Abstract
Background
Knowledge about genetic diversity and relationships among germplasms could be an invaluable aid in diospyros improvement strategies.
Methods
This study was designed to analyze the genetic diversity and relationship of local and natural varieties in Guangxi Zhuang Autonomous Region of China using start codon targeted polymorphism (SCoT) markers. The accessions of 95 diospyros germplasms belonging to four species Diospyros kaki Thunb, D. oleifera Cheng, D. kaki var. silverstris Mak, and D. lotus Linn were collected from different eco-climatic zones in Guangxi and were analyzed using SCoT markers.
Results
Results indicated that the accessions of 95 diospyros germplasms could be distinguished using SCoT markers, and were divided into three groups at similarity coefficient of 0.608; these germplasms that belong to the same species were clustered together; of these, the degree of genetic diversity of the natural D. kaki var. silverstris Mak population was richest among the four species; the geographical distance showed that the 12 natural populations of D. kaki var. silverstris Mak were divided into two groups at similarity coefficient of 0.19. Meanwhile, in order to further verify the stable and useful of SCoT markers in diospyros germplasms, SSR markers were also used in current research to analyze the genetic diversity and relationship in the same diospyros germplasms. Once again, majority of germplasms that belong to the same species were clustered together. Thus SCoT markers were stable and especially useful for analysis of the genetic diversity and relationship in diospyros germplasms.
Discussion
The molecular characterization and diversity assessment of diospyros were very important for conservation of diospyros germplasm resources, meanwhile for diospyros improvement.
Introduction
The genus diospyros (Ebenaceae) consists of approximately 400 to 500 highly heterozygous species, most of which are distributed in the subtropical and tropical regions of Asia, Africa, and central South America [1,2]. Diospyros Linn originated from China, and has over 900 cultivars [3,4]. Nine diospyros species are cultivated in China as fruit trees or rootstock, among these, D. kaki Thunb is the most economically valuable and widely cultivated species [5,6]. D. kaki Thunb in China accounting for 89.79% and 73.84% of total planting area and yield worldwide, respectively [7].
Guangxi is one of the most important production areas for diospyros, and diospyros germplasm resources are abundant and widely distributed because of the unique climate and diverse ecology [8]. Previous reports had mainly focused on planting cultivars, without local and natural varieties. Therefore, the assignment of cultivar identity is a major problem due to insufficient information, meanwhile, due to presence of synonyms and homonyms among local and natural varieties, which affect significantly the exploration, utilization, and protection of diospyros germplasm resources in Guangxi.
Investigation and analysis of genetic diversity of diospyros germplasms will provide information basis to support diospyros improvement and botanical research as well as support conservation efforts. So far, different marker techniques such as plant morphology [9], karyotype [10], isoenzyme [11] and DNA-based markers random amplified polymorphic DNA (RAPD) [12], simple sequence repeat (SSR) [13], restriction fragment length polymorphism (RFLP) [14], sequence-related amplified polymorphism (SRAP) [4], amplified fragment length polymorphism (AFLP) [15], and inverse sequence-tagged repeat (ISTR) [2] have been applied to study the genetic diversity and relationships between diospyros species and their relatives. Nevertheless, the relationships between diospyros accessions of local and natural varieties are still not completely clarified in spite of all previous efforts, probably because of the low resolution of the germplasm resources collection, conservation and exploitation.
Recently, a molecular marker technique termed start codon targeted (SCoT) polymorphism, a simple and novel DNA marker technique, was developed by Collard and Mackill [16]. SCoT marker technique is a simple and novel targeted molecular marker tool which base on the short conserved region flanking the start codon (ATG) in plant genes; it can generate more information related to biological traits than random DNA markers [16]. SCoT markers employ longer primers (18-mer) producing high polymorphism which is reproducible [17], it is suggested that primer length and annealing temperature are not the sole factors determining reproducibility [16]. As a single primer amplification molecular marker technique, SCoT markers are technically simple, not time-consuming and not laborious, and requires no prior sequence information and targeting functional regions [18]. SCoT markers have been broadly and successfully used for evaluation of genetic diversity, phylogenetics, fingerprinting, variation, and differentiation since 2009 [17–19].
However, up to now, no report has been available on the investigation and analysis of genetic diversity of diospyros germplasms using SCoT molecular markers in Guangxi. The objective of the present research was to identify the genetic diversity and relationships of diospyros germplasm of the local and natural varieties to provide theoretical basis for classification, protection, and utilization of diospyros germplasm resources in Guangxi China.
Materials and Methods
Plant material
A total of 95 Diospyros Linn and 189 D. kaki var. silvestris individuals corresponding to 12 natural populations were investigated and labeled in Guangxi Zhuang autonomous region, China (Fig 1, Table 1, and S1 Table), then young leaves were collected from the labeled individuals in the wild area, frozen in liquid nitrogen, and stored at -80°C until analysis.
Fig 1. The localities of the natural D. kaki var. silvestris Mak samples.
