Abstract
Fast calcium signaling is regulated by numerous calcium channels exhibiting high spatiotemporal profiles which are currently measured by fluorescent calcium sensors. There is still a strong need to improve the kinetics of genetically encoded calcium indicators (sensors) to capture calcium dynamics in the millisecond time frame. In this review, we summarize several major fast calcium signaling pathways and discuss the recent developments and application of genetically encoded calcium indicators to detect these pathways. A new class of genetically encoded calcium indicators designed with site-directed mutagenesis on the surface of beta-barrel fluorescent proteins to form a pentagonal bipyramidal-like calcium binding domain dramatically accelerates calcium binding kinetics. Furthermore, novel genetically encoded calcium indicators with significantly increased fluorescent lifetime change are advantageous in deep-field imaging with high light-scattering and notable morphology change.
Introduction
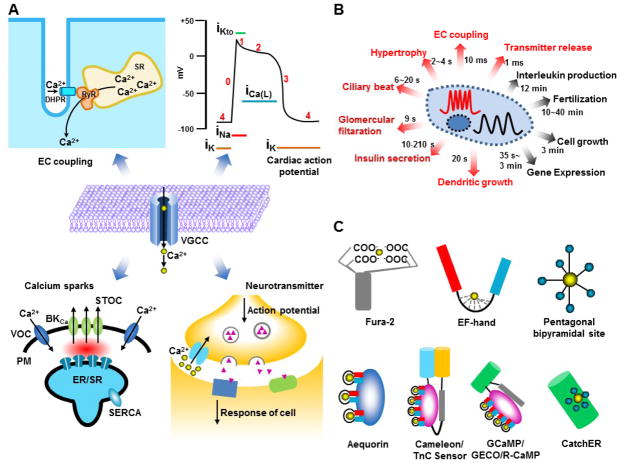
Calcium (Ca2+), a second messenger and the most ubiquitous signaling molecule, plays an important role in regulating various biological functions in living organisms (Figure 1A). The time scale of calcium ion flow varies from milliseconds in muscle contractions to days for fertilization and development (Figure 1B) [1]. Rapid calcium signaling regulates calcium channels, excitation-contraction coupling, action potential, calcium sparks, and release of neurotransmitters (Figure 1A). Voltage gated calcium channels (VGCCs) exhibit a high open and close frequency and deliver fast calcium movement through a hydrophilic path in response to plasma membrane voltage changes, allowing precise calcium signaling within milliseconds [2, 3]. During channel activation, calcium concentration is estimated to be hundreds of micromolar within several nanometers from the mouth of the channels, generating Ca2+ microdomains. A high Ca2+ gradient is generated between the microdomain and bulk cytosol [4, 5].
Figure 1.
Calcium signaling and fluorescent calcium sensors. (A) Fast calcium signaling regulated by the voltage-gated calcium channels (VGCC) includes EC coupling, cardiac action potential, calcium sparks and neurotransmitter release. (B) Different time scales of calcium signaling. (C) Representatives of calcium dye and genetically encoded calcium indicators. Fura-2 is synthesized calcium dye. Aequorin is used as genetically encoded calcium indicator. Cameleon and TnC sensor are composed by calmodulin and troponin C containing four EF-hands, respectively, flanked with FRET pair. GCaMP, GECO and R-CaMP are single fluorescent protein based sensor with calmodulin. CatchER is the single fluorescent protein with a designed calcium binding site on the surface of EGFP.
In muscle cells, electrical stimuli applied to the plasma membrane can be converted to muscle contraction by a process known as excitation-contraction coupling (EC coupling). In skeletal muscle, an action potential activates the dihydropyridine receptor (DHPR) anchored in the T tubule of the sarcolemma. DHPR then physically interacts with ryanodine receptors (RyR) expressed in the sarcoplasmic reticulum (SR) membrane to induce SR calcium release; this interaction occurs within milliseconds. After stimulation, a transient asymmetric calcium spike lasting several to tens of milliseconds occurs in the cytosol, with a fast calcium recovery phase due to SERCA pump refilling of SR calcium and buffering effects of calcium binding proteins in the cytosol [6].
The VGCC is transiently activated after the initial Na+ influx and K+ efflux in cardiac muscles, forming a plateau and a sequential slow decayed phase of membrane potential lasting for about 200 ms, much longer than that of skeletal muscle or neurons lasting for only 2–4 ms. This limits the firing rate up to several Hz, preventing the tetanus contraction of cardiac muscles. The fast calcium influx through the calcium channel triggers SR calcium release through calcium-induced calcium release (CICR) to further elevate cytosolic calcium before decreasing. The Ca2+ influx is terminated by closing of the VGCC with cytosolic calcium pumped back into the SR by the SERCA pump or extruded to the extracellular space by the sodium-calcium exchanger (NCX) [7]. A normal contracting cardiac muscle cell exhibits a train of cytosolic calcium spikes with the time to peak around tens of milliseconds, and a decay phase within hundreds of milliseconds.
