Abstract
In patients with hepatitis C virus (HCV) infection, enhanced activity of indoleamine-2,3-dioxygenase 1 (IDO) has been reported. IDO - a tryptophan-catabolizing enzyme - has been considered as both an innate defence mechanism and an important regulator of the immune response. The molecular mechanism of IDO induction in HCV infection and its role in the antiviral immune response remain unknown. Using primary human hepatocytes, we show that HCV infection stimulates IDO expression. IDO gene induction was transient and coincided with the expression of types I and III interferons (IFNs) and IFN-stimulated genes in HCV-infected hepatocytes. Overexpression of hepatic IDO prior to HCV infection markedly impaired HCV replication in hepatocytes, suggesting that IDO limits the spread of HCV within the liver. siRNA-mediated IDO knock-down revealed that IDO functions as an IFN-mediated anti-HCV effector. Hepatic IDO was most potently induced by IFN-γ, and ongoing HCV replication could significantly upregulate IDO expression. IRF1 (IFN-regulatory factor 1) and STAT1 (signal transducer and activator of transcription 1) regulated hepatic IDO expression. Hepatic IDO expression also had a significant inhibitory effect on CD4+ T-cell proliferation. Our data suggest that hepatic IDO plays a dual role during HCV infection by slowing down viral replication and also regulating host immune responses.
Key Words: Indoleamine-2,3-dioxygenase; Hepatocyte; Hepatitis C virus; Antiviral immune response
Introduction
Hepatitis C virus (HCV), a member of the Flaviviridae family, is one of the most important causes of chronic liver disease, which may lead to cirrhosis and hepatocellular carcinoma [1]. Hepatic innate immunity is the first line of defence against HCV and plays a crucial role in the initiation and modulation of adaptive immune responses [2]. Although the initiated immune response can clear HCV, the majority of acutely infected individuals develop persistent infection. Persistence of HCV has been ascribed to HCV escape mutations and impaired innate and adaptive immune responses through mechanisms that are not yet fully understood [for review, see [2,][3]].
Indoleamine-2,3-dioxygenase 1 (IDO) is an enzyme that metabolizes tryptophan to kynurenine, which can be further metabolized to various downstream catabolites such as 3-hydroxykynurenine. IDO is induced mainly by type II interferons (IFN-γ) and to a lesser extent by type I IFNs (IFN-α/β) and other inflammatory cytokines in human tissues and cell subsets [4]. IDO has been considered to be part of the host's innate defence mechanism as it can control pathogen proliferation by depleting tryptophan from the local microenvironment [5]. Nutrient deprivation is an evolutionarily ancient but important strategy of the innate host defence. IDO-mediated tryptophan depletion in cell culture models has been shown to inhibit the replication of various viruses such as herpes simplex virus, hepatitis B virus and flaviviruses [6, 7, 8].
Previous research has demonstrated that IDO plays a crucial role in immunological homeostasis [9] and pathogen persistence [10]. CD4+ T cells are very sensitive to tryptophan shortage and accumulation of kynurenine catabolites, which cause their arrest in the G1 phase of the cell cycle, anergy and even apoptosis. For example, IDO-producing dendritic cells inhibit T-cell activation and proliferation or induce Treg (T-regulatory) cells through tryptophan starvation and/or the formation of immunotoxic kynurenine catabolites such as 3-hydroxykynurenine and 3-hydroxyanthranilic acid [4]. In vitro studies have shown that HIV stimulates IDO expression in dendritic cells [11]. Moreover, during HIV infection, IDO is overexpressed in lymphoid tissue and parallels with the accumulation of Treg cells [12], suggesting that the activation of IDO facilitates HIV persistence.
An increased serum kynurenine-to-tryptophan ratio, which is an index for IDO activity, has been previously demonstrated in patients with chronic HCV infection compared to patients with resolved HCV infection and healthy individuals [13, 14, 15, 16]. Studies in HCV-infected chimpanzees showed that IDO mRNA expression was upregulated in the liver during the acute phase of infection but returned rapidly to baseline levels in chimpanzees that subsequently cleared the infection. In contrast, chimpanzees that developed persistent infection maintained a high level of hepatic IDO mRNA expression [14, 17]. However, the molecular mechanism of IDO induction in HCV infection and its impact on anti-HCV effector functions remains poorly defined.
In this study, we show that hepatic IDO expression exerts inhibitory effects on both HCV replication and immune cells. The dichotomous nature of IDO may favour persistence within the host over viral eradication.
Materials and Methods
Cells, Reagents and Antibodies
Human Huh7.5.1 cells were kindly provided by Dr. F. Chisari (The Scripps Research Institute, La Jolla, Calif., USA; MTA No. 636) [18]. Primary human hepatocytes (PHH) were provided by Stephen Strom of the University of Pittsburgh through the Liver Tissue Procurement and Distribution System (N01-DK-7-0004/HHSN26700700004C). PHH were isolated from liver resections according to standard perfusion protocols. After 7 days of culture, PHH remained attached and maintained a healthy and highly differentiated phenotype. Peripheral blood mononuclear cells were isolated from healthy blood donors (EFS-Alsace, Strasbourg, France) by Ficoll density gradient centrifugation. CD4+ T-cell positive selection was performed by magnetic sorting using CD4+ microbeads (Miltenyi Biotech). Human recombinant IFN-α and IFN-γ were obtained from Roche and InterMune, respectively. Rabbit polyclonal anti-IDO1 (Abcam), anti-β actin (Abcam), anti-IFN-regulatory factor 1 (IRF1; Santa Cruz Biotechnology), and anti-signal transducer and activator of transcription 1 (STAT1) and anti-phospho STAT1 (Tyr701) antibodies (Cell Signaling) were used for Western blot analysis. PE-conjugated mouse anti-human IFN-γR, anti-HCV core (clone C7-50, Thermo Scientific) and anti-IRF1 antibodies were used for flow cytometry analysis. The IDO inhibitor 1-DL-MT, a racemic mixture of 1-methyl-tryptophan comprising both isomers (D + L), was purchased from Sigma-Aldrich.
Viruses
The production of HCV pseudoparticles (HCVpp) bearing envelope glycoproteins of strains HCV-J (1b) and JFH1 (2a) and the production of vesicular stomatitis virus pseudoparticles have been described previously [19]. HCVpp were added to Huh7.5.1 cells for 3 h at 37°C. HCVpp infection was determined by measuring luciferase activity 72 h after infection. HCVpp entry was assessed with a luciferase assay system (Promega) using a Mithras LB940 plate reader (Berthold Technologies). The production of cell-culture-derived HCV (HCV JFH1 strain kindly provided by Dr. T. Wakita of the National Institute of Infectious Disease, Tokyo, Japan; MTA No. 1664) has been described previously [20]. For infection experiments, Huh7.5.1 cells and PHH were infected with JFH1 at a multiplicity of infection of 0.05 and 0.1, respectively, for 3 h at 37°C to allow HCV entry. HCV JFH1 infection efficiency of PHH ranged from 4 to 7 RNA copies per hepatocyte. PHH were infected with 50 µl of serum derived from a patient infected with HCV genotype 1b (viral load of 6.68 × 105 IU/ml) who had participated in an HCV clinical trial (ClinicalTrials.gov identifier NCT00638144). The cells were then washed with a heparin solution (60 µg/ml) to remove surface-bound viral particles followed by two washes in PBS. JFH1 infection was analysed in cell lysates by quantification of viral RNA at different time points after infection. HCV RNA copy numbers were determined by real-time quantitative RT-PCR (qRT-PCR), as described previously [21]. Ultraviolet (UV)-inactivated JFH1 virus was produced by placing 2 ml of JFH1 under a Vilber UV lamp (VL-115.M) for 20 min. The UV-inactivated virus was non-infectious, as determined by titration on naïve Huh7.5.1 cells.
Real-Time RT-PCR Analysis
RNA was extracted from cell samples using the RNeasy Kit (Qiagen) according to the manufacturer's instructions. Complementary DNA (cDNA) was synthesized using a cDNA reverse transcription kit (Fermentas). PCR was performed using TaqMan probe-based chemistry. The qPCR reaction mix was incubated for 5 min at 95°C and 10 min at 50°C followed by 40 PCR cycles, with each cycle comprising 95°C for 15 s and 60°C for 1 min (Rotor-Gene 6000; Corbett Life Science). Primers and internal probes for IDO1, IDO2, CXCL10 chemokine, STAT1, IRF1, melanoma differentiation-associated protein 5 (MDA5), double-stranded RNA-activated protein kinase R (PKR), IFN-α, IFN-β, IFN-λ, claudin-1, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were purchased from Applied Biosystems. GAPDH was used as a reference gene for normalization of quantitative PCR data.
