Abstract
The ensemble of antibodies found in serum and secretions represents the key adaptive component of B-cell mediated humoral immunity. The antibody repertoire is shaped by the historical record of exposure to exogenous factors such as pathogens and vaccines, as well as by endogenous host-intrinsic factors such as genetics, self-antigens, and age. Thanks to very recent technology advancements it is now becoming possible to identify and quantify the individual antibodies comprising the serological repertoire. In parallel, the advent of high throughput methods for antigen and immunosignature discovery opens up unprecedented opportunities to transform our understanding of numerous key questions in adaptive humoral immunity, including the nature and dynamics of serological memory, the role of polyspecific antibodies in health and disease and how protective responses to infections or vaccine challenge arise. Additionally, these technologies also hold great promise for therapeutic antibody and biomarker discovery in a variety of settings
Antibody Serology: Past Achievements
Serology is classically defined as the study of proteins, predominantly antibodies, found in blood and secretions such as saliva. The genesis of serology dates to the end of the 19th century and the pioneering “serum therapy” of Emil von Behring and Paul Ehrlich, followed for decades by elegant studies on the specificity of serological reactions by Karl Landsteiner. But it would take Landsteiner until the twilight of his career to formally demonstrate that an anti-serum does not comprise merely a single antibody but rather a mixture of different antibody populations of unknown complexity[1]. It was another decade before the plasma cell, which is responsible for the secretion of antibodies, was discovered, and then it was only 50 years ago, in 1965 that it was convincingly shown that antibodies are produced by B lymphocytes [2]. This latest discovery coincided with the development of new technologies in protein chemistry and the advent of molecular biology that, together, catalyzed a remarkable pace of progress in the understanding of B cell development and antibody formation. We now know that long-lived plasma cells constitute the (most likely) irreversible end-point of B cell development, show little or no evidence of proliferation and produce copious amounts of antibodies for years, and quite possibly for decades, in humans [3]. Long-lived plasma cells reside predominantly but not exclusively in the bone marrow, surviving within specialized anatomical niches with the help of anti-apoptotic signals provided by stromal cells [4]. Of note, a fraction of bone marrow plasma cells have been recently reported to lack CD19 expression and to be protected from mobilization and replacement by newly formed antibody producing cells following infection[5•], underscoring the heterogeneity of the long-lived compartment of plasma cells and, by extension, the pool of serum immunoglobulins.
The compendium of antibodies produced first by long-lived plasma cells and second by transient waves of short-lived plasma cells or plasmablasts (elicited in response to pathogen, vaccine or autoantigen stimulation) constitutes the two main components of antibody serological immunity. A third component is contributed by natural antibodies which recognize common pathogen antigens such as galactose-α-1,3-galactose (Anti-Gal) and have an innate-like protective function [6•,7]. The relative contribution of antibodies from long-lived plasma cells, transient plasmablasts and by “natural antibody”-producing cells (rodent B1 and marginal zone B cells, and perhaps the human analogues of B1 cells, although these have yet to definitively identified) to the serological immunity is poorly understood. The antibody concentration in blood is homeostatically controlled. For example, the Immunoglopbulin G (IgG) concentration in healthy adults is maintained at roughly between 7–17 mg/ml. The level of IgG produced by long-lived plasma cells changes very slowly over time [8]. Plasmablast bursts following vaccine or infection are accompanied by a dramatic increase in the antigen-specific antibody titer but generally seem to have little impact on the total concentration of immunoglobulin in blood. It is noteworthy that the fate of the large number of plasmablasts present in circulation at the peak of the plasmablast wave has never been quantitatively followed although it is well established that the majority apoptose and only a relatively small fraction mature to long lived plasma cells and are able to take residence in niches so that they can contribute to serological memory[9]. Finally, the fraction of natural antibodies in blood or secretions is not known, nor is it known how the total level of natural antibody (comprising mostly IgM but also some IgG and IgA) vary as a function of time in health or in inflammation.
Antibody Serology: Future Directions
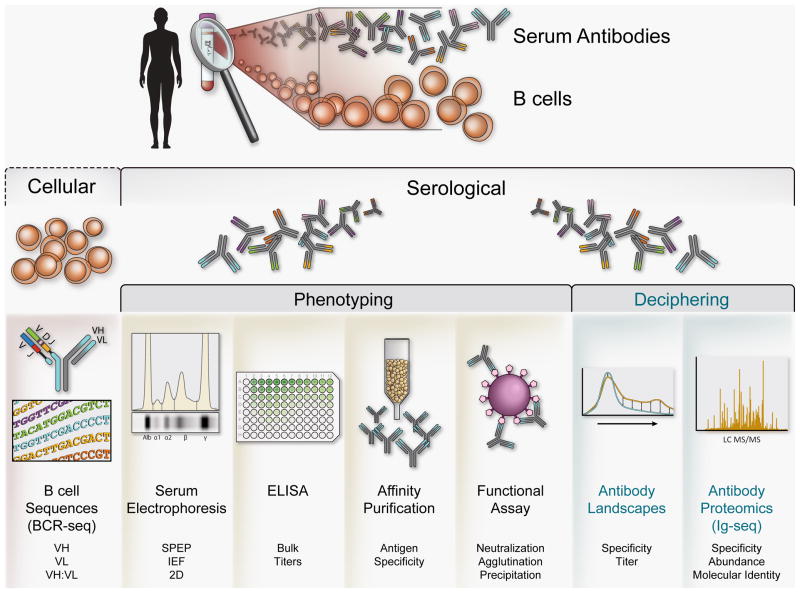
The essence of serological immunity is predicated on the existence of a diverse repertoire of antibodies, elicited over the life of the host and representing the integrated response to numerous antigenic stimuli. Due to the complexity and temporally dynamic nature of the antibody repertoire, the identification of its component immunoglobulins represents a formidable challenge. All serological studies to date have relied on the detection of an ensemble of antibodies that either could be resolved by a certain analytical technique or bound to a specified antigen (Fig. 1). Among the most useful metrics for assessing humoral immunity, the presence of neutralizing antibodies in the serum following vaccination or infection represents the best correlate for vaccine efficacy and for protection during invasive infections [10,11,12]. The limitations imposed by the inability to resolve complex serum antibody mixtures into their constituent clonal representatives and the need to have pre-established the identity of antigens of potential interest have obscured central questions of profound basic and clinical significance, some of which are outlined below:
Figure 1. Approaches for the analysis of human antibody repertoires.