Calcium sparks, elementary events of the CICR through the RyR in cardiac EC coupling, were discovered by fast fluorescence imaging [8]. The opening of the RyRs in cardiac or skeletal muscle cells produces calcium transients with 10 ms to peak and 20 ms half-decay, restricted around 2 μm. Activation of numerous RyRs produces multiple simultaneous calcium sparks, ranging from 50 to 5000 in a cell [9], which is regulated by the SR calcium content. The summation of the sparks generates the cytosolic calcium change. The counterpart of the calcium sparks are Ca2+ blinks, the transient decrement of Ca2+ in SR exhibiting similar fast kinetics and a much smaller region.
The neurotransmitter released from the presynaptic vesicles [10] triggered by presynaptic calcium channel activation will induce the postsynaptic receptors, for the synaptic transmission. Calcium microdomains and high calcium gradients are formed around the presynaptic calcium channels during the sub-millisecond calcium influx. In varying cell types, the local calcium concentration ranges from 10 μM to 200 μM for membrane fusion. Additionally, numerous calcium sensor proteins on the vesicle membrane surface are activated throughout the neurotransmitter release process [11]. Calcium dependent inactivation regulates the rapid termination of the calcium influx within 1–2 ms [12].
Need for calcium sensors with fast kinetics
The link between abnormal calcium channels and pathology requires precise tools to study them. Fluorescence microscopy with calcium sensors is indispensable in understanding live cell processes with high spatiotemporal resolution. Development of cell permeable calcium dyes has contributed greatly to our understanding of intracellular calcium signaling [1, 13]. Dyes such as Fluo-5N with fast off rates are commonly used to monitor calcium dynamics (Table 1). However, calcium dyes without targeting are not suitable for microdomain imaging. The current genetically encoded calcium indicators (GECIs) possessing the EF-hand motif show slow kinetics of signal decay (Table 1) that hamper their applications in probing fast calcium transients, especially in neuronal and skeletal muscle cells. Most calcium dyes with high affinity can only perfuse up to 80 nm from the channel, and are unable to measure tens of micromolar calcium in calcium microdomains. Imaging of calcium sparks should be deconvoluted with equations for intrinsic signals, and significant modification of the decay curve [14]. A similar convolution was applied for skeletal muscle myoplasmic calcium measurement, for which an initial peak is sometimes missing during the long-last pulse stimulation [15]. Furthermore, the complete decay of calcium in each spike was not measured in high frequency action potential stimulation, which is different from the membrane voltage changes. Rather a single transient peak was recorded for both single and multiple action potentials [16]; summation of calcium peaks is possible but not clear. This separation or summation of calcium spikes can be influenced by the choice of sensor [17, 18]. Therefore, there is a strong demand for tools to image fast transient or high-frequency calcium signaling generated by the activity of numerous calcium channels and receptors. GECIs with fast on and off rates and simple quantitation are required to monitor rapid calcium signaling in microdomains.
Table 1.
Kinetics of recent major calcium indicators.
| GECI/Dye | Rise time constant | Decay time constant | kon (M−1 s−1) | koff (s−1) |
|---|---|---|---|---|
| R-CatchER | 0.33 ms | >2.5×107 | >2100 | |
| CatchER [35] | 1.44 ms | ~3.89×106 | ~700 | |
| CH-GECO2.1 [53] | 3.70 × 105* | 0.423 | ||
| R-GECO1 [42] | 9.52 × 109* | 0.752 | ||
| TN-XL [54] | 240 ms | 430 ms | 2.3 | |
| TN-XXL [28] | 1.04 s | 0.88 s | 1.1 | |
| D1ER [24] | 3.6×106 | 256 | ||
| YC3.3 [18] | 1.41 s | 1.05 s | 12 | |
| YC3.6 [18] | 0.82 s | 0.73 s | 1.4 | |
| YC2.6 [18] | 5.24 s | 0.2 | ||
| D3cpv [18] | 0.68 s | 1.96 s | 0.5 | |
| TN-L15 [18] | 0.81 s | 1.49 s | 0.7 | |
| TN-XL [18] | 0.59 s | 0.20 s | 5 | |
| GCaMP1.6 [18] | 1.38 s | 0.45 s | 2.2 | |
| GCaMP3 [31] | 0.22 s | 4.6 | ||
| Fast-GCaMP-RS09 [31] | 36 ms | 27 | ||
| OGB-1 [18] | 0.24 s | 0.38 s | 2.6 | |
| Fluo-4 [55] | 6.0×108 | 210 | ||
| Calcium Green-5N [56] | 4.0×108 | 9259 | ||
| Mag-Fura-2 [56] | 7.5×108 | 26760 | ||
| Fluo-5F [55] | 3.0×108 | 300 |
The unit is M−ns−1. n(CH-GECO2.1)=0.58 and n(R-GECO1)=1.6.