Immunoblotting
Proteins (5-10 µg) were loaded onto a 10% SDS-PAGE gel. The separated proteins were transferred onto PVDF membranes. The membrane was incubated with primary antibody for 2 h followed by incubation with secondary antibody (IgG horse-radish peroxidase conjugated; Pierce) for 1 h at room temperature. The membrane was developed with ECF Western blotting detection reagent (Amersham Biosciences) and visualized with an image analyser (Typhoon™ Trio, GE Healthcare). Quantitative densitometric analyses were performed on digitized images using ImageQuant software (GE Healthcare).
Staining of Surface and Intracellular Antigens by Flow Cytometry
For analysis of cell surface antigens, cells were stained with fluorochrome-labelled antibodies (PE-conjugated mouse anti-human IFN-γR; BD Biosciences). For analysis of intracellular antigens, cells were permeabilized using Cytofix/Cytoperm (BD Biosciences). The cells were incubated with primary antibodies (anti-HCV core or anti-IRF1 antibody) for 45 min, followed with fluorochrome-labelled antibodies (Alexa555 or PE-conjugated secondary antibody; BD Biosciences) for 30 min. Antigen expression was analysed on an LSRII flow cytometer using DIVA software (BD Biosciences).
T-Cell Proliferation Assay
T-cell proliferation assays were performed by staining cells with CellTrace CFSE cell proliferation dye (Invitrogen). The percentages of cells in different generations were calculated using ModFit LT for Win32 (Verity Software House, Topsham). The proliferation index was calculated according to the ModFit programme and represents the sum of the cells in all generations divided by the computed number of original parent cells present at the start of the experiments. The proliferation index is proportional to the percentage of cells that have proliferated in culture and is also weighted for the number of divisions that cells have undergone.
Transient Transfection and RNA-Mediated Interference
Huh7.5.1 cells were transfected with either pcDNA3.1 encoding full-length human IDO cDNA (pcDNA IDO) [22], IRF1 cDNA (InvivoGen) or pcDNA3.1 (vector control; Invitrogen) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. PHH were transfected using Lipofectamine LTX reagent (Invitrogen) according to the manufacturer's instructions. The siRNAs directed against IRF1 consist of pools of 4 individual siRNAs (Smartpool) and were obtained from Dharmacon Research, as was the control siRNA GLO (siControl). Huh7.5.1 cells were transfected with 50 nM of either the IRF1-targeting siRNA pool or the control siRNA GLO using Dharmafect 4 siRNA-transfection reagent (Dharmacon Research) according to the manufacturer's instructions. At 48 h after transfection, Huh7.5.1 cells were stimulated with IFN-γ (50 IU/ml) for 7 h.
Measurement of IDO Activity
IDO functional activity was measured in terms of the ability of cells to catabolize exogenous tryptophan (100 µM; Sigma Aldrich) to kynurenine by reverse-phase high-performance liquid chromatography (HPLC), as detailed previously [23]. Alternatively, kynurenine was measured spectrophotometrically using the Ehrlich reagent method, as described previously [24]. The values were compared with a standard curve with defined concentrations of kynurenine (0-100 µM; Sigma Aldrich). IFN-γ-stimulated PHH or IDO-transfected Huh7.5.1 cells were washed and resuspended in HBSS buffer containing 100 µM of tryptophan. After 4 h, the supernatants were harvested and assessed for the presence of kynurenine. The IDO specific inhibitor, 1-DL-MT, was used to block IDO activity.
Co-Culture Conditions
Isolated CD4+ T cells were labelled with CFSE (10 µM) and stimulated with anti-CD3 and anti-CD28-coated beads (Invitrogen) at a bead-to-cell ratio of 1:1 in the presence of IL-2 (30 IU/ml, Invitrogen) 48 h prior to co-culture. Then, CFSE-labelled and activated CD4+ T cells were co-cultured with IDO-transfected Huh7.5.1 cells at a ratio of 1:1. After 72 h of co-culture, CD4+ T cells were collected and their proliferative responses were analysed by flow cytometry as described above. As a control, some cultures were supplemented with 1-DL-1MT (1,000 µM) at the time of co-culture.
Determination of IDO-Mediated Anti-HCV Activity
Huh7.5.1 cells were transfected with siRNAs targeting the type I IFN receptor 2 (IFNαR2), with siRNAs targeting IDO or with non-targeting control siRNAs at a final concentration of 50 nM, using a reverse transfection protocol with Oligofectamine (Invitrogen), as described previously [25]. After 72 h, the cells were infected with HCV JFH1. At 24 h after infection, the cells were either mock-treated or treated with INF-α at 10 IU/ml. At 24 h after IFN-α treatment, the cells were harvested, and total cellular RNA was extracted using the RNeasy Mini Kit (QIAGEN). For the IDO gene expression assay, cDNA was synthesized from total cellular RNA using the First Strand cDNA Synthesis Kit (Roche). The mRNA levels of IDO were measured by qRT-PCR using IDO-specific primers and probes (IDT) on an ABI 7500 real-time PCR system. Relative IDO mRNA levels were calculated by the ΔΔCT method and were normalized by 18S rRNA levels (Applied Biosystems). Copy numbers of intracellular HCV RNA were measured by qRT-PCR with the primers, probes and parameters described previously [26]. The relative amount of HCV RNA was normalized to 18S rRNA.
Statistical Analysis
Unpaired t tests were used to analyse data and compare different conditions. Data are expressed as means ± SD, and p values less than 0.05 were considered significant.
Results
PHH Express an Enzymatically Active IDO
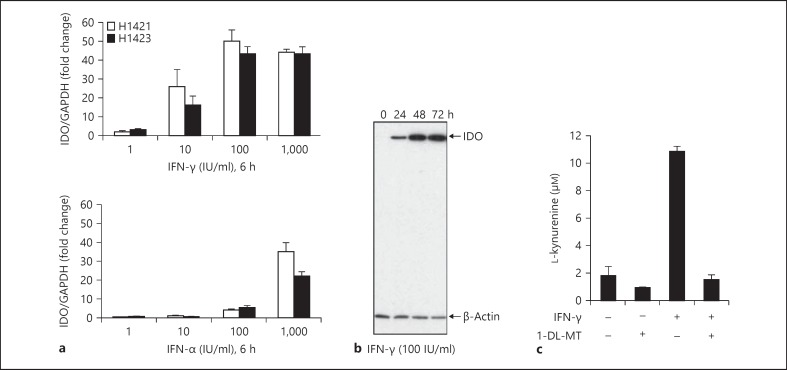
Hepatocytes are the primary target cells supporting HCV replication and provide the first line of immune defence against HCV. We thus explored the capacity of PHH to produce a functional IDO protein. PHH from two different donors were stimulated with different doses of type I and II IFNs. As shown in figure 1, IFN-γ markedly enhanced IDO mRNA expression in a dose-dependent manner, reaching a plateau at 100 IU/ml. IDO was prominently induced by IFN-γ, while IFN-α had a much weaker effect on IDO mRNA expression (fig. 1a). Western blot analysis confirmed the expression of IDO protein in PHH. IFN-γ-induced IDO protein expression was detectable throughout 3 days in PHH (fig. 1b). To confirm that IDO is functional in PHH, we monitored the degradation of exogenous tryptophan to kynurenine by HPLC. Kynurenine production was observed in IFN-γ-stimulated PHH, and the presence of the IDO inhibitor 1-DL-MT abolished IDO activity (fig. 1c), indicating that hepatic IDO protein is enzymatically active.
Fig. 1.
Expression of IDO in PHH. a PHH from 2 donors (H1421, H1423) were incubated with different doses of IFNs. IDO expression is shown as a ratio of IDO/GAPDH compared to non-stimulated PHH. b PHH were incubated with IFN-γ and harvested at the indicated time points. Levels of IDO protein expression in PHH lysates were determined by Western blotting. β-Actin levels are shown as a loading control. c PHH were stimulated with IFN-γ (100 IU/ml) for 48 h in the presence and absence of 1-DL-MT (100 µM). Then, cells were cultured for 4 h in HBSS buffer containing 100 µM of tryptophan. Kynurenine accumulation was measured in cell culture supernatants by HPLC. Data are given as mean kynurenine production ± SD from 2 independent experiments.