Table 1. Diospyros germplasm resources and their respective localities.
| NO. | Germplasms | Species | Localities | NO. | Germplasms | Species | Localities |
|---|---|---|---|---|---|---|---|
| 1 | Yueshi | D. kaki Thunb | Gongcheng county | 49 | PL1 | D. oleifera Cheng | Pingle county |
| 2 | Niuxinshi | D. kaki Thunb | Pingle county | 50 | PL2 | D. oleifera Cheng | Pingle county |
| 3 | Jixinshi | D. kaki Thunb | Longlin county | 51 | JC1 | D. oleifera Cheng | Jinchengjiang district |
| 4 | Xiaofangshi | D. kaki Thunb | Leye county | 52 | JC2 | D. oleifera Chen | Jinchengjiang district |
| 5 | Dafangshi | D. kaki Thunb | Leye county | 53 | YZ1 | D. oleifera Cheng | Yizhou city |
| 6 | Huoshi | D. kaki Thunb | Gongcheng county | 54 | YZ2 | D. oleifera Cheng | Yizhou city |
| 7 | Jingshi | D. kaki Thunb | Hengxian county | 55 | YZ3 | D. oleifera Cheng | Yizhou city |
| 8 | HZ | D. kaki Thunb | Zhongshan county | 56 | YZ4 | D. kaki Thunb | Yizhou city |
| 9 | LQ1 | D. kaki Thunb | Liangqing district | 57 | YZ5 | D. oleifera Cheng | Yizhou city |
| 10 | LQ2 | D. kaki Thunb | Liangqing district | 58 | YZ6 | D. oleifera Cheng | Yizhou city |
| 11 | LL1 | D. oleifera Cheng | Longlin county | 59 | YZ7 | D. oleifera Cheng | Yizhou city |
| 12 | LL2 | D. kaki Thunb | Longlin county | 60 | YZ8 | D. oleifera Cheng | Yizhou city |
| 13 | LL3 | D. kaki Thunb | Longlin county | 61 | YZ9 | D. oleifera Cheng | Yizhou city |
| 14 | LL4 | D. kaki Thunb | Longlin county | 62 | YZ10 | D. oleifera Cheng | Yizhou city |
| 15 | LY1 | D. kaki Thunb | Leye county | 63 | YZ11 | D. kaki Thunb | Yizhou city |
| 16 | LY2 | D. kaki Thunb | Leye county | 64 | YZ12 | D. kaki Thunb | Yizhou city |
| 17 | LY3 | D. kaki Thunb | Leye county | 65 | YZ13 | D. kaki Thunb | Yizhou city |
| 18 | LY4 | D. kaki Thunb | Leye county | 66 | QZ1 | D. oleifera Cheng | Quanzhou county |
| 19 | LY5 | D. oleifera Cheng | Leye county | 67 | QZ2 | D. oleifera Cheng | Quanzhou county |
| 20 | LY6 | D. oleifera Cheng | Leye county | 68 | QZ3 | D. oleifera Cheng | Quanzhou county |
| 21 | LY7 | D. oleifera Cheng | Leye county | 69 | QZ4 | D. oleifera Cheng | Quanzhou county |
| 22 | LY8 | D. oleifera Cheng | Leye county | 70 | QZ5 | D. oleifera Cheng | Quanzhou county |
| 23 | GN1 | D. kaki Thunb | Gangnan district | 71 | QZ6 | D. oleifera Cheng | Quanzhou county |
| 24 | GN2 | D. kaki Thunb | Gangnan district | 72 | QZ7 | D. oleifera Cheng | Quanzhou county |
| 25 | GN3 | D. kaki Thunb | Gangnan district | 73 | QZ8 | D. oleifera Cheng | Quanzhou county |
| 26 | GN4 | D. kaki Thunb | Gangnan district | 74 | QZ9 | D. oleifera Cheng | Quanzhou county |
| 27 | GN5 | D. kaki Thunb | Gangnan district | 75 | QZ10 | D. oleifera Cheng | Quanzhou county |
| 28 | GB1 | D. kaki Thunb | Gangbei district | 76 | XL1 | D. oleifera Cheng | Xilin county |
| 29 | GB2 | D. kaki Thunb | Gangbei district | 77 | XL2 | D. oleifera Cheng | Xilin county |
| 30 | GB3 | D. kak iThunb | Gangbei district | 78 | XL3 | D. oleifera Cheng | Xilin county |
| 31 | GB4 | D. kak iThunb | Gangbei district | 79 | XL4 | D. oleifera Cheng | Xilin county |
| 32 | GB5 | D. kaki Thunb | Gangbei district | 80 | XL5 | D. kaki Thunb | Xilin county |
| 33 | GB6 | D. kaki Thunb | Gangbei district | 81 | XL6 | D. oleifera Cheng | Xilin county |
| 34 | GB7 | D. kaki Thunb | Gangbei district | 82 | XL7 | D. oleifera Cheng | Xilin county |
| 35 | GB8 | D. kaki Thunb | Gangbei district | 83 | XL8 | D. oleifera Cheng | Xilin county |
| 36 | GB9 | D. oleifera Cheng | Gangbei district | 84 | XL9 | D. oleifera Cheng | Xilin county |
| 37 | GB10 | D. oleifera Cheng | Gangbei district | 85 | XL10 | D. oleifera Cheng | Xilin county |
| 38 | GB11 | D. oleifera Cheng | Gangbei district | 86 | TL1 | D. oleifera Cheng | Tianilin county |
| 39 | GB12 | D. oleifera Cheng | Gangbei district | 87 | TL2 | D. oleifera Cheng | Tianilin county |
| 40 | QB1 | D. oleifera Cheng | Qinbei district | 88 | TL3 | D. oleifera Cheng | Tianilin county |
