Abstract
High throughput RNA sequencing has accelerated discovery of the complex regulatory roles of small RNAs, but RNAs containing modified nucleosides may escape detection when those modifications interfere with reverse transcription during RNA-seq library preparation. Here we describe AlkB-facilitated RNA Methylation sequencing (ARM-Seq) which uses pre-treatment with Escherichia coli AlkB to demethylate 1-methyladenosine, 3-methylcytidine, and 1-methylguanosine, all commonly found in transfer RNAs. Comparative methylation analysis using ARM-Seq provides the first detailed, transcriptome-scale map of these modifications, and reveals an abundance of previously undetected, methylated small RNAs derived from tRNAs. ARM-Seq demonstrates that tRNA-derived small RNAs accurately recapitulate the m1A modification state for well-characterized yeast tRNAs, and generates new predictions for a large number of human tRNAs, including tRNA precursors and mitochondrial tRNAs. Thus, ARM-Seq provides broad utility for identifying previously overlooked methyl-modified RNAs, can efficiently monitor methylation state, and may reveal new roles for tRNA-derived RNAs as biomarkers or signaling molecules.
Introduction
Next-generation RNA-sequencing has provided insight into the importance of small RNAs in a wide range of biological contexts. Transfer RNAs (tRNAs) are among the most abundant RNAs in all organisms, so it is perhaps unsurprising that tRNA fragments and half molecules are often abundant constituents of small RNA sequencing libraries1–3. There is increasing evidence that these tRNA-derived RNAs can have important functions distinct from those of mature tRNAs4–8, including potential roles in disease4, 5, 9. However, tRNA-derived fragments are likely to escape sequencing-based detection when they contain nucleoside modifications similar to those in mature tRNAs. Many tRNA modifications cause pauses or stops during reverse transcription10, a critical step in most RNA-seq protocols. These so-called “hard-stop” modifications, including 1-methyladenosine (m1A), 1-methylguanosine (m1G), 2,2,-dimethylguanosine (m2,2G), and 3-methylcytidine (m3C), are more prevalent in tRNAs than other classes of RNAs, and likely play important roles in the biogenesis, stability, and functional activities of tRNA-derived small RNAs, much as they do for mature tRNAs11. For example, specific modifications can target specific tRNAs for cleavage into half-molecules12, protect tRNAs from cleavage13, 14, or alter the interaction of tRNA fragments with proteins such as Dicer or Piwi2, 3, 8.
We developed AlkB-facilitated RNA Methylation sequencing (ARM-Seq) to provide sensitive and specific detection of methyl-modified RNAs using RNA-seq. In ARM-Seq, RNA is treated with a de-alkylating enzyme, Escherichia coli AlkB, prior to the reverse transcription step in library preparation. Differential abundance analysis comparing treated to untreated samples efficiently identifies RNAs sequenced more frequently after demethylation. The known substrates of E. coli AlkB in RNA are m1A, documented in approximately half of all well-characterized tRNAs, and m3C, a less common modification documented primarily in tRNAs15–17. There is also evidence that E. coli AlkB can demethylate m1G, which is nearly as prevalent as m1A in tRNAs, although by a different mechanism18.
Analyses of budding yeast (Saccharomyces cerevisiae) and human cell lines show that ARM-Seq greatly increases the abundance and diversity of reads for small RNAs derived from tRNAs in widely divergent model organisms. ARM-Seq can be used to predict the identity and position of modified residues when compared to previous documentation17, demonstrating that most tRNA-derived fragments contain modifications found in corresponding mature tRNAs. This approach, corroborated by primer extension experiments, correctly predicts the m1A modification state for the complete set of known yeast tRNAs with 94% accuracy, including several where modifications were verified to differ from previous documentation. Furthermore, ARM-Seq provides compelling evidence for m1A modifications in a large proportion of human tRNAs where modification patterns were unknown or not documented. Thus, ARM-Seq facilitates sequencing of methyl-modified RNAs that otherwise escape detection in standard sequencing protocols, and can be used to rapidly characterize methylation patterns across diverse transcriptomes.