HCV Stimulates IDO Expression in PHH
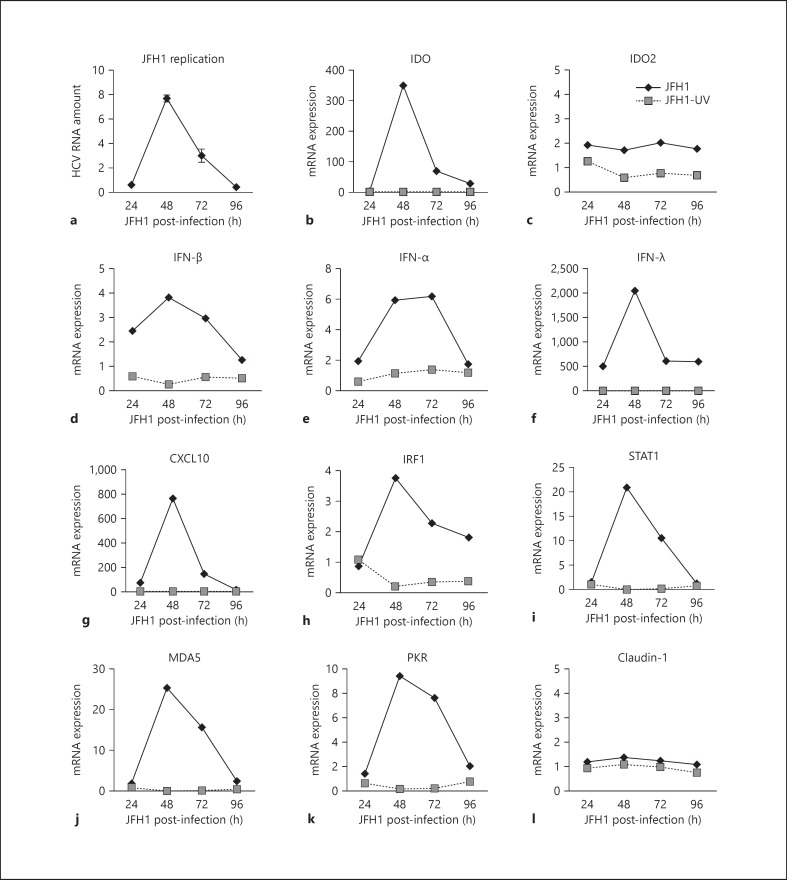
Although hepatocytes are the primary target cells of HCV in vivo, in vitro infection of PHH remains a difficult task. Here we took advantage of the development of recombinant cell-culture-derived HCV JFH1 virus [20] to evaluate IDO expression in PHH. PHH were exposed to the infectious JFH1 virus and replication-deficient JFH1. As shown in figure 2 and consistent with previous publications [27, 28], limited replication of JFH1 in PHH was detected over 24–96 h, suggesting that innate antiviral host defences produced an antiviral state restricting HCV replication and spread in PHH. Using a qRT-PCR approach we detected a robust increase in IDO mRNA expression within 48 h of infection that coincided with JFH1 replication and the expression of CXCL10, IFN-α, IFN-β, and IFN-λ, as well as the induction of IFN-stimulated genes (ISGs) such as STAT1, IRF1, MDA5, and PKR. In contrast, JFH1 infection did not stimulate the expression of IDO2, a paralogue of IDO [29], and claudin-1, a tight junction protein and a putative receptor for HCV entry [30]. Gene induction was dependent on HCV replication, as UV-inactivated non-infectious JFH1 had little if any effect on gene expression (fig. 2). These results suggest that HCV replication drives hepatic IDO expression together with proteins that exhibit significant antiviral properties.
Fig. 2.
HCV infection stimulates IDO expression in PHH. a PHH (donor H0199) were infected with JFH1 for 3 h, washed and then lysed at the time points indicated. HCV RNA levels were normalized to GAPDH mRNA levels. Results are presented as fold induction to 3-hour post-infected PHH. Gene induction was measured by real-time RT-PCR analysis in PHH exposed to JFH1 or UV-infected JFH1. Gene induction of IDO (b), IDO2 (c), IFN-β (d), IFN-α (e), IFN-λ (f), CXCL10 (g), IRF1 (h), STAT1 (i), MDA5 (j), PKR (k), and claudin-1 (l) are shown as fold change over uninfected PHH cultures.
IRF1 Regulates HCV-Induced Hepatic IDO Expression
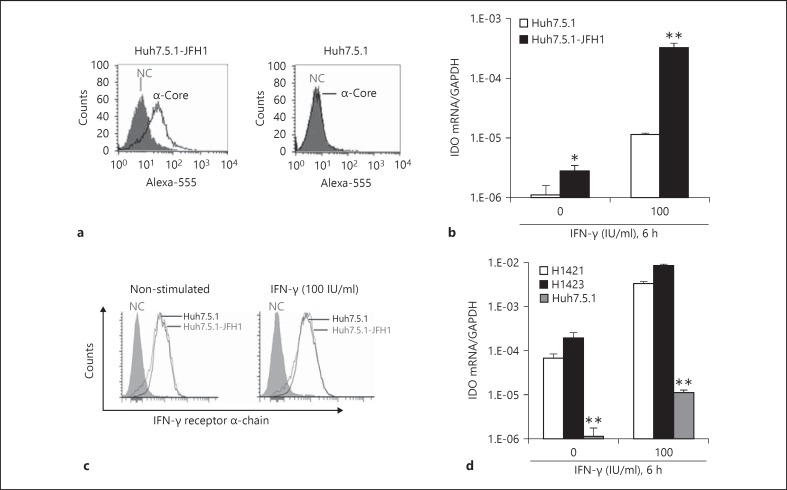
Since HCV replication is transient in primary liver cells [27, 28] (fig. 2), we used the Huh7-derived cell culture system [20] to gain a deeper insight into the molecular mechanism of HCV-induced IDO. As shown in figure 3a, at day 5 after JFH1 exposure almost all Huh7.5.1 cells were infected, as evidenced by the presence of HCV core-positive cells during flow cytometry analysis. Expression of IDO mRNA was very low or undetectable in uninfected Huh7.5.1 cells (fig. 3b). As in PHH, JFH1 infection and IFN-γ stimulated IDO expression in Huh7.5.1 cells (fig. 3b). Furthermore, IFN-γ significantly upregulated IDO expression in JFH1-infected Huh7.5.1 cells (fig. 3b) and to a similar extent as that observed previously in Huh7 cells by Larrea et al. [14]. To exclude the possibility that this enhancement was the direct result of greater IFN-γ receptor density on the cell surface of JFH1-infected Huh7.5.1 cells, we determined IFN-γ receptor α-chain expression levels using flow cytometry. As expected, there was no difference between JFH1-infected Huh7.5.1 cells and non-infected control Huh7.5.1 cells (fig. 3c). However, we failed to detect IDO at the protein level in Huh7.5.1 cells by Western blot (data not shown), probably because IDO is expressed at very low levels in these cells. Of note, levels of IDO gene transcription were extremely low in Huh7.5.1 cells compared to PHH in the presence or absence of IFN-γ (fig. 3d).
Fig. 3.
Ongoing HCV replication in Huh7.5.1 cells enhances IFN-γ-induced IDO expression. NC = Negative control. a Robust HCV infection of Huh7.5.1 cells was determined using anti-HCV core staining and flow cytometry at day 5 after inoculation. b JFH1-infected Huh7.5.1 cells and uninfected control cells were stimulated with IFN-γ and then analysed for IDO gene expression. c After 4 h of IFN-γ stimulation, JFH1-infected Huh7.5.1 cells and control cells were analysed for IFN-γ receptor α-chain expression. d IDO gene expression levels in PHH (donor H1421, H1423) and Huh7.5.1 cells in the presence or absence of IFN-γ. * p < 0.05, ** p < 0.01.
