Abstract
Clofazimine (CFZ) is an optically active, red-colored chemotherapeutic agent that is FDA – approved for the treatment of leprosy and is on the World Health Organization's list of essential medications. Interestingly, CFZ massively accumulates in macrophages where it forms crystal–like–drug inclusions (CLDIs) following oral administration of the drug in animals and humans. Analysis of the fluorescence spectra of CLDIs formed by resident tissue macrophages revealed that CFZ, when accumulated as CLDIs, undergoes a red shift in fluorescence excitation (from Ex:540-570 nm to Ex:560-600 nm) and emission (Em: 560-580 nm to Em: 640-700 nm) signal relative to the soluble and free-base crystal forms of CFZ. Using epifluorescence microscopy, CLDI(+) cells could be identified, relative to CLDI(−) cells, based on a >3-fold increment in mean fluorescence signal at excitation 640 nm and emission at 670 nm. Similarly, CLDI(+) cells could be identified by flow cytometry, based on a >100-fold increment in mean fluorescence signal using excitation lasers at 640 nm and emission detectors >600 nm. CLDI's fluorescence excitation and emission was orthogonal to that of cell viability dyes such as propidium iodide and DAPI, cellular staining dyes such as Hoechst 33342 (nucleus) and FM 1-43 (plasma membrane), as well as many other fluorescently-tagged antibodies used for immunophenotyping analyses. In vivo, >85% of CLDI(+) cells in the peritoneal exudate were F4/80(+) macrophages and >97% of CLDI(+) cells in the alveolar exudate were CD11c(+). Most importantly, the viability of cells was minimally affected by the presence of CLDIs. Accordingly, these results establish that CFZ fluorescence in CLDIs is suitable for quantitative flow cytometric phenotyping analysis and functional studies of xenobiotic sequestering macrophages.
Keywords: CFZ, intracellular crystals, immunophenotyping, spectral microscopy
INTRODUCTION
Mammalian organisms possess various passive and active transport mechanisms that mediate the transport, distribution and elimination of foreign chemical agents such as environmental pollutants, toxicants as well as pharmaceutical agents, referred to as “xenobiotics”. Many xenobiotics are eliminated from the organism through a combination of excretory and metabolic pathways but there are some that are highly stable and therefore can bio-accumulate in the organism upon prolonged exposure. While these agents could potentially disrupt the normal physiological functions of cells and organs, organisms may also possess active cellular mechanisms that allow them to adapt to bio-accumulating chemical agents. In the latter category, several weakly basic drug molecules have been shown to accumulate in macrophages and some form aggregates with cellular membranes while others precipitate as insoluble intracellular complexes. Clofazimine (CFZ) is a model xenobiotic - a weakly basic, FDA approved drug for the treatment of leprosy (1) that accumulates in macrophages, forming crystal-like-drug-inclusions (CLDIs) (2–7). Following biochemical isolation it is possible to analyze the composition of CLDIs, and we have elucidated that CLDIs are comprised of domains of pure CFZ hydrochloride crystals (CFZ-HCl), bound and separated by membranes likely of cellular origin (8).
Previously, bioaccumulation of uric acid crystals in macrophages, and the phagocytosis of silica particles, have been linked to activation of pro-inflammatory signaling pathways in macrophages (9–12). Considering that CFZ possesses immunomodulatory activity (2,13,14), we decided to determine whether CLDIs may elicit similar changes in macrophage function by developing a method by which CLDI-containing macrophages could be isolated and functionally studied. Since in solution, CFZ is weakly fluorescent and its fluorescence is pH-sensitive as per the local microenvironment (5,15), we sought to determine whether formation of CLDIs in macrophages is associated with specific shifts in the fluorescence excitation and emission spectra that could be used to detect CLDI-containing macrophages. Here, we report how CLDI formation leads to a far-red shift in CFZ's fluorescence signal, that is distinctively different from the fluorescence of soluble CFZ as well as free-base CFZ crystals (CFZ-TC). Additional experiments demonstrate how this far-red shift in fluorescence of CLDI-containing macrophages facilitates immunophenotyping and functional analysis of these xenobiotic sequestering cells.
MATERIALS AND METHODS
Animal Experiments
Mice (4 week old, male C57Bl6) were purchased from the Jackson Laboratory (Bar Harbor, ME) and acclimatized for 1 week in a specific-pathogen-free animal facility. Animal care was provided by the University of Michigan's Unit for Laboratory Animal Medicine (ULAM) and the experimental protocol was approved by the Committee on Use and Care of Animals in accordance with NIH guidelines. An oral diet containing CFZ was fed to mice as previously described (2,3,8). CFZ (C8895; Sigma-Aldrich, St. Louis, MO, USA) was dissolved in sesame oil (Roland, China, or Shirakiku, Japan) to achieve a concentration of 3 mg/ml, which was mixed with Powdered Lab Diet 5001 (PMI International, Inc., St. Louis, MO, USA) to produce a 0.03% drug to powdered feed mix. A corresponding amount of sesame oil was mixed with chow for vehicle treatment (control mice). On average, food consumption for a 25 g mouse was 3 g per day, resulting in 10 mg of bioavailable drug/kg per day. For CFZ treatment, the drug diet was administered for 8 weeks followed by regular chow for 8 weeks (washout phase).
Isolation of CLDIs from Mouse Spleen
At 8 weeks post drug feeding, mice were euthanized by exsanguination while deeply anesthetized by an intraperitoneal injection of ketamine (100 mg/kg)/xylazine (10 mg/kg) and spleens were harvested and cut open to prepare tissue homogenate in phosphate-buffered saline (PBS). The tissue homogenate was sonicated for 30 minutes and centrifuged (100 × g for 1 minute) to remove large cell debris. A solution of 10% sucrose in PBS was added to the acquired supernatant and the mixture was centrifuged (100 × g). The resulting supernatant was centrifuged (21,000 × g for 1 min) to pellet drug inclusions which were then resuspended in 2 ml of 10% sucrose. CLDIs were further purified using a 3-layer discontinuous gradient (50%, 30% and 10% sucrose in PBS) centrifugation method (3200 × g for 30 min, no brakes) (14). The CFZ content of the isolated CLDIs was determined spectrophotometrically (λ=495 nm) by procuring 100 μl of CLDIs (in triplicate) by centrifugation (21,000 × g for 1 min) and dissolution in DMSO followed by comparison with calibrated CFZ standards.
Fluorimetry
CFZ was dissolved in DMSO to achieve a concentration of 20 μM. Fluorescence excitation and emission scans were done in increments of 10 nm from 400 nm to 800 nm on a Perkin-Elmer LS-55 fluorescence spectrometer using standard cuvettes. Data were imported into Microsoft® Excel (Redmond, WA, USA) (MS-Excel) for further analysis. The fluorescence yield was background-subtracted using data obtained from solvent alone (DMSO) and was normalized to the maximum fluorescence yield measured across the spectral wavelength range tested.
Spectral Confocal Microscopy
For preparation of slides, CFZ drug crystals were dusted on a glass slide followed by the application of a glass cover slip. For slides of CLDIs, a 20 μl drop of purified CLDIs was placed on a glass slide and allowed to dry overnight in the dark. The following day, a single drop of Prolong® Gold (Life Technologies, Carlsbad, CA) was added to the CLDIs and a cover slip was applied prior to imaging. Spectral confocal microscopy was performed on a Leica Inverted SP5X confocal microscope system with 2-photon FLIM (Leica Microsystems Inc., Buffalo Grove, IL) using excitation wavelengths (λ=470–670 nm). Image analysis and quantification was performed on Leica LAS AF. Several regions of interest (ROIs) of individual crystals were used to obtain fluorescence data which were imported into MS-Excel for further analysis. All fluorescence yields were normalized to the maximum fluorescence yield measured across the spectral range tested and background subtracted using data obtained from a blank slide.
Epifluorescence Microscopy
Visualization of all samples (cells or crystals) was done on a Nikon Eclipse Ti (Nikon Instruments Inc., Melville, NY, USA). The fluorescence filters (Excitation/Emission) used were DAPI (350/405 nm, exposure=50 ms), FITC (490/510 nm, exposure=100–500 ms), Texas Red (590/610 nm, exposure<500 ms) and Cy5 (640/670 nm, exposure=15 ms). Brightfield color photographs were acquired using a Nikon DS-Fi2 camera while fluorescence photographs were acquired using a Photometrics® CoolSNAP™ MYO (Photometrics, Tucson, AZ, USA) camera. CLDIs (seen as intense red pigmentation) were counted and analyzed for physical dimensions using the Nikon Elements software (Nikon Instruments Inc., Melville, NY, USA).