Results
ARM-Seq enables detection of methylated small RNAs derived from tRNAs
We first tested the ARM-Seq methodology (Fig.1) on S. cerevisiae, where tRNAs and their modifications17 have been most extensively characterized. Initial experiments showed that demethylation conditions used for ARM-Seq specifically removed m1A and m3C modifications from target RNAs (Supplementary Figure 1). ARM-Seq more than doubled the proportion of small RNA sequencing reads from tRNA genes from 6.9% to 15.1% (Fig.2a, Supplementary Table 1). These increases corresponded almost entirely to tRNA-derived small RNAs rather than full-length mature tRNAs (Supplementary Table 2), indicating that a large proportion of tRNA-derived small RNAs in yeast contain AlkB-sensitive modifications. In contrast, the share of reads mapping to other major classes of small RNAs diminished slightly (Supplementary Table 1).
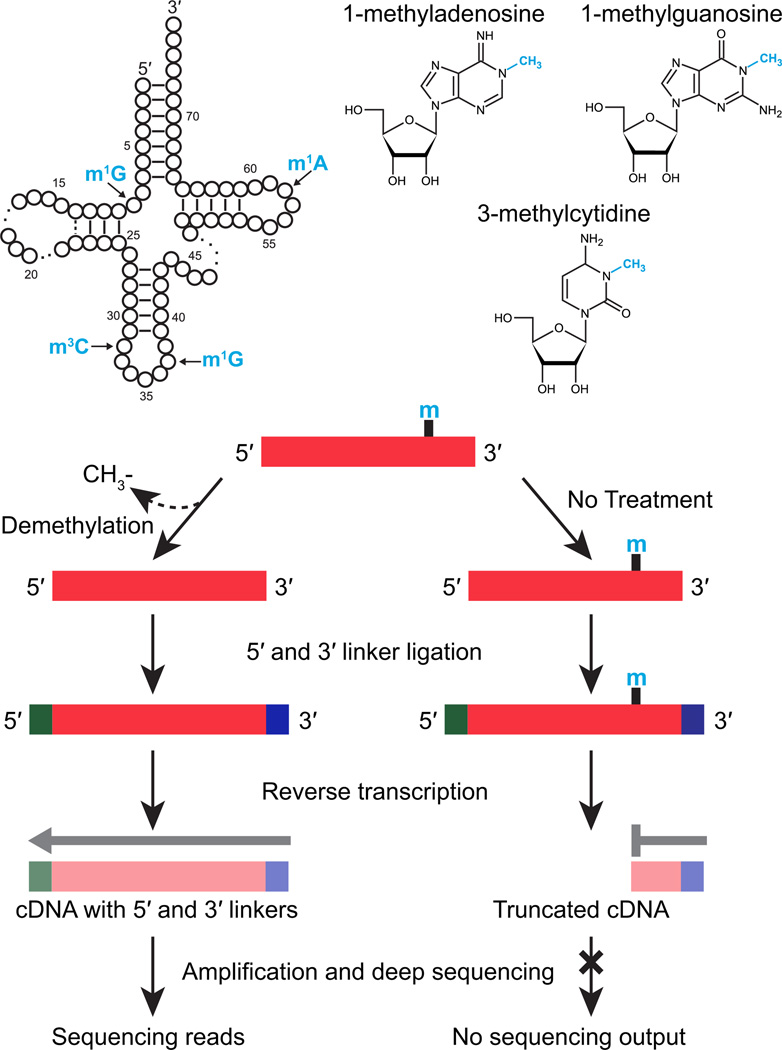
Figure 1. ARM-Seq facilitates sequencing of m1A, m3C, or m1G modified RNAs.
AlkB-facilitated RNA methylated sequence (ARM-Seq) uses enzymatic demethylation of RNA samples prior to RNA-seq library preparation to reveal RNAs containing m1A, m3C, or m1G. Widely used protocols for small RNA sequencing, including NEBNext (New England Biolabs) and TruSeq (Illumina), require ligation of sequencing adapters to both the 5′ and 3’ ends of each RNA prior to reverse transcription for library preparation. Without any additional treatments, sequencing output from these protocols will therefore represent only RNAs with appropriate end chemistry for sequencing adapter ligations (5′-monophosphate and 3′-OH, the expected end chemistry of mature tRNAs, some classes of tRNA-derived fragments, microRNAs, and snoRNAs) that produce full-length cDNAs. “Hard-stop” modifications such as m1A, m3C or m1G, which commonly occur in tRNAs, cause premature termination of cDNA synthesis, preventing PCR amplification and subsequent sequencing. Typical positions for these modifications are indicated in the schematic showing tRNA secondary structure in canonical cloverleaf form. In ARM-Seq, removal of m1A, m3C, or m1G modifications by AlkB treatment facilitates the production of full-length cDNAs from previously modified templates, producing a ratio of reads in treated versus untreated samples that can be used to identify methylated RNAs.
