Abstract
Mitotic spindle formation relies on the stochastic capture of microtubules at kinetochores. Kinetochore architecture affects the efficiency and fidelity of this process with large kinetochores expected to accelerate assembly at the expense of accuracy, and smaller kinetochores to suppress errors at the expense of efficiency. We demonstrate that upon mitotic entry, kinetochores in cultured human cells form large crescents that subsequently compact into discrete structures on opposite sides of the centromere. This compaction occurs only after the formation of end-on microtubule attachments. Live-cell microscopy reveals that centromere rotation mediated by lateral kinetochore-microtubule interactions precedes formation of end-on attachments and kinetochore compaction. Computational analyses of kinetochore expansion-compaction in the context of lateral interactions correctly predict experimentally-observed spindle assembly times with reasonable error rates. The computational model suggests that larger kinetochores reduce both errors and assembly times, which can explain the robustness of spindle assembly and the functional significance of enlarged kinetochores.
Introduction
Chromosome segregation during cell division is enacted by the mitotic ‘spindle’. Chromosomes connect to the spindle via kinetochores that capture microtubules and attach to their plus ends, the principle described as ‘search-and-capture’ (S&C)1–5. A ramification of the S&C mechanism is that kinetochore size and shape play a fundamental role in determining the efficiency and fidelity of chromosome segregation. Intuitively, larger kinetochores are expected to increase the probability of encounters between kinetochores and microtubules which would also promote errors such as attachment of sister kinetochores to the same spindle pole (syntelic) or attachment of a single kinetochore to both poles (merotelic). Cellular regulations that minimize erroneous attachments while expediting spindle assembly remain unknown.
Here we demonstrate that the shape of the kinetochore’s outer layer changes dramatically and rapidly during the normal course of mitosis. At the onset of spindle assembly, sister kinetochores expand to almost completely encircle the centromere. After the formation of end-on attachments to microtubules the enlarged kinetochores downsize into small discs on opposite sides of the centromere. Computational analyses suggest that the observed reorganization of the kinetochore architecture simultaneously enhances efficiency of microtubule capture and suppresses the number of erroneous attachments. Error reduction is due to improvements in the angular orientation of enlarged kinetochores that result from lateral interactions with microtubules prior to the formation of end-on attachments. If these lateral interactions are impeded, the number of errors increases significantly.
Results
The outer layer of unattached kinetochores encircles the centromere
Properly attached kinetochores appear as nearly diffraction-limited spots in fluorescence light microscopy (LM)6 and as ~200-nm discs positioned on opposite sides of the centromere in electron-microscopy (EM)7,8. However, the kinetochore outer layer is enlarged when cells are arrested in mitosis due to lack of microtubules9–11. Enlarged kinetochores have been observed also during prometaphase in HeLa cells12. To test the idea that kinetochore size and shape change during normal spindle assembly we detailed the kinetochore architecture at various mitotic stages.
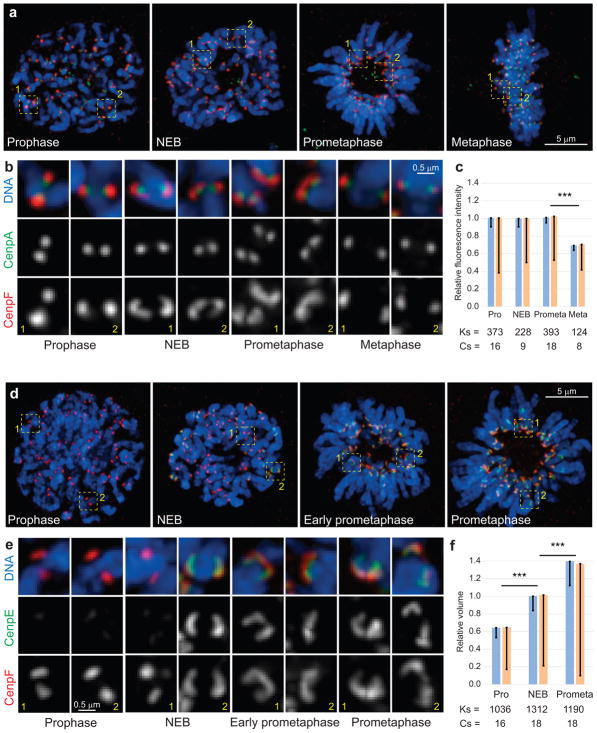
In non-transformed human cells RPE1, the outer-kinetochore protein CenpF forms compact spots during late prophase and metaphase, but partially encircles the centromere shortly after nuclear envelope breakdown (NEB, 193 of 274 kinetochores in 3 cells) and during prometaphase (267 of 550 kinetochores in 6 cells) (Fig. 1). Despite the apparent change in morphology, the amount of CenpF at the kinetochore remains constant from prophase through prometaphase (Fig. 1c). To detail changes within the outer layer we co-visualized CenpF13 and CenpE14,15, the two most peripheral kinetochore proteins capable of direct interactions with microtubules. CenpE appears at the kinetochores only after NEB (Fig. 2). We have previously demonstrated that the central region of forming spindles becomes devoid of chromosomes ~1 min after NEB in RPE1 cells16 (see Supplementary Movies 2–3). This feature allows one to identify cells that are in the first minute of spindle assembly. In these cells, CenpE is detected only at some of the kinetochores. Intriguingly, CenpF forms compact spots in CenpE-negative kinetochores but partially encircles the centromere in CenpE-positive kinetochores (Fig. 1d,e). As the amount of CenpF remains constant during prophase-prometaphase (Fig. 1c), relative volumes occupied by this protein can be compared. Volumetric analysis (Fig. 1f) suggests that the outer layer expands during spindle assembly.
Figure 1. Changes in the outer kinetochore architecture at various stages of mitosis.
(a–c) The outer layer enlarges at the onset of spindle assembly and subsequently downsizes. (a) Maximal intensity projections (include all kinetochores) depicting RPE1 cells at various stages of mitosis. (b) Examples of individual kinetochores from the boxed areas in (a), shown at higher magnification. The outer layers (red, CenpF) appear as compact spots during prophase and metaphase but expand into crescents that partially encircle the centromere during NEB and prometaphase. (c) Intensity of CenpF fluorescence remains constant during outer layer expansion and decreases during metaphase. (d–f) The outer layer both recruits additional proteins (CenpE, green) and expands (CenpF, red). (d) Maximal intensity projections (include all kinetochores) depicting RPE1 cells. (e) Examples of individual kinetochores from the boxed areas in (d), shown at higher magnification. (f) The volume occupied by CenpF increases from late prophase through prometaphase. Blue bars in (c) and (f) are calculated as mean of mean values for multiple kinetochores in individual cells (n values listed below the bars, Cs; cells). Error bars represent standard error of the mean (s.e.m.). Yellow bars are mean values calculated for all kinetochores, pooled from all cells in that class (n values listed below the bars, Ks; kinetochores). Error bars represent standard deviation (s.d.). Triple asterisks denote differences with p < 0.005 (two-tailed Student’s test) for both blue vs. blue and yellow vs. yellow bars.