What has been done in the field?
Calcium imaging has evolved from aequorin and calcium dyes to GECIs [19]. Figure 1C summarizes the structures of the major classes of calcium indicators. Calcium dyes based on BAPTA are mainly applied in bulk cytosol due to their strong binding affinity. Relatively weak affinity dyes are used to measure calcium concentration in internal calcium stores containing high calcium [20], but require plasma membrane permeabilization, an obstacle for intact live cell imaging. Novel strategies have been applied to improve the targeting capability of calcium dyes [21], but they are not as practical as GECIs. The kinetics of calcium sensors are highly dependent on the calcium binding protein used as the sensor and the target peptides used. Both calmodulin and troponin C based sensors bind calcium cooperatively between four EF-hands with a fluorescence change resulting from the calcium induced global protein conformational change [22–26]. These types of calcium sensors have been successfully applied to measure cytosolic calcium due to fast on rates [27–29]. Applying these sensors to measure rapid calcium release from the ER is limited by their slow off rate [30]. A fast GCaMP sensor has recently been reported to have a 20 fold improvement in the koff rate [31], and progress has also been made for red fluorescent sensors [32]. However, the slow off rates of these sensors may originate from the coupled EF hand calcium binding motifs and the subsequent large global conformational change [33–36]. Multiple binding processes are introduced by modification of multiple calcium binding sites in calmodulin and troponin C raising additional challenges for quantitative measurements of rapid calcium dynamics by the developed sensors.
CatchER (a GFP-based Calcium sensor for detecting high concentrations in the high calcium concentration environment such as ER)
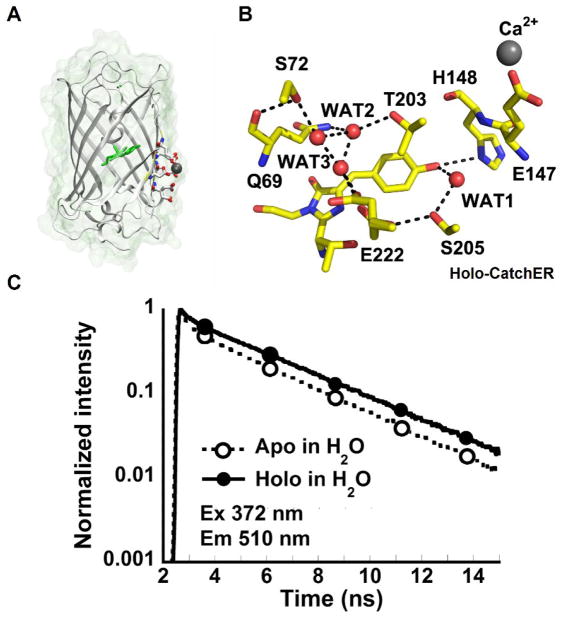
A previous calcium sensor—G1 reported by our lab was created by grafting an EF-hand motif into EGFP and exhibited ratiometric change in response to calcium but its kinetics were limited by an intermediate state [37]. The design of CatchER with a fast off rate was achieved based on key determinants for fine-tuning Ca2+ binding affinity and Ca2+ -induced conformational changes and the established chromophore properties of fluorescent proteins. We reasoned that Ca2+ sensors with fast fluorescence response can be better designed by coupling Ca2+ binding sites directly to the chromophore rather than relying on stretched protein-protein interaction to modulate chromophore conformation. First, four or five oxygen ligand atoms from protein residues (typically, carboxyl groups of D, E, N, Q) are situated in the pentagonal bipyramidal geometry [38] to form a calcium binding site similar to natural calcium binding proteins [39–42]. A single calcium binding site without cooperativity from protein coupling is important for fast off rate. Second, appropriate choice of residue charge and type can fine-tune Ca2+ binding affinity and metal selectivity [43, 44]. Third, diffusion-limited access of Ca2+ to the site requires good solvent accessibility [45]. Fourth, propagating Ca2+-induced, local conformational and electrostatic changes to the chromophore can be achieved by proper location of the charged ligand residues with respect it [46, 47]. Fifth, these changes must occur rapidly—more rapidly than the rate of conversion from neutral to anionic state ascribed to these chromophores [37, 48]. Sixth, the created binding site must not interfere with the chromophore’s synthesis and formation. We have designed CatchER by introducing five acidic residues to form a hand-like metal binding pocket in EGFP (Figure 2). The calcium binding site exhibits a pentagonal bipyramidal geometry and spreads across three antiparallel beta-sheets on the surface of EGFP. This designed calcium binding pocket is in close proximity to the hydrophobic chromophore, with hydrogen bond interaction, but facing outwards with high solvent accessibility. Calcium binding affects the hydrogen bond network between the binding residues and chromophore, triggering fluorescence change. Calcium binding to CatchER induces the ratiometric changes in the absorption spectra, as well as an increase in fluorescence emission at 510 nm upon excitation at both 395 and 488 nm. This novel designed calcium pocket lacks two binding ligands from its ideal coordination, thereby exhibits lower binding affinity and fast kinetics [35]. The X-ray crystal structures of CatchER have been resolved at 1.78–1.20 Å in Ca2+ loaded and Ca2+ free form, respectively (Figure 3A) [35]. Calcium binding altered the conformations of Thr203 and Glu222 associated with two forms of Tyr66 of the chromophore to induce the optical change [49]. From the X-ray crystal structure, Ca2+ ions were partially occupied in two locations within the extended designed binding site, suggesting the ability of Ca2+ ions to move around these possible locations may be responsible at least in part for the fast kinetics of metal-ion binding to CatchER [49]. A more dynamic observation was achieved by NMR [35].