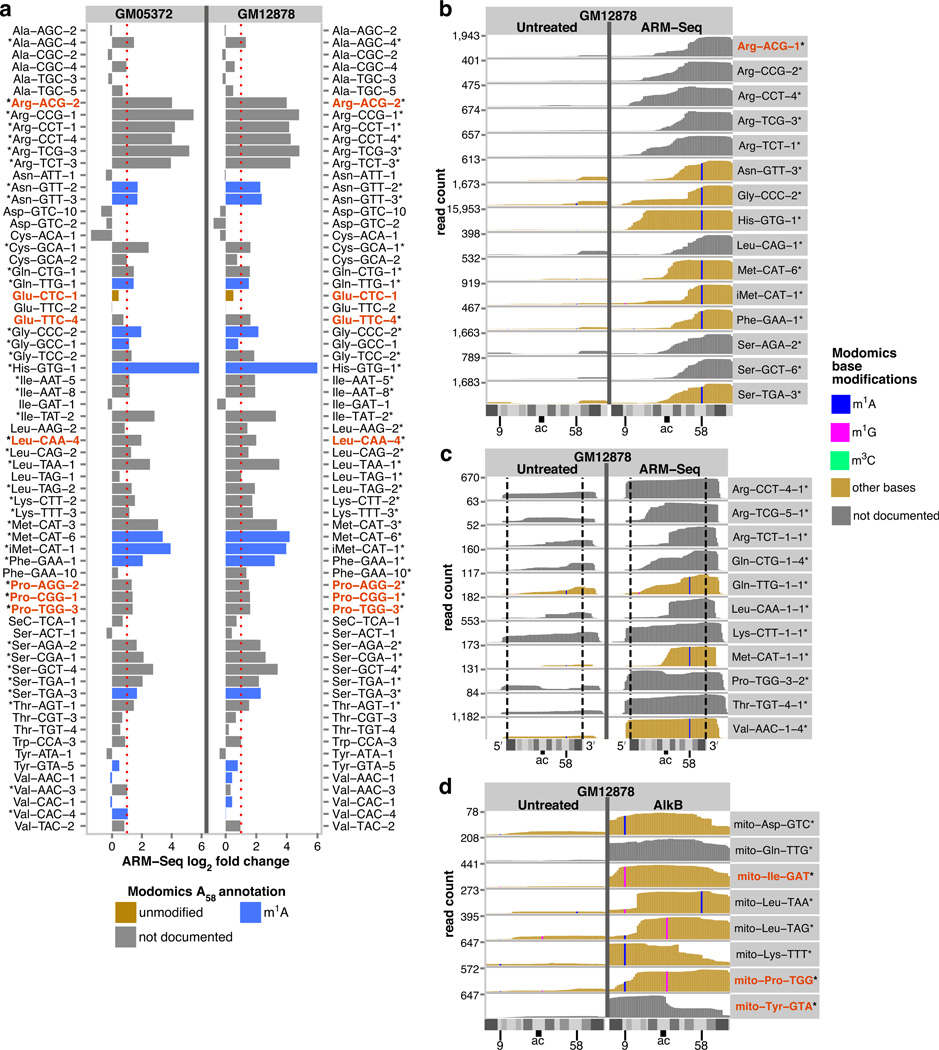
Figure 2. ARM-Seq reveals m1A-modified tRNA fragments in S. cerevisiae.
