Abstract
FBXW7 (F-box and WD40 domain protein 7) is a tumor suppressor frequently inactivated in human cancers. The precise molecular mechanisms by which FBXW7 exerts antitumor activity remain under intensive investigation and are thought to relate in part to FBXW7-mediated destruction of key cancer-relevant proteins. Enolase 1 (ENO1) possesses oncogenic activity and is often overexpressed in various human cancers, besides its critical role in glycolysis. However, the detailed regulatory mechanisms of ENO1 expression remain unclear. Here we show that the elevated expression of ENO1 was identified in FBXW7-depletion HCT116 cells through two-dimensional protein electrophoresis and mass spectrometry assays (2DE-MS). Subsequent western blotting and immunohistochemical assays confirmed that ENO1 expression reversely correlates with FBXW7 expression in several cells and colon cancer tissues. Furthermore, we show that FBXW7 physically binds to ENO1 and targets ENO1 for ubiquitin-mediated degradation. Functionally, we found that FBXW7 suppresses the ENO1-induced gene expression, lactate production, cell proliferation and migration. These findings suggest that ENO1 is a novel substrate of FBXW7, and its activity can be negatively regulated by FBXW7 at the posttranslational level. Our work provides a novel molecular insight into FBXW7-directed tumor suppression through regulation of ENO1.
Enolase 1 (ENO1) is a conserved glycolytic enzyme that catalyze the formation of phosphoenolpyruvate from 2-phosphoglycerate, which generates ATP during glycolysis.1,2 Several studies have shown that, besides its major role in glycolysis, ENO1 is a multifunctional protein participating in several physiological processes, including growth control, hypoxia tolerance and autoimmune activities.1,3–5 Remarkably, accumulating evidence suggests that ENO1 can function as an oncogenic protein by promoting cell proliferation, invasion and metastasis. ENO1 expression is frequently increased in diverse tumors, including head and neck,6 thyroid,7 breast,8 lung,9,10 prostate,11 colon and gastric cancer,12,13 glioma 14and cholangiocarcinoma.15 Moreover, overexpression of ENO1 is positively associated with progression and poor prognosis in several tumors.
The expression of ENO1 is regulated at multiple levels. Nuclear factor kappa B (NFκB) and hypoxia-inducible factor-1α (HIF-1α) are involved in the transcriptional activation of ENO1.8,16 MiR-206 may target ENO1 and inhibit the expression of ENO1.17 Activation of the Src and MEK/ERK signal pathways upregulates ENO1 expression,18 whereas retinoic acid reduces ENO1 at the protein level.19 Although a lot of efforts have been made in studying the regulatory mechanism of ENO1 activity, the possibility that other factors might regulate ENO1 expression during tumorigenesis remains to be investigated.
FBXW7 (F-box and WD40 domain protein 7) is a conserved F-box WD40 protein and functions as a substrate recognition subunit of the SCF (SKP1/CUL1/F-box protein) E3 ubiquitin ligase.20 FBXW7 targets a network of proteins with central roles in cell division, cell growth and differentiation for ubiquitination and proteasome degradation. As most FBXW7 substrates are potent oncoproteins, such as cyclin E,21,22 Notch,23 mTOR,24,25 Aurora A,26,27 etc, FBXW7 represents a major tumor suppressor. Numerous cancer-associated mutations of FBXW7 have been found in various cancers28–30 and loss of FBXW7 function results in tumorigenesis.31 However, a detailed understanding of the full set of FBXW7 substrates and the mechanisms that link FBXW7 deficiency to tumorigenesis is still lacking.
In this present study to further investigate the suppressive role of FBXW7 in tumorigenesis, we used proteomics approach to globally identify FBXW7-regulated proteins in colon cancer cells. Differential expression of ENO1 protein in normal and FBXW7-deficient cells was first identified. Further studies indicated that FBXW7 is a novel regulator of ENO1 protein. FBXW7 physically bound to and facilitated the ubiquitination and degradation of ENO1. Consequently, the biological activity of ENO1 was inhibited by FBXW7. Our observations suggest that ENO1 has an important role in FBXW7-induced growth inhibition of cancer cells.
MATERIALS AND METHODS
Cell Culture and Cell Transfection
Human colon cancer cells HCT116, DLD-1, breast cancer cell MDA-MB-468, prostate cancer cells DU145 and PC3 and HEK293T cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). The cell lines HCT116 FBXW7−/− and DLD-1 FBXW7−/− were gifts from Dr Bert Vogelstein. Cell culture was according to the manufacturer’s protocol. The cells were maintained in a humidified incubator at 37 °C in an atmosphere of 95% air and 5% carbon dioxide. Transfection of HEK293T and HCT 116 cells was performed using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s instructions.
Reagent and Antibodies
MG132 (Sigma-Aldrich, MO, USA) was dissolved in dimethyl sulfoxide (DMSO). GSK3β inhibitor VIII was from Calbiochem. Cells were treated with 10 µM MG132 or 25 µM GSK3β inhibitor VIII and incubated for 6 h. Antibodies against FBXW7 (ab109617) were purchased from Abcam. ENO1 and β-actin antibodies were from Cell Signaling Technology. The ENO1 antibody for immunohistochemistry was from Proteintech. Unless otherwise noted, all other chemicals were from Sigma.
Two-Dimensional (2-D) Electrophoresis and Mass Spectrometry (MS)
Total protein of HCT116 FBXW7+/+ and HCT116 FBXW7−/−cells was prepared and then labeled with Cy2 and Cy5, respectively. 2-D electrophoresis was performed using Bergman’s method as previously described.13 Following 2-D electrophoresis, gels were scanned into a computer. The 2-D gel profile was analyzed using the PDQuest software (Bio-Rad). Finally, the differentially expressed protein spots were identified using the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) on a mass spectrometer (Voyager-DE PRO).
Western Blotting
Cells were harvested and lysed in RIPA buffer (1% Triton X-100, 0.1% SDS, 50 mM Tris PH 7.5, 150 mM NaCl, 0.5% sodium deoxycholate, 10 mM NaF) supplemented with protease inhibitors (Roche). Lysates were clarified by centri-fugation at 14 000 g for 20 min at 4 °C. Protein concentration was then determined by the BCA Kit (Biyuntian). After incubation at 95 °C for 5 min in SDS sample buffer, protein extracts were separated by 10% SDS-PAGE and then electroblotted onto PVDF membranes. The membranes were blocked with 5% (w/v) BSA in TBST for 2 h and then incubated with antibodies anti-FBXW7 (1:1000), anti-ENO1 (1:1000) and anti-β-actin (1:2000) at 4 °C overnight. After three washes with TBST, the membranes were incubated for 1 h at room temperature in a 1:10 000 dilution of IgG-horseradish peroxidase secondary antibody (Sigma-Aldrich) and visualized with chemiluminescence (Pierce Protein Biology Products/Thermo Scientific, Rockford, IL, USA).
Coimmunoprecipitation Assay
Cells were seeded in 10-cm culture plates and treated with 10 µM MG132 for 6 h and then lysed in NP-40 lysis buffer. About 1-mg cell lysates were incubated with the indicated antibodies overnight at 4 °C on a verticle roller. The lysates were incubated with 30 µl of Protein A/G agarose beads (Calbiochem) for 4 h and then the beads were washed four times with cold PBS. The immunoprecipitated proteins were then identified by western blotting. The membranes were immunoblotted with the corresponding primary antibodies.