Figure 2.
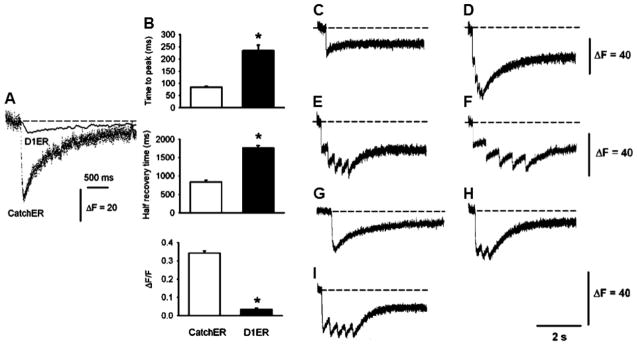
CatchER tracks Ca2+ release and uptake kinetics in response to single or repetitive muscle fiber depolarization. (A) SR Ca2+ transients were recorded in FDB fibers expressing either CatchER or D1ER elicited by 100-ms command pulses to 20 mV under whole-cell patch-clamp. CatchER’s and D1ER citrine’s fluorescence is illustrated to compare their amplitude and kinetics. D1ER citrine emission (535 nm) was chosen over the cyan fluorescence protein (485 nm) because of its larger amplitude. The D1ER excitation wavelength was set at 436 nm. D1ER’s and CatchER’s fluorescence was recorded using a spectrofluorometer and confocal microscope in the line-scan mode (see above), respectively. The dashed line indicates the baseline. (B) Time to peak, half recovery time, and response amplitude normalized to basal fluorescence were analyzed for CatchER and D1ER citrine fluorescence (ΔF/F). Asterisks indicate a statistically significant difference (*P < 0.01). Values are mean ± SEM for 19 and 13 fibers expressing CatchER or D1ER FDB, respectively. Transient changes in CatchER’s fluorescence response to various 20-mV command pulses: (C) a 10-ms pulse, (D) five 10-ms pulses at 10 Hz, (E) a 10-ms pulse at 3.3 Hz, (F) a 10-ms pulse at 1.6 Hz, (G) a 100-ms pulse, (H) 100-ms pulse at 5 Hz, and (I) 50-ms pulse at 3.3 Hz. Images provided with kind permission from Springer Science and Business Media and Tang et al., PNAS Copyright © 2011.
Figure 3.
Structure and fluorescence lifetime of CatchER (A) The X-ray crystal structure of Ca2+-CatchER (PDB ID: 4L12). The residues shown in stick are the designed calcium binding site. Residues 164-168 were shown with 50% transparency. The residue shown in green is the chromophore. The atom shown in grey is the calcium atom. The oxygen atoms and nitrogen atoms are indicated in red and blue, respectively. (B) The proton wire observed in the crystal structure of Ca2+-CatchER (PDB ID: 4L12). The H-bonds are shown in the black dash line with the cut off of 3.5. Only side chains were shown in S72, T203, S205 and E222. (C) Fluorescence decay traces of apo-CatchER (dash line) and CatchER supplemented with 10 mM Ca2+ (Holo, solid line). CatchER was excited at 372 nm and emitted at 510 nm in the time range of 15 ns. Ex and Em are short for excitation and emission, respectively. Reprinted (adapted) with permission from Zou, et al., J. Phys. Chem. B, 2014. Copyright © 2015 American Chemical Society.
Our novel GECI—CatchER has been applied to measure the ER calcium dynamics in non-excitable mammalian cells [50]. Furthermore, having fast kinetics, CatchER was able to record the SR luminal Ca2+ in flexor digitorum brevis (FDB) muscle fibers during voltage stimulation (Figure 2), and successfully determined the decreased SR calcium release in aging mice [51]. Thermostablity and brightness impaired the application of the early version of GCaMP, which was dim and experienced difficulty folding at 37°C [52]. Later it was improved using site directed mutagenesis. We have further improved the optical properties of CatchER. The new version of CatchER variant has been recently developed for optimal expression at 37°C (Reddish et al., unpublished results). Recently, we have designed red fluorescent protein based CatchER with improved pKa value (Zhuo et al., unpublished results).