(a) ARM-Seq more than doubled the fraction of yeast small RNA sequencing reads mapping to tRNAs, revealing a diversity of methylated small RNAs derived from tRNAs. The majority of these were 3′-fragments and half-molecules of tRNAs, where m1A at position 58 (m1A58) is the most prevalent hard-stop modification. Full-length tRNAs comprised less than 1% of tRNA reads in both AlkB-treated and untreated samples, consistent with a known bias in sequencing library preparation where 5′ linker ligation is impeded by recessed 5′ ends of mature tRNAs. (b) ARM-Seq read profiles show increases in 3′-fragment reads relative to untreated samples that predict the presence of m1A58 in Thr-AGT, Leu-GAG and Gln-TTG (indicated by *). By contrast, ARM-Seq profiles for Arg-CCG, Gly-CCC and His-GTG show comparable or diminished 3’ reads for untreated samples, predicting un-modified A58 in these tRNAs. (c) Primer extensions targeting the corresponding mature tRNAs demonstrate that these ARM-Seq results reflect the modification patterns of mature tRNAs, confirming the A58 modification state documented in Modomics for Thr-AGT and His-GTG, providing new information on the m1A58 modification state of Arg-CCG, Gly-CCC and Leu-GAG tRNAs, and presenting new evidence that Gln-TTG tRNAs contain m1A58. (d) As a genome-scale screen, ARM-Seq correctly predicts m1A58 modification state for yeast tRNAs with accuracy of 94% as corroborated by documentation in Modomics, or verification by primer extension (for tRNAs indicated in red), based on increases of two-fold or more (dotted red line) and P < 0.01 (indicated by *).
ARM-Seq predicts the m1A58 modification state of mature tRNAs
Next, we showed that ARM-Seq abundance ratios (RNA-seq read counts from AlkB-treated versus untreated RNA) and read profiles detected known m1A tRNA modifications as effectively as traditional primer extension experiments. Thr-AGT tRNA, which is known to contain m1A58, showed a 16-fold increase in normalized read count corresponding to fragments that include A58 (Fig.2b). Primer extensions targeting mature Thr-AGT tRNA revealed a hard-stop band corresponding to m1A58 in an untreated sample, versus much reduced band intensity in the corresponding AlkB-treated sample, consistent with demethylation of the expected m1A58 modification (Fig.2c). By contrast, ARM-Seq produced no significant effect for His-GTG (Fig.2b), a true negative where an expected unmodified A58 was also confirmed by primer extension (Fig.2c). Similar comparisons confirmed ARM-Seq predictions for three isodecoder groups with no previous modification data (Leu-GAG, Arg-CCG, Gly-CCC), and one isodecoder group (Gln-TTG) where A58 was previously documented as unmodified7, but shown to be methylated by both ARM-Seq and primer extension (Fig.2b–c).
Since ARM-Seq read profiles of tRNA-derived small RNAs correctly predicted the m1A58 modification state for the mature tRNAs tested, we examined ARM-Seq results for the complete set of yeast tRNAs. Based on our initial verified test data, we used a two-fold increase in read abundance and a DEseq2 P-value <0.01 (see Online Methods) as our threshold for identifying all significant ARM-Seq responses.
ARM-Seq correctly predicted the modification state for 22 of 26 yeast tRNAs with documented17 m1A58 modifications (Fig.2d, Supplementary Figures 2–3, Supplementary Table 2). Among the other four tRNAs, ARM-seq predicted unmodified A58 in two (Leu-TAA-1 & Lys-CTT-1), and these were confirmed by primer extension (Fig.2d, Supplementary Figure 4). The last two tRNAs expected to contain m1A58 (Ile-TAT-1, Val-CAC-1) showed visible increases in read count but were not quite significant by our cutoff criteria (Fig.2d, Supplementary Figure 2b).
Conversely, ARM-Seq produced profiles consistent with unmodified A58 for 15 of 19 tRNAs in isodecoder groups expected to lack m1A58 (Supplementary Figure 2), and correctly identified three others (Gln-TTG isodecoders) where unexpected m1A58 modifications were confirmed by primer extension (Fig.2b–c). ARM-Seq profiles for the last tRNA in this group, Ser-CGA, showed evidence for demethylation of both an expected m3C32 modification, and an unexpected m1A58.
ARM-Seq also predicted m1A58 modifications for five yeast tRNAs in isodecoder groups not represented in Modomics and unmodified A58 for three others, with primer extensions confirming m1A58 for Leu-GAG and unmodified A58 for Arg-CCG and Gly-CCC (Fig.2c–d, Supplementary Figure 2d). The final tRNA not represented in Modomics, Pro-AGG, showed evidence for partial AlkB sensitivity that was also confirmed by primer extension (Supplementary Figure 4).
Summarizing for all yeast tRNAs where m1A58 modification state was either corroborated by documentation in Modomics or verified by primer extensions, ARM-Seq correctly predicted 26 of 28 that contain m1A58 (93% sensitivity) and 18 of 19 that contain unmodified A58 (95% specificity), demonstrating a combined accuracy of 94% overall.
