Abstract
Biochemical tests have been previously developed to identify carbapenemase-producing Enterobacteriaceae, Pseudomonas spp. (Carba NP test) and Acinetobacter spp. (CarbAcineto NP test). We evaluated a modified Carba NP test to detect carbapenemase production in Enterobacteriaceae, Pseudomonas and Acinetobacter species using a single protocol with rapid results and found good reliability and speed.
Keywords: Carba NP test, carbapenemase, carbapenems, Gram negative, multidrug-resistant bacteria
Multidrug-resistant Gram-negative bacteria (GNB) are increasingly being reported worldwide. The spread of carbapenemase-producing Enterobacteriaceae, Pseudomonas and Acinetobacter species have become a global threat. The emergence of resistance to carbapenems makes the treatment for infections caused by these carbapenem-resistant strains very limited [1–3]. Different types of carbapenemases have been reported, such as Ambler class A Klebsiella pneumoniae carbapenemase (KPC) and Guiana extended spectrum (GES) β-lactamase, Ambler class B metallo-β-lactamases (MBL) and Ambler class D oxacillinase type [1].
Rapid methods for detecting carbapenemase producers have been described, such as the MALDI-TOF MS (matrix-assisted laser desorption/ionization time-of-flight mass spectrometry) carbapenemase assay [4]. Previous studies have described a rapid biochemical carbapenemase detection method based on imipenem hydrolysis, the Carba NP test, for Enterobacteriaceae[5] and Pseudomonas species [6], as well as the CarbAcineto NP test for Acinetobacter species [7]. Recently, however, several authors have published evaluations of the Carba NP and the CarbAcineto NP tests; their criticisms focussed essentially on the absence of detection of oxacillinase (OXA) type carbapenemases [8–10].
Here we describe a modified Carba NP (MCNP) test which enables the rapid detection of different carbapenemases (KPC, MBL and OXA types) from Enterobacteriaceae, Pseudomonas and Acinetobacter species using a single protocol.
One hundred ten previously characterized GNB, including 69 carbapenemase-producing GNB (Enterobacteriaceae n = 14, Pseudomonas aeruginosa n = 11 and Acinetobacter baumannii n = 44), and 41 non-carbapenemase-producing GNB, including Enterobacteriaceae (n = 24), P. aeruginosa (n = 5) and A. baumannii (n = 12), were tested in two laboratories including Unité de recherche sur les maladies infectieuses et tropicales émergentes (URMITE), Aix-Marseille University, Marseille, France, and Microbial Ecology laboratory, Béjaia University, Béjaia, Algeria (Table 1). Carbapenemase activity was assessed using phenotypic and genotypic tests, including the modified Hodge test, MALDI-TOF MS assay, PCR amplification and sequencing [4,11].
Table 1.
Test results for Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter baumannii strains
| Group | Species | Carbapenemase or other β-lactamase gene | Test result by: |
||
|---|---|---|---|---|---|
| MHT | MALDI-TOF MS | MCNP | |||
| Enterobacteriaceae | Klebsiella pneumoniae H5500M | NDM-1 | + | + | + |
| K. pneumoniae KpnaseyM | NDM-1 | + | + | + | |
| K. pneumoniae U2A 2246M | KPC-3 | + | + | + | |
| K. pneumoniaeA | KPC-3 | + | + | + | |
| K. pneumoniae 360M | KPC-2 | + | + | + | |
| K. pneumoniaeM | OXA-48 | + | + | + | |
| K. pneumoniae 413A | TEM-1/CTX-M-3/SHV-12/DHA-1 | − | − | − | |
| K. pneumoniae 473A | TEM-1/CTX-M-15/OXA-1 | − | − | − | |
| K. pneumoniae 463A | TEM-1/CTX-M-15 | − | − | − | |
| K. pneumoniae 550A | TEM-1/CTX-M-3 | − | − | − | |