| 41 | QB2 | D. oleifera Cheng | Qinbei district | 89 | WX | D. oleifera Cheng | Laibin county |
| 42 | QB3 | D. oleifera Cheng | Qinbei district | 90 | Yeshi1 | D.kaki var.silvestris | Pingle county |
| 43 | QB4 | D. kaki Thunb | Qinbei district | 91 | Yeshi2 | D.kaki var.silvestris | Pingle county |
| 44 | QB5 | D. oleifera Cheng | Qinbei district | 92 | Yeshi3 | D.kaki var.silvestris | Leye county |
| 45 | QB6 | D. oleifera Cheng | Qinbei district | 93 | Yeshi4 | D.kaki var.silvestris | Tianlin county |
| 46 | GC1 | D. oleifera Cheng | Gongcheng county | 94 | Yeshi5 | D.kaki var.silvestris | Quanzhou county |
| 47 | GC2 | D. oleifera Cheng | Gongcheng county | 95 | Junqianzi | D.lotus Linn | Gongcheng county |
| 48 | GC3 | D. oleifera Cheng | Gongcheng county |
Note: The germplasms that have not been previously reported, or whose names are not sure were temporarily named after the first letter of their localities and numbers.
Ethics Statement
The study was approved by Guangxi Zhuang autonomous region government, which was part of diospyros germplasm resources protection in China and supported by the public basic scientific research project foundation of Guangxi Zhuang Autonomous Region (Rzz200901).
DNA extraction and genotype analysis
DNA was individually extracted as described by [20–22]. PCR reactions were performed in 20 μL volumes containing 2.0 μL of 10× buffer, 30 ng of genomic DNA, 30 μmol·L-1 primers, 4.0 mmol·L-1 dNTPs, and 0.2 unit of Taq polymerase (BoRi, Hangzhou, China). SCoT-PCR Amplification was performed in a GeneAmp PCR System 9700 (Perkin-Elmer Corp., Norwalk, CT, USA) under initial denaturation for 4 min at 95°C, followed by 36 cycles of 40 s at 95°C, 40 s at 50°C and 2 min at 72°C, followed by final extension of 7 min at 72°C.
SSR primers were also used to analyze and verify the genetic diversity and relationship of local and natural varieties in Guangxi Zhuang Autonomous Region of China, which were previously described in detail [22–24]. SSR-PCR Amplification was performed under initial denaturation for 5 min at 94°C, followed by 30 cycles of 50 s at 94°C, 1 min at annealing temperature and 50s at 72°C, followed by final extension of 4 min at 72°C. The SCoT-PCR and SSR-PCR products were separated and stained, following [25].
Data analysis
The SCoT and SSR profiles for each band were scored as present (1) or absent (0) based on size comparison with the standard (2000 bp DNA Ladder Plus). Cluster analysis and dendrogram construction were carried out using NTSYS-pc 2.1e software [26]. Cluster analysis based on the genetic similarity matrix was performed using unweighted pair group method with arithmetic mean (UPGMA) method [27]. Nei’ s gene diversity index (He), Shannon’s information index (Ho), observed number of alleles (Na), and effective number of alleles (Ne) were calculated using the GenAlEx 6.5 program [28]. Nei’s unbiased genetic distances were calculated for all population pairs, and were used to construct a phylogenetic tree [29]. Principal coordinate analysis (PCoA) [30] was performed based on the variance covariance matrix calculated from the marker data. The distribution of genetic variation among the populations was analyzed using AMOVA 1.55 software, and a Mantel test in NTSYS-pc 2.10e software was used to test whether the matrix of the genetic distances correlates with the matrix of geographical distances.
Results
Analysis of validation, amplification and polymorphic of SCoT markers in diospyros germplasm resources
A total of 80 pairs of SCoT primers were selected from the previous literatures [16,17], which were used to screen for polymorphic markers using three accessions (S2 and S3 Tables) and the polymorphic markers identified were used to genotype individuals of 95 Diospyros Linn and 189 D. kaki var. silvestris. Only primers that exhibited unambiguous and reproducible band patterns were selected for further analysis. Thus, a total of 18 primers that exhibit distinct and reliable band patterns were utilized for bands scoring.