Quantitative Real-Time PCR (qRT-PCR)
The total RNA was isolated using Trizol reagent (Invitrogen, USA). Total RNA was reversely transcribed by the SuperScript II Reverse Transcriptase (Invitrogen).The mRNA expression was determined by qRT-PCR using SYBR Green PCR Master Mix (Thermo). The primers sequences were as follows:
FBXW7 forward primer 5′-CGAACTCCAGTAGTATTGT GGACCT-3′ and reverse primer 5′-TTCTTTTCATTTTTGT TGTTTTTGTATAGA-3′;
CCL20 forward primer 5′-GACTGCTGTGTTGGATACAC-3′ and reverse primer 5′-CTGAGGAGACGCACAATATA-3′; and
GAPDH forward primer 5′-GCCGCATCTTCTTTTGCGT CGC-3′ and reverse primer 5′-TCCCGTTCTCAGCCTTGAC GGT-3′;
GAPDH was used as the reference gene. The relative levels of gene expression were represented as 2−ΔCt(Ct gene – Ct reference). Experiments were repeated in triplicate.
Wound-Healing Assay
Cells were grown to confluence in six-well plates to a density of approximately 5 × 106 cells per well and denuded by dragging sterile plastic 200-µl micropipette tips through the center of the plate. Images were captured under a phase-contrast microscopy at 0 and 44 h.
Cell Proliferation Assay
The viability of cells was determined by assaying the reduction of 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT). Briefly, cells were plated onto 96-well plates with 2 × 103 cells per well. At the indicated time, the medium was replaced by 100 µl medium and incubated with 10 µl MTT (5 mg/ml) for 4 h. The particles were dissolved with DMSO, and the absorbance was determined at 492 nm.
Lactate Assay
Cells were collected and dispersed using ultrasonic disintegrator and then the lysates were centrifuged at 2000 g for 15 min at 4 °C. The supernatant was collected and total protein concentration was determined by the BCA Kit (Biyuntian). The level of lactic acid was measured using commercially available diagnostic kits according to the manufacturer’s instructions (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
Immunohistochemistry and Scoring
A total of 50 samples of colorectal cancer tissues and corresponding adjacent non-cancerous tissues were obtained from patients undergoing surgical excision of tumors in Qilu Hospital of Shandong University (Jinan, China). The samples were fixed with 10% formalin and embedded in paraffin and then were sliced into 5-µm sections. The sections were deparaffinized in xylene and rehydrated through graded alcohol. After incubation in 0.3% H2O2 and blocked with 10% goat serum, the sections were incubated overnight at 4 °C with antibodies against ENO1 (1:200) and against FBXW7 (1:300). Subsequently, the sections were treated with secondary antibody for 30 min, developed with diaminobenzidine (DAB) substrate (DAB Color Kit (Beijing Zhong-shan Golden Bridge Biotechnology)) and counterstained with hematoxylin. Staining was observed in five randomly selected fields in the high-power microscope. The staining intensity was based on the average percentage of positive cells. The scoring results were analyzed by two investigators.
Statistical Analysis
SPSS 17.0 software (SPSS, Chicago, IL, USA) was used for statistical analysis. The expression differences between two different groups were analyzed with the Student’s t-test. A Spearman correlation analysis was performed to assess the relationship between FBXW7 and ENO1 expression in colorectal cancer tissues using Cox regression analyses. The differences of cell proliferation and cell migration were assessed with the two-tailed unpaired Student’s t-test. P < 0.05 was considered statistically significant.