| K. pneumoniae 47A | TEM-1/CMY-4 | − | − | − | |
| K. pneumoniae 123A | TEM-1/CMY-4 | − | − | − | |
| K. pneumoniae 318A | CTX-M-15 | − | − | − | |
| K. pneumoniae 476A | CTX-M-15 | − | − | − | |
| K. pneumoniae 613A | CMY-4 | − | − | − | |
| K. pneumoniae 615A | CMY-4 | − | − | − | |
| Escherichia coli H5636M | NDM-1 | + | + | + | |
| E. coli 181A | NDM-5 | + | + | + | |
| E. coliM | NDM-5 | + | + | + | |
| E. coli 99A | OXA-48 | + | + | + | |
| E. coli 100A | OXA-48 | + | + | + | |
| E. coli 132A | OXA-48 | + | + | + | |
| E. coli 192A | OXA-48 | + | + | + | |
| E. coliM | OXA-48 | + | + | + | |
| E. coli 544A | TEM-1/CTX-M-15/OXA-1 | − | − | − | |
| E. coli 469A | TEM-1/CTX-M-3/OXA-1 | − | − | − | |
| E. coli 472A | TEM-1/CTX-M-3/OXA-1 | − | − | − | |
| E. coli 234A | TEM-1/CTX-M-15/CMY-4 | − | − | − | |
| E. coli 611A | TEM-1/CTX-M-15/CMY-4 | − | − | − | |
| E. coli 536A | TEM-1/CTX-M-15 | − | − | − | |
| E. coli 534A | CTX-M-15/OXA-1 | − | − | − | |
| E. coli 542A | CTX-M-15/OXA-1 | − | − | − | |
| E. coli 560A | TEM-1/CMY-4 | − | − | − | |
| E. coli 606A | TEM-1/CMY-4 | − | − | − | |
| E. coli ATCC 25922M | — | − | − | − | |
| E. coli 7624M | — | − | − | − | |
| E. cloacae 308A | TEM-1/CTX-M-15/OXA-1 | − | − | − | |
| E. cloacae 570A | TEM-1/CTX-M-15 | − | − | − | |
| Pseudomonas | P. aeruginosaM | VIM-2 | + | + | + |
| P. aeruginosaM | VIM-2 | + | + | + | |
| P. aeruginosaM | VIM-2 | + | + | + | |
| P. aeruginosaM | VIM-2 | + | + | + | |
| P. aeruginosaM | VIM-2 | + | + | + | |
| P. aeruginosaM | VIM-2 | + | + | + | |
| P. aeruginosaM | VIM-2 | + | + | + | |
| P. aeruginosaM | VIM-2 | + | + | + | |
| P. aeruginosaM | VIM-2 | + | + | + | |
| P. aeruginosaM | VIM-2 | + | + | + | |
| P. aeruginosaM | — | − | − | − | |
| P. aeruginosaM | — | − | − | − | |
| P. aeruginosaM | — | − | − | − | |
| P. aeruginosaM | — | − | − | − | |
| P. aeruginosaM | — | − | − | − | |
| P. aeruginosa UAA 2257M | IMP-1 | + | + | + | |
| Acinetobacter | A. baumanniiA | OXA-23 | + | + | + |
| A. baumanniiA | OXA-23 | + | + | + | |
| A. baumanniiA | OXA-23 | + | + | + | |
| A. baumanniiA | OXA-23 | + | + | + | |
| A. baumanniiA | OXA-23 | + | + | + | |
| A. baumanniiA | OXA-23 | + | + | + | |
| A. baumanniiM | OXA-23 | + | + | + | |
| A. baumanniiM | OXA-23 | + | + | + | |
| A. baumanniiM | OXA-23 | + | + | + | |
| A. baumanniiM | OXA-23 | + | + | + | |
| A. baumanniiM | OXA-24 | + | + | + | |
| A. baumanniiM | OXA-24 | + | + | + | |
| A. baumanniiM | OXA-24 | + | + | + | |
| A. baumanniiM | OXA-24 | + | + | + | |
| A. baumanniiM | OXA-24 | + | + | + | |
| A. baumanniiM | OXA-24 | + | + | + | |
| A. baumanniiA | OXA-24 | + | + | + | |
| A. baumanniiA | OXA-24 | + | + | + | |
| A. baumanniiA | OXA-24 | + | + | + | |
| A. baumanniiA | OXA-24 | + | + | + | |
| A. baumanniiM | OXA-58 | + | + | + | |
| A. baumanniiM | OXA-23/OXA-24 | + | + | + | |
| A. baumanniiA | OXA-23/OXA-24 | + | + | + | |
| A. baumanniiA | OXA-23/OXA-24 | + | + | + | |
| A. baumanniiA | NDM-1 | + | + | + | |
| A. baumanniiA | NDM-1 | + | + | + | |
| A. baumanniiA | NDM-1 | + | + | + | |
| A. baumanniiA | NDM-1 | + | + | + | |
| A. baumanniiA | NDM-1 | + | + | + | |
| A. baumanniiM | NDM-1 | + | + | + | |
| A. baumanniiM | NDM-1 | + | + | + | |
| A. baumanniiM | NDM-1 | + | + | + | |
| A. baumanniiM | NDM-1 | + | + | + | |
| A. baumanniiM | NDM-1 | + | + | + | |
| A. baumanniiM | OXA-23/NDM-1 | + | + | + | |
| A. baumanniiM | OXA-23/NDM-1 | + | + | + | |
| A. baumanniiM | OXA-23/NDM-1 | + | + | + | |
| A. baumanniiM | OXA-23/NDM-1 | + | + | + | |