Identification of the CLDI Signal by Flow Cytometry in RAW264.7 cells
Macrophages phagocytose CLDIs isolated from mouse spleen following 8 weeks of CFZ treatment (14). RAW 264.7 cells (TIB-71™ ATCC, Manassas, VA) cells were maintained with DMEM + 10 % fetal bovine serum (FBS) (10082; Gibco®, Invitrogen, Carlsbad, CA, USA) with 1 % penicillin/streptomycin (15140; Gibco®, Invitrogen, Carlsbad, CA, USA) at 37 °C, 5% CO2. The cells were seeded at 4 × 105 cells/well in a 6-well plate 18–20 hours prior to incubation with isolated and purified spleen CLDIs at a solution equivalent concentration of 40 μM CLDIs (14). Following 24 hours post CLDI incubation, cells were gently scraped and suspended in sterile flow cytometry tubes at a density of ~2 × 106 cells/ml of phosphate-buffered saline (PBS) + 5% FBS. The cells were analyzed on a MoFlo® Astrios™ (Beckman Coulter, Brea, CA, USA) using various laser combinations. Unless otherwise mentioned, forward and side scatter were measured using the 488 nm laser. Laser settings are referenced in the following format – Excitation Emission/Bandwidth in nm. For example, if the excitation laser used is 488 nm and the emission detector is at 576 nm with a bandwidth of 21 nm, the written format is 488 576/21. To check the activity of viability dyes and signal compensation, just prior to analysis, propidium iodide (PI) (00-6990-50; Affymetrix eBioscience, San Diego, CA, USA) or 4,6-diamidino-2-phenylindole, dihydrochloride (DAPI) (D1306; Life Technologies, Grand Island, NY, USA) was added to the cells (5 μl per 1×106 cells). All gating and analysis was done on at least 10,000 cells (DAPI(−) or PI(−) cell population for viability studies) using FlowJo (FlowJo, LLC, Ashland, OR, USA). MIFlowCyt-compatible information for this experiment has been included in the Supplementary Information document (See pages 5–11 in supplementary document). Statistical analysis of sensitivity and specificity was conducted by acquiring >6 brightfield microscopy images of sorted cell populations on standard microscopy slides and >350 cells were counted for each sorting experiment. Sensitivity and specificity were calculated as follows.
Where TP (True Positive) = CLDI(+) in CLDI(+) cell population,
FP (False Positive) = CLDI(−) in CLDI(+) cell population,
TN (True Negative) = CLDI(−) in CLDI(−) cell population,
FN (False Negative) = CLDI(+) in CLDI(−) cell population,
Peritoneal Lavage
Peritoneal lavage was done as previously reported before and after the initiation of CFZ or vehicle treatment (1 week, 2 weeks, 4 weeks, 8 weeks and 16 weeks (8 weeks drug feed + 8 weeks washout phase)) (16). Mice were euthanized as described above followed by sterilization of the outer skin with 70% ethanol. A small incision was made along the midline of the abdomen followed by abdominal skin retraction up to the thoracic boundary and the animal extremities to expose the intact peritoneal wall. A smaller incision was then made on the peritoneal wall to expose the cavity. The entire peritoneal cavity was washed with ice-cold sterile PBS + 5% FBS (5–10 ml) and collected as peritoneal exudate. The exudate was then centrifuged (100 × g for 5 min, 4 °C) and resuspended in 1.5 ml of PBS + 5% FBS. Cells were counted using a hemocytometer for viable cells using Trypan Blue and for CLDI-containing cells.
Alveolar Lavage
Mice were euthanized as described above and the trachea was surgically exposed and cannulated with an 18G luer stub and the lungs were lavaged to obtain alveolar exudate by instilling calcium- and magnesium-free Dulbecco's PBS (DPBS) containing 0.5 mM EDTA in 1-ml aliquots for a total of 6 ml. The alveolar exudate fluid was centrifuged (400 × g, 10 min, 4 °C) and resuspended in RPMI 1640 media. Viable (using Trypan Blue staining method) and CLDI containing cells were counted using a hemocytometer.
Flow Cytometry of Peritoneal and Alveolar Exudate
Peritoneal and alveolar lavage was performed as stated before to obtain the respective exudates. Purified Fc block CD16/32 (1 μl) was added for every 100,000 cells for analysis. For CLDI fluorescence signal experiments, cells were pelleted by centrifugation (100 × g for 5 min) followed by resuspension in PBS + 5% FBS (500 μl). For functional assays, antibodies - anti-F4/80-eFluor 450 (48-4801, Affymetrix eBioscience, San Diego, CA, USA), anti-CD86-FITC (553691, BD Biosciences, San Jose, CA, USA), anti-CD206-FITC (141704, BioLegend®, San Diego, CA, USA), anti-CD11c-eFluor450 (48-0114, Affymetrix eBioscience) and anti-Ly6G-eFluor450 (48-5931, Affymetrix eBioscience) were added to the cell suspension at a loading of 1 μg/100,000 cells (1:10 volumetric ratio) and incubated in the dark (30 min, 4 °C). Following incubation, the samples were diluted with PBS and pelleted by centrifugation (100 × g, 5 min). The supernatant was discarded and the pellet was re-suspended in 300 μl of PBS + 2% FBS. Just prior to analysis, PI (5 μl per 1×106 cells) was added to the cells to assess viability. Sample measurements were done on a MoFlo® Astrios™ EQ. All gating and analysis was done on at least 10,000 live cells (PI(−) cell population) using FlowJo. CLDI(+) cells were gated either using 640 671/30 or 640 795/70. A small sub-population of CD206(+) cells, which was present in both CLDI(+) F4/80(−) and CLDI(−) F4/80(+) populations, exhibited high CD206 expression based on the intensity of the fluorescence signal. These cells also had extremely high side-scatter that were measured as saturated signals. This small sub-population of cells were not considered for evaluation of change of CD206 expression in the peritoneal macrophages. MIFlowCyt-compatible information for this experiment has been included in the Supplementary Information document (See pages 5–8, 12–17 in supplementary document).
Laser-Scanning Confocal Microscopy
CLDI treated RAW264.7 cells were generated as mentioned above but on cover-slips in a 6-well plate. The cells were washed with PBS buffer following which Hoechst 33342 (Invitrogen, Carlsbad, CA) and FM® 1-43 (Molecular Probes T35356, Invitrogen) were used to stain cell nuclei and the plasma membrane, respectively. Cells were incubated with 1:1 (v/v) dye mixtures of 5 μg/ml Hoechst 33342 and 7 μM FM® 1-43 in HBSS (300 μl) for 15 min at room temperature. The confocal imaging of the live cells was performed on an Olympus Fluoview 500 (Olympus America Inc., Center Valley, PA, USA) using lasers for DAPI (405 450/50), FITC (488 525/50) and Cy5 (640 671/30) channels. Z-stack images of the cells were captured along the Z-axis (interval=0.25 μm) and analyzed using the Nikon NIS-Elements 3.2 confocal software (Nikon Instruments Inc., Melville, NY). For peritoneal macrophages stained with F4/80, a drop of the stained cell sample was mounted on a blank microscopy slide; Prolong Gold and a cover-slip were applied followed by immediate scanning.
Data Plotting and Statistical Analysis
Plots were constructed using Origin 9.0 (OriginLab Corporation, Northampton, MA, USA) and laid out in figure format using scalable vector graphics format (svg) in either Inkscape (www.inkscape.org) or GIMP (www.gimp.org). Flow cytometry plots were obtained in svg format directly from FlowJo and assembled using Inkscape. All statistical analysis was performed using Student's t-test in MS-Excel. Correlation statistics were done using a Pearson's test in Origin 9.0. Results were considered significant if p≤0.05. MIFlowCyt compatible information was prepared using established standards (17). The completed MIFlowCyt checklist along with full gating strategies, machine information and data analysis has been included in the Supporting Document. All original list mode files have been saved onto internal servers and can be obtained via email from the authors.