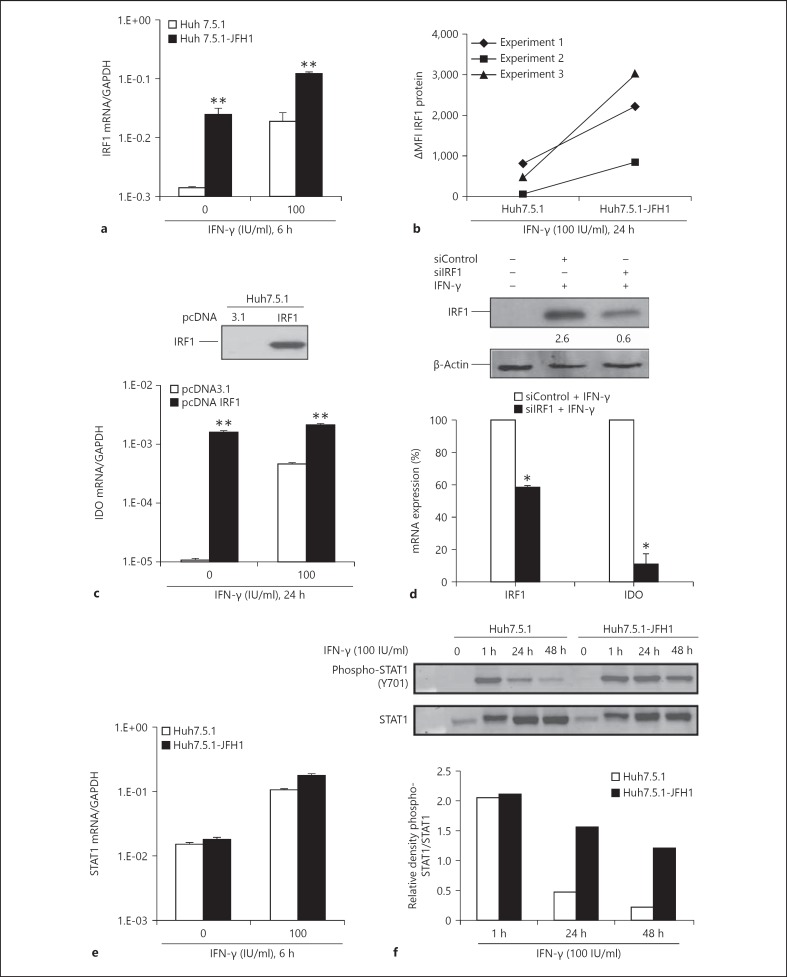
Because IRF1 is an IFN-γ-induced transcriptional activator critical for IDO regulation [31], we investigated IRF1 mRNA expression level in Huh7.5.1 cells in the presence and absence of JFH1 infection. JFH1-infected cells demonstrated an approximately 1 log higher IRF1 mRNA expression compared to uninfected control cells (fig. 4a). IFN-γ treatment of JFH1-infected cells further increased IRF1 expression level (fig. 4a). Analysis of IRF1 protein expression by flow cytometry confirmed the presence of increased amounts of IRF1 protein in JFH1-infected Huh7.5.1 cells compared to uninfected cells upon IFN-γ treatment (fig. 4b). To demonstrate that IRF1 is a critical factor in hepatic IDO expression, we transiently overexpressed the human IRF1 gene in Huh7.5.1 cells. As shown in figure 4c, IRF1 overexpression alone stimulated the expression of IDO mRNA in Huh7.5.1 cells. siRNA-mediated knock-down of IRF1 abrogated IFN-γ-induced IDO expression almost completely in Huh7.5.1 cells (fig. 4d), indicating that IRF1 regulates hepatic IDO expression.
Fig. 4.
IRF1 regulates IDO expression in HCV infection. MFI = Mean fluorescence intensity. a JFH1-infected Huh7.5.1 cells and uninfected control cells were analysed for IRF1 expression in the absence and presence of IFN-γ. IRF1 expression is shown as a ratio of IRF1/GAPDH. b IFN-γ-induced IRF1 protein expression is shown as delta mean fluorescence intensity (ΔMFI: MFI of IFN-γ-stimulated cells minus MFI of non-stimulated cells). c Huh7.5.1 cells were transfected with pcDNA IRF1 or pcDNA3.1. IRF1 expression was verified by Western blotting 48 h after transfection. IDO mRNA expression is shown in the presence or absence of IFN-γ in transfected cells. d IRF1 was silenced in Huh7.5.1 cells. Numbers represent densitometric analysis of IRF1 bands protein normalized to β-actin levels. Then, cells were stimulated with IFN-γ (50 IU/ml) for 7 h, and the relative levels of IRF1 and IDO mRNA expression were determined. Data are expressed as the percentage of gene expression for IRF1-silenced Huh7.5.1 cells in comparison to siControl (= 100%). e JFH1-infected Huh7.5.1 cells and control cells were analysed for STAT1 expression in the absence and presence of IFN-γ. STAT1 expression is shown as a ratio of STAT1/GAPDH. f After treatment with IFN-γ, cells were harvested at the indicated times. Equal amounts of total cell lysates were immunoblotted with anti-STAT1 and anti-phospho-STAT1 antibodies. Densitometry analysis shows phospho-STAT1 levels relative to STAT1 levels. * p < 0.05, ** p < 0.01.
A subgroup of genes induced by IFN-γ requires both IRF1 and STAT1 for transcriptional activation. Previous studies have shown that full activation of STAT1 is crucial for IFN-γ-induced IDO expression in murine dendritic cells [32]. We, therefore, studied levels of STAT1 expression and activation in JFH1-infected Huh7.5.1 cells. As shown in figure 4e, f, STAT1 mRNA and protein expression did not markedly differ between JFH1-infected and control Huh7.5.1 cells in the absence or presence of IFN-γ. However, we observed that IFN-γ-induced STAT1 tyrosine phosphorylation was prolonged in JFH1-infected Huh7.5.1 cells compared to uninfected Huh7.5.1 control cells, even though JFH1 infection per se was not able to promote virtually detectable STAT1 phosphorylation. Prolonged phosphorylation of STAT1 may contribute to enhanced upregulation of IFN-γ-induced IDO expression in JFH1-infected Huh7.5.1 cells.
Antiviral Activity of Hepatic IDO in HCV Infection
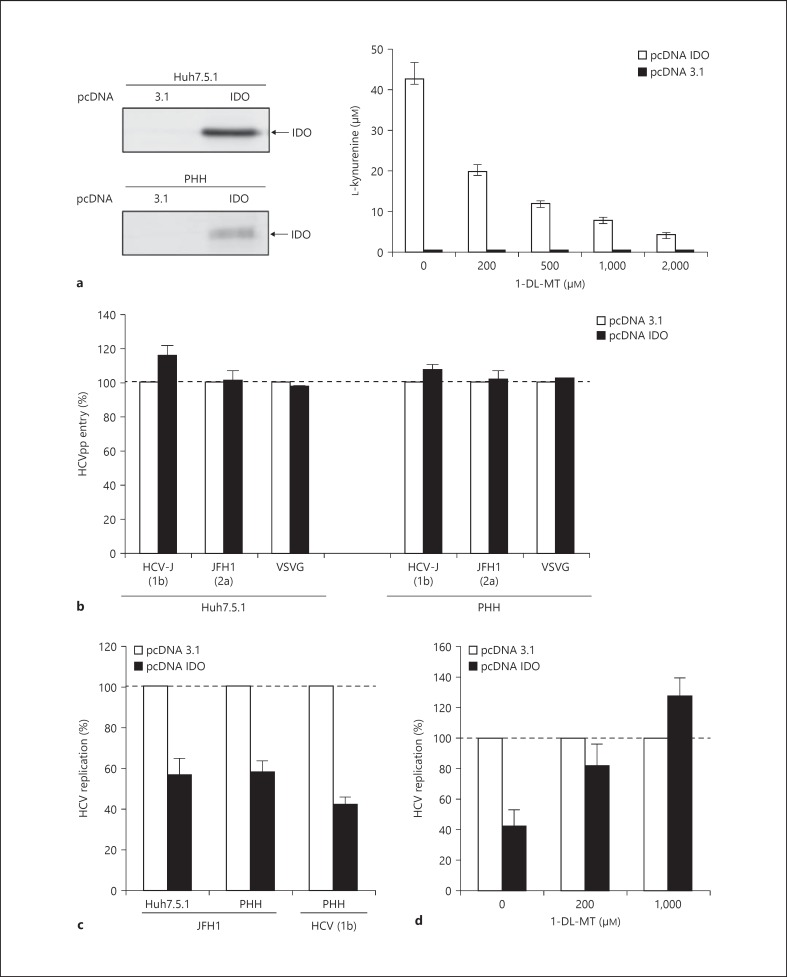
To determine whether hepatic IDO has an impact on HCV replication, we transiently overexpressed the human IDO gene in Huh7.5.1 cells and PHH prior to HCV exposure. At 48 h after transfection, IDO was expressed at the protein level in Huh7.5.1 cells and PHH (fig. 5a). Enzymatic activity of the expressed IDO protein was verified by the production of kynurenine in the cell culture supernatant (fig. 5a). In an additional experiment, we demonstrated that the IDO inhibitor 1-DL-MT inhibited IDO-mediated tryptophan-to-kynurenine conversion in a dose-dependent manner without causing cytotoxic effects to IDO-transfected Huh7.5.1 cells (fig. 5a). To assess whether IDO has an effect on HCV entry, we studied HCVpp entry into Huh7.5.1 cells and PHH. As shown in figure 5b, transient IDO overexpression did not inhibit HCVpp entry in Huh7.5.1 cells and PHH, indicating that IDO does not prevent the virus from entering the cells based on the HCVpp system. However, we cannot completely rule out an effect on entry because recent studies suggest that the HCVpp system does not completely reproduce all the steps of HCV entry [33]. Expression of IDO reduced HCV replication up to 50% in JFH1-infected Huh7.5.1 cells and PHH (fig. 5c). Similar results were observed in Huh7.5.1 cells co-transfected with a JFH1 subgenomic replicon (data not shown) or PHH exposed to serum-derived HCV (genotype 1b; fig. 5c), indicating that IDO attenuates HCV replication. To assess whether IDO's antiviral activity relies on its catalytic function, we added the IDO inhibitor 1-DL-MT to the cell cultures. As shown in figure 5d, the presence of 1-DL-MT restored HCV replication to normal level in JFH1-infected Huh7.5.1 cells. These results suggest that IDO's antiviral activity is mediated by tryptophan shortage and/or the accumulation of kynurenine.