| A. baumanniiM | OXA-23/NDM-1 | + | + | + | |
| A. baumanniiM | OXA-23/NDM-1 | + | + | + | |
| A. baumanniiA | OXA-23/NDM-1 | + | + | + | |
| A. baumanniiA | OXA-23/NDM-1 | + | + | + | |
| A. baumanniiA | OXA-23/NDM-1 | + | + | + | |
| A. baumanniiA | OXA-23/NDM-1 | + | + | + | |
| A. baumanniiA | TEM-128 | − | − | − | |
| A. baumanniiA | TEM-128 | − | − | − | |
| A. baumanniiA | TEM-128 | − | − | − | |
| A. baumanniiA | TEM-128 | − | − | − | |
| A. baumanniiA | TEM-128 | − | − | − | |
| A. baumanniiA | TEM-128 | − | − | − | |
| A. baumanniiA | TEM-128 | − | − | − | |
| A. baumanniiM | TEM-128 | − | − | − | |
| A. baumanniiM | TEM-128 | − | − | − | |
| A. baumanniiM | TEM-128 | − | − | − | |
| A. baumannii AYEM | VEB-1 | − | − | − | |
| A. baumannii SDFM | — | − | − | − | |
MALDI-TOF MS, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; MCNP, modified Carba NP test; MHT, modified Hodge test.
AStrains tested in Microbial Ecology Laboratory, Béjaia University, Béjaia, Algeria.
MStrains tested in Unité de recherche sur les maladies infectieuses et tropicales émergentes (URMITE), Aix-Marseille University, Marseille, France.
The Carba NP and the CarbAcineto NP tests are straightforward biochemical tests which identify carbapenemase production in GNB by detecting imipenem hydrolysis using phenol red solution as a colour indicator and a bacterial lysis buffer (B-PER II, Bacterial Protein Extraction Reagent) for Enterobacteriaceae and Pseudomonas species (Carba NP test) [5,6] and 5 M NaCl for Acinetobacter species (CarbAcineto NP test) [7].
In order to use a single protocol to detect the production of carbapenemases in the three types of bacteria (Enterobacteriaceae, Pseudomonas and Acinetobacter) and to accelerate the speed with which results are produced, the lysis buffer and pH of the colour indicator solution used in the Carba NP and CarbAcineto NP tests were changed.
In the MCNP test, the lysis buffers used for the Carba NP test and CarbAcineto NP test, B-PER II, Bacterial Protein Extraction Reagent and NaCl 5 M, respectively, were replaced by cetyl trimethyl ammonium bromide (CTAB) 0.02%, and the pH value of the phenol red solution was adjusted to 7.5 (instead of 7.8). In addition, two steps used in the previous protocols [5,6], centrifugation and incubation at room temperature for 30 minutes, were eliminated in our method. These modifications simplify the lysis step and produce results more quickly.
The MCNP test was performed as follows. One inoculation loop (10 μL) of the tested strain, directly recovered from a Mueller Hinton agar plate (bioMérieux, Marcy l'Étoile, France), was resuspended in 200 μL of 0.02% CTAB (Sigma-Aldrich Chimie, Saint-Quentin-Fallavier, France) and vortexed for 1 to 2 minutes. Subsequently, 100 μL of the bacterial suspension was mixed with 100 μL of diluted phenol red solution (2 mL of phenol red (Sigma-Aldrich) solution 0.5% (wt/vol) with 16.6 mL of distilled water) containing 0.1 mM ZnSO4 (pH 7.5) in the first tube, tube 1, used as negative control, and a diluted phenol red solution containing 0.1 mM ZnSO4 (pH 7.5) supplemented with 6 mg/mL of commercially available imipenem (Tienam 500; Merck Sharp & Dohme, Paris, France) in the second tube, tube 2. Tubes 1 and 2 were vortexed, then incubated at 37°C for a maximum of 2 hours.
Carbapenemase activity was revealed when the test and negative control solutions, respectively, were yellow vs. red or orange vs. red. In contrast, both solutions remained red in the case of noncarbapenemase producers (Fig. 1).
Fig. 1.