Isolated B cells are sorted into several subsets based on expressed cell markers that correspond to the developmental stage of the B cell. These populations can be further processed for high-throughput sequencing to generate the antibody repertoire encoded by B cells (cellular repertoire, left side of the figure). The corresponding serum immunoglobulins are isolated from the samples and can be analyzed by various methods including well established technologies such as 2D gels or by recently established methodologies such as high resolution shotgun proteomics (serological repertoire, left side of the figure). The methodologies for serological immunoglobulin analysis can be broadly based upon the phenotype of an antibody subpopulation (e.g., ELISA titer of antigen-specific fraction) or upon decipherment of the molecular identity and sequence determination of an antibody subpopulation (e.g., LC-MS/MS immunoglobulin sequencing, Ig-seq).
-
First and foremost, there is nearly no information on the number of sequences (clonal diversity), functions and relative concentrations of the individual antibodies in serum. Upon reflection it is surely striking that after over a century of immunology research that the clonality of serum antibodies (number of distinct antibody clonotypes having same IGHV and IGHJ segments and highly homologous CDR-H3 sequences) in blood and in secretions has not been established experimentally even to a first approximation, although a remarkably prescient estimate of antibody diversity was first proposed by Talmage based on theoretical arguments, over half a century ago [13]. Nor do we know anything about how the clonality of serum antibodies changes as a function at extreme ages and disease status, or about the concentration distribution of individual antibodies in biological fluids. Without a detailed description of the identities and relative amounts of the antibodies that comprise the serological repertoire it is not possible to establish precisely how the B cell developmental program shapes protective immunity.
Information on the serological antibody repertoire is particularly pertinent to vaccine design/evaluation and to antibody discovery from patients. In an effort to better delineate the serological response to influenza by taking into account antigenic variation among strains circulating in humans, Fonville et al. sought to establish correlations between serum mediated neutralization against panels of different hemagglutinins and their phylogenetic distance [14]. The authors further developed a visualization tool for describing complex serological data by plotting antibody-mediated immunity as a function of the antigenic relationships among viruses (landscape plot).
For rapidly evolving viral pathogens such as HIV-1 or influenza, immortalization techniques and single cell cloning coupled with high throughput micro-neutralization assays of peripheral plasmablasts or memory B cells have led to the discovery of numerous broadly neutralizing antibodies (bNAbs) displaying neutralization breadth towards many or even nearly all known clinical isolates of a particular virus [15•,16•,17,18•,19]. To provide a first-order approximation of component antibody neutralization specificities in sera, Georgiev et al.[20•] proposed matching bNAb and serological neutralization potency fingerprints towards HIV-1 isolates.
Identification of bNAbs coupled with the reconstruction of the evolutionary route that led to the development of phylogenetically-related antibodies from an un-mutated common ancestor (UCA) is essential for the design of vaccine immunogens with the potential of eliciting broad protection. However, the fact that a DNA encoding a bNAb has been isolated from peripheral B cells does not mean that the respective antibody is present in serum or that it is produced at physiologically relevant amounts (i.e. at a concentration near the equilibrium dissociation constant such that the antibody can bind antigen). This divergence was described initially by Burnet stating that there is no requirement in the clonal selection theory that every cellular receptor has a corresponding secreted globulin [21]. As a matter of fact, considering that the BCR repertoire diversity in the memory and plasmablast compartments is orders of magnitude greater than that of the serological repertoire [22••,23] it follows that the overwhelming majority of peripheral B cell-encoded antibodies are unlikely to be present in detectable amounts as soluble proteins in blood or secretions and thus could not have contributed to humoral immunity.
Second, while it is well established that a significant fraction of antibodies display polyreactivity and that these antibodies have important physiological functions in processes such as the clearance of cell debris and in pathogen recognition[24,25], there is a paucity of methods for quantifying and characterizing the polyreactive fraction of the serological response. It is noteworthy that, in addition to natural antibodies, which are typically polyreactive, IgG antibodies encoded by human memory B cells [26] and even bone marrow plasma cells and also bNAbs elicited by anti-viral immune responses in response to HIV-1 infection [27] often recognize multiple antigens. There is a clear need to understand the mechanisms that drive polyreactivity and its implications in health and disease. One possible explanation is that polyreactivity originates from B cells that were not removed from the repertoire during B-cell development. For some pathogens, notably HIV, polyreactivity may confer a selective advantage to pathogen–specific antibodies [26,28].
Third, in many instances the antigens that are recognized by serum antibodies are not known a priori. The significance of identifying antigens, antigen surrogates (i.e. antigen-mimics that can be chemically distinct from the antigens that elicited an antibody response) and immunosignatures [29] for disease diagnosis is being increasingly recognized. Additionally, mapping the serum antigen reactivity profile in a comprehensive manner is key to understanding which environmental exposures play a more dominant role in shaping humoral immunity.
Antigen and Immunosignatures Discovery
Earlier methods for antigen discovery include the use of peptide libraries either immobilized in array format or displayed on M13 phage and serological analysis of recombinant cDNA expression libraries (SEREX), which employs cDNA libraries from tumor tissues for cancer antigen discovery and finally, antigen arrays [29,30•,31,32]. More recently advances in library screening methodologies and separately, in high throughput gene and peptide synthesis technologies have led to an explosion of high-resolution methods for the discovery of autoantigens and antigen surrogates. Daugherty and coworkers developed a technology that capitalized on E.coli display [33] of very large random peptide libraries for the identification of peptide motifs that bound to antibodies from patient sera. This technique led to the discovery of a diagnostically-relevant epitope for celiac disease that exhibited a high degree of amino acid identity to deamidated gliadin, a key antigen in celiac disease. In a separate study the same group reported a 7 peptide panel of highly statistical diagnostic significance for preeclampsia [34,35]. Using massively parallel DNA synthesis Larman et al. [36] constructed libraries of >410,000 overlapping 26-mer peptides spanning all ORFs in the human genome on T7 phage. Phage was immunopreciptated with patient sera and diagnostic immunosignatures were determined by NextGen sequencing. In an alternative approach very high density printed arrays of synthetic random peptide arrays were used for immunosignature discovery and were reported to exhibit higher accuracy than tiled peptide epitope arrays [37,38]. Since peptides may not be able to capture antibodies that bind to post-translational modifications and conformational epitopes Kodadek and coworkers pioneered the use of libraries of unnatural molecules and specifically peptoids (N-substituted oligoglycine polymers) for the discovery of surrogate antigen biomarkers from patient sera [29,39]. Other powerful methods that are currently used for antigen discovery include antigen microarrays [40,41], nucleic acid programmable protein arrays (NAPPAs) produced using in-vitro transcription/translation[42], peptide arrays [43] or finally arrays based on a different targets such as DNA, peptides and recombinant/native proteins [44].