ARM-seq reveals abundant methylated RNAs derived from human tRNAs
The tRNA repertoire in humans is substantially more complex. Of 414 unique human mature tRNA sequences identified by tRNAscan-SE19, 20, just 43 match entries in Modomics. ARM-Seq demethylation increased the proportion of RNA-seq reads mapping to tRNAs from 2.9% to 10.1% in an Epstein-Barr virus transformed B-cell line (GM12878), and from 3.9% to 13.2% in a B-cell lymphoma-derived cell line (GM05372), about 3.5-fold in each case (Supplementary Figure 5, Supplementary Table 1). These increases again corresponded to detection of modified tRNA-derived small RNAs, rather than full-length mature tRNAs (Supplementary Table 3). The tRNA 3′-fragments only detectable with ARM-Seq all included A58, positively predicting 15 of the 17 (88%) human isodecoder groups expected to contain m1A58 modifications (Fig.3a–b). ARM-Seq also correctly identified the only isodecoder group expected to contain unmodified A58 (Glu-CTC; Supplementary Figure 6d). Examining all isotypes, ARM-Seq produced an unprecedented set of methylation predictions encompassing the full spectrum of human isodecoder groups (Supplementary Figure 6a–b; Supplementary Data 1).
Figure 3. ARM-Seq reveals methylated RNAs derived from human cytosolic tRNAs, tRNA precursors, and mitochondrial tRNAs.
(a) Transcriptome-scale screening using ARM-Seq provides evidence for m1A58 modification in a majority of human tRNA isotypes, showing a consistent profile of modification in two B-cell derived human cell lines (with * indicating significant responders). (b) Profiles for many tRNA-derived small RNAs revealed by ARM-Seq show little, if any detection in untreated samples, indicating high levels of modification. (c) ARM-Seq also provides the first evidence that many human pre-tRNAs are m1A58 modified at an early stage prior to removal of 5’ leader and 3’ trailer sequences from primary transcripts (demarcated by dashed lines), demonstrating the ability to resolve sequential modification and processing steps involved in tRNA maturation. The 5′-leader sequences of these precursor-derived RNAs were typically short (4–5 nt) when present, which might reflect either nucleolytic processing or dephosphorylation of triphosphorylated primary transcripts to generate 5’-monophosphate ends (required for RNA-seq library inclusion). By contrast, the 3′-trailers were often 9–10 nt or longer, frequently ending with a poly-U sequence, suggesting that these represent the 3′-ends of primary RNA polymerase III transcripts. Reads for full-length and fragmentary pre-tRNAs revealed by ARM-Seq included the T-loop region, consistent with m1A58 modifications. (d) Fragments of human mitochondrial tRNAs revealed by ARM-Seq demonstrate a capacity to also demethylate m1A9 (in mito-Asp-GTC, mito-Lys-TTT), m1G9 (mito-Ile-GAT), and m1G37 (mito-Leu-TAG, mito-Pro-TGG), enabling investigation of mitochondrial diseases related to tRNA modification and processing. tRNAs for which ARM-Seq predictions were verified by primer extension are indicated in red.
ARM-Seq identifies methyl-modified pre-tRNAs and mitochondrial tRNAs
A subset of transcripts revealed by ARM-Seq in the human samples preferentially mapped to tRNA genes rather than mature tRNA transcripts because they included genomically-encoded sequences found only in tRNA precursors (Fig.3c, Supplementary Figures S7–S11). Most tRNA base modifications are thought to occur after cleavage of 5′-leader and 3′-trailer sequences from tRNA-precursor transcripts21. Evidence demonstrating m1A58 modification of initiator methionine pre-tRNAs in yeast and exogenous pre-tRNAs in Xenopus laevis oocytes established a limited precedent for this particular modification at an earlier stage in pre-tRNA processing22, 23, but direct evidence for early m1A58 modification has been lacking for most organisms, including humans. Surprisingly, ARM-Seq identified modified precursors for most human acceptor types (Supplementary Figure 12, Supplementary Table 3), even though pre-tRNAs are less abundant and more challenging to detect than mature tRNAs. Overall, pre-tRNAs in 33 different isodecoder families from 86 different human tRNA gene loci showed significant ARM-Seq responses in at least one of the two cell lines. A large subset of these, 38 loci, showed significant ARM-Seq responses in both cell lines. Primer extensions confirmed an AlkB-sensitive block corresponding to m1A58 in a human Leu-CAA pre-tRNA (Supplementary Figure 8b). Thus, ARM-Seq provides the first evidence that many human pre-tRNAs are m1A58-modified prior to 5′-leader and 3′-trailer removal, suggesting this pattern occurs broadly among eukaryotes.