For multi-color imaging with different fluorescent sensors, a red fluorescent protein-based calcium sensor (R-CatchER) has been designed to expand the imaging wavelength (Zhuo et al., unpublished results). The red fluorescent protein based ER sensor R-LAR-GECO1 was reported recently with a weaker binding affinity of 24 μM by modifying the interaction surface between CaM and M13 of the cytosolic calcium sensor R-GECO1 [53, 54]. It is interesting progress since the binding affinity decreased around 50 folds, whereas earlier it was reported as impossible to tune the binding affinity of GCaMP significantly lower by the same mutations found in Cameleon [22, 52].
From Intensity to Lifetime
Two photon fluorescence lifetime imaging is an indispensable method for understanding molecular signaling in environments with highly dynamic morphological changes and light scattering, such as the brain [55–58]. Until now, synthesized calcium indicators, such as OGB1, have single exponential lifetime decay, allowing for quantification of calcium change with high accuracy. However, there are limited reports of calcium fluorescence lifetime imaging with GECIs. A study reported that there was no significant lifetime change before and after adding Ca2+ to Cameleon YC3.1 that used ECFP and Venus [59], which are commonly used by other FRET-based calcium sensors. The multiple component lifetime decay of CFP makes it difficult to accurately determine the lifetime change. A fluorescent protein with a single exponential lifetime decay and high photostablity would be a good alternative for the FRET pair donor. Recently, an mTFP-Cit pair based TnC sensor was reported to exhibit significant lifetime change [60]. However, the kinetic property of this sensor was not recorded.
We have recently reported that CatchER is the first protein-based calcium indicator with the single fluorescent moiety to show the direct correlation between the lifetime and calcium binding (Figure 3C). Calcium binding increases the mean fluorescence lifetime of the deprotonated form of CatchER by 44%. Compared to wild type GFP, the site-directed mutagenesis to create calcium binding ligands disrupted the proton-transfer path and altered the optical properties. Calcium binding rescues this path and affects the fluorescence intensity (Figure 3B) [61]. Our finding provides important insights for a strategy to design calcium sensors and suggests that CatchER could be a useful probe for FLIM imaging of calcium in situ.
Perspectives
Fluorescent sensor based calcium imaging has dramatically increased our knowledge of cellular functions. However, there are still pressing needs for the accurate determination of fast calcium signaling besides simulation, especially in excitable cells expressing calcium channels. For further development, accelerating the kinetics and broadening the binding affinities of GECIs will compensate current imaging limitations. Looking for different calcium binding motifs other than the EF-hand for GECIs may provide a beneficial approach in overcoming these issues. For neuronal and deep field imaging with strong light-scattering, it could be advisable to design calcium sensors with high sensitivity for fluorescence lifetime imaging. Furthermore, EGFP-based calcium sensors are generally pH sensitive to the physiological conditions, as CatchER exhibited a pKa around 6.9 in the presence of Ca2+, with a 0.7 unit decrease than apo-form. Recently reported GCaMPs displayed a more significant pKa shift from 8~10 in the apo-form to 6~7.2 in the holo-form [16]. The pH insensitive sensors such as blue fluorescent protein [53] or red fluorescent protein-based sensor (Zhuo et al, unpublished results) are superior for monitoring calcium signaling in the Golgi and lysosome. The new, advanced GECIs will significantly improve our understanding of the molecular basis of Ca2+ homeostasis in health and pathology.
Highlights.
Fast calcium signaling is mainly regulated by calcium channels.
Calcium indicators facilitate our understanding of fast calcium signaling.
It is necessary to improve the kinetics of genetically encoded calcium indicators.
CatchER with a pentagonal bipyramidal-like binding domain exhibits fast kinetics.
Fluorescent lifetime calcium imaging is advantageous for deep field imaging.