RESULTS
Fluorescence Spectral Analysis of CFZ and CLDIs
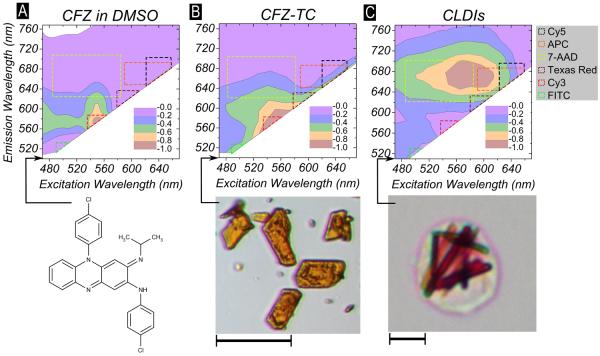
The fluorescence excitation and emission scan of CFZ dissolved in DMSO indicates that CFZ in solution is fluorescent in the range - Excitation: 540–560 nm, Emission: 560-600 nm (Figure 1A). To characterize the fluorescence of CFZ crystallized as free-base triclinic crystal (closest triclinic polymorph of neutral CFZ crystal, CCDC refcode: DAKXUI01), we used spectral confocal microscopy on individual CFZ-TC crystals dispersed on a microscopy slide. CFZ-TC has a similar fluorescence profile as soluble CFZ (Figure 1B). However, residual fluorescence of CFZ-TC is also detected at lower (Ex:480–500 nm, Em: 510–540 nm) and higher (Ex: 580–600 nm, Em: 600–640 nm) spectral ranges. To characterize and identify a specific spectral signature associated with the intracellular accumulated and crystallized CFZ–CLDIs, the spleens of 8-week drug-fed mice were harvested and CLDIs were isolated and purified. By visual inspection, CLDIs were dark red, rod shaped particles that were distinct from the orange-red CFZ-TC crystals (Figure 1B–C, inset photographs). When subjected to fluorescence spectral analysis using the spectral confocal microscope, CLDIs exhibited a red-shift in their emission fluorescent spectra with peak fluorescence activity at Ex:560-600 nm, Em: 650–690 nm (Figure 1C). To place this specific red-shift in the context of other commonly used dyes, CFZ-TC exhibited a fluorescence profile similar to Fluorescein Isothiocyanate (FITC), Cyanine 3 (Cy3), Rhodamine and Texas Red (TR) whereas CLDIs had a red-shifted fluorescence profile similar to TR, Cyanine 5 (Cy5), 7-Aminoactinomycin D, (7-AAD), Allophycocyanin (APC) and other commercial Cy5 derivatives.
Figure 1.
Fluorimetric analysis of (A) CFZ dissolved in DMSO (20 μM) done using conventional solution fluorimetry (bottom – chemical formula of CFZ) and (B) CFZ-TC (bottom – representative picture, scale bar = 10 μm) and (C) isolated CLDIs from spleen (bottom – representative picture, scale bar = 10 μm) acquired using spectral confocal microscopy. The excitation wavelength (nm, Ex) and emission wavelength (nm, Em) are shown on the X-axis and Y-axis, respectively. The normalized fluorescence yield is shown by a contour plot that was normalized to the maximum measured fluorescence yield. Both solubilized CFZ in DMSO and CFZ-TC had peak fluorescence yield at Ex:540-570 nm, Em: 560–580 nm while CLDIs had peak fluorescence yield at Ex:560-600 nm, Em: 640–700 nm. The excitation/emission combination for detection of commonly used fluorophores: FITC, Cy3, Texas Red, 7-AAD, APC and Cy5 are also shown as dotted bounded shapes super-imposed on the contour plots.
Identification of CLDI-Containing Cell Subpopulations with Standard Flow Cytometer Configurations
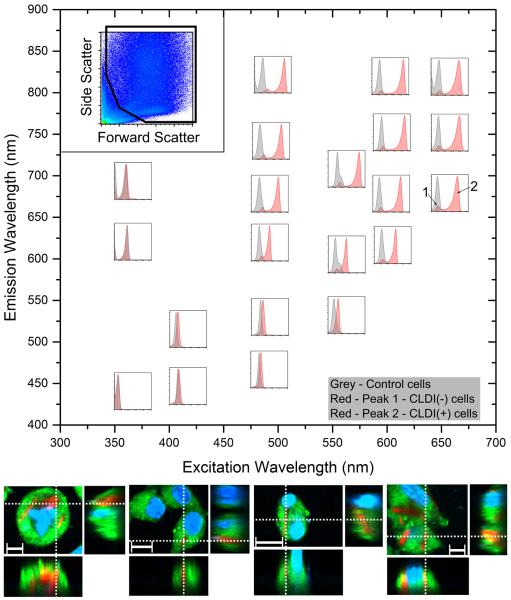
To design a single-cell analysis technique to study the pharmacology of xenobiotic-sequestering macrophages and the impact of CLDIs on cellular functions, we proceeded to determine whether flow cytometery could be used to distinguish between cells that contain and do not contain CLDIs. Samples were analyzed using standard excitation lasers and detector configurations, to determine the specific fluorescence cytometry settings that can be used for analyzing CLDI containing cell subpopulations and to establish any spectral overlap detected in the other channels (Figure 2). The use of ultraviolet (355 nm) and violet (405 nm) excitation lasers resulted in the detection of single populations at all emission detector settings, corresponding to the background autofluorescence signal of unlabeled cells. Similarly, a single cell population was detected when samples were excited with the blue (488 nm) or green (561 nm) excitation lasers at emission detection <600 nm. However, at emission detection >600 nm, two distinct cell populations were observed, one which corresponded to the background fluorescence signal of CLDI-free cells (labeled 1), and the other which corresponded to the positive fluorescence signal of the CLDI containing cells (labeled 2). Furthermore, the use of the red (592 nm) and far-red (640 nm) excitation laser resulted in the detection of two cell sub-populations corresponding to the CLDI-free and CLDI-containing cells, at all tested emission detector settings. The laser and detector settings that detected the subpopulation of CLDI-containing cells corresponded to the CLDI fluorescence spectra, which were independently measured using the spectral confocal microscope (Figure 1C).
Figure 2.
Flow cytometric analysis of RAW264.7 incubated with CLDIs using a variety of laser combinations on a MoFlo Astrios. (top left inset) The forward scatter versus the side scatter used for further analysis. Various sub-plots are arranged based on excitation and emission wavelength combination in the main graph (X-axis = excitation wavelength, Y-axis = emission wavelength). A single population is observed with RAW264.7 cells alone (shown in grey histogram in each sub-plot). A single population is observed with RAW264.7 cells with CLDIs when excited with UV laser (excitation=355 nm, all emission detector settings), violet laser (excitation=405 nm, all emission detector settings), blue laser (excitation=488 nm, emission=513 nm, 576 nm) and orange laser (excitation=561 nm, emission=579 nm). In all other laser combinations, two populations were observed (labeled 1 and 2). (bottom) Confocal microscopy of RAW264.7 incubated with CLDIs confirmed the phagocytosis of CLDIs using the fluorescence of CLDIs in the Cy5 channel while cells were stained with membrane stain FM 1–43 (green) and nuclear counterstain Hoechst 33342 (blue) (Scale bar = 10 μm, dotted lines show the visualization planes for the three orthogonal views).
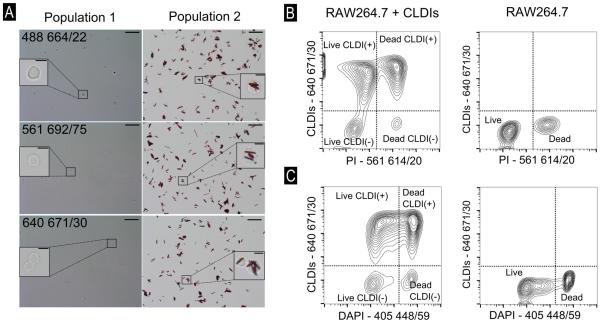
To confirm these observations, a fluorescence activated cells sorting experiment was performed using 488 664/22, 561 692/75 and 640 671/30 [See italicized statement in Methods and Supplementary Information containing MIFlowCyt information on description of laser settings] along with laser-scanning confocal microscopy of incubated cells. When RAW264.7 cells incubated with CLDIs and labeled with fluorescent cellular staining dyes – Hoechst 33342 (nucleus) and FM 1-43 (plasma membrane) were imaged, the intracellular accumulation of CLDIs was confirmed via their far-red fluorescence (Figure 2, bottom right inset) distinct from the green fluorescence of FM 1-43 and blue fluorescence of Hoechst 33342. Additionally, as per brightfield microscopy, the sorted cell population with the lower fluorescence signal corresponded to CLDI-free cells whereas the sorted cell population within the high fluorescence signal corresponded to CLDI-containing cells (Figure 3A). The sorting sensitivity was 98.34%, 98.82% and 99.01% at 488 664/22, 561 692/75 and 640 671/30, respectively, whereas sorting specificity was 86.30%, 90.80% and 93.98%, respectively (See Methods for formula). The detection of CLDIs in subsequent flow cytometry experiments was done using either 640 671/30 or 640 795/70 whereas for microscopy, CLDIs were detected using the red laser (Excitation: 640 nm, Emission: 670 nm).