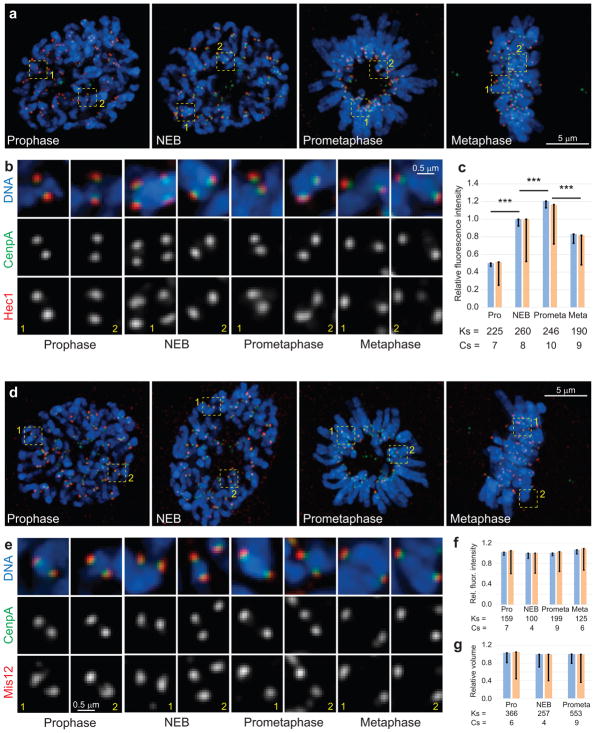
Figure 2. The kinetochore core remains relatively compact throughout mitosis.
(a,d) Maximal intensity projections (include all kinetochores) depicting RPE1 cells at various stages of mitosis. (b,e) Examples of individual kinetochores from the boxed areas in (a) and (d) shown at higher magnification. The shape of Hec1 (b, red) and Mis12 (d, red) distributions remains relatively compact. Notice that the amount of Hec1 increases significantly (c) while the amount of Mis12 and the volume occupied by this protein remain constant (f,g). Also notice that the shape of inner kinetochore remains compact throughout mitosis (green, CenpA-GFP). Blue bars in (c), (f), and (g) are calculated as mean of mean values for multiple kinetochores in individual cells (n values listed below the bars. Cs; cells). Error bars represent s.e.m. Yellow bars are mean values calculated for all kinetochores pooled from all cells in that class (n values listed below the bars, Ks; kinetochores). Triple asterisks denote differences with p < 0.005 (two-tailed Student’s test) for both blue vs. blue and yellow vs. yellow bars.
Unlike CenpF, proteins that reside deeper inside the kinetochore (Hec1, Mis12)6 appear as compact spots throughout mitosis (Fig. 2) even though the amount of Hec1 increases during prometaphase (Fig. 2c) while the amount of Mis12 remains constant (Fig. 2c). The volume occupied by Mis12 also remains constant (Fig. 2g). Thus, changes in the kinetochore architecture occur primarily in the outer layers of the organelle.
To evaluate the shape of the outer layer at a higher resolution, we employed correlative LM/EM. Serial-section analyses of three cells at NEB reveal highly variable kinetochore morphology. Although the exact location of kinetochores is determined via correlation of LM and EM images (Fig. 3a), most of the kinetochores lack distinct plates. In one cell all kinetochores appear as ill-defined spots of fibrous material (Fig. 3b). In the other two cells, ~30% of kinetochores (53/185) display trilaminar plates that largely encircle the centromere. The gap between the plates of sister kinetochores is typically 100–200 nm on one side and larger on the other side of the centromere (Fig. 3d). The rest of kinetochores appear as ill-defined spots or partially assembled plates embedded in a small cloud of fibrous material (Fig. 3c). These observations suggest that at the onset of spindle assembly, the outer plate rapidly expands from a compact cloud to a large crescent on the surface of the centromere.
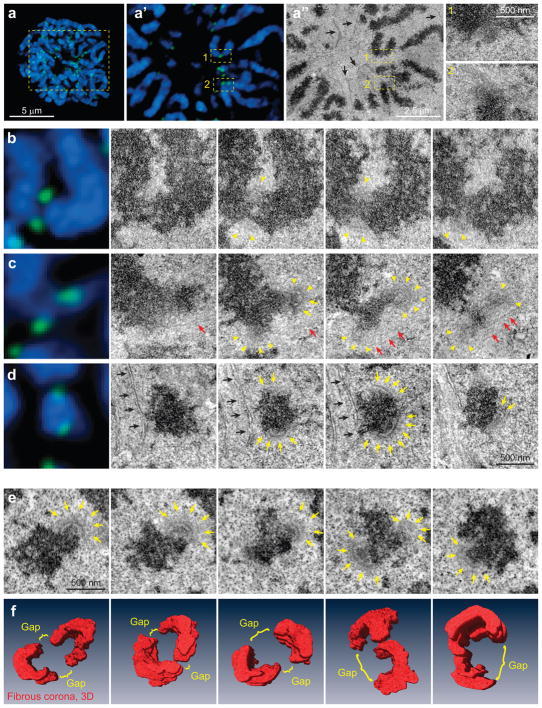
Figure 3. Kinetochore morphology at the onset of spindle assembly.
(a) Correlative LM/EM analysis of kinetochore morphology at NEB. Maximal intensity projection of the entire cell and an individual focal plane are shown. Via correlation of LM (a′) and EM (a″) images, positions of individual kinetochores are identified. Boxed areas are shown at higher magnification. Kinetochore 1 lacks the distinct trilaminar plate. Black arrows mark partially-disassembled nuclear envelope. (b–d) Examples of kinetochore morphology in RPE cells at NEB. Each chromosome is shown in LM (left) and serial EM sections. Approximately 50% of kinetochores are morphologically indistinct (b). The other 50% display partially- (c), or fully assembled plates that largely encircle the centromere (d). Arrowheads denote fibrous corona, arrows point at distinct trilaminar plates. (e–f) Morphology of ‘virgin’ kinetochores in a cell fixed less than 2 min after NEB in the absence of microtubules (see Figure S2 for whole-cell view and preparation details). (e) Complete series of sections through sister kinetochores. Yellow arrows mark trilaminar plates, red arrows point at microtubules, black arrows mark remnants of nuclear envelope. (f) Examples of 3-D reconstructions of the outer layer (first image corresponds to the kinetochores shown in (e)). The outer layers largely encircle the centromere.
Microtubules in proximity of partially-assembled plates indicate that microtubule-mediated forces may play a role in shaping the kinetochore outer layer17 (Fig. 3c). To characterize the architecture of ‘virgin’ kinetochores that have not interacted with microtubules, we treated cells in late prophase with nocodazole and fixed them immediately after detection of NEB (Supplementary Fig. 1). Since nocodazole completely depolymerizes microtubules within 2 min17, this experimental approach produces kinetochores whose architecture is unaffected by interactions with microtubules or prolonged mitotic arrest.
CenpF immunofluorescence suggests that the outer layers of virgin kinetochores consistently form crescents that encircle the centromere (367/500 kinetochores in 10 cells). In most cases, sister kinetochores appear to fuse together on one side of the centromere with a gap on the other side of the centromere, where sister kinetochores are separated by chromatin (Supplementary Fig. 1d). Serial-section EM confirms that the outer layers of sister kinetochores partially encircle the centromere (>80 kinetochores in the three cells). 3D reconstructions of six typical centromeres suggest that the outer layers of virgin sister kinetochores cover >50% of the centromere circumference but remain separated by ~100–200 nm (Fig. 3f).
The outer layer compacts after formation of end-on microtubule attachment
The observation that the kinetochore outer layers transition from encircling a large part of the centromere after NEB to ~200-nm discs on the opposite sides of the centromere at metaphase1,2 prompted us to search for the event that triggers kinetochore compaction.
Visual inspection of fluorescently labelled prometaphase kinetochores and microtubules suggests that the majority of compact kinetochores are attached to the ends of microtubule bundles. In contrast, enlarged kinetochores either lack microtubule attachments or interact with microtubules laterally (Supplementary Fig. 2). Because unambiguous discrimination of end-on vs. lateral interactions is difficult in LM, we treated RPE1 cells with the CenpE (kinesin-7) inhibitor GSK-92329518. CenpE drives congression of chromosomes from the vicinity of the pole to the spindle equator15,19,20 and mediates conversion from lateral to end-on attachments21. Partial inhibition of CenpE with 15-nM GSK-923295 results in the accumulation of mono-oriented chromosomes and prolongation of mitosis to >1 hr in >80% of cells. Immunofluorescence demonstrates that outer (CenpF), core (Hec1, Mis12), and inner (CenpA-GFP) kinetochore proteins appear as compact spots on congressed chromosomes in GSK-923295-treated cells. In contrast, both CenpF and Hec1 form crescents on chromosomes that reside near the spindle poles (Fig. 4a,b).