RESULTS
FBXW7 Downregulates ENO1 Protein Level
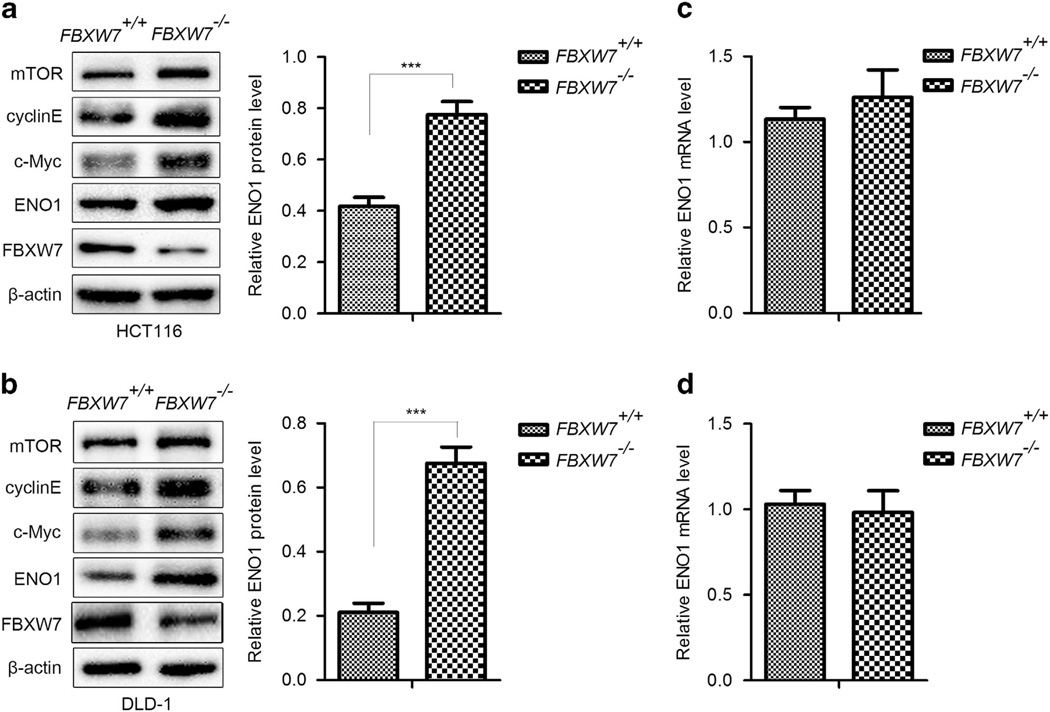
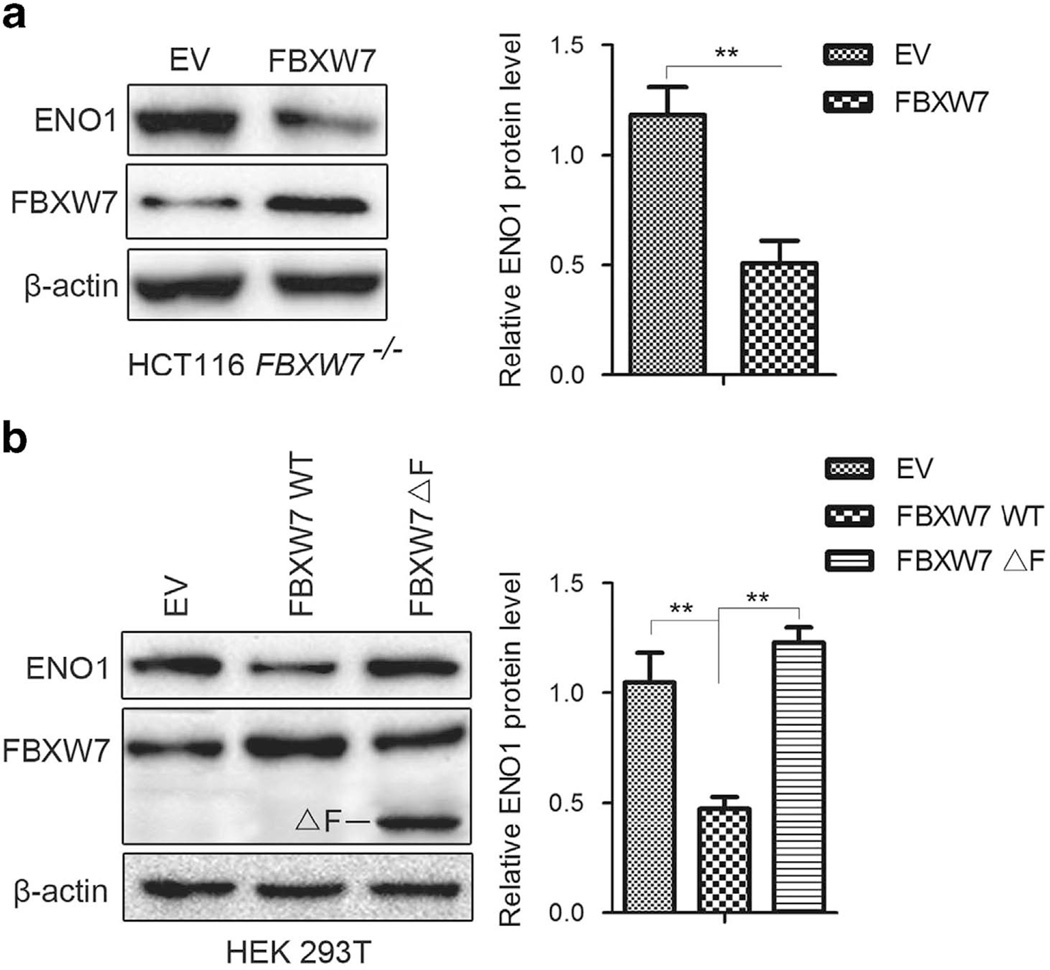
FBXW7 exerts its antitumorigenesis roles mainly through downregulating oncoprotein levels. To identify new substrates recognized by FBXW7, we performed 2-D gel electrophoresis (2-DE) and MS in the human colorectal cancer cell line HCT116 with homozygous deletion of the FBXW7 gene (HCT116 FBXW7−/−), compared with its wild-type (WT) counterpart. Several differentially expressed proteins were separated and identified, of which ENO1 protein was elevated significantly in HCT116 FBXW7−/− cells compared with WT cells. The increased expression of ENO1 protein and other identified protein, including cyclinE, c-Myc and mTOR, were validated by western blotting analysis in HCT116 and DLD-1 cells with depletion of FBXW7 (Figures 1a and b). Further, FBXW7 expression was silenced by two different short hairpin RNA (shRNA) constructs (shFBXW7A and shFBXW7B) in other cell lines, such as MDA-MB-468, DU145 and PC3 cells. Elevated ENO1 expression was observed in these FBXW7-silenced cells (Supplementary Figure 1A – C). To further validate our observation, re-introduction of WT FBXW7 expression into HCT116 FBXW7−/−cells reversed the ENO1 expression induced by FBXW7 depletion (Figure 2a). Similar results were observed in 293T cells with exogenous expression of WT FBXW7 while exogenous expression of FBXW7ΔF (without the F-box that interacts with Skp1) failed to decrease the ENO1 protein level (Figure 2b). Consistent with a posttranslational mode of regulation, no changes in ENO1 mRNA levels were observed after depletion of FBXW7 in HCT116 and DLD-1 cells (Figures 1c and d) or silenced expression of FBXW7 in other cell lines (Supplementary Figure 1D – F). Together, these results suggest that FBXW7 can downregulate ENO1 protein expression at the posttranscriptional level.
Figure 1.
Knockout of FBXW7 upregulates the protein level of ENO1. (a and b) Western blotting assay on the whole cell lysates of HCT116 (a) and DLD-1 (b) FBXW7−/− and FBXW7+/+ cells using the indicated antibody. The graph shows quantitative analysis. (c and d) qRT-PCR analysis to detect the mRNA level of ENO1 in HCT116 (c) and DLD-1 (d) FBXW7−/− and FBXW7+/+ cells. All results are representative of three independent experiments. ***P < 0.001 based on Student’s t-test.
Figure 2.
Overexpression of wild-type FBXW7 downregulates the protein level of ENO1. Wild-type FBXW7 was re-introduced into HCT116 FBXW7−/− and HEK293T cells. At the same time, FBXW7 ΔF (F-box deletion) was also introduced into HEK293T cells. Western blotting assay on the whole cell lysates of HCT116 FBXW7−/− (a) and HEK293T cells (b) using the anti-ENO1 and anti-FBXW7 antibodies. The graph shows quantitative analysis. All results are representative of three independent experiments. **P < 0.01 based on Student’s t-test. EV: empty vector.
FBXW7 Promotes Proteasomal Degradation of the ENO1 Protein
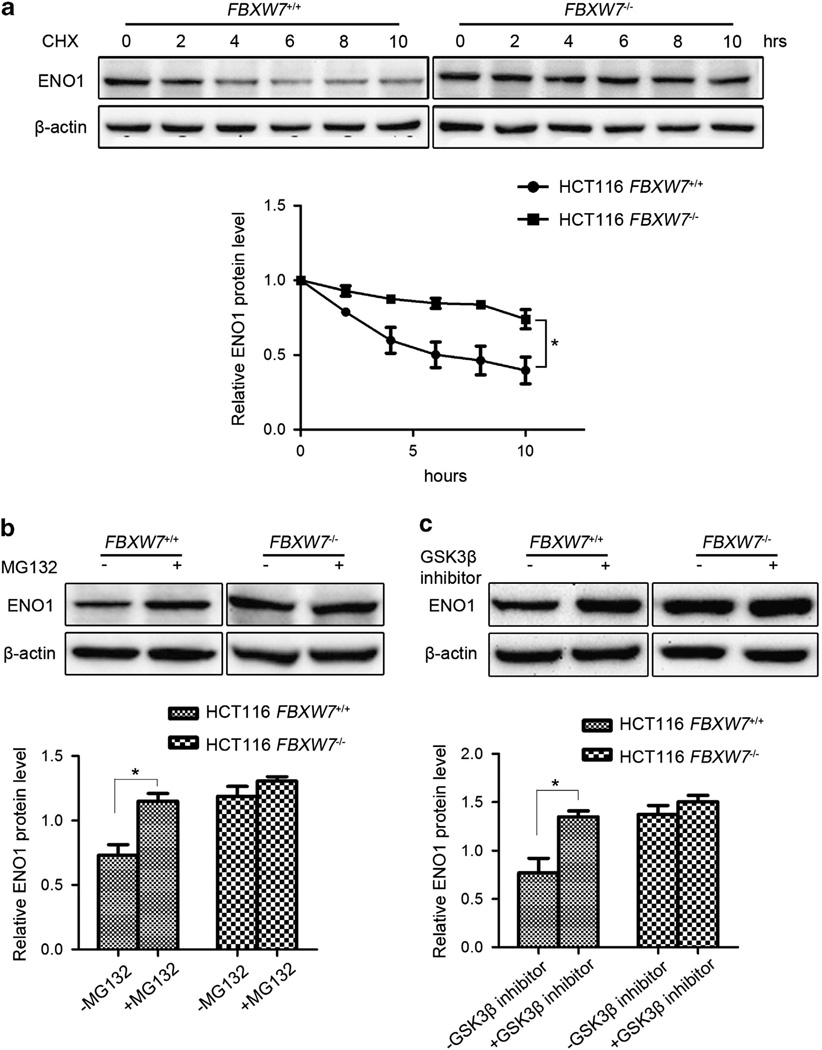
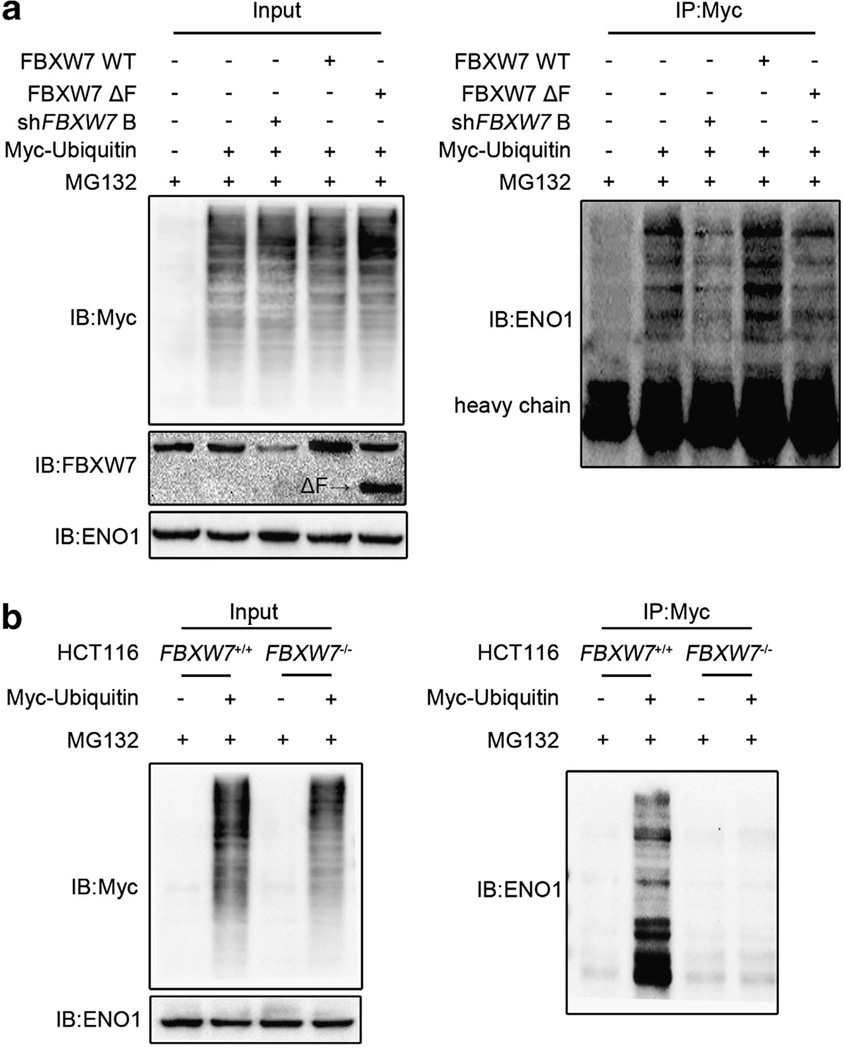