Figure 3.
(A) Representative images of brightfield microscopy of sorted cell populations – labeled 1 and 2 in Figure 2 at 488 664/22, 561 692/75 and 640 671/30, scale bar = 50 μm. Inset photographs show digitally zoomed-in regions (scale bars=10 μm). Flow cytometric compensation to obtain accurate readout of RAW264.7 cells with CLDIs for viability using (B) propidium iodide (PI) and (C) DAPI. Y-axis − 640 671/30. X-axis – corresponding compensated fluorescence axes for determining PI and DAPI fluorescence. Four populations were observed with the use of PI and DAPI to verify that viability of cells could be studied with appropriate compensation.
Functional Flow Cytometric Analysis of Live vs Dead, CLDI-Containing Cells
In flow cytometry experiments, a fundamental functional assay involves discriminating live and dead cells using standard fluorescent labels. Accordingly, to determine whether CLDI-containing cells were viable, RAW264.7 cells loaded with CLDIs were further incubated with membrane impermeant cell viability dyes – PI or DAPI prior to flow cytometric analysis. Using the laser-detector settings for PI – 561 614/20, two populations were detected using CLDI-incubated RAW264.7 cells. We conducted a compensation of the spectral overlap that was observed at 561 614/20 to verify that PI can be used in the presence of CLDIs (at 640 671/30) without signal overlap (Figure 3B). Incubating control (CLDI-free) cells with PI yielded two distinct cell populations, corresponding to live (low fluorescence signal) and dead (high fluorescence signal) cells (Figure 3B). In contrast, after CLDI-containing cell samples were incubated with PI, four populations were observed, corresponding to viable CLDI-free (Live CLDI(−)) and viable CLDI-containing cells (Live CLDI(+)), and dead CLDI-free (Dead CLDI(−)) and dead CLDI-containing cells ((Dead CLDI(+)) (Figure 3B, Figure S2). In the same manner, flow cytometric analysis revealed that DAPI could be used to determine the viability of cells containing CLDIs (Figure 3C). With DAPI, four distinct populations were observed (without compensation), corresponding to live (weak UV signal) and dead (strong UV signal), CLDI containing (marked as CLDI(+)) and CLDI free (marked as CLDI(−)) cells. Thus, both PI and DAPI were suitable as fluorescent markers for discriminating between live and dead cell subpopulations of CLDI-containing cells.
In vivo Flow Cytometry Detection of CLDI-containing cells in Peritoneal and Alveolar Exudates from CFZ-treated mice
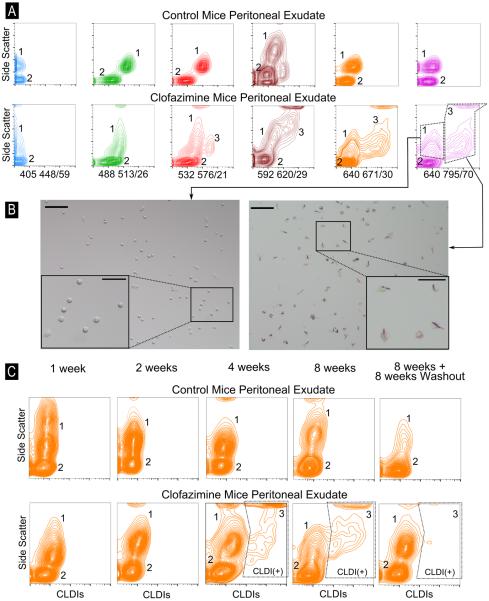
To measure the presence of CLDIs in peritoneal exudates obtained from CFZ-treated mice, we performed a quantitative, flow cytometric analysis on peritoneal exudate of mice following 8-week drug administration. While the control mouse peritoneal exudate showed two distinct populations (labeled 1 and 2 - Figure 4A) at all tested laser settings, the CFZ mouse peritoneal exudate showed two populations when assayed with the 405 448/59 or 488 513/26 laser-detector combination. With the use of 532 576/21, 592 620/29, 640 671/30 and 640 795/70, however, three distinct populations were observed (labeled 1, 2 and 3 - Figure 4A). The mean fluorescence intensity of population 3 was increased >100-fold relative to population 1 at 640 671/30 and 640 795/70. To confirm that the third population was CLDI(+) cells, we sorted the cell populations labeled 1 and 3 using the 640 795/70 laser-detector combination and plated them in cell culture media overnight. Population 1 was confirmed to be CLDI(−) cells whereas population 3 was indeed CLDI(+) cells (Figure 4B).
Figure 4.
(A) Flow cytometric analysis of peritoneal exudate in control mice and CFZ-treated mice done using various laser combinations. Two distinct populations were observed in the control peritoneal exudate (labeled as 1 and 2) and at 405 448/59 and 488 513/26 in the CFZ peritoneal exudate while three populations were observed at 532 576/21, 592 620/29, 640 671/30 and 640 795/70 and two populations were observed The Y-axis shows the side-scatter profile of cells with high-scatter associated with peritoneal macrophages while low side-scatter is associated with B-lymphocytes. (B) The populations labeled 1 and 3 at 640 795/70 in (A) were sorted, plated with complete growth media and visualized using standard epifluorescence microscopy. Population 3 shows a remarkably high proportion of cells containing CLDIs; population 2 are cells containing no CLDIs (scale bar = 100 μm). Inset photographs show digitally zoomed in regions (scale bar = 50 μm). (C) Flow cytometric analysis of peritoneal exudate at various time-points post drug feeding. Y-axis in each plot is the side-scatter profile while the X-axis is the 640 795/70 laser-detector combination. CLDI(+) cells are observed starting at 4 weeks of CFZ treatment.
Next we proceeded to determine whether the time course of CLDI accumulation in macrophages as determined by flow cytometry corresponded to the expected time course of CLDI accumulation following drug treatment (2,3). Following drug treatment of mice for varying time periods, peritoneal exudates were obtained and analyzed by flow cytometry as well as light microscopy, revealing that CLDIs were not present at 1 week and 2 weeks, but could be detected starting at 4 weeks (Figure 4C).