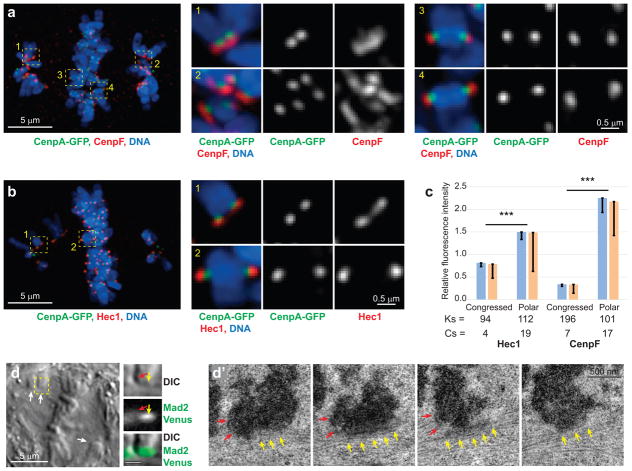
Figure 4. Kinetochore outer layer compaction occurs upon the formation of end-on microtubule attachments.
(a,b) Kinetochores are enlarged on polar, but compact on congressed, chromosomes. Whole-cell images are maximal-intensity projections that include all kinetochores in the cell. Individual kinetochores are shown as maximal-intensity projections. CenpF delineates the outer layer (a) and Hec1 – the kinetochore cores (b). (c) Amounts of both CenpF and Hec1 are increased on polar chromosomes. Blue bars are mean kinetochore intensity calculated as mean of mean values for multiple kinetochores in individual cells (n values are listed below the bars. Cs: cells). Error bars represent s.e.m. Yellow bars are mean values calculated for all Ks kinetochores pooled from all cells in that class (n values listed below the bars. Ks; kinetochores). Error bars represent s.d. Triple asterisks denote differences with p < 0.005 (two-tailed Student’s test) for both blue vs. blue and yellow vs. yellow bars. (d, d′) Sister kinetochores on congressing chromosomes display distinctly different morphologies. (d) A single-plane DIC image of the cell near the centre of mitotic spindle. Arrows denote congressing chromosomes. The centromere region of one congressing chromosome (yellow box) is also shown at higher magnification in DIC and Mad2-Venus fluorescence (bar = 1 μm). Arrows point at the approximate positions of the leading (yellow) and trailing (red) kinetochores. (d′) Consecutive 80-nm electron microscopy sections through the centromere of the chromosome shown in (d). Notice the size difference between the trailing end-on attached kinetochore (red arrows) vs. the leading kinetochore (yellow arrows) that lacks end-on attachment but interacts with microtubules laterally. The cells are treated with 15-nM GSK 923295, a cell-permeable inhibitor of CenpE to slow down chromosome congression and conversion from lateral to end-on microtubule attachments.
In GSK-923295-treated RPE cells with one allele of Mad2 replaced with Mad2-Venus22, at least one kinetochore on each mono-oriented chromosome remains Mad2-positive. Correlative LM/EM analysis of 31 Mad2-positive kinetochores in 4 cells demonstrates that these kinetochores lack end-on microtubule attachments but interact laterally with the walls of adjacent microtubules. The outer layer of these kinetochores is enlarged to 400–500 nm and is either spread alongside an adjacent microtubule bundle (Fig. 4d) or partially encircles the centromere (see Fig. 3d). In contrast, end-on attached kinetochores in the same cells are ~200 nm discs (Fig. 4d′). This difference in the architecture of end-on attached vs. laterally interacting kinetochores suggests that kinetochore compaction occurs only after the formation of end-on microtubule attachment and independently in sister kinetochores.
Intensity measurements demonstrate that kinetochores of congressed chromosomes in GSK-923295-treated cells contain as much Hec1 as in untreated metaphase cells (cf., Fig. 4c and 1f) while the amount of CenpF decreases to ~50% of the normal metaphase level (cf., Fig. 4c and 1c, p < 0.001). In contrast, the amount of both CenpF and Hec1 on polar chromosomes is higher than seen during any stage of normal mitosis (cf. Fig. 4c, 1c, and 1f, all p <0.001 in each comparison). This difference suggests that kinetochores enlarge if end-on attachment is delayed. Similar increases in fluorescence intensity and morphological enlargement of the outer layer are observed when cells are arrested in the absence of microtubules (Supplementary Fig. 3).
Large compactable kinetochores support rapid and low-error microtubule capture
Our observations that the outer layers of sister kinetochores rapidly expand to encircle the centromere at the onset of spindle assembly and subsequently compact into small discs upon formation of end-on microtubule attachments are consistent with previous reports in various types of mammalian9,12,23 and fly cells24,25. However, in the context of the S&C mechanism, kinetochore enlargement at the onset of mitosis appears counterproductive because large kinetochores are expected to increase attachment errors. Indeed, kinetochore enlargement during mitotic arrest is postulated to drive the drastic increase in attachment errors in nocodazole wash-out experiments26.
To explore potential effects of kinetochore enlargement on the number of erroneous attachments we employed computational modelling. In minimalistic stochastic models of spindle assembly, randomly located chromosomes are expected to form stable end-on microtubule attachments instantaneously when a growing microtubule plus end runs into a kinetochore. Microtubule capture by sister kinetochores is uncorrelated and the kinetochore that has already attached to a microtubule can acquire an additional connection if it is hit by another microtubule. Thus, after the initial formation of monotelic attachment, the next capture is by chance correct or erroneous (results in syntelic or merotelic attachments)27–30.
A major limitation of these simplistic models is that they predict unrealistically long times for spindle assembly due to a low probability of encounters between microtubules and 200-nm small kinetochores27. Our experimental observations suggest that the size of the capture target was underestimated in these models, which prompted us to replace small discoid kinetochores with large crescents (Supplementary Fig. 4a) that become compact after microtubule attachment. The duration of compaction (τcomp, Supplementary Fig. 4b) is not directly revealed in our experimental analyses. Therefore we explored various compaction times and found that this parameter does not significantly affect the time of spindle assembly or the number of errors (Supplementary Fig. 4d,d′). Enlarged kinetochores accelerate the time of spindle assembly to 5–11 minutes (Supplementary Fig. 4e), which is in agreement with the observation that the metaphase plate forms in ~8 min in RPE1 cells16. However, the number of erroneous attachments predicted by this simulation is large (>30%, Supplementary Fig. 4e).
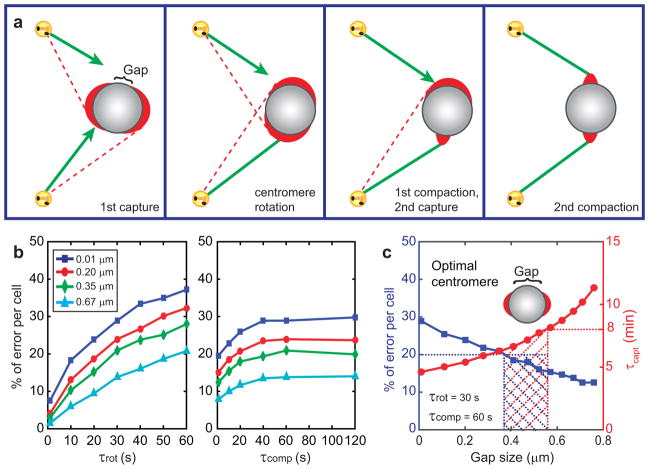
We have previously hypothesized that the number of erroneous attachments is reduced by the rotation of the chromosome that occurs immediately upon the initial capture28. This rotation orients the centromere so that its axis (the line connecting the centres of sister kinetochores) becomes roughly parallel to the captured microtubule (Fig. 5a). As a result, the unattached sister kinetochore is less likely to capture microtubules from the same spindle pole as its sister, which suppresses attachment errors28.