Genotypes of 95 diospyros germplasm Accessions were analyzed using the 18 screened primers. Up to 241 unambiguous and reproducible bands were generated, of those, 233 (96.68%) were polymorphic; only eight fragments were present in all the 95 accessions. The sizes of the amplified bands ranged from 250 bp to 2500 bp, of those, most of the band sizes ranged from 500 bp to 2000 bp (S1 Fig). The number of bands amplified by the selected primers ranged from 7 to 16 per primer, with an average of 13.39 bands per primer.
Genetic diversity analysis of diospyros germplasm resources using SCoT markers
The coefficients of similarity between the germplasms were calculated using Ntedit software. These results showed that the lowest similarity coefficient was observed between YZ4 and QZ4, which was 0.485. The highest similarity coefficient was observed between GN3 and GB7, which was 0.946. The data indicated that the genetic relationship between YZ4 and QZ4 was the farthest while the genetic relationship between GN3 and GB7 was the nearest.
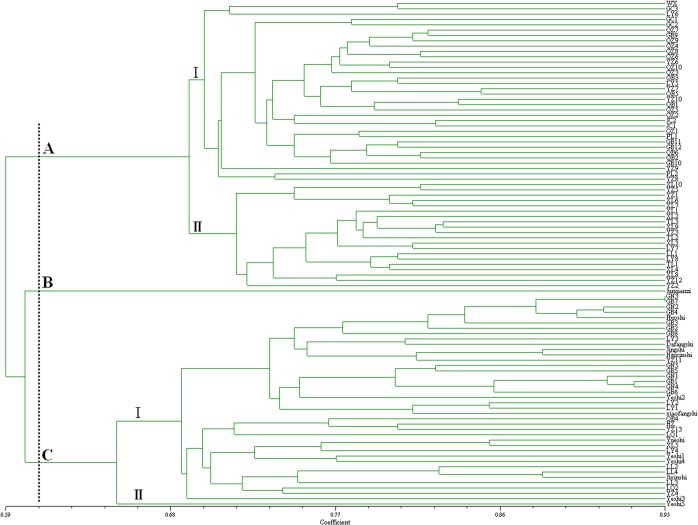
Based on corresponding similarity coefficients among the tested accessions of 95 diospyros germplasms, a dendrogram was constructed (Fig 2). The cluster analysis demonstrated that the genetic relationship of the diospyros germplasms accessions genotypes was complex. The 95 accessions fell under divided into three major groups at a genetic distance threshold of 0.608. Group A included 54 accessions, all of which belonged to D. oleifera Cheng. Group B contained only one accession belonged to D. lotus Linn. Group C included 40 accessions, 35 of those belonged to Diospyros kaki Thunb and 5 belonged to D. kaki var. silvestris Mak. These results indicated that D. kaki Thunb, D. oleifera Cheng, and D. lotus Linn could be distinguished from each other by the18 SCoT primers selected, and the genetic relationship of D. kaki Thunb and D. kaki var. silverstris Mak seemed closer than those of other species.
Fig 2. UPGMA dendrogram of the accessions of 95 diospyros germplasms based on SCoT molecular markers.
Principal component Analysis (PCA) based on the genetic similarity matrix of SCoT
Principal component Analysis (PCA) based on the genetic similarity matrix of SCoT was conducted and the two-dimensional clustering was obtained (S2 Fig), 95 accessions of diospyros germplasms were divided into four group, the accessions of group A belonged to D. oleifera Cheng, group B belonged to D. lotus Linn, group C belonged to Diospyros kaki Thunb and D. kaki var. silverstris Mak, and group D belonged to D. kaki var. silverstris Mak only. The result of principal component analysis was corresponding to clustering analysis.
Genetic diversity analysis of diospyros germplasm resources using SSR markers
In order to further verify the stable and useful of SCoT markers in diospyros germplasms, 15 SSR markers screened that showed polymorphisms selected from previous research were also used to analyze the genetic diversity and relationship in the same diospyros germplasms accessions. Results indicated 159 unambiguous and reproducible bands were generated, of those, 154 were polymorphic. The number of bands amplified by the selected primers ranged from 7 to 14 per primer, with an average of 10.60 bands per primer. The sizes of the amplified bands ranged from 80 bp to 500 bp, of those, most of the band sizes rangeed from 80 bp to 230 bp (S3 Fig and S4 Table). The coefficients of similarity between the germplasms were also calculated using Ntedit software, once again, results showed that the lowest similarity coefficient was observed between YZ4 and QZ4.