As FBXW7 is an E3 ligase, we first evaluated whether FBXW7 could regulate ENO1 protein stability. The turnover of ENO1 protein in the HCT116 FBXW7+/+ and FBXW7−/− cells was measured, respectively, by cycloheximide (CHX) chase assay. The result showed that the half-life of ENO1 was significantly extended when FBXW7 expression is depleted, indicating that FBXW7 decreased ENO1 protein level by promoting its degradation (Figure 3a). Then we determined whether the degradation of ENO1 by FBXW7 is through the proteasome-dependent pathway. HCT116 FBXW7+/+ and FBXW7−/− cells were treated with the proteasome inhibitor MG132. Indeed proteasome inhibition caused a significant increase in ENO1 levels in HCT116 FBXW7+/+ cells but not in HCT116 FBXW7−/− cells (Figure 3b). As FBXW7-mediated degradation of most substrates occurs in a GSK3β-dependent manner, we examined whether FBXW7 targets ENO1 also in a GSK3β-dependent manner. Similar with the treatment of MG132, Gsk3β inhibitor treatment resulted in a significant increase in ENO1 levels in HCT116 FBXW7+/+ cells but not in HCT116 FBXW7−/− cells (Figure 3c). To test whether FBXW7 ubiquitinates ENO1, HEK293T cells were transfected with Myc-tagged ubiquitin (Myc-ub) alone, in combination with WT FBXW7, FBXW7ΔF or shFBXW7 B. Immunoprecipitation with Myc-ub followed by ENO1 immunoblotting showed that overexpression of WT FBXW7 but not of FBXW7ΔF increased the ENO1 ubiquitination while knockdown expression of FBXW7 reduced the ENO1 ubiquitination (Figure 4a). Additionally, we examined the ENO1 ubiquitination in HCT116 FBXW7+/+ and FBXW7−/− cells and also found that the ENO1 ubiquitination is decreased in FBXW7−/− HCT116 cells (Figure 4b). Together, these data indicate that FBXW7 acts as an E3 ligase and promotes ENO1 degradation through ubiquitin/proteosome pathway in a GSK3β-dependent manner.
Figure 3.
FBXW7 facilitates ENO1 turnover in proteasome-dependent pathway. (a) HCT116 FBXW7+/+ and FBXW7−/− cells were treated with 50 µg/ml CHX for 0, 2, 4, 6, 8 and 10 h. Western blotting analysis was carried out to detect the endogenous level of ENO1 using the anti-ENO1 antibody. The graph shows quantitative analysis of the CHX chase data. (b) HCT116 FBXW7+/+ and FBXW7−/− cells were treated with or without 10 µM MG132 for 6 h. Western blotting analysis was carried out using the anti-ENO1 antibody. The graph shows quantitative analysis. (c) HCT116 FBXW7+/+ and FBXW7−/− cells were treated with or without 25 µM GSK3β inhibitor VIII for 6 h. Western blotting analysis was carried out using the anti-ENO1 antibody. The graph shows quantitative analysis. All results are representative of three independent experiments. *P < 0.05 based on Student’s t-test.
Figure 4.
FBXW7 promotes the ubiquitination of ENO1. (a) HEK293T cells were co-transfected with Myc-ubiquitin and wild-type FBXW7, FBXW7ΔF or shFBXW7 B and then treated with MG132 for 6 h. Cell lysates were immunoprecipitated with anti-Myc antibody and analyzed by immunoblotting with the anti-ENO1 antibody. Left panel shows the input levels of the indicated proteins. (b) HCT116 FBXW7+/+ and FBXW7−/− cells were transfected with Myc-ubiquitin and treated with MG132 for 6 h. Cell extracts were immunoprecipitated with the anti-Myc antibody and analyzed by immunoblotting with the anti-ENO1 antibody. Left panel shows the input levels of the indicated proteins. All results are representative of three independent experiments.
