobiotic mouse model. Infect Immun. 1979. February;23(2):403–11. . Pubmed Central PMCID: 414179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Finfer S, Bellomo R, Lipman J, French C, Dobb G, Myburgh J. Adult-population incidence of severe sepsis in Australian and New Zealand intensive care units. Intensive care medicine. 2004. April;30(4):589–96. . [DOI] [PubMed] [Google Scholar]
- 3. Katouli M, Bark T, Ljungqvist O, Svenberg T, Mollby R. Composition and diversity of intestinal coliform flora influence bacterial translocation in rats after hemorrhagic stress. Infect Immun. 1994. November;62(11):4768–74. . Pubmed Central PMCID: 303185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Katouli M, Nettebladt CG, Muratov V, Ljungqvist O, Bark T, Svenberg T, et al. Selective translocation of coliform bacteria adhering to caecal epithelium of rats during catabolic stress. J Med Microbiol. 1997. July;46(7):571–8. . [DOI] [PubMed] [Google Scholar]
- 5. Bark T, Katouli M, Ljungqvist O, Mollby R, Svenberg T. Bacterial translocation after non-lethal hemorrhage in the rat. Circulatory shock. 1993. September;41(1):60–5. . [PubMed] [Google Scholar]
- 6. Katouli M, Ramos NL, Nettelbladt CG, Ljungdahl M, Robinson W, Ison HM, et al. Host species-specific translocation of Escherichia coli . Eur J Clin Microbiol Infect Dis. 2009. September;28(9):1095–103. 10.1007/s10096-009-0754-0 [DOI] [PubMed] [Google Scholar]
- 7. Nettelbladt CG, Katouli M, Bark T, Svenberg T, Mollby R, Ljungqvist O. Evidence of bacterial translocation in fatal hemorrhagic pancreatitis. The Journal of trauma. 2000. February;48(2):314–5. . [DOI] [PubMed] [Google Scholar]
- 8. Ljungdahl M, Lundholm M, Katouli M, Rasmussen I, Engstrand L, Haglund U. Bacterial translocation in experimental shock is dependent on the strains in the intestinal flora. Scandinavian journal of gastroenterology. 2000. April;35(4):389–97. . [DOI] [PubMed] [Google Scholar]
- 9. Pang X, Hua X, Yang Q, Ding D, Che C, Cui L, et al. Inter-species transplantation of gut microbiota from human to pigs. ISME J. 2007. June;1(2):156–62. . [DOI] [PubMed] [Google Scholar]
- 10. Girardeau JP, Lalioui L, Said AM, De Champs C, Le Bouguenec C. Extended virulence genotype of pathogenic Escherichia coli isolates carrying the afa-8 operon: evidence of similarities between isolates from humans and animals with extraintestinal infections. J Clin Microbiol. 2003. January;41(1):218–26. . Pubmed Central PMCID: 149575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramos NL, Lamprokostopoulou A, Chapman TA, Chin JC, Romling U, Brauner A, et al. Virulence characteristics of translocating Escherichia coli and the interleukin-8 response to infection. Microb Pathog. 2011. February;50(2):81–6. 10.1016/j.micpath.2010.11.003 [DOI] [PubMed] [Google Scholar]
- 12. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012. May;19(5):455–77. Pubmed Central PMCID: 3342519. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006. June;60(5):1136–51. . Pubmed Central PMCID: 1557465. Epub 2006/05/13. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000. October;16(10):944–5. . Epub 2000/12/20. eng. [DOI] [PubMed] [Google Scholar]
- 15. Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011. August 8;12(1):402 . Epub 2011/08/10. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011. April 1;27(7):1009–10. Pubmed Central PMCID: 3065679. Epub 2011/02/01. eng. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. ACT: the Artemis Comparison Tool. Bioinformatics. 2005. August 15;21(16):3422–3. . Epub 2005/06/25. eng. [DOI] [PubMed] [Google Scholar]
- 18. Wurpel DJ, Beatson SA, Totsika M, Petty NK, Schembri MA. Chaperone-Usher Fimbriae of Escherichia coli. PLoS One. 2013;8(1):e52835 Epub 2013/02/06. eng. 10.1371/journal.pone.0052835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009. January;5(1):e1000344 Pubmed Central PMCID: 2617782. Epub 2009/01/24. eng. 10.1371/journal.pgen.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bingen E, Picard B, Brahimi N, Mathy S, Desjardins P, Elion J, et al. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J Infect Dis. 1998. March;177(3):642–50. . [DOI] [PubMed] [Google Scholar]
- 21. Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000. January;181(1):261–72. . [DOI] [PubMed] [Google Scholar]