Finally, for very complex conformational antigens on proteins that cannot be immobilized on arrays (e.g. integral membrane proteins) or contain complex post-translational modifications or for which a peptide or synthetic surrogate is not available, novel solution phase assays are being developed for diagnostic purposes. For example radio-binding assays (RBA) [45] using in vitro transcription/translation RBAs have been used for detection of autoantibodies. In one notable example Joseph and co-workers, studied the association of scleroderma with an humoral immune response to cancer. They found an association of genetic alterations the RNA polymerase III subunit (RPC1) encoding gene (POLR3A) in patients with autoantibodies to RPC1 suggesting that POLR3A mutations triggered cellular immunity and cross-reactive humoral immune responses [46]. Alternatively, light-emitting recombinant antigens (LIPS) [47] have been developed and used for the profiling of 16 autoantigens in SLE patients [48]. Solution phase assays are useful for diagnostic purposes but are difficult to implement for antigen discovery since thousands of soluble proteins having a native conformation would need to first be produced and validated
The Serological Antibody Repertoire
Until recently determining the sequence and relative concentration of the antibodies in the serum repertoire was considered a nearly impossible task: biological fluids contain many thousands of different antibodies all of which are chemically very similar, having an overall high degree of sequence identity, and whose concentrations can vary by several orders of magnitude in a dynamic fashion. Starting in 2008 LC-MS/MS instrumentation began to be used to detect immunoglobulin tryptic fragments in serum samples [49,50]. However, the Ig-derived peptides detected in these earlier studies, were overwhelmingly derived from the framework regions and did not provide sufficient information to piece together complete antibody sequences. By restricting the diversity of the antigen-specific antibody pool from the serum of immunized rabbits using antigen-affinity chromatography under stringent elution conditions, Polakiewicz and coworkers succeeded in using MS to identify overlapping antibody peptides that could help assemble a complete V gene. Combinatorial pairing of separate VH and VL sequences deduced from LC-MS/MS was then used then used to produce several antibodies displaying high affinity for antigen, first from rabbits and subsequently from humans [51,52]. In an alternate approach, dating to 2011, our lab invented a technology for determining the serological repertoire to a specified antigen [53] by combining: (i) NextGen V gene sequencing from peripheral memory B cells and plasmablasts to first create an archive of the antibodies encoded by an individual; (ii) enrichment of the pool of antigen-specific antibodies by affinity chromatography; (iii) determining the CDR-H3 peptides and other informative peptides as needed (e.g. CDR-H2) by LC-MS/MS bottom up proteomics; and (iv) the use of stringent informatics filters to assign the informative mass spectra to the entire VH gene with the help of the sequenced antibody gene archive from (i) above [54•,55]. In parallel we developed methods for the massive sequencing of the natively paired VH:VL repertoire, first in 10s of thousands and later on, in millions of peripheral B cells[56,57••] In this manner, the mass spectrometric identification of a unique peptide in a serum antibody could be mapped into the paired VH:VL repertoire archive to reveal the complete sequence of that antibody which then could be produced and studied in vitro. Thanks to the exquisite sensitivity of modern MS instrumentation, individual serum antibodies can be detected semi-quantitatively at levels as low as 0.4 ng/ml[22••]. Given that for an antibody to bind to antigen it has to be present at a concentration at or above its equilibrium dissociation constant, which is estimated to have a ceiling of around 0.1 nM or [58] the approach outlined above has more than adequate resolution for the detection of the repertoire of physiologically relevant antibodies in a sample.
As with any analytical technique, the experimental pipeline described above has certain limitations. First, a fraction of antibodies in the repertoire may evade discovery due to constraints inherent in LC-MS/MS proteomics. Such fault may arise when proteolysis fails to produce unique peptides for some sequences, physicochemical properties such as exceptional length limit peptide observation, or MS/MS spectral identification is ambiguous or incorrect. Additionally, unanticipated post-translational modifications or an incomplete sequence database may prevent assignment of some spectra. We estimate that the fraction of CDR-H3 peptide spectra that cannot be assigned is <20%, based on semiautomated inspection of the MS/MS data. Antibodies that evade detection using the proteomic approach outlined above may potentially be identified through use of alternative proteases and instrument fragmentation methods[59]Second, bottom up proteomic approaches reveal the antibody clonotypes (i.e. the family of antibodies having near identical CDR-H3s and V and J segments and likely derived from the same precursor B cell) in the repertoire. Somatic variants of antibodies having the same CDR-H3 are determined based on the NextGen sequencing data but cannot be unambiguously quantified proteomically. Third, the analysis of the antibody repertoire is both technically demanding and expensive, requiring access to VH:VL paired repertoire sequencing, high-end (Thermo-Electron Obritrap or equivalent mass accuracy) LC-MS/MS instrumentation and finally the requisite informatic and data storage infrastructure. Nonetheless, the above technical limitations notwithstanding, we now have the capability to delineate the serum antibody repertoire with medium-high resolution and with excellent sensitivity.
What’s Next? Addressing Fundamental Questions in Humoral Immunity
What kind of information can we glean by utilizing these new tools? We are at the dawn of a new era in the molecular-level understanding of the serological response and numerous questions wait to be answered. Examples of some major outstanding issues we expect to be addressed in the near future are listed below:
The size of the serological antibody repertoire: We have argued based on the amount of total IgG in healthy humans and the fact that physiologically relevant antibodies must exist at concentrations at or above KD, that a possible upper bound for the number of distinct clonotypes in human blood can be estimated to be ≥104 [23]. Preliminary MS analyses by our lab place the size of the IgG repertoire for a middle age adult at <20,000 distinct clonotypes consistent with previous numbers presented by Talmage during the late 50’s [13]. To put this number in perspective, the serological memory repertoire to vaccines we have analyzed so far such as tetanus[22••] TIV seasonal influenza, inactivated polio and pneumonococcal polysaccharide typically comprises between 50–400 antibody clonotypes each. In turn these results suggest that as much as 10–15% of the blood IgG pool corresponds or is “taken up” by vaccine-induced antibodies. How clonotypic diversity of the IgG pool changes as a function of age especially in the elderly and during early development and the relationship between overall antibody diversity and susceptibility to infection will need to be determined. Also it will be interesting to determine whether vaccination with a new vaccine impacts the established serological memory antibody repertoire to an earlier vaccine.