Fig. 5.
Impact of hepatic IDO expression on HCV infection. a Huh7.5.1 cells and PHH (H0204, H0216, H0208) were transfected with pcDNA IDO or pcDNA3.1. IDO protein expression was verified by Western blotting. Functional activity of the expressed IDO protein in Huh7.5.1 cells was verified by the accumulation of kynurenine in the supernatant in the presence and absence of the specific IDO inhibitor 1-DL-MT using the Ehrlich reagent method. b At 48 h after transfection, Huh7.5.1 cells and PHH (donor H0204, H0216, H0208) were assessed for HCVpp entry (genotype 2a: JFH1, genotype 1b: HCV-J). VSVG = Vesicular stomatitis virus G glycoprotein. Data are expressed as the percentage of HCVpp entry into IDO-transfected PHH compared to pcDNA3.1-transfected control cells (= 100%). Vesicular stomatitis virus G glycoprotein-pseudotyped virions were included as a control. c At 48 h after transfection, Huh7.5.1 cells and PHH were incubated with JFH1 or HCV serum (genotype 1b). At 2 days after JFH1 infection, viral replication in IDO-transfected cells was measured. HCV RNA copies were normalized to GAPDH. Data are shown as mean percentage of HCV replication compared to pcDNA3.1-transfected cells (= 100%). d Rescued HCV JFH1 replication in IDO-transfected Huh7.5.1 cells. JFH1 replication in IDO-transfected Huh7.5.1 cells was determined in the presence and absence of the specific IDO inhibitor 1-DL-MT. Data are shown as the mean ± SD of triplicate determinations and are representative of 3 independent experiments.
IDO Mediates Antiviral Effects of IFNs
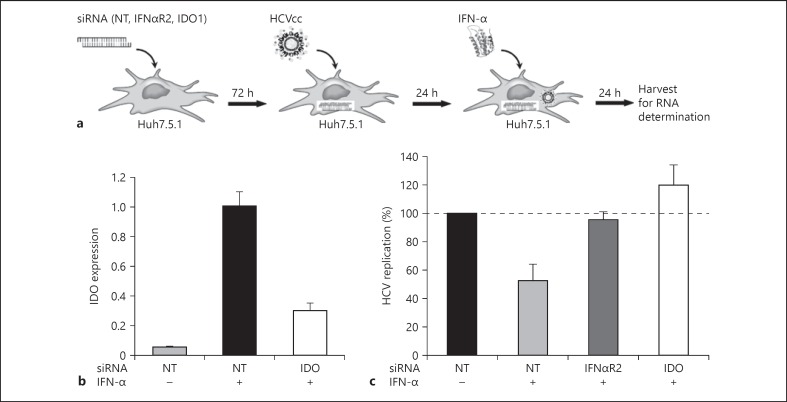
To investigate whether IDO participates in innate immune responses to HCV, we performed IDO siRNA knock-down experiments in Huh7.5.1 cells. As shown in figure 6a, siRNA-treated cells were subsequently challenged with JFH1 virus and a suboptimal dose of IFN-α (10 IU/ml). The treatment of cells with suboptimal doses of IFN-α was necessary since higher doses of IFN-α would minimize the siRNA effect. Efficient siRNA transfection and knock-down of IDO in Huh7.5.1 cells was monitored at day 5 after transfection by qRT-PCR. We observed at this time point a target gene knock-down of 70% - a value still considered to be highly efficient (fig. 6b). As shown in figure 6c, IFN-α at suboptimal dose decreased HCV replication by 50% in Huh7.5.1 cells. The downregulation of HCV replication was IFN-α dependent, as evidenced by the rescue of HCV replication in Huh7.5.1 cells following IFNαR2 siRNA knock-down. IDO siRNA knock-down also reversed IFN-α-mediated suppression of HCV replication, indicating that IDO is an ISG with anti-HCV activity.
Fig. 6.
IDO function as an ISG effector with anti-HCV activity. NT = Non-targeting control. a Flowchart describing the 3 main parts of the experiment. Huh7.5.1 cells were transfected with siRNAs against IFNαR2, IDO or NT siRNAs. After 72 h, cells were infected with HCV JFH1 virus and, after an additional 24 h, IFN-α (10 IU/ml) was added. Cells were harvested 24 h later, and HCV replication was measured by qRT-PCR. b Knock-down efficiencies of IDO. IDO mRNA levels were quantified by qRT-PCR at day 5 after siRNA transfection. c Intracellular HCV RNA amounts. Data are shown as mean percentage of HCV replication compared to cells treated with non-targeting control siRNAs (= 100%). Values represent the mean ± SD of triplicate measurements.
Hepatic IDO Modulates CD4+ T-Cell Proliferation
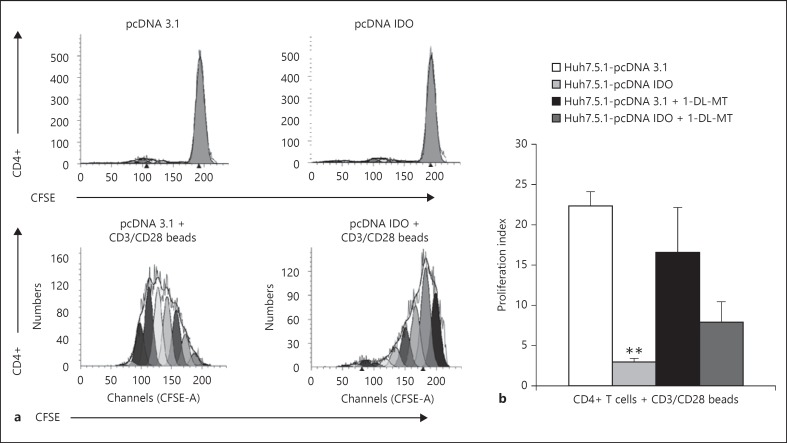
IDO-expressing dendritic cells have a crucial role in the downregulation of immune responses [4]. To investigate whether hepatic IDO has similar effects on immune cells, we performed co-culture experiments in which naïve or activated CD4+ T lymphocytes were cultured in the presence of IDO-transfected Huh7.5.1 cells. As shown in figure 7a, the presence of IDO-transfected Huh7.5.1 cells or control cells transfected with the empty vector did not have an effect on naïve CD4+ T cells. In contrast, the presence of IDO-expressing Huh7.5.1 cells significantly reduced the proliferation of anti-CD3/CD28 stimulated CD4+ T lymphocytes compared to control cells (fig. 7a, b). These results suggest that IDO expressed in hepatocytes plays an important role in the regulation of hepatic immune responses. The addition of the IDO inhibitor 1-DL-MT during the co-culture experiment partially rescued CD4+ T-cell proliferation (fig. 7b). The incomplete reversal of suppression by 1-DL-MT might be due to the fact that 1-DL-MT-mediated blockage was incomplete, as evidenced by the production of kynurenine of IDO-transfected Huh7.5.1 cells in the presence of 1,000 µM of 1-DL-MT (fig. 5a).
Fig. 7.