Figure 5. Effects of kinetochore enlargement-compaction on the efficiency and fidelity of capture-driven spindle assembly.
(a) Cartoon of the events envisioned in this model. Kinetochores are shown as red crescents. Green lines represent properly attached microtubules, red lines - potential erroneous attachments. (b) Error frequencies predicted at various gap sizes for various rotation and compaction times. (c) The frequency of errors and duration of spindle assembly for different gap sizes at specific rotation and compaction times. Shaded area represents the range of parameters that result in all chromosomes attaching in <8 min with <20% of errors. Predicted optimal geometry of the centromere is shown as inset in (c). Notice that the predicted optimal geometry is not similar to the centromere architecture observed in prometaphase cells (Figure 3).
Simulations that consider both rotation and compaction of initially large crescent-shaped kinetochores suggest that rapid rotation significantly reduce the errors. Such rapid rotations (~30 s) have been directly observed in live cells16. Similarly, the errors are suppressed if kinetochores compact in <60 s (Fig. 5b). At the conservatively pessimistic parameters of τrot = 30s and τcomp = 60s, the model predicts similar capture times but lower error rates (12–30%, Fig. 5c) than simulations without the rotation. However, due to inverse relationship between the speed of spindle assembly and the number of errors, efficient mitosis seems to require a specific and highly uniform geometry of the centromere (Fig. 5c). For example, spindle assembly in ≤8 min at the error rate ≤20% is predicted for a narrow range of gaps between sister kinetochores (0.35–0.55 μm, Fig. 5c). This prediction is ill compatible with the robustness of cell division31 and the observed variability in the centromere architecture (Fig. 3f). Specifically, the 0.1–0.2 μm gaps observed on many virgin centromeres (Fig. 3f) would lead to >25% errors.
Lateral interactions that precede formation of end-on attachments increase robustness of spindle assembly
Difficulties in reconciling simulations with experimental observations prompted us to reevaluate a fundamental assumption embedded in all existing computational models of spindle assembly – that the formation of microtubule attachment is a single-step process. In-vivo observations reveal that formation of end-on attachments is preceded by lateral interactions between the kinetochores and the walls of microtubules4,16,32,33. These interactions dominate during early prometaphase when centromere axes become partially aligned with the spindle axis16,33. The extent of this angular alignment is similar in cells rendered incapable of forming of end-on attachments34 by depletions of the NDC80 component Nuf216. Thus the angular alignment is driven primarily, if not exclusively, by lateral interactions between kinetochores and microtubules.
To gain further insights into the role of lateral interactions during incorporation of a chromosome into the spindle, we used flattened RPE1 cells that express fluorescent fusion of the checkpoint protein Mad222. Correlative LM/EM demonstrates that lateral interactions do not remove Mad2 from the kinetochores (Fig. 4), which provides a readout for the formation of end-on attachment. The prolonged mitosis in flattened cells, with some chromosomes attaching to microtubules only during late prometaphase35, enables us to follow the behaviour of individual chromosomes during their incorporation into the spindle. Time-lapse recordings of 26 chromosomes in 17 cells demonstrate that centromeres rotate to roughly align with the spindle axis while Mad2-Venus is still present on both sister kinetochores (Fig. 6). The disappearance of Mad2 from the kinetochores occurs 5–15 min after the centromere becomes roughly aligned with the spindle axis and initiates congression to the spindle equator (Fig. 6, Supplementary Video 1). This behaviour supports the idea that centromere rotation precedes formation of end-on attachments.
Figure 6. Centromere rotation on the surface of the spindle precedes formation of end-on microtubule attachment.
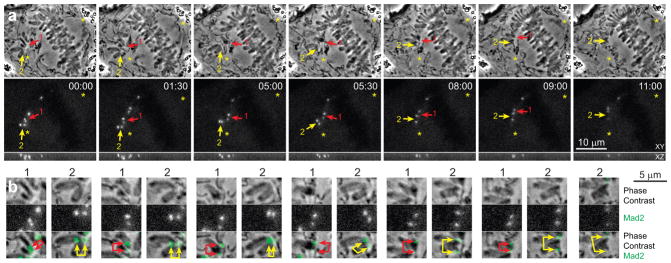
(a) Selected frames from a multi-mode time-lapse recording of a RPE1 cell flattened to 3-μm (See Supplementary Video 1 for full recording). Top row shows phase contrast (medial slice) and the bottom row – Mad2-Venus fluorescence (maximal intensity projections). Both kinetochores on monooriented chromosomes are Mad2-positive (arrows) indicating the absence of end-on attachments. Rapid rotation that orients centromere axes roughly parallel to the spindle axis precedes chromosome congression and release of Mad2 from the kinetochores. (b) Higher-magnification view of the centromere marked 1 (red arrows) and 2 (yellow arrows). Notice that the centromeres become stretched only during congression shortly before the release of Mad2 from the kinetochores. The cell is in a chamber that restricts mitotic rounding to ~3 μm.
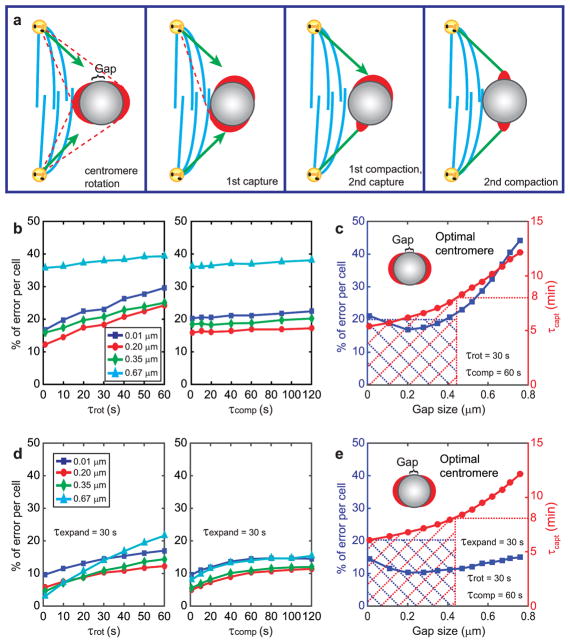
Based on these observations, we modified the model to incorporate a two-step process in which centromere rotation, mediated by lateral kinetochore-microtubule interactions, precedes microtubule end-on capture (Fig. 7a). We postulate that the centromere rotates until the interacting microtubule reaches the edge of the kinetochore at the gap (Supplementary Fig. 5a). Lateral interactions are expected to initiate within seconds after NEB16.
Figure 7. Computational models that consider centromere rotation due to lateral interactions with microtubules predict experimentally-observed parameters of spindle assembly.
(a) Cartoon of the events envisioned in the model. Blue lines represent the central part of the nascent spindle with high density of microtubules and devoid of chromosomes. Kinetochores can glide alongside of these microtubules resulting in a rotation of the centromere. Green lines represent properly end-on attached microtubules, red lines - potential erroneous attachments. (b–c) Results of the simulation that considers a single centromere rotation that takes place prior to end-on microtubule attachment and is driven by lateral interactions. (b) Error frequencies predicted for various gap sizes at various rotation and compaction times. (c) Error frequencies and durations of spindle assembly for various gap sizes at conservatively estimated rotation and compaction times. (d–e) Results of the all-inclusive simulation that considers expansion and compaction of the kinetochore as well as two subsequent rotations: one driven by lateral interactions (as in b–c) and one that results from the end-on microtubule attachment (as in Fig. 5b–c). (d) Error frequencies predicted for various gap sizes at 30-s expansion time and various rotation and compaction times. (c) Error frequencies and durations of spindle assembly for various gap sizes at specific expansion, rotation, and compaction times. Shaded areas in (c) and (e) mark the range of parameters that result in all chromosomes attaching in <8 min with <20% of errors. Predicted optimal geometry of the centromere is shown as insets in (c) and (e). Notice the similarity of the predicted optimal geometry with the experimental observations (Figure 3).