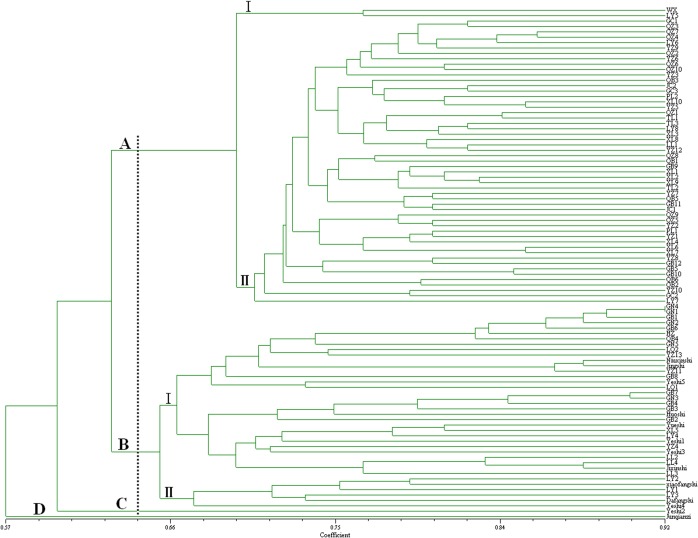
A dendrogram analyzed using SSR markers was also constructed (Fig 3). The 95 accessions fell under divided into four major groups at a genetic distance threshold of 0.642. Group A contained 55 accessions of D. oleifera Cheng, Group B contained 34 accessions of Diospyros kaki Thunb and 5 accessions of D. kaki var. silverstris Mak. Group C contained only one accession belonged to D. kaki var. silverstris Mak. Group D contained one accession belonged to D. lotus Linn. These results indicated that D. kaki Thunb, D. oleifera Cheng, and D. lotus Linn could be distinguished from each other by the15 SSR primers selected, group A belonged to D. oleifera Cheng, group B belonged to Diospyros kaki Thunb and Group B belonged to D. lotus Linn. The genetic relationship of D. kaki Thunb and D. kaki var. silverstris Mak seemed closer than that of other species; however, genetic variation between wild persimmon germplasm belonged to different sources was larger. Nevertheless, the results of SSR molecular markers were basically consistent with that of molecular markers SCoT analysis.
Fig 3. UPGMA dendrogram of the accessions of 95 diospyros germplasms based on SSR molecular markers.
Principal component Analysis (PCA) based on the genetic similarity matrix of SSR analysis was conducted and the two-dimensional clustering was obtained (S4 Fig), 95 accessions of diospyros germplasms were divided into four group, the accessions of group A belonged to D. oleifera Cheng, group B belonged to Diospyros kaki Thunb and D. kaki var. silverstris Mak, group C belonged to D. kaki var. silverstris Mak and group D belonged to D. lotus Linn. The results of principal component analysis were corresponding to clustering analysis.
Genetic diversity analysis of natural D. kaki var. silverstris Mak populations
The genotypes of 189 individuals from 12 natural D. kaki var. silverstris Mak populations were analyzed using 13 selected primers. Altogether, 159 unambiguous and reproducible bands were generated, of those, 155 (97.48%) were polymorphic, with sizes ranging from 300 bp to 1800 bp (S3 Table). The number of bands ranged from 9 to 17 per primer, with an average of 12.23 bands per primer. These results showed that the natural D. kaki var. silverstris Mak distributed in Guangxi were rich in polymorphisms.
The genetic diversity among Zhongshan populations was determined to be the highest, with He and Ho values of 0.178 and 0.269, respectively. By contrast, the genetic diversity among Youjiang populations was found to be the lowest, with He and Ho values of 0.122 and 0.192, respectively. The genetic diversity values of these populations were ranked as Zhongshan > Leye > Luzhai > Quanzhou > Tianlin > Xilin > Hengxian > Longlin > Qintang > Wuxuan > Huanjiang > Youjiang from high to low. The number of alleles (Na) and effective number of alleles (Ne) showed the same results (Table 2).
Table 2. Results of analysis of the genetic diversity of the 12 natural D. kaki var. silverstris Mak. populations.
| Population | size | Observed number of allele/Na | Effective number of allele/Ne | Shannon’s information index/Ho | Nei’s gene diversity/He | Percentage of polymorphic loci (PPL)/% |
|---|---|---|---|---|---|---|
| LL | 16.000 | 1.528 | 1.224 | 0.239 | 0.146 | 64.15 |
| XL | 15.000 | 1.226 | 1.247 | 0.233 | 0.150 | 53.46 |
| TL | 19.000 | 1.415 | 1.258 | 0.247 | 0.157 | 62.89 |
| YJ | 15.000 | 1.239 | 1.199 | 0.192 | 0.122 | 48.43 |
| QZ | 14.000 | 1.201 | 1.278 | 0.240 | 0.160 | 49.69 |
| HJ | 16.000 | 1.170 | 1.218 | 0.196 | 0.128 | 45.28 |
| ZS | 14.000 | 1.321 | 1.307 | 0.269 | 0.178 | 57.23 |
| LZ | 16.000 | 1.277 | 1.272 | 0.245 | 0.160 | 57.23 |
| WX | 15.000 | 1.164 | 1.224 | 0.201 | 0.131 | 48.43 |
| QT | 18.000 | 1.289 | 1.232 | 0.222 | 0.142 | 54.09 |
| LQ | 16.000 | 1.333 | 1.249 | 0.232 | 0.149 | 57.86 |
| LY | 15.000 | 1.434 | 1.260 | 0.257 | 0.162 | 63.52 |
| mean | 15.750 | 1.300 | 1.247 | 0.231 | 0.149 | 55.19 |
| total | 189.000 | 1.975 | 1.424 | 0.417 | 0.266 | 97.48 |
Data from the AMOVA molecular detection prove that inter-populations have 54.12% genetic variation (Table 3), the results showed that the genetic differentiation of D. kaki var. silverstris Mak was more highly affected in inter-population groups. Gene flow, estimated by Gst [Nm = 0.5 (1—Gst)/Gst)], was 0.585, indicated that genetic differentiation exists between the natural D. kaki var. silverstris Mak populations and that species variation was also affected within the population. The SCoT data were analyzed through PCoA. The accumulative contribution rate of the first and second coordinates was 49.96%, indicated the main original information. The 189 individuals in the two-dimension diagram occupy a large space and were widely distributed, showed that its genetic background was broad and extremely diverse. Some populations overlap and interact, showed that the permeation of genes and communication took place among the natural D. kaki var. silverstris Mak populations (S5 Fig). The geographic distance and genetic distance between the wild D. kaki var. silverstris Mak populations were calculated through Mantel test analysis, results showed that genetic distance had positive relationship with geographical distance (r = 0.5559; S5 Table).