Concentration distribution of circulating antibodies: In biological fluids individual antibodies are present at widely different concentrations. For example we have found that out of the 80–120 antibody clonotypes that typically constitute the serum titer to the tetanus toxoid vaccine a single clonotype can account for as much as 15–30% of antigen-specific response [22••]. We have observed a similar polarization of the serological repertoire, manifest by the presence of one or a few highly abundant antibodies with a number of vaccine antigens. Obviously, such antibodies must be the product of large plasma cell expansions. The mechanisms that dictate the extent of expansion of certain B cell clones following antigen stimulation are not clear. Since highly abundant antibodies display comparable binding affinity as much less prevalent clones, differences in the degree of plasma cell expansions observed in peripheral blood at day 7 (the typical peak of plasmablast response in serum) clearly cannot be solely due to antigen affinity. Of note, extremely highly abundant antibody clones (clinically defined as globulin level >30 mg/ml) are observed in about 3% of asymptomatic adults over 50 yr old and in multiple myeloma patients[60]. There is a documented reciprocal change with age in the antibody titer to extrinsic and intrinsic antigens [61], but whether this is related to the presence of age-associated highly expanded serum antibodies remains to be determined.
The relationship and contribution of the different isotype repertoires to humoral immunity: In addition to IgG, human blood contains an appreciable level of IgM and IgA. IgD is present at a level 1/100 of that of IgA and IgE is found at an even lower concentration [62]. The relationships among the isotypic repertoires in serum need to be elucidated. For example to what degree is the antigen-specific IgG repertoire clonally related to the respective IgA repertoire? Additionally, there might be isotypic “layers” to human B cell memory, as recently described in the mouse, wherein particular memory B-cell subsets are predisposed to differentiate into plasma cells while others re-enter germinal center reactions [63]. Are features such as degree of somatic hypermutation, V gene usage, CDR-H3 length etc comparable among the class switched repertoires? While some of these issues are beginning to be explored for peripheral B cells through the use of NextGen sequencing approaches [64,65] there is presently no information for the repertoire at the protein level. A related immunological puzzle which could be clarified using Ig-seq proteomics is the extent to which peripheral blood IgM+IgD+CD27+ circulating B cells are germinal center-derived memory B cells or are instead related to marginal zone B cells found in human spleen, and whether, analogous to the mouse, they constitute a distinct B-cell lineage harboring a pre-diversified repertoire that is responsible for predominating anti-polysaccharide responses against encapsulated bacteria such as S. pneumonia. Finally an equally intriguing question is the degree to which the antigen-specific repertoires in blood and in secretions are clonally related and whether they are directed to the same epitopes.
Significance of antibody post-translational modifications: Post-translational modifications impact all aspects of antibody function including antigen-binding, stability to aggregation and degradation, interaction with Fc binding proteins and effector functions and half-life[66•]. Antibody post-translational modifications have been studied in the context of quality control of therapeutic antibodies, a key issue for the biopharmaceutical industry. How post-translational modifications influence the selection and function of circulating antibodies is not known. Certainly, oxidation must be quite prevalent in circulating antibodies since they originate from oxidatively stressed plasma cells and then are exposed to the high redox potential of the serum for many days. It is quite plausible that surface Ig on B cells is also subject to oxidation and perhaps oxidized residues may contribute to antigen affinity and B cell selection during the germinal center reaction. Fc glycosylation is well established to impact the effect of antibodies on the innate immune system via the modulation of Fc receptor interactions[67]. While glycosylation heterogeneity in monoclonal antibodies has been studied extensively [66•] recent advances now enable deep analyses of human glycosylation in complex mixtures. For example, Pučić et al. used hydrophilic interaction chromatography and mass spectrometry to investigate correlation between gender, age and heredity on the relative abundance of serum IgG N-glycan structures for a cohort of 2298 individuals[68]. Though this work focused solely on the characterization of the pooled N-glycans from each sample, MS based methods may enable the future study of serum Ig glycosylation at the individual antibody level[69].
The polyspecific antibody repertoire: As mentioned above, the immunological significance of antibody polyreactivity or polyspecificity is being increasingly appreciated [24,70]. However, polyspecificity as a feature of the serum antibody response has not been investigated in a systematic way. Part of the problem is inherent to the diversity of the antibody repertoire of normal serum. Perhaps this is an area where lessons and tools from the biotherapeutics industry which has developed a number of widely used metrics and probes to evaluate polyspecificity in the context of therapeutic antibody development can be applied to molecular immunology[71]., Cell-based studies have shown that up to 50% of B cells found in cord blood of human newborns, and approximately 20% of peripheral blood B cells circulating in human adults encode polyreactive antibodies[72,73]. Furthermore, although significantly reduced by putative checkpoint mechanisms, the terminal repertoire of bone marrow plasma cells still encompasses ≥10% polyreactive antibodies [74] which suggests that a similar observation awaits discovery at the level of the serum repertoire (Fig. 1).
Conclusions
We are on the verge of a new and exciting research era in serology. For the first time, newly developed technologies provide us with the means to interrogate the adaptive immune response at unprecedented resolution to determine both the spectrum of antigens towards which the serological response is directed and the antibodies that comprise the respective polyclonal antibody pools towards these antigens. Together with new tools for the analysis of B cell subsets and BCR repertoires we can now begin to envision the possibility of a fairly complete description of the nature of antibody responses in health and disease. Needless to say numerous challenges remain: First off, the molecular level dissection of humoral immunity is technically demanding and requires a combination of tools that are currently available only in a very small set of laboratories. It will be interesting to see how and whether these techniques may be “industrialized” to become available for clinical use and for use by the wider research community. Second, these studies result in very large and complex data files and a major effort will be required for data storage, annotation, storage and meta-analysis. Third, numerous experimental challenges still need to be addressed: how to develop appropriate probes for the quantification and for characterizing the composition of the polyspecific antibody repertoire, how to best characterize the antigenic determinants in complex pathogens (e.g. S. aureus and other bacterial pathogens, parasites etc) how to efficiently identify the neutralizing antibodies present in serum (as contrasted to relying on B cell cloning which as was argued above may not necessarily yield the biologically relevant antibodies in a patient) and many others. Finally and perhaps most importantly the deluge of data that can be generated by the tools described above and by other systems immunology approaches should by no means overshadow the need for first formulating testable hypotheses and well posed questions that in turn can provide definitive insights into the working of the immune system. Critical thinking, appreciation of the biological context and, for disease biology a clear understanding of the clinical need must be the driving force behind any large scale science including in this case modern serology.