Simulations that consider centromere rotations prior to end-on attachment predict that larger kinetochores increase both the speed and accuracy of the spindle assembly. Unlike in all previous models, the relationship between the speed and accuracy of spindle assembly is not inverse and the <20% of error rate is predicted for a much wider range of gap sizes than in the ‘attachment-rotation’ scenario (cf. Fig. 7c and 5c). Incorporation of both types of centromere rotation (one driven by lateral interactions and one resulting from end-on attachment) into the model decreases the number of errors to ~10–15%, and the error rate becomes largely insensitive to the kinetochore size. With any gap size <0.5 μm, the assembly time is <8 min, and the error rate is <15% (Fig. 7e). Interestingly, the lowest error rate (Fig. 7e) is predicted when the gap between sister kinetochores is the commonly observed ~0.2-μm (Figs. 2,3). Thus, lateral interactions that precede formation of end-on attachments increase robustness of spindle assembly.
Deviant geometry of the nascent spindle increases chromosome mis-segregation
Our model suggest that the kinetochore architecture is adapted for spindle assembly that involves partial angular alignment of centromeres which normally occurs on the surface of a nascent spindle prior to the formation of end-on attachments16. If pre-alignment is impeded, enlarged kinetochores would lead to an increase in the number of errors (Fig. 5c). This prompted us to evaluate the organization of the early-prometaphase spindle in cells that mis-segregate chromosomes.
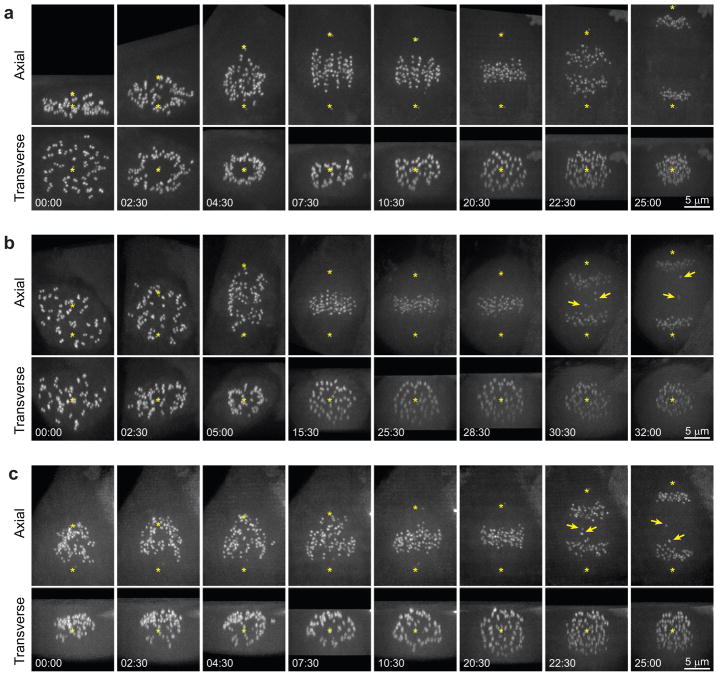
The largest frequency of centromere rotations is observed during early prometaphase16 when the centromeres reside on the surface of a nascent spindle (Fig. 8a). This pattern forms in the vast majority of RPE1 cells (>90% [47/50]). We find that those untreated RPE1 cells that fail to form the clear zone (3/50) are prone to display lagging chromosomes during anaphase (Fig. 8b). To experimentally impede formation of the clear zone, we treated RPE1 cells with 3-μM nocodazole (for <30 min), located cells that just entered mitosis, washed out the drug, and followed formation of the spindle by 3-D time-lapse microscopy. Under these conditions, the clear zone consistently fails to form (n=21) and the centromeres are intermixed with microtubules, which impedes angular alignment of the centromeres. 43% of the nocodazole-treated cells (9/21) display lagging chromosomes during anaphase-telophase (Fig. 8c). Therefore, a lack of a clear zone in the centre of the nascent spindle correlates with an increased frequency of errors.
Figure 8. Abnormal geometry of the nascent spindle during early prometaphase correlates with erroneous chromosome segregation.
(a) Typical pattern of spindle formation (RPE1 cell). Notice that all centromeres (CenpA-GFP) reside on the surface of the nascent spindle for the first 6–8 min of prometaphase. (b) Example of an untreated RPE1 cell where the formation of the clear central zone fails for unknown reasons. The centromeres are interspersed from the onset of spindle assembly. Although all chromosomes congress onto the metaphase plate with only a minor delay, several chromosomes lag behind during anaphase (arrows) which is indicative of erroneous kinetochore attachments. (c) Spindle assembly after nocodazole washout. Formation of the clear central zone does not occur and chromosomes are lagging during anaphase (arrows). Asterisks mark mother centrioles (labelled with centrin-GFP). To ensure the detection of the clear zone each time-point is rotated and shown as maximal intensity projections in precisely axial and transverse orientations. See Supplementary Figure 6 and Videos 3–5 for conventional views of these cells. Time in minutes: seconds from NEB (a, b) or completion of nocodazole washout (c).
DISCUSSION
Direct observations of microtubule capture by kinetochores4,32 have established S&C5 as the basic principle of spindle assembly. Multiple mechanisms such as the spatially selective stabilization of microtubules by RanGTP36, the rotation of chromosomes28, the formation of a specific spatial arrangement during early prometaphase16, and the sweeping movements of growing microtubules37 promote microtubule capture and thus accelerate spindle assembly. However, the inverse relationship between the efficiency and accuracy of S&C-driven spindle formation inherent in all previous computational models is incompatible with the well-established robustness of mitotic regulations.
Our observation that the kinetochore outer layer expands at the onset of mitosis, demonstrates that previous computational models underestimated the size of microtubule-capturing target. By introducing adaptable kinetochore geometry and pre-alignment of centromeres due to rapid lateral kinetochore-microtubule interactions prior to the formation of end-on microtubule attachments4,16,32,33,38, we have constructed the first model that predicts realistically-rapid spindle assembly with error rates that are sufficiently low to be handled by error-correction mechanisms26,39,40. A non-trivial prediction of our model is that kinetochore expansion during the phase of spindle assembly when lateral interactions dominate, creates a synergistic relationship between the efficiency and fidelity of spindle assembly. This can explain the robustness of the process in spite of the inevitable size variation of individual kinetochores in real cells. A corollary of this prediction is that conditions that impede domination of lateral interactions during the initial stages of spindle assembly increase the number of erroneous attachments.
Centromere alignment via lateral interactions primarily takes place when centromeres reside on the surface of the nascent spindle16,33. Therefore, conditions that affect the formation of the hollow spindle during early prometaphase are deleterious for chromosome segregation. Consistent with this notion, attachment errors are consistently observed in cells depleted for chromokinesins41,42, condition that has been shown to disrupt formation of the ring16. Further, chromosomes tend to mis-segregate when microtubules and centromeres become intermixed during early prometaphase due to ‘reversible’ drug treatments (Fig. 8) or transient deviations from the proper geometry in cells with abnormal centrosomal activity43–46. Subtle and transient changes in the geometric constraints during the initial stages of spindle assembly may still have devastating consequences for genomic stability even when the architecture of the mature metaphase spindles is not directly affected.