Table 3. Analysis of molecular variance (AMOVA) within and among natural D. kaki var. silvestris Mak. populations.
| Source of variance | d.f | SSD | MSD | Variance component | Percentage(%) | P* |
|---|---|---|---|---|---|---|
| Among populations (AP) | 11 | 2198.555 | 199.869 | 11.814 | 45.88 | < 0.001 |
| Within populations (WP) | 177 | 2466.979 | 13.938 | 13.938 | 54.12 | < 0.001 |
| Total | 188 | 4665.534 |
Note: MSD,expected mean squares
*Number of permutation = 1,000
A dendrogram was constructed based on the results of Nei’s genetic distance using the UPGMA method (Fig 4). Twelve natural D. kaki var. silverstris Mak populations in Guangxi were divided into two clusters at similarity coefficient of 0.19. One cluster contained populations from Longlin, Xilin, Tianlin, Youjiang, and Leye; whereas the other contained populations from Quanzhou, Huanjiang, Zhongshan, Luzhai, Wuxuan, Qintang, and Hengxian. The genetic distance between the Wuxuan and Qintang populations was 0.091, and the genetic relationship was the closest among the populations. However, the genetic distance between the Youjiang and Quanzhou populations was 0.292, showed the farthest genetic distance. The clustering result supported the distribution of the two principal coordinates by PCoA.
Fig 4. UPGMA dendrogram of the natural D. kaki var. silvestris Mak populations based on Nei’s genetic distance.
Discussion
Guangxi is located in low latitude ranging from 50 m to 1500 m. diospyros has been cultivated in Guangxi for over 400 years, and possesses abundant germplasm resources. The diospyros germplasm resources in Guangxi are not fully utilized because of poor identification and insufficient information. Many diospyros germplasms around the region that had low commodity value were neglected, and were usually cut down or top grafted by local farmers. The genetic diversity of diospyros germplasm resources decreased with the passage of time, and many rural local varieties that had great value in germplasm preservation and breeding died due to the lack of protection. Most diospyros germplasms in China were local varieties, meanwhile, another diospyros germplasms were developed through bud mutation, nevertheless, which were sporadic [3,31]. Thus, it is necessary to investigate and analyze the genetic diversity of diospyros germplasms to provide theoretical basis for classification, protection and utilization of diospyros germplasm resources in Guangxi China.
Molecular marker technology has been widely applied in research on diospyros germplasm resources [13,32,33]. Some studies suggested that the genetic diversity of diospyros germplasms revealed by SCoT markers was consistent with phenotypic analysis [34]. D. kaki Thunb, D. oleifera Cheng, and D. lotus Linn germplasms were used in current experiment to be completely distinguishable through UPGMA analysis. The D. kaki var. silvestris Mak germplasms are closer to D. kaki Thunb than the other species. D. kaki var. silvestris Mak is the native species of D. kaki Thunb [35]. Meanwhile, in order to further verify the stable and useful of SCoT markers in diospyros germplasms, SSR markers were also used in current research to analyze the genetic diversity and relationship in the same diospyros germplasms. The results of SSR molecular markers were basically consistent with that of molecular markers SCoT analysis. Thus SCoT markers were stable and especially useful for analysis of the genetic diversity and relationship in diospyros germplasms.