FBXW7 Physically Binds to ENO1
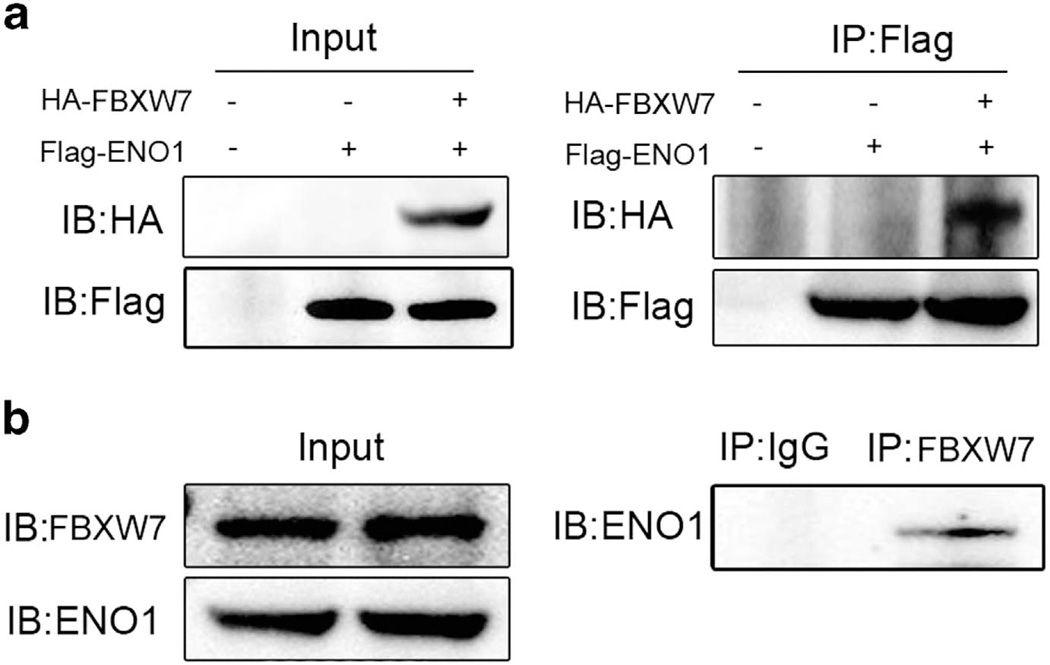
Next, we performed coimmunoprecipitation assay to determine whether ENO1 interacts with FBXW7. HEK293T cells were transiently transfected with Flag-tagged ENO1 alone or in combination with HA-tagged FBXW7. Lysates were immunoprecipitated with anti-Flag antibody and then probed for FBXW7 by anti-HA antibody. Western blotting analysis revealed that HA-tagged FBXW7 coimmunoprecipitated with Flag-tagged ENO1, indicating a physical interaction between the two proteins (Figure 5a). Moreover, to determine whether endogenous FBXW7 could bind to ENO1, lysates of HCT116 FBXW7+/+ cells were immunoprecipitated with anti-FBXW7 antibody and probed for ENO1 by anti-ENO1 antibody. Western blotting revealed that FBXW7 coprecipitated with endogenous ENO1 (Figure 5b). These results demonstrate that FBXW7 binds to ENO1 and targets ENO1 for proteasomal degradation.
Figure 5.
FBXW7 binds with ENO1 physically. (a) HEK293T cells were cotransfected with Flag-ENO1 and HA-FBXW7. Cell lysates were immuno-precipitated with anti-Flag and subjected to subsequent immunoblotting with anti-HA. (b) Cell lysates of HCT116 were immunoprecipitated with the FBXW7 antibody, followed by immunoblotting with the anti-ENO1 antibody. All results are representative of three independent experiments.
FBXW7 Negatively Regulates Biological Activities of ENO1
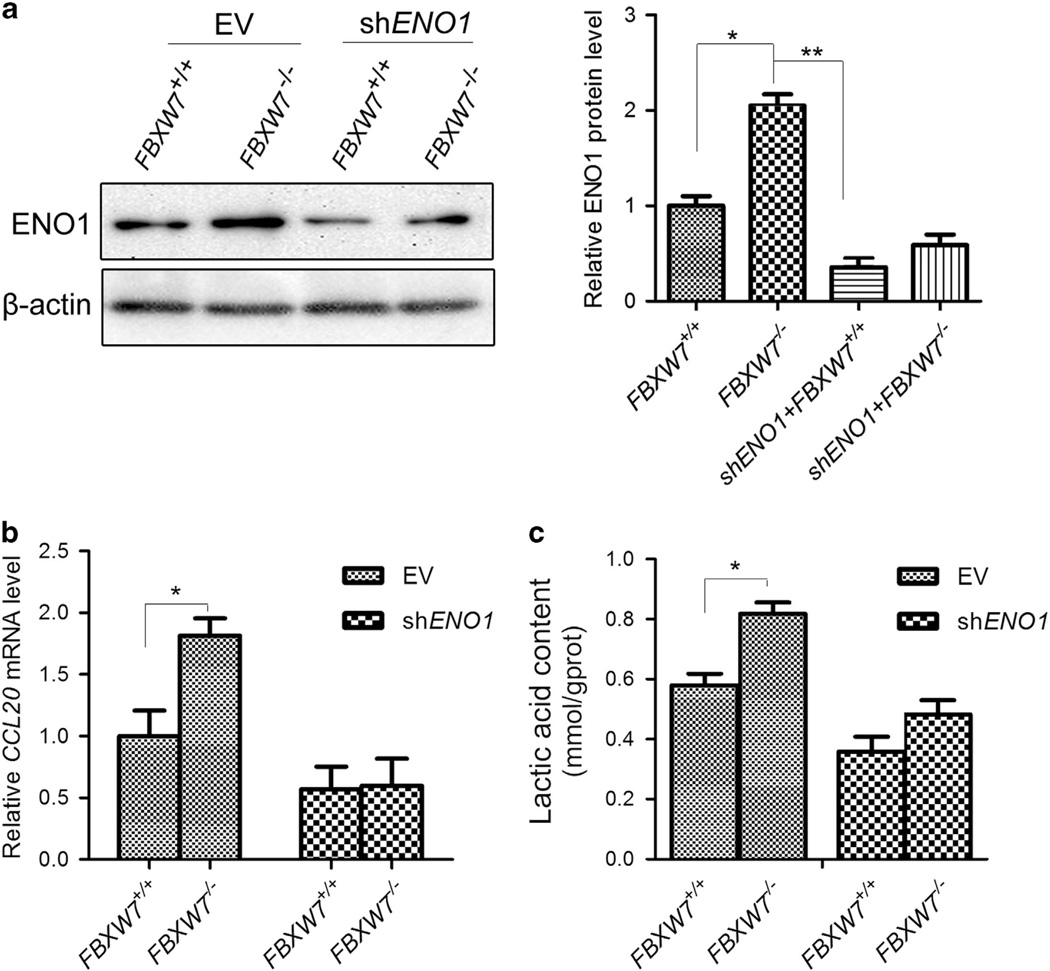
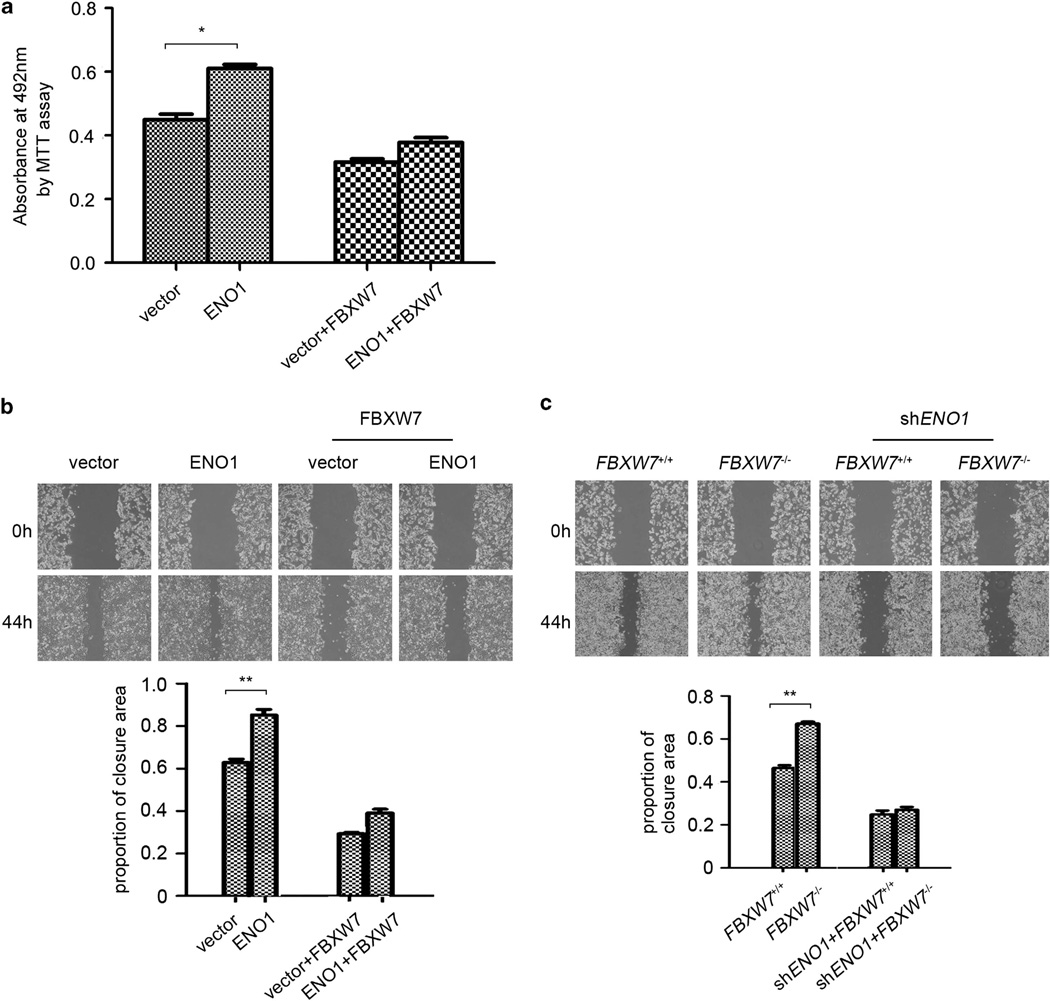
To determine the biological implications of FBXW7 in regulating ENO1 turnover, we monitored the effects of FBXW7 on ENO1-mediated cellular activities. We first analyzed whether FBXW7 regulated ENO1 target gene expression. As it is reported that chemokine CCL20 is a downstream target of ENO1 and can be upregulated by overexpression of ENO1 at the transcriptional level,6 we examined the effect of FBXW7 on ENO1-mediated CCL20 expression. ENO1 expression was silenced by shRNA construct, respectively, in HCT116 FBXW7+/+ and FBXW7−/− cells. The levels of ENO1 in the different cell groups were first verified by western blotting (Figure 6a). Then the CCL20 expression in these cells was measured by qRT-PCR. The results showed that depleting FBXW7 expression in HCT116 cells significantly increased CCL20 expression while silencing ENO1 blocked FBXW7 loss-induced CCL20 overexpression (Figure 