Hepatic IDO expression inhibits CD4+ T cell proliferation. a Flow cytometry analysis of CD4 and CFSE staining following CD4+T/Huh7.5.1 cell co-culture. Huh7.5.1 cells were transfected with pcDNA IDO or pcDNA3.1. At 48 h after transfection, cells were added to CD3/CD28-activated CD4+ T cells or control CD4+ T cells. CFSE fluorescence of undivided non-stimulated and CD3/CD28-activated CD4+ T cells undergoing multiple rounds of cell division following Huh7.5.1 co-culture is shown. b Pooled data from 3 different co-culture experiments are shown as mean proliferation index ± SD. Data were compiled from 3 different healthy CD4+ T-cell donors. Inhibition of CD4+ T-cell proliferation by IDO-transfected Huh7.5.1 cells was statistically significant. ** p < 0.01. The presence of 1-DL-MT (1,000 µM) for the duration of the co-culture partially restored IDO-mediated CD4+ T-cell suppression.
Discussion
The present study shows that IDO is transiently expressed in HCV-infected hepatocytes and coincides with an increase in IFNs and ISGs, suggesting that IDO participates in innate anti-HCV immune mechanisms. Indeed, we found that HCV replication is less efficient in Huh7.5.1 cells overexpressing IDO. Furthermore, we were able to recapitulate these findings in PHH, although the level of IDO transfection and HCV JFH1 infection in these cells was very low compared to Huh7.5.1 cells. Our findings are in line with recent studies showing that overexpression of IDO prior to infection with West Nile virus, a mosquito-borne flavivirus, significantly inhibited West Nile virus replication via tryptophan starvation [8]. In addition, Mao et al. [7] demonstrated that IDO inhibited hepatitis B virus replication via tryptophan deprivation. Since tryptophan shortage restricts mammalian cell growth and viral replication is dependent on the host cell metabolic machinery, a tryptophan-deficient intracellular environment is presumably less favourable for HCV replication. Type I IFN production by the infected hepatocyte serves as the first line of host defence against HCV, and current evidence suggests that IFN-mediated anti-HCV activity is performed by the combined action of multiple ISGs [28, 34]. To investigate whether IDO is an ISG with anti-HCV activity, cells with silenced IDO gene expression were exposed to IFN-α at a suboptimal dose to avoid minimizing siRNA efficiency. Knock-down of IDO rescued HCV replication in Huh7.5.1 in the context of IFN-α treatment, suggesting that IDO contributes to the suppression of HCV replication mediated by IFNs. Our findings are in line with in vivo studies that showed a transient upregulation of hepatic IDO during the acute phase of HCV infection in chimpanzees that spontaneously cleared HCV infection [14].
HCV infection triggered IRF1 expression in hepatocytes and Huh7.5.1 cells. Gene expression profiling of JFH-1-infected Huh7 cells by microarray analysis also demonstrated an increase in IRF1 expression [35]. In addition, IRF1 mRNA expression levels have been shown to be upregulated in liver biopsy samples from HCV-infected patients [36]. IRF1 is a pleiotropic transcription factor with antiviral, antiproliferative and proapoptotic properties. IRF1 regulates the expression of several ISGs such as PKR and 2′,5′-oligoadenylate synthetase [31]. Previous studies have suggested that IRF1 is one of the key cellular factors that regulate intracellular HCV replication [37]. Of note, we observed a substantial increase in the level of HCV replication in JFH1-infected Huh7.5.1 cells following partial IRF1 gene silencing (data not shown). It has been previously shown that the IDO promoter contains a putative binding site for IRF1 [38]. Overexpression of IRF1 alone was sufficient to upregulate IDO expression in Huh7.5.1 cells. On the other hand, partial silencing of the IRF1 gene in Huh7.5.1 cells abolished IFN-γ-induced IDO expression. These findings suggest that IDO expression in HCV-infected cells is driven by IRF1. IRF1 can be induced by many stimuli, including IFNs (-α, -β, -γ), TNF-α, IL-6, and viral infections. Positive feedback regulation of IFN-β expression by IRF1 has been also observed, suggesting a complex gene network in the regulation of the IFN pathway by IRF1 [31]. The double-stranded RNA replication intermediate of HCV is recognized in the infected hepatocyte through the combined action of multiple pathways, including PKR, RIG-I (retinoic acid-inducible gene I)-like helicases and TLR-3 (Toll-like receptor 3) pathways, which ultimately leads to the induction of type I IFNs or proinflammatory cytokines [2]. Thus, it is possible that HCV-induced IFN production triggers IRF1 production in hepatocytes. However, a detailed analysis of the underlying mechanism in this study is hampered by the fact that Huh7.5.1 cell lines are highly adapted, with defects in RIG-I and TLR3 pathway signalling [39] that allow high levels of HCV replication by impairing innate immunity.
IDO mRNA expression was most potently induced by IFN-γ in hepatocytes and Huh7.5.1 cells. IFN-γ-induced IDO was enzymatically active in hepatocytes, as evidenced by the production of kynurenine, the main metabolite of tryptophan. However, we were unable to detect IDO protein expression in IFN-γ-stimulated Huh7.5.1 cells by Western blot, suggesting that IDO is expressed at low levels in these cells. The absence of a detectable IDO protein signal in Huh7.5.1 cells may also be explained by the very low transcription rate of the IDO gene in these cells compared to PHH. Ongoing HCV replication in Huh7.5.1 cells significantly upregulated IFN-γ-induced IDO expression. Similar effects on IFN-γ-induced IDO expression have been previously reported in Huh7 cells by Larrea et al. [14]. In their study, IFN-γ-induced IDO mRNA expression was analysed for up to 40 h after IFN-γ treatment. Interestingly, IDO mRNA expression levels remained significantly upregulated until this time point compared to uninfected control cells, suggesting that IFN-γ induces a sustained IDO expression in HCV-infected hepatocytes. Co-operative binding of IRF1 and STAT1 to the IDO promoter appears to play an important role in the regulation of IDO gene expression [32]. IFN-γ mediated IRF-1 expression is driven primarily by phosphorylated STAT1 homodimers [40]. Analysis of phospho-STAT1 levels demonstrated prolonged STAT1 phosphorylation in JFH1-infected Huh7.5.1 cells in the presence of IFN-γ. Delayed dephosphorylation of activated STAT1 in JFH1-infected Huh7.5.1 cells may sustain IRF1 expression as well as IDO. Although further studies are necessary to support this hypothesis, prolonged phosphorylation in the presence of IFN-γ in virus-infected cells has been described for the Sindbis virus, an enveloped plus-strand, mosquito-borne ARN virus [41] and, most recently, for cytomegalovirus [42]. Finally, adenovirus has been shown to prolong phosphorylation of STAT1 induced by IFN-α in infected cells by inhibiting STAT1 dephosphorylation [43].
IFN-γ production is mainly attributed to natural killer cells and activated T cells which play key roles in innate and adaptive immune responses, respectively. Previous studies showed that co-culture of IFN-γ-producing T cells with HCV-infected Huh7 cells resulted in 100-fold higher IDO levels than in experiments using exogenous IFN-γ alone [14]. However, whether induced hepatic IDO exerts immunoregulatory effects upon innate and adaptive immune cells was not investigated. In the present study, we found that hepatic IDO expression significantly inhibited the proliferation of activated CD4+ T cells, suggesting that HCV-infected hepatocytes in an IFN-γ rich environment are capable of exerting a negative feedback loop on CD4+ T cells. Further studies are necessary to address the underlying mechanism of hepatic IDO-mediated inhibition. A shortage in tryptophan supply combined with the production of kynurenine pathway metabolites have been associated with the emergence of a regulatory phenotype in naïve CD4+CD25- T cells, affecting CD4+ as well as CD8+ T cell function [44]. Downstream kynurenine pathway metabolites such as 3-hydroxyanthranilic acid have also been described as causing dysfunction of activated T cells, T cell anergy and even apoptosis [4]. Whether hepatocytes possess the enzymatic machinery to further metabolize kynurenine into immunomodulatory downstream metabolites remains unknown. Functional exhaustion and, ultimately, deletion of HCV-specific CD4+ T cells is a hallmark of chronic HCV infection [3]. Sustained hepatic IDO expression within the liver environment may play a role in dampening the HCV-specific T-cell response. Of note, there are two other enzymes involved in tryptophan metabolism: TDO and IDO2, a paralogue of IDO [29]. IDO2 gene expression is not induced by IFNs, and its function is still unclear. TDO is constitutively expressed in the liver and plays a crucial role in systemic tryptophan homoeostasis [45]. Cells expressing recombinant human TDO have been shown to inhibit T-cell proliferation and production of IFN-γ [46], suggesting that hepatic TDO might contribute to the tolerogenic properties of the liver. However, since TDO does not respond to inflammatory signals and cannot be blocked by the IDO-specific inhibitor 1-DL-MT, our in vitro data point to a prominent role for IDO in mediating antiviral and immune regulatory effects in HCV infection.