Acknowledgments
We thank Mani Salarian, Fantashia Goolsby, Kenneth Huang, and Cassandra Miller for their critical review. We appreciate encouragements from Charles Louis, Aldebaran Hofer, Tullio Pozzan, Roger Tsien, Osvaldo Delbono, and Francesco Zorzato for developing calcium sensors and pioneer work by past members Jin Zou, April Ellis, Yun Huang, Yusheng Jiang, Malcom Delgado and Angela Holder, and collaborators including Giovanni Gadda, Kyril Solntsev, Vincent Rehder, Moica Lurtz, and Irene Weber and Chen Zhang. This work is supported, in part, by NIH grants GM070555, GM081749 and EB007268 and a Brain & Behavior (BB) seed grant to J.J.Y.; a BB fellowship to S.T. and Y.Z; and NIH supplemental grant to F.R..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rudolf R, et al. Looking forward to seeing calcium. Nat Rev Mol Cell Biol. 2003;4(7):579–86. doi: 10.1038/nrm1153. [DOI] [PubMed] [Google Scholar]
- 2.Yang J, et al. Molecular determinants of Ca2+ selectivity and ion permeation in L-type Ca2+ channels. Nature. 1993;366(6451):158–61. doi: 10.1038/366158a0. [DOI] [PubMed] [Google Scholar]
- 3.Ellinor PT, et al. Ca2+ channel selectivity at a single locus for high-affinity Ca2+ interactions. Neuron. 1995;15(5):1121–32. doi: 10.1016/0896-6273(95)90100-0. [DOI] [PubMed] [Google Scholar]
- 4.Muller A, et al. Endogenous Ca2+ buffer concentration and Ca2+ microdomains in hippocampal neurons. J Neurosci. 2005;25(3):558–65. doi: 10.1523/JNEUROSCI.3799-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parekh AB. Ca2+ microdomains near plasma membrane Ca2+ channels: impact on cell function. J Physiol. 2008;586(13):3043–54. doi: 10.1113/jphysiol.2008.153460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melzer W. Skeletal muscle fibers: Inactivated or depleted after long depolarizations? J Gen Physiol. 2013;141(5):517–20. doi: 10.1085/jgp.201310997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brini M, Carafoli E. The plasma membrane Ca2+ ATPase and the plasma membrane sodium calcium exchanger cooperate in the regulation of cell calcium. Cold Spring Harb Perspect Biol. 2011;3(2) doi: 10.1101/cshperspect.a004168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng H, Lederer WJ, Cannell MB. Calcium sparks: elementary events underlying excitation-contraction coupling in heart muscle. Science. 1993;262(5134):740–4. doi: 10.1126/science.8235594. [DOI] [PubMed] [Google Scholar]
- 9.Banyasz T, et al. A new approach to the detection and statistical classification of Ca2+ sparks. Biophys J. 2007;92(12):4458–65. doi: 10.1529/biophysj.106.103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasai H, Takahashi N, Tokumaru H. Distinct initial SNARE configurations underlying the diversity of exocytosis. Physiol Rev. 2012;92(4):1915–64. doi: 10.1152/physrev.00007.2012. [DOI] [PubMed] [Google Scholar]
- 11.de Jong AP, Fioravante D. Translating neuronal activity at the synapse: presynaptic calcium sensors in short-term plasticity. Front Cell Neurosci. 2014;8:356. doi: 10.3389/fncel.2014.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budde T, Meuth S, Pape HC. Calcium-dependent inactivation of neuronal calcium channels. Nat Rev Neurosci. 2002;3(11):873–83. doi: 10.1038/nrn959. [DOI] [PubMed] [Google Scholar]
- 13.Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260(6):3440–50. [PubMed] [Google Scholar]
- 14.Cheng H, Lederer WJ. Calcium sparks. Physiol Rev. 2008;88(4):1491–545. doi: 10.1152/physrev.00030.2007. [DOI] [PubMed] [Google Scholar]
- 15.Royer L, Pouvreau S, Rios E. Evolution and modulation of intracellular calcium release during long-lasting, depleting depolarization in mouse muscle. J Physiol. 2008;586(Pt 19):4609–29. doi: 10.1113/jphysiol.2008.157990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499(7458):295–300. doi: 10.1038/nature12354. The authors of this study create the GCaMP6 family of ultrasensitive GECIs for application in neural cells. These probes are able to measure calcium transients in dendritic spines over weeks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada Y, et al. Quantitative comparison of genetically encoded Ca2+ indicators in cortical pyramidal cells and cerebellar Purkinje cells. Front Cell Neurosci. 2011;5:18. doi: 10.3389/fncel.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendel T, et al. Fluorescence changes of genetic calcium indicators and OGB-1 correlated with neural activity and calcium in vivo and in vitro. J Neurosci. 2008;28(29):7399–411. doi: 10.1523/JNEUROSCI.1038-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizzuto R, et al. Rapid changes of mitochondrial Ca2+ revealed by specifically targeted recombinant aequorin. Nature. 1992;358(6384):325–7. doi: 10.1038/358325a0. [DOI] [PubMed] [Google Scholar]