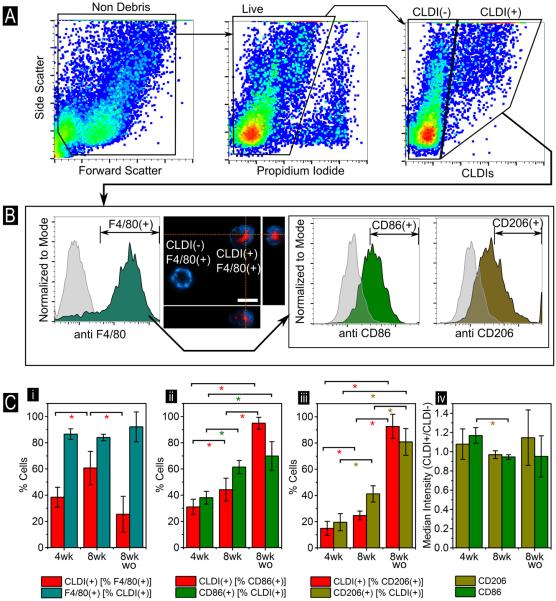
To determine whether flow cytometry could be used to characterize the molecular phenotype of CLDI-containing macrophage sub-populations, multi-stain flow cytometry was conducted on peritoneal exudates of CFZ-treated animals. For this purpose, cells obtained from peritoneal exudates were incubated with other commonly employed macrophage-targeted antibodies possessing fluorescent signals distinct from that of CLDIs (405 448/59 and 488 513/26; Figure 1). Since CLDI accumulation has been reported to be macrophage targeted (3,18–20), we chose the murine pan-macrophage marker F4/80 as well as other macrophage phenotype surface markers – CD86 and CD206 (Figure 5). The schematic for quantification of the gating strategy is shown in Figure 5A,B and Figure S1. The analysis revealed that in peritoneal exudates, CLDIs were primarily localized to F4/80(+) peritoneal macrophages (PMΦ) (>85% of total CLDI(+) cells at all measured time points (Figure 5B, no statistically significant changes across the times measured). This observation was also confirmed via laser-scanning confocal microscopy using the far-red fluorescence region for CLDIs and the violet laser to visualize F4/80 extracellular membrane staining (Figure 5B). The accumulation, however, was both treatment and time dependent with respect to F4/80(+) PMΦ as ~38% of F4/80(+) PMΦ were CLDI(+) after 4 weeks of CFZ treatment which increased to ~61% after 8 weeks (p<0.05) of treatment and declined to ~25% at 8 weeks of washout (p<0.05; Figure 5C(i)). Within F4/80(+) CD86(+) cells, CLDI accumulation increased in a time-dependent fashion, from ~31% after 4 weeks of CFZ treatment to ~95% at 8 weeks + 8 weeks washout (p<0.05, Figure 5C(ii)). As a proportion of F4/80(+) CD206(+) cells, CLDI(+)% increased from ~15% after 4 weeks of CFZ treatment to ~93% after 8 weeks + 8 weeks washout (p<0.05, Figure 5C(iii)). Collectively, these findings suggest that CLDI accumulation was not restricted to the CD86(+) or CD206(+) macrophage phenotype (Figure 5B). In a related trend, % CLDI(+) cells that could be identified as F4/80(+) CD86(+) increased from ~38% at 4 weeks post-drug feeding to ~70% at 8 weeks + 8 weeks washout (p<0.05) whereas % CLDI(+) cells that could be identified as F4/80(+) CD206(+) increased from ~20% at 4 weeks post-drug feeding to ~81% at 8 weeks + 8 weeks washout (p<0.05). While the accumulation of CLDIs in CD86(+) cells was higher relative to CD206(+) cells at 4 weeks and 8 weeks post-drug feeding (p<0.05), the accumulation was not statistically deviated towards any phenotype at 8 weeks + 8 weeks washout (p>0.05). In contrast, the ratiometric CD86 median surface expression in CLDI(+) relative to CLDI(−) decreased from ~1.17 at 4 weeks post-drug feeding to 0.95 (p<0.05, Figure 5C(iv)) at 8 weeks post-drug feeding. The ratiometric CD206 median surface expression in CLDI(+) relative to CLDI(−) CD206 remained between ~0.97 and ~1.15 for CD206 with no statistically significant difference at the three measured time points (Figure 5C(iv)). Since the fold-change in the median surface expression is much less than the benchmark of two-fold change in expression to ascertain significant change in expression of the two surface markers – the changes, as a result of being CLDI(+) reflect relatively minor expressional differences between the two populations. Finally, the viability of F4/80(+) cells, as measured via PI(−) cells, was unaffected by the presence of CLDIs (viability of CLDI(+) F4/80(+) = 87±6%, CLDI(−) F4/80(+) = 88±12%, p=0.84 between the two groups, n=4). The viability measured via flow cytometry also agreed with the sample viability of CLDI+ cells in the peritoneal exudate measured by Trypan blue staining (87±9%, n=4).
Figure 5.
(A) Flow cytometry analysis of peritoneal exudate in CFZ mice (Schematic for quantification shown in Figure S1). Gating was done via viability using PI (compensated 561 614/10 settings) and then for CLDI-associated signal at 640 795/70. (B) The peritoneal exudate was also dual labeled with antibodies for the pan-macrophage marker F4/80 (detected using 405 448/59) followed by either CD86 or CD206 (488 513/26). Gating for CLDI(+) followed by F4/80(+) and CD86(+) or CD206(+) is shown. CLDI+ cells were significantly F4/80+ with a medium expression of CD86 and CD206. Gating for antibody positivity was done via the corresponding isotype (grey background histogram). Confocal microscopy shows the membrane staining of the F4/80 surface marker in blue while CLDIs are shown in red (Scale bar = 10 μm, dotted lines show the visualization planes for the three orthogonal views). (C) Cumulative statistics on CLDI(+) cells that are (i) F4/80+, (ii) CD86+ and (iii) CD206+ in CFZ mice at four weeks (4wk), eight weeks (8wk) and eight weeks + eight weeks washout (8wk wo) post drug feeding. Error bars show standard deviation (n=4 mice). (iv) Median expression of CD86 and CD206 in CLDI+ F4/80+ relative to CLDI- F4/80+ were also compared at all time points. * indicates p<0.05 (Student's t-test).
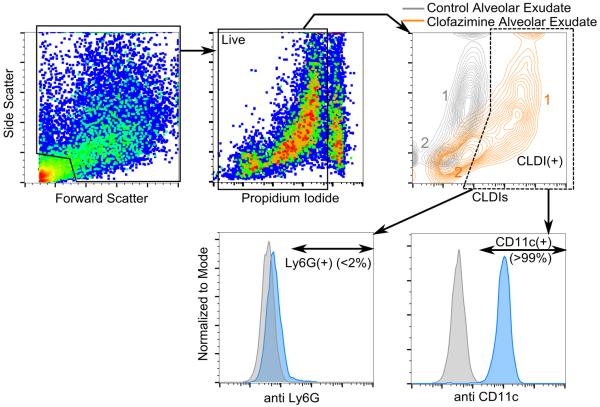
To follow up our analysis on peritoneal exudate, we performed a flow cytometric analysis on alveolar exudate of drug fed mice at 8 weeks to confirm the macrophage-specific accumulation of CLDIs in the lung (2,4,18) (Figure 6). Both the control and CFZ mice alveolar exudates had two distinct populations. However, population 2, that had high side-scatter, had a distinctly higher fluorescence at the settings used to detect CLDIs (640 671/30), marking them out as CLDI(+) cells. Upon further testing with fluorescent antibodies (at 405 448/59), specific for alveolar macrophages (CD11c) and neutrophils (Ly6G) (21), CLDI(+) cells were determined to be overwhelmingly CD11c(+) (>99%) whereas <2% of Ly6G(+) cells were detected as CLDI+.
Figure 6.
Flow cytometric analysis of alveolar exudate at 8 weeks post drug feeding. Propidium iodide (PI) fluorescence was detected using the compensated 561 614/20 laser-detector setting where CLDI fluorescence was detected using the 640 671/30 setting. Gating was done on live cells followed by CLDI(+) cells. The population labeled 2 in orange color showed significant CLDI signal marking them out as CLDI(+) cells. The alveolar exudate was also labeled with antibodies for the macrophage surface marker CD11c (detected using 405 448/59) or the neutrophil surface marker Ly6G (detected using 405 448/59). CLDI(+) cells were overwhelmingly CD11c(+) (>99%, 6 pooled mice lavage sample, repeated twice) and Ly6G(−) (6 pooled mice lavage sample, repeated twice). Gating for antibody positivity was done using the corresponding isotype (grey background histogram).
Correlation of CLDI amount with Mean Cellular Fluorescence Intensity (MFI)
In fluorescence activated cell sorting experiments, attempts to relate the MFI of sorted cell populations with the total number of CLDIs per cell did not yield interpretable data. Therefore, to determine whether the fluorescence signal per cell was related to the number of CLDIs in each cell, we incubated RAW264.7 macrophage cells with CLDIs, and acquired fluorescence images with conventional epifluorescence microscopy using the Cy5 filter for CLDIs (Figure S3). The normalized background corrected MFI of cells was strongly correlated (Pearson's r=0.70, p<0.0001) with the number of CLDIs/cell. As expected, cell MFI was uncorrelated to intracellular CLDI geometry and morphology (Figure S4, Pearson's r = −0.2 to 0.3, p-value < 0.005). The resulting fluorescence signal readily distinguished CLDI(+) cell relative to a CLDI(−) cells (p-values < 0.005, Table S1). Cells containing 1 vs 2 CLDIs were also distinguishable (p < 0.05). However, cells containing > 2 CLDIs could not be distinguished from cells containing fewer CLDIs, in a statistically relevant manner. Given that each CLDI contains ~25 femtomoles CFZ (24.3 ± 13.5, n=8 mice), we infer that this method could be used to quantify CFZ sequestration in CLDIs, up to 50 femtomoles of CFZ (corresponding to 2 CLDIs per cell).