In conclusion, we identified a dichotomous role of hepatic IDO in HCV infection. The dominant nature of this role - anti-HCV activity or immune counter-regulation - in HCV infection remains to be investigated. In the early phase of HCV infection, the upregulation of IDO may contribute to the host's innate antiviral defence mechanism by slowing down HCV replication. At later time points, when HCV-specific T cells are activated and accumulate in the liver, a sustained IFN-γ-induced upregulation of IDO in HCV-infected hepatocytes may counter-regulate the anti-HCV T-cell response and favour tolerance. In addition, IDO expression might protect hepatocytes against a possible overshooting T-cell response or might be secondary to the progressive inflammation events of the liver, which is a hallmark of persistent HCV infection. Hepatic IDO might be beneficial in acute infection and deleterious in chronic infection. However, the question arises as to whether it is possible to potentiate the contribution of IDO to anti-HCV activity and reduce its immunosuppressive effect in the chronic phase of infection. In a model of bacterial infection, Müller et al. [47] demonstrated that IDO-mediated antimicrobial and immunoregulatory effects require different threshold levels of tryptophan. A 10- to 40-fold greater tryptophan depletion was required to mediate an immunoregulatory effect compared to the antibacterial effect [47]. This observation, together with the fact that IDO-mediated regulation of T-cell responses is closely linked to toxic effects of kynurenine and other immunosuppressive metabolites of the kynurenine pathway [4] may provide the opportunity to modulate the IDO pathway towards favouring the antiviral effect. Thus, the attenuation of IDO activity with small molecule inhibitors such as 1MT, or the inhibition of toxic kynurenine accumulation via the activation of other downstream enzymes that are involved in the production of harmless by- or end-products, may tip the balance toward IDO-mediated antiviral effects with less IDO-mediated immune suppression. However, to answer these questions an IDO knock-out mouse susceptible to HCV has to be developed. Since IDO inhibitor drugs have already entered phase II clinical trials in cancer patients (www.clinicaltrials.gov. NCT01685255, NCT01604889), knowledge of the dominant role of IDO in acute and chronic HCV infection may pave the way for novel therapeutic intervention.
Acknowledgments
We would like to thank T. Baumert, INSERM U1110, for supporting the study and providing access to the L3 laboratory facility. We also wish to thank S. Strom and his colleagues at the University of Pittsburgh for providing the PHH through the Liver Tissue Procurement and Distribution System (N01-DK-7-0004/HHSN26700700004C) and the team of P. Bachellier, Pôle des Pathologies Digestives, Hépatiques et Transplantation, Hôpitaux Universitaires de Strasbourg, France for providing liver resections for PHH isolation. We are grateful to S. Durand, INSERM U1110, Strasbourg for excellent technical assistance in the isolation of PHH and to S. Raghuraman, Inflammation and Infection Research Centre, University of New South Wales, Sydney, Australia for helpful discussion. Finally, we thank C. Sodroski, Liver Diseases Branch, NIDDK, NIH for proofreading the manuscript. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH and the Agence Nationale de la Recherche sur le Sida (2010-242 to H.B.).
References
- 1.Liang TJ, Heller T. Pathogenesis of hepatitis C-associated hepatocellular carcinoma. Gastroenterology. 2004;127:S62–S71. doi: 10.1053/j.gastro.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 2.Horner SM, Gale M. Regulation of hepatic innate immunity by hepatitis C virus. Nat Med. 2013;19:879–888. doi: 10.1038/nm.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rehermann B. Pathogenesis of chronic viral hepatitis: differential roles of T cells and NK cells. Nat Med. 2013;19:859–868. doi: 10.1038/nm.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol. 2004;4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- 5.Pfefferkorn ER. Interferon gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proc Natl Acad Sci U S A. 1984;81:908–912. doi: 10.1073/pnas.81.3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams O, Besken K, Oberdorfer C, MacKenzie CR, Takikawa O, Daubener W. Role of indoleamine-2,3-dioxygenase in alpha/beta and gamma interferon-mediated antiviral effects against herpes simplex virus infections. J Virol. 2004;78:2632–2636. doi: 10.1128/JVI.78.5.2632-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao R, Zhang J, Jiang D, Cai D, Levy JM, Cuconati A, Block TM, Guo JT, Guo H. Indoleamine 2,3-dioxygenase mediates the antiviral effect of gamma interferon against hepatitis B virus in human hepatocyte-derived cells. J Virol. 2010;85:1048–1057. doi: 10.1128/JVI.01998-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeung AW, Wu W, Freewan M, Stocker R, King NJ, Thomas SR. Flavivirus infection induces indoleamine 2,3-dioxygenase in human monocyte-derived macrophages via tumor necrosis factor and NF-κB. J Leuk Biol. 2012;91:657–666. doi: 10.1189/jlb.1011532. [DOI] [PubMed] [Google Scholar]
- 9.Curti A, Trabanelli S, Salvestrini V, Baccarani M, Lemoli RM. The role of indoleamine 2,3-dioxygenase in the induction of immune tolerance: focus on hematology. Blood. 2009;113:2394–2401. doi: 10.1182/blood-2008-07-144485. [DOI] [PubMed] [Google Scholar]
- 10.Barth H, Raghuraman S. Persistent infection diseases say - IDO. Role of indoleamine-2,3-dioxygenase in disease pathogenesis and implications for therapy. Crit Rev Microbiol. 2014;40:360–368. doi: 10.3109/1040841X.2012.742037. [DOI] [PubMed] [Google Scholar]
- 11.Boasso A, Herbeuval JP, Hardy AW, Anderson SA, Dolan MJ, Fuchs D, Shearer GM. HIV inhibits CD4+ T-cell proliferation by inducing indoleamine 2,3-dioxygenase in plasmacytoid dendritic cells. Blood. 2007;109:3351–3359. doi: 10.1182/blood-2006-07-034785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nilsson J, Boasso A, Velilla PA, Zhang R, Vaccari M, Franchini G, Shearer GM, Andersson J, Chougnet C. HIV-1-driven regulatory T-cell accumulation in lymphoid tissues is associated with disease progression in HIV/AIDS. Blood. 2006;108:3808–3817. doi: 10.1182/blood-2006-05-021576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cozzi A, Zignego AL, Carpendo R, Biagiotti T, Aldinucci A, Monti M, Giannini C, Rosselli M, Laffi G, Moroni F. Low serum tryptophan levels, reduced macrophage IDO activity and high frequency of psychopathology in HCV patients. J Viral Hepat. 2006;13:402–408. doi: 10.1111/j.1365-2893.2005.00706.x. [DOI] [PubMed] [Google Scholar]
- 14.Larrea E, Riezu-Boj JI, Gil-Guerrero L, Casares N, Aldabe R, Sarobe P, Civeira MP, Heeney JL, Rollier C, Verstrepen B, Wakita T, Borras-Cuesta F, Lasarte JJ, Prieto J. Upregulation of indoleamine 2,3-dioxygenase in hepatitis C virus infection. J Virol. 2007;81:3662–3666. doi: 10.1128/JVI.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zignego AL, Cozzi A, Carpenedo R, Giannini C, Rosselli M, Biagioli T, Aldinucci A, Laffi G, Moroni F. HCV patients, psychopathology and tryptophan metabolism: analysis of the effects of pegylated interferon plus ribavirin treatment. Dig Liver Dis. 2007;39((suppl 1)):S107–S111. doi: 10.1016/s1590-8658(07)80021-1. [DOI] [PubMed] [Google Scholar]
- 16.Higashitani K, Kanto T, Kuroda S, Yoshio S, Matsubara T, Kakita N, Oze T, Miyazaki M, Sakakibara M, Hiramatsu N, Mita E, Imai Y, Kasahara A, Okuno A, Takikawa O, Hayashi N, Takehara T. Association of enhanced activity of indoleamine 2,3-dioxygenase in dendritic cells with the induction of regulatory T cells in chronic hepatitis C infection. J Gastroenterol. 2013;48:660–670. doi: 10.1007/s00535-012-0667-z. [DOI] [PubMed] [Google Scholar]