Highlights.
New technologies have revolutionized serological antibody analysis
NextGen sequencing and shotgun proteomics help identify and quantify serum mAb’s
Clonality and dynamics of serum mAb repertoire in various states can be addressed
New tools available for identifying novel antigens that elicit serological response
Acknowledgments
Work on immune profiling in our lab was supported by grants HDTRA1-12-C-0105 from DTRA, RO1CA106006 and 5U19AI057234-09 from NIH and by the Clayton Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Landsteiner K, van der Scheer J. On cross reactions of egg albumin sera. J Exp Med. 1940;71:445–454. doi: 10.1084/jem.71.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper MD. The early history of B cells. Nat Rev Immunol. 2015;15:191–197. doi: 10.1038/nri3801. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida T, Mei HE, Dörner T, Hiepe F, Radbruch A, Fillatreau S, Hoyer BF. Memory B and memory plasma cells. Immunol Rev. 2010;237:117–139. doi: 10.1111/j.1600-065X.2010.00938.x. [DOI] [PubMed] [Google Scholar]
- 4.Kometani K, Kurosaki T. Differentiation and maintenance of long-lived plasma cells. Curr Opin Immunol. 2015;33c:64–69. doi: 10.1016/j.coi.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 5•.Mei HE, Wirries I, Frolich D, Brisslert M, Giesecke C, Grun JR, Alexander T, Schmidt S, Luda K, Kuhl AA, Engelmann R, Durr M, Scheel T, Bokarewa M, Perka C, Radbruch A, Dörner T. A unique population of IgG-expressing plasma cells lacking CD19 is enriched in human bone marrow. Blood. 2015;125:1739–1748. doi: 10.1182/blood-2014-02-555169. On mature B cells, CD19 is a co-receptor of the B cell receptor and regulates its signaling. This study characterizes distinct subsets of human plasma cells (PC) differing by CD19 expression. Using a combination of FACS-sorting, single cell sequencing, EliSpot and statistical analysis, the researchers demonstrated that the CD19-B cell population in the BM are independent from being replaced by new PC subset and comprise a lower level of somatic hypermutation. The authors concluded that this PC subset are differentiated in early life. [DOI] [PubMed] [Google Scholar]
- 6•.Panda S, Ding JL. Natural antibodies bridge innate and adaptive immunity. J Immunol. 2015;194:13–20. doi: 10.4049/jimmunol.1400844. Natural Abs, mostly isotypes IgM, IgG3, and IgA, were discovered nearly half a century ago. Despite knowledge about the role of the polyreactive natural IgM there is a lack of clarity about the physiological role of natural IgG and natural IgA because they appear incapable of recognizing Ags on their own and are perceived as nonreactive. In this review, the authors describe the findings on natural Abs, focusing on the role of natural IgM, recent findings on natural IgG, which collaborates with serum lectins, and how this immune complex links innate immunity to adaptive immunity. [DOI] [PubMed] [Google Scholar]
- 7.Galili U. Anti-Gal: an abundant human natural antibody of multiple pathogeneses and clinical benefits. Immunology. 2013;140:1–11. doi: 10.1111/imm.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amanna IJ, Carlson NE, Slifka MK. Duration of humoral immunity to common viral and vaccine antigens. N Engl J Med. 2007;357:1903–1915. doi: 10.1056/NEJMoa066092. [DOI] [PubMed] [Google Scholar]
- 9.Radbruch A, Muehlinghaus G, Luger EO, Inamine A, Smith KG, Dorner T, Hiepe F. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat Rev Immunol. 2006;6:741–750. doi: 10.1038/nri1886. [DOI] [PubMed] [Google Scholar]
- 10.Plotkin SA. Vaccines: correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47:401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 11.Plotkin SA. Correlates of Protection Induced by Vaccination. Clin Vaccine Immunol. 2010:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rappuoli R. Bridging the knowledge gaps in vaccine design. Nat Biotechnol. 2007;25:1361–1366. doi: 10.1038/nbt1207-1361. [DOI] [PubMed] [Google Scholar]
- 13.Talmage DW. Immunological Specificity: Unique combinations of selected natural globulins provide an alternative to the classical concept. Science. 1959;129:1643–1648. doi: 10.1126/science.129.3364.1643. [DOI] [PubMed] [Google Scholar]
- 14.Fonville JM, Wilks SH, James SL, Fox A, Ventresca M, Aban M, Xue L, Jones TC, Le NM, Pham QT, Tran ND, Wong Y, Mosterin A, Katzelnick LC, Labonte D, Le TT, van der Net G, Skepner E, Russell CA, Kaplan TD, Rimmelzwaan GF, Masurel N, de Jong JC, Palache A, Beyer WE, Le QM, Nguyen TH, Wertheim HF, Hurt AC, Osterhaus AD, Barr IG, Fouchier RA, Horby PW, Smith DJ. Antibody landscapes after influenza virus infection or vaccination. Science. 2014;346:996–1000. doi: 10.1126/science.1256427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. The high variability of viral glycoproteins and sophisticated viral escape mechanisms presents a great challenge to the development of antibody-based therapies or vaccines that could confer broad and long-lasting protection. In this review the authors describe viral escape mechanisms and neutralization. The review focuses on new approaches that were developed during the past five years that enabled the high efficiency investigation of human memory B cells and plasma cells and to identify and isolate several broadly neutralizing antiviral antibodies against highly variable pathogens such as HIV-1 and influenza virus. [DOI] [PubMed] [Google Scholar]