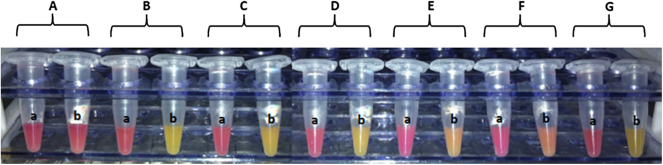
Modified Carba NP test results for Enterobacteriaceae, Pseudomonas and Acinetobacter species. (A) Escherichia coli ATCC 25922. (B) NDM-5-positive E. coli. (C) KPC-2-positive Klebsiella pneumoniae 360. (D) IMP-1-positive Pseudomonas aeruginosa UAA 2257. (E) OXA-23-positive Acinetobacter baumannii. (F) OXA-24-positive A. baumannii. (G) NDM-1-positive A. baumannii. (a) Tube containing phenol red solution 0.1 mM ZnSO4 (pH 7.5) and cetyl trimethyl ammonium bromide (CTAB) 0.02%. (b) Tube containing phenol red solution 0.1 mM ZnSO4 (pH 7.5) supplemented with 6 mg/mL of imipenem and CTAB 0.02%.
The results showed that the MCNP method detected all carbapenemases produced by carbapenem-resistant strains with 100% sensitivity and 100% specificity. Positive results were observed at different times for the different carbapenemases types (MBL, KPC and OXA-48 at 10 to 30 minutes vs. 1 to 2 hours for OXA type). The most interesting aspect of this method is that the colour changed from red to orange or yellow (positive result) even before incubation in some cases (NDM-5-producing Escherichia coli, NDM-1-producing Klebsiella pneumoniae (Kpnasey) and imipenem-producing P. aeruginosa UAA2257). Moreover, a higher inoculums (two inoculation loops (10 μL)) is recommended for Acinetobacter species tests.
Currently, the MCNP test is routinely used in Timone Hospital, Marseille, France. It was performed when antibiotic susceptibility testing revealed a resistance to ertapenem and susceptibility or resistance to imipenem. The suspicion of carbapenemase producers, in particular OXA-48, was based on this phenotype. Between November 2014 and May 2015, a total of 233 strains were tested. Among them, 35 positives strains with carbapenemase producers were detected (Table 2). These positives strains were isolated from 25 different patients. These results confirm the efficiency of the MCNP test with high sensitivity, given the detection of all strains producing OXA-48-type carbapenemases. Also, two carbapenemase-producing A. baumannii were detected, thus confirming the advantages of the MCNP test.
Table 2.
Results of MCNP test applied for carbapenem-resistant strains isolated in La Timone Hospital, Marseille, France
| Date | Sample source | Strain | Antibiotic susceptibility testing results |
MCNP result | Carbapenemases gene detected | |
|---|---|---|---|---|---|---|
| ETP | IMP | |||||
| 10/11/2014 | Urine | Klebsiella pneumoniae | R | R | + | OXA-48 |
| 20/11/2014 | Rectal swab | Escherichia coli | R | S | + | OXA-48 |
| 25/11/2014 | Bronchoalveolar lavage fluid | K. pneumoniae | R | S | + | OXA-48 |
| 05/12/2014 | Bronchoalveolar lavage fluid | K. pneumoniae | R | R | + | OXA-48 |
| 08/12/2014 | Rectal swab | K. pneumoniae | R | S | + | OXA-48 |
| 09/12/2014 | Blood culture | K. pneumoniae | R | R | + | OXA-48 |
| 11/12/2014 | Urine | Enterobacter cloacae | R | S | + | OXA-48 |
| 21/12/2014 | Stools | K. pneumoniae | R | S | + | OXA-48 |
| 31/12/2014 | Spittle | K. pneumoniae | R | R | + | OXA-48 |
| 12/01/2015 | Urine | E. coli | R | S | + | OXA-48 |
| 24/01/2015 | Armpit swab | K. pneumoniae | R | S | + | OXA-48 |
| 02/02/2015 | Rectal swab | K. pneumoniae | R | R | + | NDM |
| 06/02/2015 | Urine | E. coli | R | S | + | OXA-48 |
| 17/02/2015 | Urine | E. coli | R | S | + | OXA-48 |
| 19/02/2015 | Spittle | K. pneumoniae | R | S | + | OXA-48 |
| 20/02/2015 | Rectal swab | K. pneumoniae | R | S | + | OXA-48 |
| 23/02/2015 | Urine | K. pneumoniae | R | S | + | OXA-48 |
| 04/03/2015 | Rectal swab | E. cloacae | R | R | + | OXA-48 |
| 04/03/2015 | Rectal swab | K. pneumoniae | R | R | + | OXA-48 |
| 16/03/2015 | Rectal swab | K. pneumoniae | R | S | + | OXA-48 |
| 23/03/2015 | Bronchial aspirate | K. pneumoniae | R | S | + | OXA-48 |
| 30/03/2015 | Armpit swab | K. pneumoniae | R | S | + | OXA-48 |
| 31/03/2015 | Urine | K. pneumoniae | R | S | + | OXA-48 |
| 13/04/2015 | Sinus | Serratia marcescens | R | R | + | OXA-48 |
| 13/04/2015 | Blood culture | E. cloacae | R | S | + | OXA-48 |
| 18/04/2015 | Blood culture | E. coli | R | I | + | OXA-48 |