DISCUSSION
The pharmacological activity of CFZ, its pharmacokinetics, and its effectiveness as an antimycobacterial agent have been intensively studied for several decades (1,22–28). Remarkably, the optical properties of CFZ have been largely unrecognized, especially in the context of its intracellular bioaccumulation behavior. While CFZ's accumulation in CLDIs has been clinically observed, the phenotypic characteristics and the mechanisms that underlie such massive intracellular bioaccumulation have not been studied (2,7,18,19,26,29). Seeking to understand whether the intracellular bioaccumulation of CFZ affects the optical properties of the drug, we determined whether there are spectral shifts in fluorescence excitation and the emission spectrum of CLDIs in relation to the soluble form of the drug. As a reference, CFZ was readily crystallized as a free base – CFZ-TC (CCDC ref code: DAKXUI01) and the spectrum of the CFZ-TC crystals were analyzed and compared to that of CLDIs. Using a confocal spectral microscope imaging system, fluorescence spectral analysis was carried out on CLDIs, and compared to the fluorescence spectra of pure crystalline forms of CFZ resulting in a red shift in the fluorescence emission spectra in CLDIs. This fluorescence shift led to a highly specific signal that was largely confined to channels that are used to detect far-red probes such as Cyanine 5-like fluorescence either in Cyanine 5 (Cy5) or its various commercial conjugates in standard epifluorescence microscopes filter setups and flow cytometers. Surprisingly, while CFZ and CFZ-TC have strong fluorescence in the green to mid-red fluorescence range, CLDIs exhibited a pronounced, red-shifted fluorescence emission profile, spanning the red to far-red range of the visible spectrum. Unlike CLDIs, both CFZ and CFZ-TC exhibited some fluorescence in the blue-green wavelengths of the electromagnetic spectrum.
To assess the utility of the far-red fluorescence activity of CLDIs, we proceeded to detect the fluorescence signal with a flow cytometry approach using different lasers on a macrophage-derived cell line that is known to internalize CLDIs through phagocytosis. Inside cells, the fluorescence of CLDIs was specific to the red and far-red fluorescence spectral channels, corresponding to the fluorescence spectra obtained with the spectral confocal microscope. The accuracy and sensitivity of the flow cytometer for detecting CLDI+ cells from a mixed population of cells was calculated to be >98% sensitive and >93% specific. Due to minimal spectral overlap in the ultraviolet, violet, blue and green fluorescence channels, CLDIs can be readily used as a marker for xenobiotic sequestering macrophages, for multi-color flow cytometric analysis with orthogonal fluorescent probes and cellular staining dyes for microscopic analysis and image cytometry (30).
Proceeding to analyze the phenotypic characteristics of xenobiotic sequestering PMΦ and alveolar macrophages (AMΦ) that bioaccumulate CFZ upon prolonged oral feeding in mice, we confirmed that the laser detector combination of 640 671/30 or 640 795/70 could be used to identify CLDI(+) cells. Three populations were observed (Figure 4) with the peritoneal exudate. Populations 1 and 2 were high side-scatter and low side-scatter respectively but had low fluorescence. Earlier reports have characterized that peritoneal exudates contain primarily PMΦ and B-cells (16,31). Based on the scatter analysis, population 1 was most likely PMΦ whereas population 2 contained B-cells. Population 3 had high side-scatter as well as high fluorescence indicating that they were CLDI(+) cells as well as most-likely a PMΦ cell population. This was confirmed by sorting the fluorescent and non-fluorescent cell populations, plating the corresponding populations on tissue culture dishes, and visually inspecting them for the presence or absence of CLDIs. In the alveolar exudate, two distinct populations were observed for both control and CFZ mice samples. The high side-scatter population was identified to contain CLDIs due to the high fluorescence signal detected with that population in the CFZ mice samples, identifying it as a potential AMΦ population as well.
Murine resident PMΦ as well as macrophages of others organs have high F4/80 plasma membrane expression (32). F4/80 is encoded by the gene Emr1, a member of the adhesion-GPCR family of proteins that play notable roles in immune modulation and related functions (33) (www.genecards.org). Based on the expression of other surface markers, these macrophages can be classified into two specific functional categories - classically activated macrophages (M1) and alternatively activated macrophages (M2) (34,35,12,36–38). M1 macrophages noticeably express high levels of the cell surface marker CD86 that is critical for host defense but can also be harmful to the host (36,37). M2 macrophages, express high levels of CD206 and are key mediators of host inflammation via the their wound healing, tissue remodeling and angiogenic functions (34,35). Most importantly, both CD86 and CD206 are exclusive to M1 and M2 type macrophages, respectively serving as important biomarkers for inflammation-related events. With the use of the CLDI specific signal, we confirmed that the majority of the CLDI(+) cells were indeed F4/80+ PMΦ (>85%) and the presence of CLDIs had a minor effect on polarizing the macrophages towards the M2 phenotype (34).
Analyzing the alveolar exudate, CLDI accumulation was specifically targeted towards CD11c(+) cells (>99%) while Ly6G(+) cells had negligibly low CLDI accumulation. Alveolar exudates from healthy humans (39) and mice (40) typically contain AMΦ, neutrophils and lymphocytes. CD11c is an integrin protein, encoded by the gene ITGAX, with useful functions in cell-cell interactions during inflammation as well as in macrophage adhesion and chemotaxis (www.genecards.org). CD11c is also an important surface cell marker for evaluating murine AMΦ (CD11c(+)) (21,41,42). Ly6G is a murine glycosylphosphatidylinositol (GPI)-anchored protein with unknown functions in cellular physiology (43). It is also a neutrophil-specific surface marker that has been used to examine inflammation models in various tissues (21,44,45). Using these specific immune markers for macrophages and neutrophils, CLDI accumulation was confirmed to be mostly present in AMΦ.
Like other fluorescent probes that emit in the far red or infrared regions of the electromagnetic spectrum (46,47), the intrinsic fluorescence of CLDIs exhibit favorable optical properties (47), with minimal background fluorescence in the far-red and near infra-red region allowing accurate quantitative analysis with the flow cytometer or fluorescence microscope (48). Accordingly, these results demonstrate that CFZ, when stabilized as CLDIs, has excellent properties as a fluorescent probe for characterizing the specific macrophage subpopulations involved in xenobiotic disposition. As with other fluorescence-based measurement method, this fluorescence signal was saturatable: while there was a significant correlation between the fluorescence signal of the cells and the number of CLDIs per cell for cells containing 0, 1 or 2 CLDIs, the correlation was lost for cells containing 3 or more CLDIs. Nevertheless, the specific fluorescence signal of CLDIs could serve as an all-or-none probe for detecting and sorting xenobiotic sequestering macrophages, and as a tracking probe to follow the function of these cells during inflammation and wound healing. Considering that recent studies have pointed out the therapeutic efficacy of CFZ for drug-resistant tuberculosis, the potential application of CLDIs as cellular targeted probes for assessment of antimycobacterial activity is promising (49–54). In terms of the functional significance of these xenobiotic sequestering macrophages, other poorly soluble drugs, endogenous metabolites and toxins have been reported to bio-accumulate within macrophages. Such bio-accumulating agents are often associated with a macrophage activation phenotype, favoring the M1 population which was not observed in the case of the CLDI-containing cells (9–12). Using fluorescence activated cell sorter, the isolation of CLDI(+) cell subpopulations will enable a precisely quantified approach towards understanding the mechanisms of xenobiotic sequestration.