- 16•.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. In this study the authors provided data regarding the understanding of the interaction between HIV-1 virus evolution and maturing broad neutralizing antibody lineages in early infection. By providing data related to the viral and antibody evolution leading to induction of a lineage of HIV-1 broadly neutralizing antibodies, they provided insights into strategies to elicit similar antibodies by vaccination. The authors reported the isolation of the CH103 CD4-binding site broad neutralizing antibody clonal lineage from an African patient and show that the CH103 broad neutralizing antibody lineage is less mutated than most other CD4-binding site broad neutralizing antibody. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O’Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med. 2014;20:143–151. doi: 10.1038/nm.3443. In this study, the researches utilize anti–HA stalk broad neutralizing antibodies that neutralize a panel of H1 or of both H1 and H5 influenza viruses, to show that the anti-stalk bNAbs required Fc-FcγR interactions for maximum bNAb-mediated neutralization of influenza virus in vivo, whereas the strain-specific anti–HA head mAbs did not. The anti-stalk bNAbs functioned after viral entry in vivo and were superior inducers of antibody-dependent cellular cytotoxicity (ADCC), suggesting a mechanism for their FcγR-dependent function in vivo. The authors underlined the potential of engineering the human IgG1 Fc to enhance binding to its activation receptors and augment antibody effector function that may results in the development of future therapies, including vaccination protocols to elicit bNAbs and passive antibody therapies using bNAbs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunol Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, Bailer RT, McKee K, Nason M, O’Dell S, Ofek G, Pancera M, Srivatsan S, Shapiro L, Connors M, Migueles SA, Morris L, Nishimura Y, Martin MA, Mascola JR, Kwong PD. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science. 2013;340:751–756. doi: 10.1126/science.1233989. In this paper the authors hypothesized that when the immune system is presented with a diverse set of HIV-1 isolates, it generates monoclonal antibodies with specific neutralization fingerprints and neutralization patterns of a polyclonal serum could be viewed as the combined effect of the neutralization fingerprints of the constituent monoclonal antibodies. They demonstrated the neutralization fingerprints for 30 neutralizing antibodies on a panel of 34 diverse HIV-1 strains and showed that similarity in neutralization fingerprint correlated with similarity in epitope. Furthermore, they used these fingerprints to delineate specificities of polyclonal sera from 24 HIV-1-infected donors. Collectively, they suggested that epitope delineation based on neutralization fingerprints may provide a transformative strategy for screening sera or characterizing antibody specificities induced upon infection or vaccination against HIV-1 as well as other viruses. [DOI] [PubMed] [Google Scholar]
- 21.Burnet FM. The Clonal Selection Theory of Acquired Immunity. Vanderbilt University Press; 1959. p. 95. [Google Scholar]
- 22••.Lavinder JJ, Wine Y, Giesecke C, Ippolito GC, Horton AP, Lungu OI, Hoi KH, DeKosky BJ, Murrin EM, Wirth MM, Ellington AD, Doerner T, Marcotte EM, Boutz DR, Georgiou G. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc Natl Acad Sci USA. 2014;111:2259–2264. doi: 10.1073/pnas.1317793111. Most vaccines confer immunity by eliciting long-term production of antibodies that bind to and neutralize the vaccine antigen. Remarkably, very little is known regarding the identities, sequence diversity, relative concentrations, or binding functionalities of the mAbs that comprise the serum repertoire elicited by vaccination. In this study the authors delineated the constituent antibodies of the human serum IgG repertoire after vaccination and examined their relationship to the antibody V gene repertoire encoded by circulating B cells. The results detail the molecular composition and characteristics of the vaccine-specific serum antibody repertoire and demonstrate differences between the end-point response (the serum antibodies) and the peripheral B cells responding to the vaccine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavinder JJ, Horton AP, Georgiou G, Ippolito GC. Next-generation sequencing and protein mass spectrometry for the comprehensive analysis of human cellular and serum antibody repertoires. Curr Opin Chem Biol. 2015;24c:112–120. doi: 10.1016/j.cbpa.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Dimitrov JD, Planchais C, Roumenina LT, Vassilev TL, Kaveri SV, Lacroix-Desmazes S. Antibody polyreactivity in health and disease: statu variabilis. J Immunol. 2013;191:993–999. doi: 10.4049/jimmunol.1300880. [DOI] [PubMed] [Google Scholar]
- 25.Zhou ZH, Zhang Y, Hu YF, Wahl LM, Cisar JO, Notkins AL. The broad antibacterial activity of the natural antibody repertoire is due to polyreactive antibodies. Cell Host Microbe. 2007;1:51–61. doi: 10.1016/j.chom.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, Wardemann H. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26:205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu M, Yang G, Wiehe K, Nicely NI, Vandergrift NA, Rountree W, Bonsignori M, Alam SM, Gao J, Haynes BF, Kelsoe G. Polyreactivity and autoreactivity among HIV-1 antibodies. J Virol. 2015;89:784–798. doi: 10.1128/JVI.02378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mouquet H, Nussenzweig MC. Polyreactive antibodies in adaptive immune responses to viruses. Cell Mol Life Sci. 2012;69:1435–1445. doi: 10.1007/s00018-011-0872-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kodadek T. Chemical tools to monitor and manipulate the adaptive immune system. Chem Biol. 2014;21:1066–1074. doi: 10.1016/j.chembiol.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Weiss-Ottolenghi Y, Gershoni JM. Profiling the IgOme: meeting the challenge. FEBS Lett. 2014;588:318–325. doi: 10.1016/j.febslet.2013.11.005. This paper reviews various methodologies that enable researchers to better understand the entire repertoire of antibodies in our serum (IgOme). They discuss various aspect of the development of the adaptive immune system and how “serum memory” is affected by immunization, boosts, encounters with pathogens, physiology and old age. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sokolove J, Lindstrom TM, Robinson WH. Development and deployment of antigen arrays for investigation of B-cell fine specificity in autoimmune disease. Front Biosci (Elite Ed) 2012;4:320–330. doi: 10.2741/379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kügler J, Zantow J, Meyer T, Hust M. Oligopeptide M13 Phage Display in Pathogen Research. Viruses. 2013;5:2531–2545. doi: 10.3390/v5102531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daugherty PS, Chen G, Olsen MJ, Iverson BL, Georgiou G. Antibody affinity maturation using bacterial surface display. Protein Eng. 1998;11:825–832. doi: 10.1093/protein/11.9.825. [DOI] [PubMed] [Google Scholar]