ARM-Seq also efficiently revealed modifications in human mitochondrial tRNAs. Eight of 22 human mitochondrial tRNAs are currently documented17, showing m1A9, m1G9, m1G37, and m1A58 as the most frequent hard-stop modifications. More extensively characterized bovine mitochondrial tRNAs show at least one difference in modification relative to humans for seven of these (all except initiator methionine), underscoring the need for specific characterization of human mitochondrial tRNAs17, 24. ARM-Seq produced significant increases identifying modified RNAs derived from 12 mitochondrial tRNAs in GM12878 cells, eight of which also showed significant responses in the GM05372 samples (Fig.3d, Supplementary Figure 7, Supplementary Table 3). In contrast to human cytosolic tRNAs, where ARM-Seq responses were attributable exclusively to m1A58 modification state, ARM-Seq profiles for human mitochondrial tRNAs provide evidence for m1A9 (in mito-Asp-GTC, mito-Lys-TTT, and mito-Pro-TGG), m1G9 (in mito-Ile-GAT and mito-Tyr-GTA), m1G37 (in mito-Leu-TAG and mito-Pro-TGG), and m1A58 (in mito-Leu-TAA). Primer extensions confirmed AlkB-mediated demethylation of m1A9 for mito-Pro-TGG, m1G9 in mito-Ile-GAT, and a previously undocumented m1G9 in mito-Tyr-GTA (Supplementary Figure 8b).
Discussion
ARM-Seq results presented here show that a large fraction of small RNAs in both budding yeast and human cells contain base modifications that reflect their biogenesis from modified tRNAs. Recently developed protocols provide tools to profile 6-methyladenosine (m6A), pseudouridine, and 5-methylcytidine (m5C) modified RNAs using high-throughput sequencing, revealing new and unexpected targets for these modifications25–29. ARM-Seq adds the capacity to profile m1A, m3C or m1G modified RNAs, which are otherwise recalcitrant to sequencing, revealing a complex landscape of modified tRNA fragments in two evolutionarily divergent organisms. Sequences of the most abundant of these are listed (Supplementary Table 4), with all 1634 read profiles available for individual examination (Supplementary Data 1).
The power of ARM-Seq as a screen for m1A, m3C and m1G modified RNAs can be maximized by leveraging prior knowledge from databases such as Modomics, and complementary experimental approaches such as primer extension and mass-spectrometry to identify the specific nature and location of modified residues. ARM-Seq demonstrates remarkable accuracy in predicting previously documented tRNA modification patterns, and perfect agreement with corresponding primer extensions for unexpected modifications. Furthermore, results showing that many human pre-tRNAs are m1A-modified demonstrate that ARM-Seq can dissect complex sequential steps of RNA processing and modification, with potential application for identifying modification-based regulatory checkpoints. ARM-Seq profiles revealing m1A and m1G-modified mitochondrial tRNAs also suggest uses investigating mitochondrial genetic diseases, where defects in mitochondrial tRNAs often play central roles30.
Our results, including untreated samples, do not show the same evidence for nucleotide misincorporation at expected hard-stop modifications that has been reported in other studies31–34. Although signature mismatches in sequencing data can identify modified or edited residues, ARM-Seq is almost certainly more sensitive and quantitative for detection of modified RNAs because it does not depend on low-frequency reverse transcription errors that are poorly understood, and possibly context-dependent.
ARM-Seq should facilitate the study of tRNA processing and modification in a wide range of biological settings, including investigation of novel model organisms, as well as comparative analyses of different developmental stages, tissue types, and disease states. Such studies may illuminate new facets of tRNA biology, for example by revealing tissue-specific functions for distinct tRNA variants35, or important regulatory functions for novel tRNA-derived small RNAs5. These typically overlooked small RNAs outnumbered microRNAs by four-fold or more (Supplementary Figure 5), which underscores their potential involvement in cellular signaling and regulation, as well as in disease states including neurodegeneration, cancer and viral infections4, 5, 9. Whether base modifications play central roles in these activities, and whether modifications have obscured detection of members of other classes of RNAs, such as mRNAs or long non-coding RNAs, are among the many potential lines of research now accessible with this methodology.