- 22. Pupo GM, Karaolis DK, Lan R, Reeves PR. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997. July;65(7):2685–92. . Pubmed Central PMCID: 175379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Duriez P, Clermont O, Bonacorsi S, Bingen E, Chaventre A, Elion J, et al. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology. 2001. June;147(Pt 6):1671–6. . [DOI] [PubMed] [Google Scholar]
- 24. Cascales E, Cambillau C. Structural biology of type VI secretion systems. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2012. April 19;367(1592):1102–11. Pubmed Central PMCID: 3297440. 10.1098/rstb.2011.0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cieza RJ, Hu J, Ross BN, Sbrana E, Torres AG. IbeA, the invasin of Adherent-Invasive Escherichia coli (AIEC) mediates interaction with intestinal epithelia and macrophages. Infect Immun. 2015. February 23 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zou Y, He L, Chi F, Jong A, Huang SH. Involvement of Escherichia coli K1 ibeT in bacterial adhesion that is associated with the entry into human brain microvascular endothelial cells. Med Microbiol Immunol. 2008. December;197(4):337–44. . [DOI] [PubMed] [Google Scholar]
- 27. Pavelka MS Jr., Wright LF, Silver RP. Identification of two genes, kpsM and kpsT, in region 3 of the polysialic acid gene cluster of Escherichia coli K1. J Bacteriol. 1991. August;173(15):4603–10. . Pubmed Central PMCID: 208135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martinez-Medina M, Garcia-Gil J, Barnich N, Wieler LH, Ewers C. Adherent-invasive Escherichia coli phenotype displayed by intestinal pathogenic E. coli strains from cats, dogs, and swine. Appl Environ Microbiol. 2011. August 15;77(16):5813–7. Pubmed Central PMCID: 3165260. 10.1128/AEM.02614-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, et al. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999. February;67(2):546–53. . Pubmed Central PMCID: 96353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 2007. September;1(5):403–18. . [DOI] [PubMed] [Google Scholar]
- 31. Archer CT, Kim JF, Jeong H, Park JH, Vickers CE, Lee SY, et al. The genome sequence of E. coli W (ATCC 9637): comparative genome analysis and an improved genome-scale reconstruction of E. coli . BMC Genomics. 2011;12:9 Pubmed Central PMCID: 3032704. 10.1186/1471-2164-12-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Richards VP, Lefebure T, Pavinski Bitar PD, Dogan B, Simpson KW, Schukken YH, et al. Genome Based Phylogeny and Comparative Genomic Analysis of Intra-Mammary Pathogenic Escherichia coli . PLoS One. 2015;10(3):e0119799 Pubmed Central PMCID: 4373696. 10.1371/journal.pone.0119799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu Y, Yang H, Li J, Zhang P, Wu B, Zhu B, et al. Putative type VI secretion systems of Vibrio parahaemolyticus contribute to adhesion to cultured cell monolayers. Arch Microbiol. 2012. October;194(10):827–35. . [DOI] [PubMed] [Google Scholar]
- 34. Liu L, Hao S, Lan R, Wang G, Xiao D, Sun H, et al. The type VI secretion system modulates flagellar gene expression and secretion in Citrobacter freundii and contributes to the adhesion and cytotoxicity to host cells. Infect Immun. 2015. April 13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chi F, Wang Y, Gallaher TK, Wu CH, Jong A, Huang SH. Identification of IbeR as a stationary-phase regulator in meningitic Escherichia coli K1 that carries a loss-of-function mutation in rpoS. Journal of biomedicine & biotechnology. 2009;2009:520283 . Pubmed Central PMCID: 2655632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Flechard M, Cortes MA, Reperant M, Germon P. New role for the ibeA gene in H2O2 stress resistance of Escherichia coli . J Bacteriol. 2012. September;194(17):4550–60. Pubmed Central PMCID: 3415484. 10.1128/JB.00089-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Proft T, Baker EN. Pili in Gram-negative and Gram-positive bacteria—structure, assembly and their role in disease. Cell Mol Life Sci. 2009. February;66(4):613–35. 10.1007/s00018-008-8477-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009. June 18;5(6):580–92. 10.1016/j.chom.2009.05.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All genome files are available from the National Center for Biotechnology Information (accession numbers PRJNA284559-PRJNA284564).