6b). Given the function of ENO1 as a glycolysis enzyme, we then examined whether FBXW7 alters the cellular lactic acid concentration in an ENO1-dependent manner. The results showed that knockdown of ENO1 significantly impaired the increase of the cellular lactic acid level induced by loss of FBXW7 expression (Figure 6c). Next, we explored whether FBXW7 affects the biological function of ENO1. MTT and wound-healing assay showed that overexpression of FBXW7 reduced the promoting effect of ENO1 overexpression on cell growth and migration (Figure 7a and b). Similarly, ENO1 knockdown also alleviated FBXW7 loss-induced enhancement of cell migration (Figure 7c). Taken together, these results suggest that FBXW7 negatively regulated ENO1-mediated cell biological function.
Figure 6.
FBXW7 inhibits ENO1-induced CCL20 gene expression and lactate production. (a) The protein levels of ENO1 in different cells were examined by western blotting analysis. The graph shows quantitative analysis. (b) qRT-PCR to measure the effect of FBXW7 on ENO1-induced CCL20 expression in HCT116 cells. (c) Lactate concentration detection by Lactate Assay Kit to assess the effect of ENO1 on FBXW7 loss-induced lactate generation in HCT116 cells. All results are representative of three independent experiments. *P < 0.05, **P < 0.01 based on Student’s t-test.
Figure 7.
FBXW7 inhibits ENO1-induced cell proliferation and migration. (a) MTT analysis was used to detect the influence of FBXW7 on ENO1-induced cell proliferation of HCT116 cells. (b and c) Wound-healing assay to study the influence of FBXW7 on ENO1-induced migration ability of HCT116 cells (b) and the influence of ENO1 on FBXW7 loss-induced migration ability of HCT116 cells (c). The graph shows quantitative analysis. All results are representative of three independent experiments. *P < 0.05; **P < 0.01 based on Student’s t-test.
FBXW7 Negatively Correlates with the ENO1 Expression in Colon Cancer Tissues
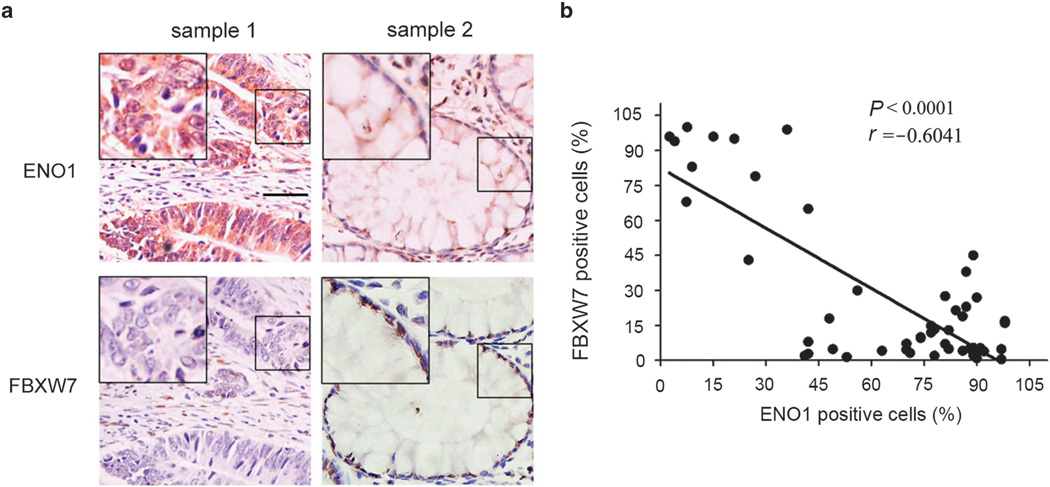
To explore any clinical significance of the observation, we then analyzed the expression of FBXW7 and ENO1 in colon cancer tissues by immunohistochemistry. The negative correlation between FBXW7 and ENO1 was revealed in these tissues (Figures 8a and b). This result is consistent with and further supports our above analyses that FBXW7 acts as a new negative regulator of ENO1.