METHODS
Methods and any associated references are available in the online version of the paper.
Note: Supplementary Information is available in the online version of the paper
METHODS
Cell Culture, chemical treatments, and live-cell microscopy
Human non-transformed hTERT-RPE1 cell line was purchased from Clontech in 2001 at passage number 118.5. Stocks of these cells at passage numbers 120–122 were generated in the Khodjakov lab and kept in liquid nitrogen. A stable clone (RPE1-18), that co-expresses CenpA-eGFP and centrin1-eGFP (both introduced via lentivirus)47, was used in most of the experiments described here. Experiments that required visualization of fluorescent Mad2 were conducted in the RPE1Mad2/Mad2-Venus cell line provided by Dr. Jonathan Pines, University of Cambridge48. All cell lines were grown in antibiotic-free DMEM supplemented with 10% FCS (Invitrogen) at 37°C, 5%CO2. For live-cell imaging, cells were grown on glass coverslips (#1½) and mounted in Rose chambers containing CO2-independent media (Invitrogen) supplemented with 10% FCS. In-house tests for mycoplasma (high-concentration Hoechst staining) are negative.
Microtubule depolymerization was induced by nocodazole (Sigma) at 3 μM. Motor activity of CenpE was inhibited with GSK-92329549 purchased from Haoyuan Chemexpress (Shanghai).
Multi-mode 3-D time-lapse recordings were obtained on a Nikon TE-2000E PFS microscope with 100X Plan Apo, N.A. 1.4 oil immersion objective lens. Fluorescence images were captured in a spinning-disc confocal mode (GSU-10, Yokogawa) on a back-illuminated Cascade 512B EM CCD camera (Photometrics). DIC images were recorded on a Photometrics CoolSnap CF camera mounted on a different port of the same microscope. Full 3-D volumes were recorded at each time point at 250-nm Z-steps (48–62 planes depending on cell thickness).
To visualize formation of the clear zone we first tracked 3-D positions of mother centrioles and then rotated the 3D volume at each time point to fix position of one mother centriole and orientation of the spindle axis50. This processing allowed us to observe chromosome movement in the precisely transverse and axial views. Mother centrioles were tracked FIJI with the standard tracking plugin. 3-D coordinates of the centrioles and the images were then imported into MatLab. The image volume was padded with black (0 value) voxels to prevent cropping during rotation. The rotation was done in two sequential steps, first in XY and then in Z via a custom MatLab script. Rotated and aligned images were transferred back to FIJI. Maximal intensity projections of the entire rotated volume were generated along the spindle axis (transverse view) and orthogonally to the spindle axis (axial view). Each view presented in Fig. 8 contains both centriole pairs and all kinetochores.
Incorporation of individual chromosomes into the spindle was observed under conditions that prevented cell rounding during mitosis. A coverslip with 3-μm microfabricated feet was placed on top of the coverslip with the growing cells. The contact between coverslips was maintained with negative pressure using a vacuum pump51.
Fixed-cell immunofluorescence
Cells were pre-extracted in warm PEM buffer (100-mM PIPES, pH 6.9, 2.5-mM EGTA, 5-mM MgCl2) supplemented with 0.5% Triton X-100 for 1 min and fixed with 1%–2% glutaraldehyde for 10 min in PEM. Microtubules were visualized with a monoclonal anti-α-tubulin antibody (DM1a, Sigma; 1:200 dilution). Kinetochores were delineated with the following antibodies: rabbit αCenpF (Novus Biologicals, NB500-101; 1:400 dilution), mouse αCenpE antibody (Abcam, ab5093; 1:200 dilution), mouse αHec1 (Abcam, ab3613; 1:200 dilution), and rabbit αMis12 (kindly provided by Dr. Ian Cheeseman; 1:400 dilution)52. Hoechst 33343 (1 μg/ml) was used to stain DNA (chromosomes). Inner kinetochores were visualized via CenpA-GFP fluorescence.
Wide-field images were recorded on a DeltaVision imaging system (Applied Precision) with a 100X NA1.35 lens (Olympus). The images were captured with a CH-350 CCD camera (Photometrics) at a 69-nm X-Y pixel size and 200-nm Z-steps. All images were deconvolved with the SoftWoRx 5.0 deconvolution software (Applied Precision) and objective lens-specific point spread functions.
Amira software (FEI) was used for surface rendering. Segmentation threshold for fluorescence images (Fig. S2) was set at 25% of maximal intensity for each dataset.
Quantification of fluorescence intensity and kinetochore volume
All measurements were conducted in ImageJ/FIJI and calculations - in MS Excel. Integrated fluorescence intensity was measured within a 3-D volume centred on a single kinetochore (whenever possible) or a small group of kinetochores (if their individual signals were not fully resolvable). The dimensions of the volume were set individually to include the entire object of interest (300–2500 voxels, 10×10×3 – 25×25×4 volumes). Background intensity for each measurement was measured in the same-dimensions volume positioned as close as possible to the object of interest. Kinetochore intensity was calculated by subtracting background intensity and dividing the result by the number of kinetochores in the volume. Intensities of multiple kinetochores (usually ~20) were measured in each cell. Mean fluorescence intensity per kinetochore was calculated for individual cells and then the mean value of per-cell averages was calculated. Alternatively, all kinetochores measured under a particular experimental condition were pooled together and the mean value was calculated for this pooled population. Results of both calculations are presented in the figures (Ks = total number of kinetochores; Cs = total number of cells). All values are normalized so that the mean intensity at NEB equals to 1.
Kinetochore volumes were measured via “3D Object Counter” routine included in the standard distribution of FIJI. As the amount of CenpF and Mis12 remains constant during late prophase-prometaphase, relative volumes occupied by kinetochores can be segmented at a constant threshold. Threshold values for segmentation were set at 20% of maximal signal intensity for CenpF and 25% for Hec1 datasets. These thresholds were empirically determined to yield maximal numbers of kinetochores per cell with minimal contamination by false objects after segmentation. As in intensity calculations, both mean values were calculated for per cell averages and for the pooled populations (both values are presented in the Figures). All values are normalized so that the mean volume at NEB equals to 1.
Mean values were compared in two-tailed Student’s test.
Correlative Electron Microscopy
Cells were fixed in 2.5% glutaraldehyde (Sigma) in PBS (pH 7.4–7.6). Differential interference contrast and fluorescence images were acquired at 0.2-μm Z steps through the entire cell volume shortly after fixation. Post-fixation, embedding, and sectioning were done as previously described53. Serial 80-nm thin sections were imaged at 80 kV on either a Zeiss 910 (Carl Zeiss) or JEOL 1400. Correlation of conspicuous morphological features between differential interference contrast and EM images was used to match the orientation and Z positions for individual focal planes and determine exact kinetochore positions.
Computational modelling
Microtubule dynamics during spindle assembly
We consider that spindle assembly takes place in the spherical volume that was occupied by the nucleus prior to NEB. Implicitly, we assume, following Wollman and coworkers (2005), that the RanGTP gradient focuses the MTs into the nuclear sphere. This accelerates the search a few-fold but has no effect on the error rate. Two centrosomes are placed at the opposite poles of the sphere at −Rcell and +Rcell positions. Each centrosome nucleates NMT microtubules that search the space isotropically. Each microtubule is represented by a rod with zero thickness that undergoes dynamic instability. The plus end of a microtubule grows steadily until a catastrophe occurs leading to microtubule shortening. The frequency of catastrophe, as well as the growth and shrinkage rates, is constant (the search with zero and small finite stochastic catastrophe frequencies have been tested and the results were not sensitive to this variation of the model). We use the optimal zero rescue frequency54. Microtubule dynamics are simulated by the Monte Carlo algorithm: a random number is generated between 0 and 1 with equal probability. At each computational step (with time increment Δt = 1 sec) the microtubule switches to shortening if this random number is less than [1−exp(−fcatΔt)]. New microtubules grow in random directions and do not turn. In all cases, if a microtubule plus end extends beyond the nuclear sphere’s boundary or encounters a chromosome arm, this microtubule undergoes catastrophe and shrinks all the way back to the centrosome.