- 34.Ballew JT, Murray JA, Collin P, Mäki M, Kagnoff MF, Kaukinen K, Daugherty PS. Antibody biomarker discovery through in vitro directed evolution of consensus recognition epitopes. Proc Natl Acad Sci USA. 2013;110:19330–19335. doi: 10.1073/pnas.1314792110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elliott SE, Parchim NF, Liu C, Xia Y, Kellems RE, Soffici AR, Daugherty PS. Characterization of antibody specificities associated with preeclampsia. Hypertension. 2014;63:1086–1093. doi: 10.1161/HYPERTENSIONAHA.113.02362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larman HB, Zhao Z, Laserson U, Li MZ, Ciccia A, Gakidis MAM, Church GM, Kesari S, Leproust EM, Solimini NL, Elledge SJ. Autoantigen discovery with a synthetic human peptidome. Nat Biotechnol. 2011;29:535–541. doi: 10.1038/nbt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stafford P, Cichacz Z, Woodbury NW, Johnston SA. Immunosignature system for diagnosis of cancer. Proc Natl Acad Sci USA. 2014;111:3072–3080. doi: 10.1073/pnas.1409432111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Navalkar KA, Johnston SA, Stafford P. Peptide based diagnostics: Are random-sequence peptides more useful than tiling proteome sequences? J Immunol Methods. 2015;417:10–21. doi: 10.1016/j.jim.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Doran TM, Simanski S, Kodadek T. Discovery of Native Autoantigens via Antigen Surrogate Technology: Application to Type 1 Diabetes. ACS Chem Biol. 2015;10:401–412. doi: 10.1021/cb5007618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeste A, Quintana FJ. Antigen microarrays for the study of autoimmune diseases. Clin Chem. 2013;59:1036–1044. doi: 10.1373/clinchem.2012.194423. [DOI] [PubMed] [Google Scholar]
- 41.Kloppot P, Selle M, Kohler C, Stentzel S, Fuchs S, Liebscher V, Muller E, Kale D, Ohlsen K, Broker BM, Zipfel PF, Kahl BC, Ehricht R, Hecker M, Engelmann S. Microarray based identification of human antibodies against Staphylococcus aureus antigens. Proteomics Clin Appl. 2015 doi: 10.1002/prca.201400123. [DOI] [PubMed] [Google Scholar]
- 42.Miersch S, Bian X, Wallstrom G, Sibani S, Logvinenko T, Wasserfall CH, Schatz D, Atkinson M, Qiu J, LaBaer J. Serological autoantibody profiling of type 1 diabetes by protein arrays. J Proteomics. 2013;94:486–496. doi: 10.1016/j.jprot.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 43.Ayoglu B, Haggmark A, Khademi M, Olsson T, Uhlen M, Schwenk JM, Nilsson P. Autoantibody profiling in multiple sclerosis using arrays of human protein fragments. Mol Cell Proteomics. 2013;12:2657–2672. doi: 10.1074/mcp.M112.026757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price JV, Haddon DJ, Kemmer D, Delepine G, Mandelbaum G, Jarrell JA, Gupta R, Balboni I, Chakravarty EF, Sokolove J, Shum AK, Anderson MS, Cheng MH, Robinson WH, Browne SK, Holland SM, Baechler EC, Utz PJ. Protein microarray analysis reveals BAFF-binding autoantibodies in systemic lupus erythematosus. J Clin Invest. 2013;123:5135–5145. doi: 10.1172/JCI70231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shum AK, Alimohammadi M, Tan CL, Cheng MH, Metzger TC, Law CS, Lwin W, Perheentupa J, Bour-Jordan H, Carel JC, Husebye ES, De Luca F, Janson C, Sargur R, Dubois N, Kajosaari M, Wolters PJ, Chapman HA, Kampe O, Anderson MS. BPIFB1 is a lung-specific autoantigen associated with interstitial lung disease. Sci Transl Med. 2013;5:206ra139. doi: 10.1126/scitranslmed.3006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joseph CG, Darrah E, Shah AA, Skora AD, Casciola-Rosen LA, Wigley FM, Boin F, Fava A, Thoburn C, Kinde I, Jiao Y, Papadopoulos N, Kinzler KW, Vogelstein B, Rosen A. Association of the autoimmune disease scleroderma with an immunologic response to cancer. Science. 2014;343:152–157. doi: 10.1126/science.1246886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ustinova J, Zusinaite E, Utt M, Metskula K, Reimand K, Huchaiah V, Merits A, Uibo R. Development of a luciferase-based system for the detection of ZnT8 autoantibodies. J Immunol Methods. 2014;405:67–73. doi: 10.1016/j.jim.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 48.Ching KH, Burbelo PD, Tipton C, Wei C, Petri M, Sanz I, Iadarola MJ. Two Major Autoantibody Clusters in Systemic Lupus Erythematosus. PLoS ONE. 2012;7:e32001. doi: 10.1371/journal.pone.0032001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Obermeier B, Mentele R, Malotka J, Kellermann J, Kumpfel T, Wekerle H, Lottspeich F, Hohlfeld R, Dornmair K. Matching of oligoclonal immunoglobulin transcriptomes and proteomes of cerebrospinal fluid in multiple sclerosis. Nat Med. 2008;14:688–693. doi: 10.1038/nm1714. [DOI] [PubMed] [Google Scholar]
- 50.VanDuijn MM, Dekker LJ, Zeneyedpour L, Smitt PA, Luider TM. Immune responses are characterized by specific shared immunoglobulin peptides that can be detected by proteomic techniques. J Biol Chem. 2010;285:29247–29253. doi: 10.1074/jbc.M110.139071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheung WC, Beausoleil SA, Zhang X, Sato S, Schieferl SM, Wieler JS, Beaudet JG, Ramenani RK, Popova L, Comb MJ, Rush J, Polakiewicz RD. A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat Biotechnol. 2012;30:447–452. doi: 10.1038/nbt.2167. [DOI] [PubMed] [Google Scholar]
- 52.Sato S, Beausoleil SA, Popova L, Beaudet JG, Ramenani RK, Zhang X, Wieler JS, Schieferl SM, Cheung WC, Polakiewicz RD. Proteomics-directed cloning of circulating antiviral human monoclonal antibodies. Nat Biotechnol. 2012;30:1039–1043. doi: 10.1038/nbt.2406. [DOI] [PubMed] [Google Scholar]
- 53.Reddy S, Ge X, Lavinder J, Boutz D, Ellington AD, Marcotte EM, Georgiou G. Rapid Isolation of Monoclonal Antibodies from Animals. US Patent. 2011