The climate in Guangxi greatly varies from the south to the north, but various natural D.kaki var. silvestris Mak populations from different regions were clustered together. Twelve populations separately gather into two big clusters. One cluster includes populations from Leye, Xilin, Tianlin, and Youjiang, which were in northwestern Guangxi, the part of Yunnan-Guizhou plateau with mountainous terrain and high altitude. The other clusters included Quanzhou of northeastern Guangxi, Huanjiang of northern Guangxi, Zhongshan of eastern Guangxi, Luzai, Wuxuan, and Qintang of central Guangxi, and Hengxian of southern Guangxi. Population genetic structures were often influenced by many factors, including habitat fragmentation, gene flow, breeding system, and seed dispersal mechanism [36,37]. When gene flow (Nm) < l, the population was more susceptible to genetic drift [38,39]. The average Nm value of natural D. kaki var. silvestris Mak is 0.585, which indicated that gene exchange between the populations was restricted, and the genetic differentiation in each population was severe.
The level of population genetic diversity was related to the size of the population; bigger populations generally have higher levels of genetic diversity [39]. The results of current study indicated that the genetic diversity of the Zhongshan population was the highest and that of the Youjiang population was the lowest. These results showed that the habitats of natural D. kaki var. silvestris Mak populations in Youjiang, Huanjiang, Wuxuan, and Qintang had been seriously damaged, and that the number of individuals has reduced.
In conclusion, SCoT molecular markers help assess genetic diversity among accessions or natural populations from different areas. The diospyros germplasm resources in Guangxi possess broad genetic background and have rich diversity, but some rural varieties and natural populations are in danger of being lost. Effective measures to protect the genetic diversity of diospyros germplasm resources are necessary. These results of current research will help in the classification, preservation, and utilization of diospyros resources.
Supporting Information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The research was supported by the public basic scientific research project foundation of Guangxi Zhuang Autonomous Region (Rzz200901), National Natural Science Foundation of China (No. 31471849) and Fund on Basic Scientific Research Project of Nonprofit Central Research Institutions. No. SSCRI 1630062014002. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Yonemori K (1997) Diospyros industry and research activities in Japan. Acta Hort 436: 21–32. [Google Scholar]
- 2. Du XY, Zhang MS, Luo ZR, Zhang QL (2009) Identification and GeneticRelationships of diospyros kakiThunb. and Related SpeciesUsing ISTR Analysis. Acta Horticulturae Sinica 36 (4): 481–486. [Google Scholar]
- 3. Wang RZ, Yang Y, Li GC (1997) Chinese diospyros germplasm resources. Acta Hortic 436: 43–50. [Google Scholar]
- 4. Guo DL, Luo ZR (2006) Genetic relationships of some PCNA persimmons (Diospyros kaki Thunb.) from China and Japan revealed by SRAP analysis. Genet Resour Crop Ev. 53: 1597–1603. [Google Scholar]
- 5. Wang RZ (1987) Persimmmon. Chin For press, Beijing: 619–636. [Google Scholar]
- 6. Luo ZR, Cai LH, Hu CG (1996) Research development of persimon germplasm resources of Diospyros kaki and their Utilization. J. Huazhong Agric Univ. 15 (4): 381–388. [Google Scholar]
- 7.FAO (2012) FAOSTAT Database. http://faostat.fao.org/
- 8.Deng LB (2013) Research on the genetic Diversity and angular leaf spot disease resistance of persimmon germplasm resources in Guangxi. Doctoral dissertation of Guangxi univ. Nanning 15–25.
- 9. Wang R, Yang Y, Li GC (1997) Chinese persimmon germplasmresources. Acta Hortic 436: 43–50. [Google Scholar]
- 10. Yang Y, Wang RZ, Li GC, Wang W (1999) Study on Chromosome Numbers of diospyros and Their Varieties. Acta Agriculturae Boreali-occidentalis Sinica 8 (3): 64–67. [Google Scholar]
- 11. Kim TC, Ko KC (1997) Taxonomic studies of persimmon (Diospyros Kaki Thumb.) by multivariate and isozyme analysis. Acta Horticulturae 436: 85–92. [Google Scholar]
- 12. Luo ZR, Yonemori K, Sugiura A (1995) Evaluation of RAPD analysis for cultivar identification of persimmons (Diospyros kaki). Journal of the Japanese Society for Horticultural Science 64(3): 535–541. [Google Scholar]
- 13. Naval MDM, Zuriaga E, Pecchioli S, Llácer G, Giordani E, Badenes ML (2010) Analysis of genetic diversity among persimmon cultivars using microsatellite markers. Tree Genet. Genomes 6: 677–687. [Google Scholar]
- 14. Hu DC, Zhang QL, Luo ZR (2008) Phylogenetic analysis in some Diospyros spp. (Ebenaceae) and Japanese persimmon using chloroplast DNA PCR-RFLP markers. Scientia Horticulturae 117: 32–38. [Google Scholar]