Methods
Purification of E. coli AlkB
AlkB was purified after growth of E. coli BL21(DE3)pLysS (12 liters) bearing plasmid JEE1167-B in the AVA421 vector36, 37, and 2 hours IPTG induction at 37 °C to express His6-3C-AlkB fusion protein. Crude lysates were made by sonication, and protein was purified by batch treatment on TALON resin, tag cleavage with His6-3C protease, and re-application to TALON resin. Unbound protein was concentrated (Amicon Ultra-15 centifugal filter unit), purified using a Hi-Load 16/60 Superdex 200 gel filtration column, and then stored as concentrated protein (15.4 mg/mL, 0.77 ml) in buffer containing 20 mM Tris-HCl pH 8.0, 50% glycerol, 0.2 M NaCl, and 2 mM dithiothreitol at −20 °C, or at −80 °C. Freezing the enzyme did not impair activity.
Growth of yeast cells and RNA isolation
S. cerevisiae cells (strain BY4741) were grown in liquid YPD medium at 30°C to OD600 1–2, and 300 OD-ml cells were harvested and quick frozen at −80 °C. Bulk RNA was prepared from cell pellets using hot phenol36, typically yielding 2 mg RNA. Bulk RNA from three independently inoculated cultures was processed separately in subsequent treatments.
Growth of human cell lines and RNA isolation
Cell pellets of human B-lymphocyte derived cell lines GM05372 and GM12878 were purchased from Coriell Institute and shipped frozen after a PBS wash. Cell lines were authenticated using microsatellite analysis, and verified as free of mycoplasma infection by Coriell Institute. Upon arrival, cells were immediately placed at −80 °C for storage prior to RNA extraction. Isolation of total RNA from 108 human cells was performed using Direct-Zol™ RNA MiniPrep Kit (Zymo Research) with TRI Reagent (Molecular Research Center, Inc.), typically yielding 400–450 µg of total RNA. Total RNA samples from each of the two human cell lines were then split into three technical replicates for subsequent treatments.
Treatment of RNA with AlkB
AlkB treatment of RNA was performed in 200 µl reaction mixtures containing 50 mM HEPES KOH, pH 8, 75 µM ferrous ammonium sulfate pH 5, 1 mM α-ketoglutarate, 2 mM sodium ascorbate, 50 µg/ml BSA, 50 µg AlkB, and 50 µg bulk RNA at 37 °C for 100 minutes. AlkB reaction buffer was prepared fresh prior to each use. Reactions were stopped by addition of 200 µl buffer containing 11 mM EDTA and 200 mM ammonium acetate, followed by phenol extraction, ethanol precipitation, and resuspension of the washed pellet in water. Control reactions for untreated samples were performed similarly, using AlkB storage buffer instead of AlkB.
Primer extension
For primer extension, ~0.7 pmol 5′-32P-phosphorylated primer was annealed to 0.2 µg bulk RNA in 5 µl H2O by heating for 3 min at 95 °C, followed by cooling to 50 °C and incubation for 1 h. Annealed primer was extended using 64 U Superscript III (Invitrogen) in a 10 µl reaction containing first strand buffer (50 mM Tris-HCl (pH 8.3, 25°C), 75 mM KCl, 3 mM MgCl2) and 1 mM each dNTP for 1 h at 50°C, stopped by addition of 10 µl formamide loading dye and freezing on dry ice. Primer extension products were resolved by electrophoresis on a 15% polyacrylamide gel containing 4 M urea, followed by visualization of the dried gel on a phosphoimager cassette. Sequences of oligonucleotides used for primer extension are listed in Supplementary Table 5.
Size selection and preparation of RNA sequencing libraries
50 µg of control or AlkB-treated RNA was processed using the MirVana miRNA Isolation Kit (Life Technologies), according to manufacturer’s instructions, to select for RNA < 200nt. RNA was concentrated to 25 µg using RNA Clean and Concentrate-25 (Zymo Research), and 10 µg was treated with DNase I (New England BioLabs). Following column cleanup of the RNA, 1 µg was used as input for NEBNext Small RNA Library Prep Kit for Illumina (New England BioLabs).