The values for the number of microtubules generated by each centrosome [NMT] and the four parameters of dynamic instability [vg, vs, fcat, fres] used in the simulations are presented in Supplementary Table 1. The radius of the nuclear/spindle sphere is set to match the geometry of mitosis in RPE1 cells47. Effects of the microtubule dynamic instability parameters have been previously explored and discussed54. In the current simulations, conservative values from the range explored in reference54 are used for vg and vs. The number of microtubules in the current simulations (600) is approximately twofold higher than in previous models (250)54. This change is introduced to account for the difference in spindle assembly time between HT-29 (15 min54) and RPE1 (8 min47) cells. The model indeed predicts slower absolute assembly time if the number of microtubules is lowered. However, the differences between the predictions in the three considered scenarios (see below) are not affected by the number of microtubules: relative differences in the assembly time as well as the predicted number of errors remain the same.
Nascent spindle
A second set of stable microtubules runs along the spindle axis and overlaps in the central part of the spindle. Based on microscopy data this dense microtubule array forms shortly after NEB (1–2 min) and persists through prometaphase47. The centromeres become positioned on the surface of the nascent spindle shortly (~2 min) after NEB (Fig. 7 and ref.45) and laterally interact with microtubule walls. In contrast to end-on attachments, lateral interactions can occur along the entire length of microtubule and there is no evidence that these interactions require microtubules to undergo plus-end dynamic instability. Therefore, the nascent spindle in our simulations comprises stable microtubules whose plus ends do not contribute to capture. Geometry of the nascent spindle is derived from previously published data47.
Microtubule capture
A microtubule plus end is instantly captured and stabilized upon encountering a kinetochore. Upon capture, a new dynamic microtubule is nucleated at the same pole to replace the stabilized one.
Chromosomes and kinetochores
Chromosomes are modelled as solid 3-D cylinders with RCH radius and Ich length (Supplementary Table 1). The initial distribution of chromosomes in the nuclear sphere and their orientation are random. Prior to capture, sister kinetochores are modelled as crescent-shaped objects (see Supplementary Table 1 and Figure S3 for dimensions) wrapped around the central part (equator) of the chromosome. The width wKT and length hKT of kinetochores are kept constant. Upon capturing a microtubule, the kinetochore crescent condenses into a small cylindrical object in τcomp time. This kinetochore geometry reflects our experimental observations (Figures 1–4).
To complete computations with a reasonable time and avoid difficulty in tracking steric inter-chromosomal interactions, single chromosomes positioned at a fixed distance away from the pole-pole axis are considered in individual simulations. The simulations are then repeated for multiple chromosome positions and random orientations. To obtain the average value of the capture time (τcapt), we multiplied the capture time of a single chromosome by the logarithm of the total number of chromosomes NCH (Supplementary Table 1).
Tested models
Four different scenarios are considered:
Completely random distribution and orientation of chromosomes and kinetochores that remain unaltered upon microtubule capture. Kinetochores are shaped as crescents at the onset of spindle assembly and compact from crescents to discs in τcomp after capture.
Microtubule capture leads to rotation of the chromosome and alignment of the centromere axis (line connecting centres of sister kinetochores) along the captured microtubule in time τrot. Kinetochores are shaped as crescents at the onset of spindle assembly and compact from crescents to discs in τcomp after capture. Because the chromosome arms are largely normal to the pole-pole axis, the rotation primarily occurs around their longitudinal axis.
Rapid lateral interactions with the stable microtubules of nascent spindle result in a rapid decrease of the angle between the centromere axis and spindle axis (line connecting the centrosomes) in time τrot. The angle of chromosome rotation is limited by the ability of kinetochores to maintain direct contact with stable microtubules, which in turn depends on the size of the kinetochore crescent. Kinetochores are shaped as crescents at the onset of spindle assembly and compact from crescents to discs in τcomp after capture. Compaction initiates only after end-on microtubule capture and completes in τcomp.
At the beginning of the search, the crescents are small, with the initial gap size of 0.76 μm. In the next 30 s, the crescents grow linearly so that the gap decreases to its final size (0.01 to 0.76 μm in various simulations). The time of crescent growth is constant, irrespective of the final gap size. Lateral interactions decrease the angle between the centromere axis and the spindle axis as in scenario iii; respective rate of rotation is very fast, a few seconds, in this case. Then, the end-on microtubule capture leads to additional rotation of the chromosome and alignment of the centromere axis (line connecting centres of sister kinetochores) along the captured microtubule in time τrot. Kinetochores compact from crescents to discs in τcomp after end-on microtubule capture.
In the most simplistic simulations (scenario i), microtubule capture is not expected to change the position or orientation of the chromosome55–57. Rotation of the centromere considered in scenarios (ii)54 and (iii) inevitably shifts the chromosome from its original position. However, in 80% of the cases the mean value of the displacement caused by the brief rapid movement during the initial interaction between kinetochores and microtubules is <1 μm in RPE1 cells47. Such a small translation does not significantly affect the probability of subsequent microtubule capture. Larger translations are rare and therefore not considered in the current model.
All simulations are carried out for various gap sizes between the crescent sister kinetochores (Supplementary Table 1). Computational data presented in the manuscript are obtained from running simulations for each set of the parameters at least 1000 times.
Computer code
The numerical codes are implemented with C programming language. Numerical experiments are performed on an IBM quad core Intel CPU server. The code of the simulation is available as Supplementary Data.
Supplementary Material
Acknowledgments
This work was supported by NIH grant GM059363 to A.K and NSF grant DMS-1118206 to A.M. The Electron Microscopy was enabled by the use of the Wadsworth Center’s Electron Microscopy Core Facility. We thank Dr. Jonathan Pines (University of Cambridge, UK) for his generous donation of Mad2-Venus cells and Dr. Sizheng Li (Air Worldwide) for assistance with the intensity quantifications.