- 20.Hofer AM, Machen TE. Technique for in situ measurement of calcium in intracellular inositol 1,4,5-trisphosphate-sensitive stores using the fluorescent indicator mag-fura-2. Proc Natl Acad Sci U S A. 1993;90(7):2598–602. doi: 10.1073/pnas.90.7.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tour O, et al. Calcium Green FlAsH as a genetically targeted small-molecule calcium indicator. Nat Chem Biol. 2007;3(7):423–31. doi: 10.1038/nchembio.2007.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyawaki A, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388(6645):882–7. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 23.Heim N, Griesbeck O. Genetically encoded indicators of cellular calcium dynamics based on troponin C and green fluorescent protein. J Biol Chem. 2004;279(14):14280–6. doi: 10.1074/jbc.M312751200. [DOI] [PubMed] [Google Scholar]
- 24.Palmer AE, et al. Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proc Natl Acad Sci U S A. 2004;101(50):17404–9. doi: 10.1073/pnas.0408030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat Biotechnol. 2001;19(2):137–41. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 26.Akerboom J, et al. Crystal structures of the GCaMP calcium sensor reveal the mechanism of fluorescence signal change and aid rational design. J Biol Chem. 2009;284(10):6455–64. doi: 10.1074/jbc.M807657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerr R, et al. Optical imaging of calcium transients in neurons and pharyngeal muscle of C. elegans. Neuron. 2000;26(3):583–94. doi: 10.1016/s0896-6273(00)81196-4. [DOI] [PubMed] [Google Scholar]
- 28.Mank M, et al. A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat Methods. 2008;5(9):805–11. doi: 10.1038/nmeth.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6(12):875–81. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jimenez-Moreno R, et al. Sarcoplasmic reticulum Ca2+ depletion in adult skeletal muscle fibres measured with the biosensor D1ER. Pflugers Arch. 2010;459(5):725–35. doi: 10.1007/s00424-009-0778-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun XR, et al. Fast GCaMPs for improved tracking of neuronal activity. Nat Commun. 2013;4:2170. doi: 10.1038/ncomms3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Inoue M, et al. Rational design of a high-affinity, fast, red calcium indicator R-CaMP2. Nat Methods. 2015;12(1):64–70. doi: 10.1038/nmeth.3185. This research highlights the design of RCaMP2 utilizing the CaMKK binding sequence, instead of the M13 peptide, that produced faster measurements of calcium transients compared to R-CaMP1.07. R-CaMP2 has a Hill coefficient near 1, making it significant compared to other GECIs of its kind with fast kinetics. [DOI] [PubMed] [Google Scholar]
- 33.Augustine GJ. How does calcium trigger neurotransmitter release? Curr Opin Neurobiol. 2001;11(3):320–6. doi: 10.1016/s0959-4388(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 34.Kirberger M, et al. Integration of Diverse Research Methods to Analyze and Engineer Ca-Binding Proteins: From Prediction to Production. Curr Bioinform. 2010;5(1):68–80. doi: 10.2174/157489310790596358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang S, et al. Design and application of a class of sensors to monitor Ca2+ dynamics in high Ca2+ concentration cellular compartments. Proc Natl Acad Sci U S A. 2011;108(39):16265–70. doi: 10.1073/pnas.1103015108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drake SK, Falke JJ. Kinetic tuning of the EF-hand calcium binding motif: the gateway residue independently adjusts (i) barrier height and (ii) equilibrium. Biochemistry. 1996;35(6):1753–60. doi: 10.1021/bi952335c. [DOI] [PubMed] [Google Scholar]
- 37.Zou J, et al. Developing sensors for real-time measurement of high Ca2+ concentrations. Biochemistry. 2007;46(43):12275–88. doi: 10.1021/bi7007307. [DOI] [PubMed] [Google Scholar]
- 38.Yang W, et al. Structural analysis, identification, and design of calcium-binding sites in proteins. Proteins. 2002;47(3):344–56. doi: 10.1002/prot.10093. [DOI] [PubMed] [Google Scholar]
- 39.Yang W, et al. Rational design of a calcium-binding protein. J Am Chem Soc. 2003;125(20):6165–71. doi: 10.1021/ja034724x. [DOI] [PubMed] [Google Scholar]
- 40.Kirberger M, et al. Statistical analysis of structural characteristics of protein Ca2+-binding sites. J Biol Inorg Chem. 2008;13(7):1169–81. doi: 10.1007/s00775-008-0402-7. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, et al. Towards predicting Ca2+-binding sites with different coordination numbers in proteins with atomic resolution. Proteins. 2009;75(4):787–98. doi: 10.1002/prot.22285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, et al. Analysis and prediction of calcium-binding pockets from apo-protein structures exhibiting calcium-induced localized conformational changes. Protein Sci. 2010;19(6):1180–90. doi: 10.1002/pro.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones LM, et al. Rational design of a novel calcium-binding site adjacent to the ligand-binding site on CD2 increases its CD48 affinity. Protein Sci. 2008;17(3):439–49. doi: 10.1110/ps.073328208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maniccia AW, et al. Using protein design to dissect the effect of charged residues on metal binding and protein stability. Biochemistry. 2006;45(18):5848–56. doi: 10.1021/bi052508q. [DOI] [PubMed] [Google Scholar]
- 45.Barondeau DP, et al. Structural chemistry of a green fluorescent protein Zn2+ biosensor. J Am Chem Soc. 2002;124(14):3522–4. doi: 10.1021/ja0176954. [DOI] [PubMed] [Google Scholar]