| 18/04/2015 | Rectal swab | K. pneumoniae | R | S | + | OXA-48 |
| 18/04/2015 | Blood culture | E. coli | R | I | + | OXA-48 |
| 04/05/2015 | Urine | K. pneumoniae | R | R | + | OXA-48 |
| 30/04/2015 | Urine | K. pneumoniae | R | I | + | NDM-1 |
| 07/05/2015 | Bronchial aspirate | K. pneumoniae | R | I | + | OXA-48 |
| 08/05/2015 | Blood culture | Acinetobacter baumannii | NT | R | + | OXA-23 |
| 11/05/2015 | Urine | A. baumannii | NT | R | + | OXA-23 |
| 18/05/2015 | Rectal swab | K. pneumoniae | R | I | + | OXA-48 |
ETP, ertapenem; IMP, imipenem; MCNP, modified Carba NP test; NT, not tested; R, resistant; S, susceptible; I, Intermediate.
In conclusion, the advantages of the MCNP test are the detection of different carbapenemase types from Enterobacteriaceae, Pseudomonas and Acinetobacter species using a single protocol, as well as the short time to results, particularly in the case of MBL-producing Enterobacteriaceae and Pseudomonas species. In addition, the effectiveness of this test on a large series of bacteria may allow us to identify the production of carbapenemase enzymes even before identification of the bacterial strain.
Interestingly, as well as using this test in developed countries such as France (URMITE laboratories, La Timone Hospital), given the simplicity and the low cost of the MCNP test, it could be used by any laboratory, including laboratories in developing countries. In Algeria, this test has been used in the Microbial Ecology Laboratory of Béjaia University since May 2014, and it will soon be introduced to laboratories in Algerian hospitals.
Conflict of interest
None declared.
Acknowledgements
We thank L. Hadjadj, A. Bergal, M. A. El-Gawad El-Sayed and N. Mathlouthi for their contributions. Supported in part by CNRS and IHU Méditerranée Infection.
References
- 1.Dortet L., Poirel L., Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int. 2014;2014:249856. doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zarrilli R., Pournaras S., Giannouli M., Tsakris A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents. 2013;41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Kempf M., Rolain J.M. Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: clinical impact and therapeutic options. Int J Antimicrob Agents. 2012;39:105–114. doi: 10.1016/j.ijantimicag.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Kempf M., Bakour S., Flaudrops C. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One. 2012;7:e31676. doi: 10.1371/journal.pone.0031676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nordmann P., Poirel L., Dortet L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2012;18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dortet L., Poirel L., Nordmann P. Rapid detection of carbapenemase-producing Pseudomonas spp. J Clin Microbiol. 2012;50:3773–3776. doi: 10.1128/JCM.01597-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dortet L., Poirel L., Errera C., Nordmann P. CarbAcineto NP test for rapid detection of carbapenemase-producing Acinetobacter spp. J Clin Microbiol. 2014;52:2359–2364. doi: 10.1128/JCM.00594-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tijet N., Boyd D., Patel S.N., Mulvey M.R., Melano R.G. Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57:4578–4580. doi: 10.1128/AAC.00878-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong P.M., McCorrister S.J., Unger M.S. MALDI-TOF MS detection of carbapenemase activity in clinical isolates of Enterobacteriaceae spp., Pseudomonas aeruginosa, and Acinetobacter baumannii compared against the Carba-NP assay. J Microbiol Methods. 2015;111:21–23. doi: 10.1016/j.mimet.2015.01.024. [DOI] [PubMed] [Google Scholar]
- 10.Osterblad M., Hakanen A.J., Jalava J. Evaluation of the Carba NP test for carbapenemase detection. Antimicrob Agents Chemother. 2014;58:7553–7556. doi: 10.1128/AAC.02761-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee K., Kim C.K., Yong D. Improved performance of the modified Hodge test with MacConkey agar for screening carbapenemase-producing Gram-negative bacilli. J Microbiol Methods. 2010;83:149–152. doi: 10.1016/j.mimet.2010.08.010. [DOI] [PubMed] [Google Scholar]