- 54•.Wine Y, Lavinder JJ, Boutz DR, Miklos AE, Hughes RA, Hoi KH, Jung ST, Horton AP, Murrin EM, Ellington AD, Marcotte EM, Georgiou G. Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response. Proc Natl Acad Sci USA. 2013;110:2993–2998. doi: 10.1073/pnas.1213737110. This paper described how the monoclonal antibody composition was delineated using a methodology that integrates proteomic analysis of the serum- derived antigen-specific polyclonal antibody pool, NextGen sequencing of the immunoglobulin heavy chain variable region (VH gene) repertoire with advanced bioinformatics tools. After booster immunization in a rabbit, it was demonstrated that the antigen- specific serum immune response is oligoclonal, comprising antibodies encoding 34 different CDRH3s that group into 30 distinct antibody VH clonotypes. Of these 34 CDRH3s, 12 account for ~60% of the antigen-specific CDRH3 peptide mass spectral counts. Proteomically identified antibodies were synthesized and shown to display subnanomolar affinities. The ability to deconvolute the polyclonal serum response is likely to be of key importance for analyzing antibody responses after vaccination and help answer fundamental questions related to the adaptive immune responses in health and disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boutz DR, Horton AP, Wine Y, Lavinder JJ, Georgiou G, Marcotte EM. Proteomic Identification of Monoclonal Antibodies from Serum. Anal Chem. 2014;86:4758–4766. doi: 10.1021/ac4037679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeKosky BJ, Ippolito GC, Deschner RP, Lavinder JJ, Wine Y, Rawlings BM, Varadarajan N, Giesecke C, Dörner T, Andrews SF, Wilson PC, Hunicke-Smith SP, Willson CG, Ellington AD, Georgiou G. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol. 2013;31:166–169. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57••.DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med. 2015;21:86–91. doi: 10.1038/nm.3743. Here the authors describe a technology that enables sequencing of the native paired VH-VL repertoire from millions of B cells within a few hours of experimental effort and using equipment that can be built inexpensively by most laboratories. Because the variable domains of antibody heavy and light chains (VH and VL, respectively) are encoded by different mRNA transcripts, until recently it was only possible to determine the paired VH-VL sequences for moderate numbers of cells, far fewer than the diversity contained reasonable samples size. Massive VH-VL repertoire analyses of three human donors provided new immunological insights including (i) the identity, frequency and pairing propensity of shared, or ‘public’, VL genes, (ii) the detection of allelic inclusion (an implicated autoimmune mechanism) in healthy individuals and (iii) the occurrence of antibodies with features, in terms of gene usage and CDR3 length, associated with broadly neutralizing antibodies to rapidly evolving viruses such as HIV-1 and influenza. [DOI] [PubMed] [Google Scholar]
- 58.Foote J, Eisen HN. Breaking the affinity ceiling for antibodies and T cell receptors. Proc Natl Acad Sci U S A. 2000;97:10679–10681. doi: 10.1073/pnas.97.20.10679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swaney DL, Wenger CD, Coon JJ. Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J Proteome Res. 2010;9:1323–1329. doi: 10.1021/pr900863u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sigurdardottir E, Turesson I, Lund S, et al. THe role of diagnosis and clinical follow-up of monoclonal gammopathy of undetermined significance on survival in multiple myeloma. JAMA Oncology. 2015 doi: 10.1001/jamaoncol.2015.23. Published advanced online. [DOI] [PubMed] [Google Scholar]
- 61.Rowley M, Buchanan H, Mackay I. Reciprocal change with age in antibody to extrinsic and intrinsic antigens. The Lancet. 292:24–26. doi: 10.1016/s0140-6736(68)92893-6. [DOI] [PubMed] [Google Scholar]
- 62.Murphy K, Travers P, Walport M, Janeway C. Janeway’s immunobiology. New York: Garland Science; 2012. [Google Scholar]
- 63.Kurosaki T, Kometani K, Ise W. Memory B cells. Nat Rev Immunol. 2015;15:149–159. doi: 10.1038/nri3802. [DOI] [PubMed] [Google Scholar]
- 64.Briney BS, Willis JR, Finn JA, McKinney BA, Crowe JE., Jr Tissue-specific expressed antibody variable gene repertoires. PLoS One. 2014;9:e100839. doi: 10.1371/journal.pone.0100839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat Biotech. 2014;32:158–168. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66•.Hmiel LK, Brorson KA, Boyne MT., 2nd Post-translational structural modifications of immunoglobulin G and their effect on biological activity. Anal Bioanal Chem. 2015;407:79–94. doi: 10.1007/s00216-014-8108-x. [DOI] [PubMed] [Google Scholar]
- 67.Pincetic A, Bournazos S, DiLillo DJ, Maamary J, Wang TT, Dahan R, Fiebiger BM, Ravetch JV. Type I and type II Fc receptors regulate innate and adaptive immunity. Nat Immunol. 2014;15:707–716. doi: 10.1038/ni.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pucic M, Knezevic A, Vidic J, Adamczyk B, Novokmet M, Polasek O, Gornik O, Supraha-Goreta S, Wormald MR, Redzic I, Campbell H, Wright A, Hastie ND, Wilson JF, Rudan I, Wuhrer M, Rudd PM, Josic D, Lauc G. High throughput isolation and glycosylation analysis of IgG-variability and heritability of the IgG glycome in three isolated human populations. Mol Cell Proteomics. 2011;10:M111.010090. doi: 10.1074/mcp.M111.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thaysen-Andersen M, Packer NH. Advances in LC-MS/MS-based glycoproteomics: getting closer to system-wide site-specific mapping of the N- and O-glycoproteome. Biochim Biophys Acta. 2014;1844:1437–1452. doi: 10.1016/j.bbapap.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 70.Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 Vaccine Development and Therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu Y, Roach W, Sun T, Jain T, Prinz B, Yu TY, Torrey J, Thomas J, Bobrowicz P, Vásquez M, Wittrup KD, Krauland E. Addressing polyspecificity of antibodies selected from an in vitro yeast presentation system: a FACS-based, high-throughput selection and analytical tool. Protein Eng Des Sel. 2013;26:663–670. doi: 10.1093/protein/gzt047. [DOI] [PubMed] [Google Scholar]
- 72.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 73.Chen ZJ, Wheeler CJ, Shi W, Wu AJ, Yarboro CH, Gallagher M, Notkins AL. Polyreactive antigen-binding B cells are the predominant cell type in the newborn B cell repertoire. Eur J Immunol. 1998;28:989–994. doi: 10.1002/(SICI)1521-4141(199803)28:03<989::AID-IMMU989>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 74.Scheid JF, Mouquet H, Kofer J, Yurasov S, Nussenzweig MC, Wardemann H. Differential regulation of self-reactivity discriminates between IgG+ human circulating memory B cells and bone marrow plasma cells. P Natl Acad Sci USA. 2011;108:18044–18048. doi: 10.1073/pnas.1113395108. [DOI] [PMC free article] [PubMed] [Google Scholar]