- 17.Rollier CS, Paranhos-Baccala G, Verschoor EJ, Verstrepen BE, Drexhage JA, Fagrouch Z, Berland JL, Komurian-Pradel F, Duverger B, Himoudi N, Staib C, Meyr M, Whelan M, Whelan JA, Adams VC, Larrea E, Riezu JI, Lasarte JJ, Bartosch B, Cosset FL, Spaan WJ, Diepolder HM, Pape GR, Sutter G, Inchauspe G, Heeney JL. Vaccine-induced early control of hepatitis C virus infection in chimpanzees fails to impact on hepatic PD-1 and chronicity. Hepatology. 2007;45:602–613. doi: 10.1002/hep.21573. [DOI] [PubMed] [Google Scholar]
- 18.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambotin M, Baumert TF, Barth H. Distinct intracellular trafficking of hepatitis C virus in myeloid and plasmacytoid dendritic cells. J Virol. 2010;84:8964–8969. doi: 10.1128/JVI.00517-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas SR, Terentis AC, Cai H, Takikawa O, Levina A, Lay PA, Freewan M, Stocker R. Post-translational regulation of human indoleamine 2,3-dioxygenase activity by nitric oxide. J Biol Chem. 2007;282:23778–23787. doi: 10.1074/jbc.M700669200. [DOI] [PubMed] [Google Scholar]
- 23.Laich A, Neurauter G, Widner B, Fuchs D. More rapid method for simultaneous measurement of tryptophan and kynurenine by HPLC. Clin Chem. 2002;48:579–581. [PubMed] [Google Scholar]
- 24.Braun D, Longman RS, Albert ML. A two-step induction of indoleamine 2,3 dioxygenase (IDO) activity during dendritic-cell maturation. Blood. 2005;106:2375–2381. doi: 10.1182/blood-2005-03-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q, Pene V, Krishnamurthy S, Cha H, Liang TJ. Hepatitis C virus infection activates an innate pathway involving IKK-α in lipogenesis and viral assembly. Nat Med. 2013;19:722–729. doi: 10.1038/nm.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas E, Gonzalez VD, Li Q, Modi AA, Chen W, Noureddin M, Rotman Y, Liang TJ. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology. 2012;142:978–988. doi: 10.1053/j.gastro.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metz P, Dazert E, Ruggieri A, Mazur J, Kaderali L, Kaul A, Zeuge U, Windisch MP, Trippler M, Lohmann V, Binder M, Frese M, Bartenschlager R. Identification of type I and type II interferon-induced effectors controlling hepatitis C virus replication. Hepatology. 2012;56:2082–2093. doi: 10.1002/hep.25908. [DOI] [PubMed] [Google Scholar]
- 29.Ball HJ, Sanchez-Perez A, Weiser S, Austin CJ, Astelbauer F, Miu J, McQuillan JA, Stocker R, Jermiin LS, Hunt NH. Characterization of an indoleamine 2,3-dioxygenase-like protein found in humans and mice. Gene. 2007;396:203–213. doi: 10.1016/j.gene.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 30.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen H, Hiscott J, Pitha PM. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 1998;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 32.Jeong YI, Kim SW, Jung ID, Lee JS, Chang JH, Lee CM, Chun SH, Yoon MS, Kim GT, Ryu SW, Kim JS, Shin YK, Lee WS, Shin HK, Lee JD, Park YM. Curcumin suppresses the induction of indoleamine 2,3-dioxygenase by blocking the Janus-activated kinase-protein kinase Cδ-STAT1 signaling pathway in interferon-γ-stimulated murine dendritic cells. J Biol Chem. 2009;284:3700–3708. doi: 10.1074/jbc.M807328200. [DOI] [PubMed] [Google Scholar]
- 33.Lindenbach BD, Rice CM. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol. 2013;11:688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blackham S, Baillie A, Al-Hababi F, Remlinger K, You S, Hamatake R, McGarvey MJ. Gene expression profiling indicates the roles of host oxidative stress, apoptosis, lipid metabolism, and intracellular transport genes in the replication of hepatitis C virus. J Virol. 2010;84:5404–5414. doi: 10.1128/JVI.02529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abbate I, Romano M, Longo R, Cappiello G, Lo Iacono O, Di Marco V, Paparella C, Spano A, Capobianchi MR. Endogenous levels of mRNA for IFNs and IFN-related genes in hepatic biopsies of chronic HCV-infected and non-alcoholic steatohepatitis patients. J Med Virol. 2003;70:581–587. doi: 10.1002/jmv.10433. [DOI] [PubMed] [Google Scholar]
- 37.Kanazawa N, Kurosaki M, Sakamoto N, Enomoto N, Itsui Y, Yamashiro T, Tanabe Y, Maekawa S, Nakagawa M, Chen CH, Kakinuma S, Oshima S, Nakamura T, Kato T, Wakita T, Watanabe M. Regulation of hepatitis C virus replication by interferon regulatory factor 1. J Virol. 2004;78:9713–9720. doi: 10.1128/JVI.78.18.9713-9720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chon SY, Hassanain HH, Gupta SL. Cooperative role of interferon regulatory factor 1 and p91 (STAT1) response elements in interferon-γ-inducible expression of human indoleamine 2,3-dioxygenase gene. J Biol Chem. 1996;271:17247–17252. doi: 10.1074/jbc.271.29.17247. [DOI] [PubMed] [Google Scholar]
- 39.Bartenschlager R, Pietschmann T. Efficient hepatitis C virus cell culture systems: what a difference the host cell makes. Proc Natl Acad Sci U S A. 2005;102:9739–9740. doi: 10.1073/pnas.0504296102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramsauer K, Farlik M, Zupkovitz G, Seiser C, Kroger A, Hauser H, Decker T. Distinct modes of action applied by transcription factors STAT1 and IRF1 to initiate transcription of the IFN-γ-inducible gbp2 gene. Proc Natl Acad Sci U S A. 2007;104:2849–2854. doi: 10.1073/pnas.0610944104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burdeinick-Kerr R, Govindarajan D, Griffin DE. Noncytolytic clearance of Sindbis virus infection from neurons by gamma interferon is dependent on Jak/STAT signaling. J Virol. 2009;83:3429–3435. doi: 10.1128/JVI.02381-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trilling M, Le VT, Rashidi-Alavijeh J, Katschinski B, Scheller J, Rose-John S, Androsiac GE, Jonjic S, Poli V, Pfeffer K, Hengel H. ‘Activated’ STAT proteins: a paradoxical consequence of inhibited Jak-STAT signaling in cytomegalovirus-infected cells. J Immunol. 2014;192:447–458. doi: 10.4049/jimmunol.1203516. [DOI] [PubMed] [Google Scholar]
- 43.Sohn SY, Hearing P. Adenovirus sequesters phosphorylated STAT1 at viral replication centers and inhibits stat dephosphorylation. J Virol. 2011;85:7555–7562. doi: 10.1128/JVI.00513-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fallarino F, Grohmann U, You S, McGrath BC, Cavener DR, Vacca C, Orabona C, Bianchi R, Belladonna ML, Volpi C, Santamaria P, Fioretti MC, Puccetti P. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor ζ-chain and induce a regulatory phenotype in naive T cells. J Immunol. 2006;176:6752–6761. doi: 10.4049/jimmunol.176.11.6752. [DOI] [PubMed] [Google Scholar]
- 45.Forouhar F, Anderson JL, Mowat CG, Vorobiev SM, Hussain A, Abashidze M, Bruckmann C, Thackray SJ, Seetharaman J, Tucker T, Xiao R, Ma LC, Zhao L, Acton TB, Montelione GT, Chapman SK, Tong L. Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase. Proc Natl Acad Sci U S A. 2007;104:473–478. doi: 10.1073/pnas.0610007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt SK, Müller A, Heseler K, Woite C, Spekker K, MacKenzie CR, Däubener W. Antimicrobial and immunoregulatory properties of human tryptophan 2,3-dioxygenase. Eur J Immunol. 2009;39:2755–2764. doi: 10.1002/eji.200939535. [DOI] [PubMed] [Google Scholar]
- 47.Müller A, Heseler K, Schmidt SK, Spekker K, Mackenzie CR, Däubener W. The missing link between indoleamine 2,3-dioxygenase mediated antibacterial and immunoregulatory effects. J Cell Mol Med. 2009;13:1125–1135. doi: 10.1111/j.1582-4934.2008.00542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]