Figure 8.
ENO1 expression is reversely correlated with FBXW7 expression in colon cancer tissues. Immunohistochemical assay was performed to detect FBXW7 and ENO1 expression in colon cancer tissues. (a) The representive staining of FBXW7 and ENO1. (b) The correlations between the expression of ENO1 and FBXW7. Scale bar indicates 50 µm.
DISCUSSION
FBXW7 represents a major tumor suppressor and has been firmly associated with tumor progression. As a physiological E3 ligase, the most common mechanism that FBXW7 exerts its tumor-suppressor function is targeting a network of onco-proteins, including cyclinE,21,32 Aurora A,27,33,34 mTOR,24 c-Myb,35 etc., for degradation by the proteasome. Although an increasing number of FBXW7 substrates have been identified recently, a detailed understanding of a full set of FBXW7 substrates and how FBXW7 inactivation causes disease remains largely unknown. In this study, to further elucidate the functional targets of FBXW7, we screened differentially expressed proteins induced by loss of FBXW7 in HCT116 cells using quantitative proteomics techniques. Among the proteins whose expression was altered by deficiency of FBXW7, ENO1 was first identified and validated with further western blotting.
In addition to its well-documented enzymatic role in glycolysis, ENO1 performs a variety of important cellular functions and has a critical role in physiological and pathophysiological process especially in carcinogenesis.1 ENO1 has been considered to be a diagnostic marker for many kind of tumors,9,14,36,37 and overexpression of ENO1 is a common scenario in several cancers and is closely connected with tumor progression and poor clinical out-comes.10,13,38 As ENO1 has an important role in tumorigenesis and tumor development, it will be important to further characterize the mechanisms through which ENO1 is regulated. Many reports have demonstrated that ENO1 can be regulated at different levels by multiple factors. For example, ENO1 can be transcriptionally activated by NFκB,8 HIF-1α4 and estrogen receptor (ER) α,2,8 while it also can be downregulated by retinoic acid at the protein level.39 Moreover, Yu et al2 observed that estradiol prolongs ENO1 protein half-life and increases ENO1 protein accumulation via a posttranscriptional mechanism. However, the posttranslational mechanisms that is involved in ENO1 regulation were not clearly investigated.
In the present study, our proteomics analysis revealed that homologous depletion of FBXW7 in HCT116 cells led to an obvious increase of ENO1 protein, which suggested an involvement of FBXW7 in the regulation of ENO1 abundance. Further experiments confirmed that ENO1 is a new target of FBXW7 and is downregulated by FBXW7 for proteasome degradation. To our knowledge, this is the first study to report that ENO1 protein is regulated by the FBXW7 E3 ligase at the posttranslational level. We provided several lines of evidence to support our finding. First, FBXW7 knockdown increases the ENO1 protein level, whereas FBXW7 overexpression decreases ENO1 protein expression. Supportively, significantly negative correlation between FBXW7 and ENO1 was found in human colorectal cancer tissues. Second, FBXW7 knockout significantly prolongs the ENO1 half-life. Meanwhile, proteasome inhibitor MG132 blocks FBXW7 loss-induced ENO1 overexpression. Third, FBXW7 interacts with ENO1 and promotes ubiquitination of ENO1. Furthermore, we also investigated the effect of FBXW7 on the biological role of ENO1 and finally found that FBXW7 suppresses the ENO1 function of promoting the CCL20 gene expression, lactate generation and colon cancer cell proliferation and migration.
In summary, the data presented herein suggest that FBXW7 negatively regulates the activity of ENO1 by facilitating its ubiquitin-mediated proteasomal degradation. Our current findings not only identify a new target of FBXW7 but also uncover a novel regulatory mechanism of ENO1, which may help us further understand the roles of FBXW7 and ENO1 in cancer development.
Supplementary Material
ACKNOWLEDGMENTS
We thank B Vogelstein for providing us with the HCT116 FBXW7−/− and DLD-1 FBXW7−/− cell lines. This work was supported by the National Natural Science Foundation of China Nos. 81172528, 31271461 and 81472583 and the Taishan Scholar Program of Shandong Province (to GW); by the National Institutes of Health, National Cancer Institute Grant R01 CA116481, and the Low Dose Scientific Focus Area, Office of Biological and Environmental Research, US Department of Energy (DE-AC02-05CH11231) (to JHM); by National Natural Science Foundation of China No. 81402193 (to WYS); and by the National Natural Science Foundation of China No. 81470127 and China Postdoctoral Science Foundation Funded Project Nos. 2011M501136 and 2012T50616 (to ZPJ).
Footnotes
Supplementary Information accompanies the paper on the Laboratory Investigation website (http://www.laboratoryinvestigation.org)
DISCLOSURE/CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Kang HJ, Jung SK, Kim SJ, et al. Structure of human alpha-enolase (hENO1), a multifunctional glycolytic enzyme. Acta Crystallogr D Biol Crystallogr. 2008;64:651–657. doi: 10.1107/S0907444908008561. [DOI] [PubMed] [Google Scholar]
- 2.Yu L, Shi J, Cheng S, et al. Estrogen promotes prostate cancer cell migration via paracrine release of ENO1 from stromal cells. Mol Endocrinol. 2012;26:1521–1530. doi: 10.1210/me.2012-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Migneco G, Whitaker-Menezes D, Chiavarina B, et al. Glycolytic cancer associated fibroblasts promote breast cancer tumor growth, without a measurable increase in angiogenesis: evidence for stromal-epithelial metabolic coupling. Cell Cycle. 2010;9:2412–2422. doi: 10.4161/cc.9.12.11989. [DOI] [PubMed] [Google Scholar]
- 4.Gao J, Zhao R, Xue Y, et al. Role of enolase-1 in response to hypoxia in breast cancer: exploring the mechanisms of action. Oncol Rep. 2013;29:1322–1332. doi: 10.3892/or.2013.2269. [DOI] [PubMed] [Google Scholar]
- 5.Bae S, Kim H, Lee N, et al. Alpha-enolase expressed on the surfaces of monocytes and macrophages induces robust synovial inflammation in rheumatoid arthritis. J Immunol. 2012;189:365–372. doi: 10.4049/jimmunol.1102073. [DOI] [PubMed] [Google Scholar]
- 6.Tsai ST, Chien IH, Shen WH, et al. ENO1, a potential prognostic head and neck cancer marker, promotes transformation partly via chemokine CCL20 induction. Eur J Cancer. 2010;46:1712–1723. doi: 10.1016/j.ejca.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 7.Kashat L, So AK, Masui O, et al. Secretome-based identification and characterization of potential biomarkers in thyroid cancer. J Proteome Res. 2010;9:5757–5769. doi: 10.1021/pr100529t. [DOI] [PubMed] [Google Scholar]
- 8.Tu SH, Chang CC, Chen CS, et al. Increased expression of enolase alpha in human breast cancer confers tamoxifen resistance in human breast cancer cells. Breast Cancer Res Treat. 2010;121:539–553. doi: 10.1007/s10549-009-0492-0. [DOI] [PubMed] [Google Scholar]