CONCLUSION
In summary, the analysis of macrophage populations in vitro and ex vivo was made possible due to the unique fluorescence spectral characteristics of CLDIs relative to neutral free base CFZ precipitates as well as the CFZ molecule in soluble form. The specific fluorescence of CLDIs was utilized in single-cell flow cytometry to study macrophage-related intracellular drug accumulation wherein we were able to measure macrophage functions including viability and expression of different cell surface markers, by exploiting the unique fluorescence spectrum of CLDIs and the availability of many other orthogonal fluorescent probes that can be detected with standard flow cytometry parameters, with minimal spectral overlap from the CLDI signal. We also confirmed that the presence of CLDIs has a minor polarizing effect towards the M2 macrophage phenotype. More importantly, the unique fluorescence shift of CFZ molecules from the freely soluble green-blue fluorescent state to the solid far-red CLDI fluorescent state should facilitate analysis of the molecular mechanisms driving drug accumulation and CLDI formation, using a variety of molecular, pharmacological, genetic, or systems biology approaches. These studies will be instrumental for furthering understanding of the role of xenobiotic sequestering macrophages in the response of the organism to bio-accumulating drugs or environmental toxicants.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institute of General Medical Sciences (NIGMS; R01GM078200 to GRR), University of Michigan's M-Cubed initiative (http://mcubed.umich.edu) and the Michigan Institute for Clinical & Health Research (MICHR) (to GRR and RKK). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the National Institutes of Health. We thank Dr. Joel Swanson (Department of Microbiology & Immunology, University of Michigan) for help with data analysis and Dave Adams, Michael Dellheim and Karen Peterson (Flow Cytometry Core, University of Michigan) for assistance with flow cytometry and with MIFlowCyt-compatible document preparation.
Footnotes
Gus R. Rosania is a consultant for Bristol Myers-Squibb. There are no other competing financial interests.
LITERATURE CITED
- 1.Cholo MC, Steel HC, Fourie PB, Germishuizen WA, Anderson R. Clofazimine: current status and future prospects. J. Antimicrob. Chemother. 2012;67:290–8. doi: 10.1093/jac/dkr444. [DOI] [PubMed] [Google Scholar]
- 2.Baik J, Stringer KA, Mane G, Rosania GR. Multiscale distribution and bioaccumulation analysis of clofazimine reveals a massive immune system-mediated xenobiotic sequestration response. Antimicrob. Agents Chemother. 2013;57:1218–30. doi: 10.1128/AAC.01731-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baik J, Rosania GR. Macrophages sequester clofazimine in an intracellular liquid crystal-like supramolecular organization. PLoS One. 2012;7:e47494. doi: 10.1371/journal.pone.0047494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conalty ML, Jina AG. Reticuloendothel. Syst. Immune Phenom. Springer; 1971. The Antileprosy Agent Clofazimine (B.663) in Macrophages: Light, Electron Microscopy and Function Studies; pp. 323–331. [Google Scholar]
- 5.O'Connor R, O'Sullivan JF, O'Kennedy R. The Pharmacology, Metabolism and Chemistry of Clofazimine. Drug Metab. Rev. 1995;27:591–614. doi: 10.3109/03602539508994208. [DOI] [PubMed] [Google Scholar]
- 6.Vandeputte D, Jacob W, Van Grieken R, Boddingius J. Study of intracellular deposition of the anti-leprosy drug clofazimine in mouse spleen using laser microprobe mass analysis. Biol. Mass Spectrom. 1993;22:221–5. doi: 10.1002/bms.1200220402. [DOI] [PubMed] [Google Scholar]
- 7.McDougall AC, Horsfall WR, Hede JE, Chaplin AJ. Splenic infarction and tissue accumulation of crystals associated with the use of clofazimine (Lamprene; B663) in the treatment of pyoderma gangrenosum. Br. J. Dermatol. 1980;102:227–230. doi: 10.1111/j.1365-2133.1980.tb05697.x. [DOI] [PubMed] [Google Scholar]
- 8.Keswani RK, Baik J, Yeomans L, Hitzman C, Johnson A, Pawate A, Kenis PJA, Rodríguez-Hornedo N, Stringer KA, Rosania GR. Mol. Pharm. 2015. Chemical Analysis of Drug Biocrystals: A Role for Counterion Transport Pathways in Intracellular Drug Disposition. 10.1021/acs.molpharmaceut.5b00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 2008;9:847–56. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herd HL, Bartlett KT, Gustafson J a., McGill LD, Ghandehari H. Macrophage silica nanoparticle response is phenotypically dependent. Biomaterials. 2015;53:574–582. doi: 10.1016/j.biomaterials.2015.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinon F, Pétrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–41. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 12.Nadra I, Mason JC, Philippidis P, Florey O, Smythe CDW, McCarthy GM, Landis RC, Haskard DO. Proinflammatory activation of macrophages by basic calcium phosphate crystals via protein kinase C and MAP kinase pathways: a vicious cycle of inflammation and arterial calcification? Circ. Res. 2005;96:1248–56. doi: 10.1161/01.RES.0000171451.88616.c2. [DOI] [PubMed] [Google Scholar]
- 13.Rensburg CEJ Van, Anderson R, O'Sullivan JF. Riminophenazine compounds : pharmacology potential and anti-neoplastic. Crit. Rev. Oncol. 1997;25:55–67. doi: 10.1016/s1040-8428(96)00229-6. [DOI] [PubMed] [Google Scholar]
- 14.Yoon GS, Sud S, Keswani RK, Standiford TJ, Stringer KA, Rosania GR. Mol. Pharm. 2015. Phagocytosed Clofazimine Biocrystals can Modulate Innate Immune Signaling by Inhibiting TNFα and Boosting IL-1RA Secretion. 10.1021/acs.molpharmaceut.5b00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrison N, Marley G. The mode of action of clofazimine DNA binding studies. Int. J. Lepr. 1976;44:133–134. [PubMed] [Google Scholar]
- 16.Zhang X, Goncalves R, Mosser DM. The Isolation and Characterization of Murine Macrophages. Curr. Protoc. Immunol. 2008;9:1–18. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JA, Spidlen J, Boyce K, Cai J, Crosbie N, Dalphin M, Furlong J, Gasparetto M, Goldberg M, Goralczyk EM, Hyun B, Jansen K, Kollmann T, Kong M, Leif R, McWeeney S, Moloshok TD, Moore W, Nolan G, Nolan J, Nikolich-Zugich J, Parrish D, Purcell B, Qian Y, Selvaraj B, Smith C, Tchuvatkina O, Wertheimer A, Wilkinson P, Wilson C, Wood J, Zigon R, Scheuermann RH, Brinkman RR. MIFlowCyt: The minimum information about a flow cytometry experiment. Cytom. Part A. 2008;73:926–930. doi: 10.1002/cyto.a.20623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conalty ML, Jackson RD. Uptake by Reticulo-Endothelial Cells of the Riminophenazine B.663. Br. J. Exp. Pathol. 1962;43:650–654. [PMC free article] [PubMed] [Google Scholar]
- 19.Sandler ED, Ng VL, Hadley WK. Clofazimine Crystals in Alveolar Macrophages From a Patient with the Acquired Immnuodeficiency Syndrome. Arch. Pathol. Lab. Med. 1992;116:541–543. [PubMed] [Google Scholar]
- 20.Harbeck RJ, Worthen GS, Lebo TD, Peloquin CA. Clofazimine crystals in the cytoplasm of pulmonary macrophages. Ann. Pharmacother. 1999;33:250. doi: 10.1345/aph.18170. [DOI] [PubMed] [Google Scholar]
- 21.Mutlu M, Budinger GRS, Perlman H. Flow Cytometric Analysis of Macrophages and Dendritic Cell Subsets in the Mouse Lung. Am. J. Respir. Cell Mol. Biol. 2013;49:503–510. doi: 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Rensburg CEJ, Joont GK, O'Sullivan JF, Anderson R. Antimicrobial activities of clofazimine and B669 are mediated by lysophospholipids. Antimicrob. Agents Chemother. 1992;36:2729–2735. doi: 10.1128/aac.36.12.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy VM, O'Sullivan JF, Gangadharam PRJ. Antimycobacterial activities of riminophenazines. J. Antimicrob. Chemother. 1999;43:615–623. doi: 10.1093/jac/43.5.615. [DOI] [PubMed] [Google Scholar]