- 46.Li S, et al. Rational design of a conformation-switchable Ca2+- and Tb3+-binding protein without the use of multiple coupled metal-binding sites. FEBS J. 2008;275(20):5048–61. doi: 10.1111/j.1742-4658.2008.06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bizzarri AR, Cannistraro S. Statistical analysis of intensity fluctuations in single molecule SERS spectra. Phys Chem Chem Phys. 2007;9(39):5315–9. doi: 10.1039/b706008d. [DOI] [PubMed] [Google Scholar]
- 48.Abbruzzetti S, et al. Kinetics of acid-induced spectral changes in the GFPmut2 chromophore. J Am Chem Soc. 2005;127(2):626–35. doi: 10.1021/ja045400r. [DOI] [PubMed] [Google Scholar]
- 49••.Zhang Y, et al. Structural basis for a hand-like site in the calcium sensor CatchER with fast kinetics. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 12):2309–19. doi: 10.1107/S0907444913021306. The authors of this study determined the crystal structures of CatchER at 1.66, 1.20, and 1.78 Å for the free, calcium bound, and gadolinium bound forms respectively. One calcium ion moves around in the binding site contributing to the fast kinetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lopez-Sanjurjo CI, et al. Lysosomes shape Ins(1,4,5)P3-evoked Ca2+ signals by selectively sequestering Ca2+ released from the endoplasmic reticulum. J Cell Sci. 2013;126(Pt 1):289–300. doi: 10.1242/jcs.116103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang ZM, et al. Residual sarcoplasmic reticulum Ca2+ concentration after Ca2+ release in skeletal myofibers from young adult and old mice. Pflugers Arch. 2012;463(4):615–24. doi: 10.1007/s00424-012-1073-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohkura M, et al. Genetically encoded bright Ca2+ probe applicable for dynamic Ca2+ imaging of dendritic spines. Anal Chem. 2005;77(18):5861–9. doi: 10.1021/ac0506837. [DOI] [PubMed] [Google Scholar]
- 53.Zhao Y, et al. An expanded palette of genetically encoded Ca2+ indicators. Science. 2011;333(6051):1888–91. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54•.Wu J, et al. Red fluorescent genetically encoded Ca2+ indicators for use in mitochondria and endoplasmic reticulum. Biochem J. 2014;464(1):13–22. doi: 10.1042/BJ20140931. The authors report the design of low affinity calcium probes LAR-GECO1 and LAR-GECO1.2 with Kd values of 24 μM and 12 μM respectively. These probes allow for simultaneous measurements of the mitochondria and the ER when combined with GFP-based probes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yasuda R. Imaging spatiotemporal dynamics of neuronal signaling using fluorescence resonance energy transfer and fluorescence lifetime imaging microscopy. Curr Opin Neurobiol. 2006;16(5):551–61. doi: 10.1016/j.conb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 56.Yasuda R, et al. Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat Neurosci. 2006;9(2):283–91. doi: 10.1038/nn1635. [DOI] [PubMed] [Google Scholar]
- 57.Lee SJ, et al. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature. 2009;458(7236):299–304. doi: 10.1038/nature07842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murakoshi H, Wang H, Yasuda R. Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature. 2011;472(7341):100–4. doi: 10.1038/nature09823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Habuchi S, et al. Resonance energy transfer in a calcium concentration-dependent cameleon protein. Biophys J. 2002;83(6):3499–506. doi: 10.1016/S0006-3495(02)75349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laine R, et al. Fluorescence lifetime readouts of Troponin-C-based calcium FRET sensors: a quantitative comparison of CFP and mTFP1 as donor fluorophores. PLoS One. 2012;7(11):e49200. doi: 10.1371/journal.pone.0049200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61••.Zhuo Y, et al. Effect of Ca on the Steady-State and Time-Resolved Emission Properties of the Genetically Encoded Fluorescent Sensor CatchER. J Phys Chem B. 2014 doi: 10.1021/jp501707n. The authors discover calcium binding in CatchER increases the mean fluorescence lifetime during indirect stimulation of the anionic chromophore by 44%. CatchER is the first single fluorophore GECI with a direct link between calcium binding and increased fluorescence lifetime. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carlson HJ, Campbell RE. Circular permutated red fluorescent proteins and calcium ion indicators based on mCherry. Protein Eng Des Sel. 2013;26(12):763–72. doi: 10.1093/protein/gzt052. [DOI] [PubMed] [Google Scholar]
- 63.McCombs JE, Palmer AE. Measuring calcium dynamics in living cells with genetically encodable calcium indicators. Methods. 2008;46(3):152–9. doi: 10.1016/j.ymeth.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scott R, Rusakov DA. Main determinants of presynaptic Ca2+ dynamics at individual mossy fiber-CA3 pyramidal cell synapses. J Neurosci. 2006;26(26):7071–81. doi: 10.1523/JNEUROSCI.0946-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Naraghi M. T-jump study of calcium binding kinetics of calcium chelators. Cell Calcium. 1997;22(4):255–68. doi: 10.1016/s0143-4160(97)90064-6. [DOI] [PubMed] [Google Scholar]