- 9.Yu L, Shen J, Mannoor K, et al. Identification of ENO1 as a potential sputum biomarker for early-stage lung cancer by shotgun proteomics. Clin Lung Cancer. 2014;15:372–378. doi: 10.1016/j.cllc.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pernemalm M, De Petris L, Branca RM, et al. Quantitative proteomics profiling of primary lung adenocarcinoma tumors reveals functional perturbations in tumor metabolism. J Proteome Res. 2013;12:3934–3943. doi: 10.1021/pr4002096. [DOI] [PubMed] [Google Scholar]
- 11.Duijvesz D, Burnum-Johnson KE, Gritsenko MA, et al. Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer. PLoS One. 2013;8:e82589. doi: 10.1371/journal.pone.0082589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang W, Li X, Rao S, et al. Constructing disease-specific gene networks using pair-wise relevance metric: application to colon cancer identifies interleukin 8, desmin and enolase 1 as the central elements. BMC Syst Biol. 2008;2:72. doi: 10.1186/1752-0509-2-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai Z, Ye Y, Liang B, et al. Proteomics-based identification of a group of apoptosis-related proteins and biomarkers in gastric cancer. Int J Oncol. 2011;38:375–383. doi: 10.3892/ijo.2010.873. [DOI] [PubMed] [Google Scholar]
- 14.Song Y, Luo Q, Long H, et al. Alpha-enolase as a potential cancer prognostic marker promotes cell growth, migration, and invasion in glioma. Mol Cancer. 2014;13:65. doi: 10.1186/1476-4598-13-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yonglitthipagon P, Pairojkul C, Bhudhisawasdi V, et al. Proteomics-based identification of alpha-enolase as a potential prognostic marker in cholangiocarcinoma. Clin Biochem. 2012;45:827–834. doi: 10.1016/j.clinbiochem.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Hamaguchi T, Iizuka N, Tsunedomi R, et al. Glycolysis module activated by hypoxia-inducible factor 1alpha is related to the aggressive phenotype of hepatocellular carcinoma. Int J Oncol. 2008;33:725–731. [PubMed] [Google Scholar]
- 17.Cui Y, Xie S, Luan J, et al. Quantitative proteomics and protein network analysis of A549 lung cancer cells affected by miR-206. Biosci Trends. 2013;7:259–263. [PubMed] [Google Scholar]
- 18.Chen S, Duan G, Zhang R, et al. Helicobacter pylori cytotoxin-associated gene A protein upregulates alpha-enolase expression via Src/MEK/ERK pathway: implication for progression of gastric cancer. Int J Oncol. 2014;45:764–770. doi: 10.3892/ijo.2014.2444. [DOI] [PubMed] [Google Scholar]
- 19.Trojanowicz B, Winkler A, Hammje K, et al. Retinoic acid-mediated down-regulation of ENO1/MBP-1 gene products caused decreased invasiveness of the follicular thyroid carcinoma cell lines. J Mol Endocrinol. 2009;42:249–260. doi: 10.1677/JME-08-0118. [DOI] [PubMed] [Google Scholar]
- 20.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 21.Van Vlierberghe P, Homminga I, Zuurbier L, et al. Cooperative genetic defects in TLX3 rearranged pediatric T-ALL. Leukemia. 2008;22:762–770. doi: 10.1038/sj.leu.2405082. [DOI] [PubMed] [Google Scholar]
- 22.Klotz K, Cepeda D, Tan Y, et al. SCF(Fbxw7/hCdc4) targets cyclin E2 for ubiquitin-dependent proteolysis. Exp Cell Res. 2009;315:1832–1839. doi: 10.1016/j.yexcr.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 23.Snyder JL, Kearns CA, Appel B. Fbxw7 regulates Notch to control specification of neural precursors for oligodendrocyte fate. Neural Dev. 2012;7:15. doi: 10.1186/1749-8104-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao JH, Kim IJ, Wu D, et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321:1499–1502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Liu Y, Lu J, et al. Rapamycin inhibits FBXW7 loss-induced epithelial-mesenchymal transition and cancer stem cell-like characteristics in colorectal cancer cells. Biochem Biophys Res Commun. 2013;434:352–356. doi: 10.1016/j.bbrc.2013.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao JH, Perez-Losada J, Wu D, et al. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 27.Kwon YW, Kim IJ, Wu D, et al. Pten regulates Aurora-A and cooperates with Fbxw7 in modulating radiation-induced tumor development. Mol Cancer Res. 2012;10:834–844. doi: 10.1158/1541-7786.MCR-12-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park MJ, Taki T, Oda M, et al. FBXW7 and NOTCH1 mutations in childhood T cell acute lymphoblastic leukaemia and T cell non-Hodgkin lymphoma. Br J Haematol. 2009;145:198–206. doi: 10.1111/j.1365-2141.2009.07607.x. [DOI] [PubMed] [Google Scholar]
- 29.Grim JE. Fbxw7 hotspot mutations and human colon cancer: mechanistic insights from new mouse models. Gut. 2013;63:707–709. doi: 10.1136/gutjnl-2013-305144. [DOI] [PubMed] [Google Scholar]
- 30.Davis H, Lewis A, Spencer-Dene B, et al. FBXW7 mutations typically found in human cancers are distinct from null alleles and disrupt lung development. J Pathol. 2011;224:180–189. doi: 10.1002/path.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akhoondi S, Sun D, von der Lehr N, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 32.Sancho R, Jandke A, Davis H, et al. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology. 2010;139:929–941. doi: 10.1053/j.gastro.2010.05.078. [DOI] [PubMed] [Google Scholar]
- 33.Finkin S, Aylon Y, Anzi S, et al. Fbw7 regulates the activity of endo-reduplication mediators and the p53 pathway to prevent drug-induced polyploidy. Oncogene. 2008;27:4411–4421. doi: 10.1038/onc.2008.77. [DOI] [PubMed] [Google Scholar]
- 34.Otto T, Horn S, Brockmann M, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Kanei-Ishii C, Nomura T, Takagi T, et al. Fbxw7 acts as an E3 ubiquitin ligase that targets c-Myb for nemo-like kinase (NLK)-induced degradation. J Biol Chem. 2008;283:30540–30548. doi: 10.1074/jbc.M804340200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, Zhou M, Zhao R, et al. Identification of candidate biomarkers for the early detection of nasopharyngeal carcinoma by quantitative proteomic analysis. J Proteomics. 2014;109C:162–175. doi: 10.1016/j.jprot.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 37.Takashima M, Kuramitsu Y, Yokoyama Y, et al. Overexpression of alpha enolase in hepatitis C virus-related hepatocellular carcinoma: association with tumor progression as determined by proteomic analysis. Proteomics. 2005;5:1686–1692. doi: 10.1002/pmic.200401022. [DOI] [PubMed] [Google Scholar]
- 38.Cappello P, Rolla S, Chiarle R, et al. Vaccination with ENO1 DNA prolongs survival of genetically engineered mice with pancreatic cancer. Gastroenterology. 2013;144:1098–1106. doi: 10.1053/j.gastro.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 39.Trojanowicz B, Sekulla C, Lorenz K, et al. Proteomic approach reveals novel targets for retinoic acid-mediated therapy of thyroid carcinoma. Mol Cell Endocrinol. 2010;325:110–117. doi: 10.1016/j.mce.2010.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.