- 24.Degang Y, Nakamura K, Akama T, Ishido Y. Leprosy as a model of immunity. Future Microbiol. 2014;9:43–54. doi: 10.2217/fmb.13.140. [DOI] [PubMed] [Google Scholar]
- 25.Scollard DM, Adams LB, Gillis TP, Krahenbuhl JL, Truman RW, Williams DL. The continuing challenges of leprosy. Clin. Microbiol. Rev. 2006;19:338–81. doi: 10.1128/CMR.19.2.338-381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barry VC, Belton JG, Conalty ML, Denneny JM, Edward DW, O'Sullivan JF, Twomey D, Winder F. A New Series of Phenazines (Rimino-Compounds) With High Antituberculosis Activity. Nature. 1957;179:1013–1015. doi: 10.1038/1791013a0. [DOI] [PubMed] [Google Scholar]
- 27.Dey T, Brigden G, Cox H, Shubber Z, Cooke G, Ford N. Outcomes of clofazimine for the treatment of drug-resistant tuberculosis: a systematic review and meta-analysis. J. Antimicrob. Chemother. 2013;68:284–293. doi: 10.1093/jac/dks389. [DOI] [PubMed] [Google Scholar]
- 28.Tolentino JG, Rodriguez JN, Abalos RM. Controlled Long-Term Therapy of Leprosy with B663 (Lamprene, Clofazimine) compared with DDS. Int J Lepr. Other Mycobact. Dis. 1974;42:416–418. [PubMed] [Google Scholar]
- 29.Belaube P, Devaux J, Pizzi M, Boutboul R, Privat Y. Small Bowel Deposition of Crystals Associated with the Use of Clofazimine (Lamprene) in the Treatment of Prurigo Nodularis. Int J Lepr. Other Mycobact. Dis. 1983;51:328–330. [PubMed] [Google Scholar]
- 30.Kling J. Measure for Measure. Nature. 2015;518:439–443. doi: 10.1038/518439a. [DOI] [PubMed] [Google Scholar]
- 31.Ray A, Dittel BN. Isolation of Mouse Peritoneal Cavity Cells. J. Vis. Exp. 2010;35:e1488. doi: 10.3791/1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Austyn JM, Gordon S. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur. J. Immunol. 1981;11:805–15. doi: 10.1002/eji.1830111013. [DOI] [PubMed] [Google Scholar]
- 33.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma'ayan A, Chua W-J, Hansen TH, Turley SJ, Merad M, Randolph GJ. Consortium the IG. Gene Expression profiles and transcriptional regulatory pathways underlying mouse tissue macrophage identity and diversity. Nat. Immunol. 2013;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi KM, Kashyap PC, Dutta N, Stoltz G, Ordog T, Donohue TS, Bauer AJ, et al. Linden DR, Szurszewski JH, Gibbons SJ, Farrugia G. CD206-Positive M2 Macrophages That Express Heme Oxygenase-1 Protect Against Diabetic Gastroparesis in Mice. Gastroenterology. 2010;138:2399–2409. doi: 10.1053/j.gastro.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellón T, Martínez V, Lucendo B, del Peso G, Castro MJ, Aroeira LS, Rodríguez-Sanz A, Ossorio M, Sánchez-Villanueva R, Selgas R, Bajo MA. Alternative activation of macrophages in human peritoneum: implications for peritoneal fibrosis. Nephrol. Dial. Transplant. 2011;26:2995–3005. doi: 10.1093/ndt/gfq771. [DOI] [PubMed] [Google Scholar]
- 36.Liu G, Xia X-P, Gong S-L, Zhao Y. The Macrophage Heterogeneity : Difference Between Mouse Peritoneal Exudate and Splenic F4 / 80 + Macrophages. J. Cell. Physiol. 2006;352:341–352. doi: 10.1002/jcp.20732. [DOI] [PubMed] [Google Scholar]
- 37.Edwards JP, Zhang X, Frauwirth KA, Mosser DM. Biochemical and functional characterization of three activated macrophage populations. J. Leukoc. Biol. 2006;80:1298–1307. doi: 10.1189/jlb.0406249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Classen A, Lloberas J, Celada A. Macrophages and Dendritic Cells. In: Reiner N, editor. Methods Mol. Biol. Vol 531. Humana Press; 2009. pp. 29–43. [Google Scholar]
- 39.Hunninghake GW, Gadek JE, Kawanami O, Ferrans VJ, Crystal RG. Inflammatory and immune processes in the human lung in health and disease: evaluation by bronchoalveolar lavage. Am. J. Pathol. 1979;97:149–206. [PMC free article] [PubMed] [Google Scholar]
- 40.Van Rijt LS, Kuipers H, Vos N, Hijdra D, Hoogsteden HC, Lambrecht BN. A rapid flow cytometric method for determining the cellular composition of bronchoalveolar lavage fluid cells in mouse models of asthma. J. Immunol. Methods. 2004;288:111–121. doi: 10.1016/j.jim.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Duan M, Li WC, Vlahos R, Maxwell MJ, Anderson GP, Hibbs ML. Distinct macrophage subpopulations characterize acute infection and chronic inflammatory lung disease. J. Immunol. 2012;189:946–55. doi: 10.4049/jimmunol.1200660. [DOI] [PubMed] [Google Scholar]
- 42.Landsman L, Jung S. Lung macrophages serve as obligatory intermediate between blood monocytes and alveolar macrophages. J. Immunol. 2007;179:3488–3494. doi: 10.4049/jimmunol.179.6.3488. [DOI] [PubMed] [Google Scholar]
- 43.Wang JX, Bair AM, King SL, Shnayder R, Huang YF, Shieh CC, Soberman RJ, Fuhlbrigge RC, Nigrovic PA. Ly6G ligation blocks recruitment of neutrophils via a β2-integrin- dependent mechanism. Blood. 2012;120:1489–1498. doi: 10.1182/blood-2012-01-404046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens SL, Bao J, Hollis J, Lessov NS, Clark WM, Stenzel-Poore MP. The use of flow cytometry to evaluate temporal changes in inflammatory cells following focal cerebral ischemia in mice. Brain Res. 2002;932:110–119. doi: 10.1016/s0006-8993(02)02292-8. [DOI] [PubMed] [Google Scholar]
- 45.Peters NC, Egen JG, Secundino N, Debrabant A, Kimblin N, Kamhawi S, Lawyer P, Fay MP, Germain RN, Sacks D. In Vivo Imaging Reveals an Essential Role for Neutrophils in Leishmaniasis Transmitted by Sand Flies. Science (80-.) 2008;321:970–975. doi: 10.1126/science.1159194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, Lukyanov S, Zaraisky AG, Davidson MW, Chudakov DM. Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. Bright far-red fluorescent protein for whole-body imaging. Nat. Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 48.Rao J, Dragulescu-Andrasi A, Yao H. Fluorescence imaging in vivo: recent advances. Curr. Opin. Biotechnol. 2007;18:17–25. doi: 10.1016/j.copbio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Grosset JH, Tyagi S, Almeida DV, Converse PJ, Li SY, Ammerman NC, Bishai WR, Enarson D, Trébucq A. Assessment of clofazimine activity in a second-line regimen for tuberculosis in mice. Am. J. Respir. Crit. Care Med. 2013;188:608–612. doi: 10.1164/rccm.201304-0753OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanson RV, Adamson J, Moodley C, Ngcobo B, Ammerman NC, Dorasamy A, Moodley S, Mgaga Z, Tapley A, Bester LA, Singh S, Grosset JH, Almeida DV. Pharmacokinetics and pharmacodynamics of clofazimine in the mouse model of tuberculosis. Antimicrob. Agents Chemother. 2015 doi: 10.1128/AAC.00260-15. AAC.00260–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tyagi S, Ammerman NC, Li S-Y, Adamson J, Converse PJ, Swanson RV, Almeida DV, Grosset JH. Proc. Natl. Acad. Sci. U. S. A. 2015. Clofazimine shortens the duration of the first-line treatment regimen for experimental chemotherapy of tuberculosis; pp. 2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Irwin SM, Gruppo V, Brooks E, Gilliland J, Scherman M, Reichlen MJ, Leistikow R, Kramnik I, Nuermberger EL, Voskuil MI, Lenaerts AJ. Limited activity of clofazimine as a single drug in a mouse model of tuberculosis exhibiting caseous necrotic granulomas. Antimicrob. Agents Chemother. 2014;58:4026–4034. doi: 10.1128/AAC.02565-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lenaerts A, Barry CE, III, Dartois V. Heterogeneity in tuberculosis pathology, microenvironments and therapeutic responses. Immunol. Rev. 2015;264:288–307. doi: 10.1111/imr.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams K, Minkowski A, Amoabeng O, Peloquin CA, Taylor D, Andries K, Wallis RS, Mdluli KE, Nuermberger EL. Sterilizing activities of novel combinations lacking first- andsecond-line drugs in a murine model of tuberculosis. Antimicrob. Agents Chemother. 2012;56:3114–3120. doi: 10.1128/AAC.00384-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.