- 15. Yonemori K, Honsho C, Kitajima A, Aradhya M, Giordani E, Bellini E, et al. (2008) Relationship of European persimmon (Diospyros kaki Thunb.) cultivars to Asian cultivars, characterized using AFLPs. Genet Resour Crop Evol 55: 81–89. [Google Scholar]
- 16. Collard BCY, Mackill DJ (2009) Start codon targeted (SCoT) polymorphism: a simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol. Biol. Rep. 27: 86–93. [Google Scholar]
- 17. Mulpuri S, Muddanuru T, Francis G (2013) Start codon targeted (SCoT) polymorphism in toxic and non-toxic accessions of Jatropha curcas L. and development of a codominant SCAR marker. Plan. Sci 207: 117–127. [DOI] [PubMed] [Google Scholar]
- 18. Xiong FQ, Zhong RC, Han ZQ (2011) Start codon targeted polymorphism for evaluation of functional genetic variation and relationships in cultivated peanut (Arachis hypogaea L.) genotypes. Mol Biol Rep 38: 3487–3494. doi: 10.1007/s11033-010-0459-6 [DOI] [PubMed] [Google Scholar]
- 19. Amirmoradi B, Talebi R, Karami E (2012) Comparison of genetic variation and differentiation among annual Cicer species using start codon targeted (SCoT) polymorphism, DAMD-PCR, and ISSR markers. Plant Syst Evol 298: 1679–1688. [Google Scholar]
- 20. Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15. [Google Scholar]
- 21. Soriano JM, Pecchioli S, Romero C, Vilanova S, Llácer G, Giordani E, et al. (2006) Development of microsatellite markers in polyploid persimmon (Diospyros kaki Lf) from an enriched genomic library. Mol Ecol Notes 6: 368–370. [Google Scholar]
- 22. He XH, Li YR, Guo YZ, Tang ZP, Li RB (2005) Genetic analysis of 23 mango cultivar collection in Guangxi province revealed by ISSR. Mol. Plant Breed. 3: 829–834. [Google Scholar]
- 23. Guo DL, Luo ZR (2004) Establishment of SSR technique of Diospyros kaki and D, lotus[J]. Journal of Agricultural Biotechnology 12(4): 386–389. [Google Scholar]
- 24. Zhang YF, Zhang QL, Yang Y, Luo ZR (2009) Deleopment of Japanese persimmon core collection by genetic distance sampling based on SSR markers. Biotechnol. & Biotechnol. Eq. 23(4): 1474–1478. [Google Scholar]
- 25. Liang QZ, Wen DQ, Xie JH, Liu LQ, Wei YZ, Wang YC and Shi SY (2014) A rapid and effective method for silver staining of PCR products separated in polyacrylamide gels. Electrophoresis 35(17): 2520–2523. doi: 10.1002/elps.201400182 [DOI] [PubMed] [Google Scholar]
- 26. Rohlf FJ (2000) NTSYS-pc: Numerical taxonomy and multivariate analysis system, version 2.1 Exeter Software, Setauket, New York. [Google Scholar]
- 27.Sneath PHA, Sokal RR (1973) Numerical taxonomy. The principles and practice of numerical classification. pp. xv + 573 pp
- 28. Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research- an update. Bioinforma 28: 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nei M (1972) Genetic distance between populations. Am Naturalist 106: 283–292. [Google Scholar]
- 30. Gower JC (1966) Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53: 325–338. [Google Scholar]
- 31. Yang Y, Ruan XF, Wang RZ, Li GC (2005) A dvances in research of germplasm resources and breeding of Dispyros kaki L. J. Northwest For Univ 20: 133–137. [Google Scholar]
- 32. Ai CX, Qin ZH, Tao JH, Wang CJ (2011) SSR fingerprints and genetic variations of the 32 persimmon major cultivars. Acta Bot. Borea1 3l: 2185–219l. [Google Scholar]
- 33. Wu S, Fu JM, Wuyun TN, Liang YQ (2012) Study on EST-SSR primer design and genetic deversity of Diospyros L. J. Central South Univ. For. Tech. 32: 153–157. [Google Scholar]
- 34. Deng LB, He XH, Li TW, Hu Y (2012) Investigation and analysis on the genetic diversity of diospyros germplasms in plateau of northwest Guangxi. Acta Hortic. Sin 39: 215–224. [Google Scholar]
- 35. Gong BC, Wang RZ, Yang Y (2011) High quality practical technology of diospyros cultivation China For press, Beijing: 67–68. [Google Scholar]
- 36. Fischer M, Matthies D (1998) RAPD variation in relation to population size and plant fitness in the rare Gentianella germanica (Gentianaceae). Am. J. Bot. 85: 811–819. [PubMed] [Google Scholar]
- 37. Ge XJ, Zhang LB, Yuan YM, Hao G (2005) Strong genetic differentiation of the East-Himalayan Megacodon stylophorus (Gentianaceae) detected by intersimple sequence repeats (ISSR). Biodivers Conserv. 14: 849–861 [Google Scholar]
- 38. Wright S (1951) The genetical structure of populations. Ann. Eug. 15: 323–354. [DOI] [PubMed] [Google Scholar]
- 39. He XH, Pan H, Deng LB, Pan JC, Li F et al. (2010) Genetic diversity of natural Myrica rubra Sieb.et Zucc populations in Guangxi revealed by ISSR markers. Sci. Agric. sin. 9: 626–632. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.