Footnotes
AUTHOR CONTRIBUTIONS
AK and BFM designed the experiments, VM, NY, CBO, and IT performed the experiments. JGA, BFM, AK, and IT conducted correlative LM/EM. AM and RP developed the computational models. RP designed MatLab code and performed computer simulations. The manuscript was written by AK, BFM, and AM.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.McIntosh JR, Molodtsov MI, Ataullakhanov FI. Biophysics of mitosis. Q Rev Biophys. 2012;45:147–207. doi: 10.1017/S0033583512000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walczak CE, Cai S, Khodjakov A. Mechanisms of chromosome behaviour during mitosis. Nature Reviews Molecular Cell Biology. 2010;11:91–102. doi: 10.1038/nrm2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayden JH, Bowser SS, Rieder CL. Kinetochores capture astral microtubules during chromosome attachment to the mitotic spindle: direct visualization in live newt lung cells. The Journal of Cell Biology. 1990;111:1039–1045. doi: 10.1083/jcb.111.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka K, et al. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- 5.Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- 6.Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinkley BR, Stubblefield E. The fine structure of the kinetochore of a mammalian cell in vitro. Chromosoma. 1966;19:28–43. doi: 10.1007/BF00332792. [DOI] [PubMed] [Google Scholar]
- 8.McEwen BF, Ding Y, Heagle AB. Relevance of kinetochore size and microtubule-binding capacity for stable chromosome attachment during mitosis in PtK 1 cells. Chromosome Research. 1998;6:123–132. doi: 10.1023/a:1009239013215. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK 1 kinetochores. Molecular Biology of the Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McEwen BF, Arena JT, Frank J, Rieder CL. Structure of the colcemid-treated PtK1 kinetochore outer plate as determined by high voltage electron microscopic tomography. The Journal of Cell Biology. 1993;120:301–312. doi: 10.1083/jcb.120.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cimini D, et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. The Journal of Cell Biology. 2001;153:517–528. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thrower DA, Jordan MA, Wilson L. Modulation of CENP-E organization at kinetochores by spindle microtubule attachment. Cell Motility & the Cytoskeleton. 1996;35:121–133. doi: 10.1002/(SICI)1097-0169(1996)35:2<121::AID-CM5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 13.Feng J, Huang H, Yen TJ. CENP-F is a novel microtubule-binding protein that is essential for kinetochore attachments and affects the duration of the mitotic checkpoint delay. Chromosoma. 2006;115:320–9. doi: 10.1007/s00412-006-0049-5. [DOI] [PubMed] [Google Scholar]
- 14.Cooke CA, Schaar B, Yen TJ, Earnshaw WC. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma. 1997;106:446–455. doi: 10.1007/s004120050266. [DOI] [PubMed] [Google Scholar]
- 15.Kim Y, Heuser JE, Waterman-Storer CM, Cleveland DW. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. The Journal of Cell Biology. 2008;181:411–419. doi: 10.1083/jcb.200802189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magidson V, et al. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loncarek J, et al. The centromere geometry essential for keeping mitosis error free is controlled by spindle forces. Nature. 2007;450:745–749. doi: 10.1038/nature06344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood KW, et al. Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5839–44. doi: 10.1073/pnas.0915068107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapoor TM, et al. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai S, O’Connell CB, Khodjakov A, Walczak CE. Chromosome congression in the absence of kinetochore fibers. Nature Cell Biology. 2009;11:832–838. doi: 10.1038/ncb1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shrestha RL, Draviam VM. Lateral to end-on conversion of chromosome-microtubule attachment requires kinesins CENP-E and MCAK. Current biology. 2013;23:1514–26. doi: 10.1016/j.cub.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collin P, Nashchekina O, Walker R, Pines J. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat Cell Biol. 2013 doi: 10.1038/ncb2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roos UP. Light and electron microscopy of rat kangaroo cells in mitosis. II. Kinetochore structure and function. Chromosoma. 1973;41:195–220. doi: 10.1007/BF00319696. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein LS. Kinetochore structure and its role in chromosome orientation during the first meiotic division in male D. melanogaster. Cell. 1981;25:591–602. doi: 10.1016/0092-8674(81)90167-7. [DOI] [PubMed] [Google Scholar]
- 25.Church K, Lin HP. Kinetochore microtubules and chromosome movement during prometaphase in Drosophila melanogaster spermatocytes studied in life and with the electron microscope. Chromosoma. 1985;92:273–282. doi: 10.1007/BF00329810. [DOI] [PubMed] [Google Scholar]
- 26.Cimini D, Moree B, Canman JC, Salmon ED. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. Journal of Cell Science. 2003;116:4213–4225. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- 27.Wollman R, et al. Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly. Current Biology. 2005;15:828–832. doi: 10.1016/j.cub.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 28.Paul R, et al. Computer simulations predict that chromosome movements and rotations accelerate mitotic spindle assembly without compromising accuracy. Proceedings of National Academy of Sciences USA. 2009;106:15708–15713. doi: 10.1073/pnas.0908261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holy TE, Leibler S. Dynamic instability of microtubules as an efficient way to search in space. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:5682–5. doi: 10.1073/pnas.91.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:4404–8. doi: 10.1073/pnas.82.13.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones JT, Myers JW, Ferrell JE, Meyer T. Probing the precision of the mitotic clock with a live-cell fluorescent biosensor. Nat Biotechnol. 2004;22:306–12. doi: 10.1038/nbt941. [DOI] [PubMed] [Google Scholar]
- 32.Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. The Journal of Cell Biology. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitajima TS, Ohsugi M, Ellenberg J. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell. 2011;146:568–81. doi: 10.1016/j.cell.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 34.DeLuca JG, et al. Hec1 and Nuf2 Are Core Components of the Kinetochore Outer Plate Essential for Organizing Microtubule Attachment Sites. Molecular Biology of the Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lancaster OM, et al. Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation. Dev Cell. 2013;25:270–83. doi: 10.1016/j.devcel.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 36.O’Connell CB, Loncarek J, Kalab P, Khodjakov A. Relative contributions of chromatin and kinetochores to mitotic spindle assembly. The Journal of Cell Biology. 2009;187:43–51. doi: 10.1083/jcb.200903076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalinina I, et al. Pivoting of microtubules around the spindle pole accelerates kinetochore capture. Nat Cell Biol. 2013;15:82–7. doi: 10.1038/ncb2640. [DOI] [PubMed] [Google Scholar]
- 38.Merdes A, De May J. The mechanism of kinetochore-spindle attachment and polewards movement analyzed in PtK2 cells at the prophase-prometaphase transition. European The Journal of Cell Biology. 1990;53:313–325. [PubMed] [Google Scholar]
- 39.Nicholson JM, Cimini D. How mitotic errors contribute to karyotypic diversity in cancer. Adv Cancer Res. 2011;112:43–75. doi: 10.1016/B978-0-12-387688-1.00003-X. [DOI] [PubMed] [Google Scholar]
- 40.Silkworth WT, Cimini D. Transient defects of mitotic spindle geometry and chromosome segregation errors. Cell Div. 2012;7:19. doi: 10.1186/1747-1028-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazumdar M, Sundareshan S, Misteli T. Human chromokinesin KIF4A functions in chromosome condensation and segregation. The Journal of Cell Biology. 2004;166:613–620. doi: 10.1083/jcb.200401142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wandke C, et al. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. The Journal of Cell Biology. 2012;198:847–63. doi: 10.1083/jcb.201110060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore-microtubule dynamics underlie chromosomal instability. Current Biology. 2009;19:1937–1942. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kleylein-Sohn J, et al. Acentrosomal spindle organization renders cancer cells dependent on the kinesin HSET. J Cell Sci. 2012;125:5391–402. doi: 10.1242/jcs.107474. [DOI] [PubMed] [Google Scholar]
- 47.Magidson V, et al. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collin P, Nashchekina O, Walker R, Pines J. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat Cell Biol. 2013 doi: 10.1038/ncb2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood KW, et al. Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5839–44. doi: 10.1073/pnas.0915068107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sikirzhytski V, et al. Direct kinetochore-spindle pole connections are not required for chromosome segregation. The Journal of Cell Biology. 2014;206:231–243. doi: 10.1083/jcb.201401090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Berre M, Aubertin J, Piel M. Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr Biol (Camb) 2012;4:1406–14. doi: 10.1039/c2ib20056b. [DOI] [PubMed] [Google Scholar]
- 52.Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. The Journal of Cell Biology. 2006;173:9–17. doi: 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rieder CL, Cassels G. Correlative light and electron microscopy of mitotic cells in monolayer cultures. Methods in Cell Biology. 1999;61:297–315. doi: 10.1016/s0091-679x(08)61987-1. [DOI] [PubMed] [Google Scholar]
- 54.Paul R, et al. Computer simulations predict that chromosome movements and rotations accelerate mitotic spindle assembly without compromising accuracy. Proceedings of National Academy of Sciences USA. 2009;106:15708–15713. doi: 10.1073/pnas.0908261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wollman R, et al. Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly. Current Biology. 2005;15:828–832. doi: 10.1016/j.cub.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 56.Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:4404–8. doi: 10.1073/pnas.82.13.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holy TE, Leibler S. Dynamic instability of microtubules as an efficient way to search in space. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:5682–5. doi: 10.1073/pnas.91.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.