Libraries were size selected on 2% SizeSelect agarose E-Gels using the 50 bp E-gel ladder (Life Technologies Corporation) as a marker to select for bands corresponding to libraries of RNA between 18–120 nt. Dilutions from column cleaned and concentrated libraries were assessed by BioAnalyzer traces using Agilent High Sensitivity DNA kit (Agilent Technologies). Sequencing of the libraries was performed at the University of California, Davis DNA Technologies and Expression Analysis Core using Illumina MiSeq paired-end sequencing. Fastq files for all sequencing runs are deposited in the NCBI Sequence Read Archive under project number SRP056032.
Mapping of sequencing reads
Reads were trimmed, removing barcoding indices and adapter sequences, and paired-end reads were merged using a custom Python script (Seqprep, J. St. John, http://github.com/jstjohn/SeqPrep). Only merged reads corresponding to RNAs at least 15 nucleotides long were analyzed further. Reads were mapped to reference genomes (Homo sapiens 2009 assembly hg19, GRCh37 or S. cerevisiae April 2011 assembly sacCer3) plus the set of mature tRNA sequences from tRNAscan-SE tRNA gene predictions for each of these genomes19. Mature tRNA sequences were generated to account for post-transcriptional processing steps: predicted introns were removed, a CCA sequence was added to the 3′ ends of all tRNAs, and a G nucleotide was added to the 5′-end of histidine tRNAs. Each of these mature tRNA sequences were padded on both ends with 20 “N” bases to allow mapping of reads with additional end sequences. Reads were mapped to the reference genomes plus the non-redundant set of predicted mature tRNA sequences using Bowtie 238, returning up to 100 alignments per read with default parameters. For analyses summarizing the composition of RNA-seq reads by RNA class, multiple mapping was not allowed and only the Bowtie 2 primary alignment was used (selected arbitrarily by the program when multiple features produced equal mapping scores). Each sample produced approximately one million mappable reads using this procedure. The proportional composition of these reads by RNA class was relatively uniform across technical replicates for the human samples, and somewhat more variable between biological replicates of the yeast samples that were derived from independently expanded cultures (Supplementary Table 1).
For differential expression analysis of reads mapped to either individual gene loci or mature tRNA sequences using DESeq2 analyses (described below), all best matches according to the Bowtie 2 scoring function were used. Reads showing equal mapping scores to tRNA gene loci (which represent unprocessed pre-tRNA transcripts) and predicted mature tRNA sequences were mapped exclusively to mature tRNAs. Thus, reads with equivalent mapping scores to multiple gene loci (encoding tRNAs that are identical after maturation) were mapped instead to a single mature tRNA sequence. In addition, reads mapped by this procedure to tRNA gene loci all contain features of tRNA precursors that are not found in mature tRNAs (e.g., intronic sequences, 3′-trailers, or 5′-leaders). These pre-tRNA features often distinguish one tRNA gene locus from another even when the mature tRNA encoded is identical. Plots of read coverage profiles for tRNAs were produced using read counts that were normalized according to size factors calculated from DESeq2 analyses (see below).
Differential expression analysis
Read counts were tabulated for all reads and assigned to mature tRNAs or genomic features where mapping produced at least 10 nucleotides of sequence overlap. Non-overlapping RNA sequences mapped to the same annotated genomic features were labeled and counted separately (for example non-overlapping RNAs mapped to a genomic feature annotated as HERVH-int were labeled HERVH-int.1, HERVH-int.2, …). Read counts for all features that exceeded a minimum threshold of 20 reads were used as input to the DESeq2 R package with default parameters39. DESeq2 takes into account variability between replicates, and normalizes read counts to account for differences in sequencing depth between samples, reporting ARM-Seq fold changes relative to untreated samples along with associated P-values that are adjusted for multiple hypothesis testing. We used a two-fold increase in read abundance with a DEseq2 P-value <0.01 as our threshold for identifying all significant ARM-Seq responses. A doubling of read counts in ARM-Seq versus untreated samples indicated the presence of AlkB-sensitive modifications in at least half of the detected RNA molecules derived from a given tRNA, while larger increases indicate an even greater proportion of modified molecules. With the exception of Supplementary Table 1, which presents raw read counts and a proportional breakdown of read mappings by RNA class that is unaffected by normalization, all read counts reported in results and Figures_reflect normalization using DESeq2 size factors.
New tRNA naming convention
tRNA transcripts and individual gene loci are labeled using a new systematic naming convention that is designed to be more stable and informative (T. Lowe and P. Chan, unpublished data). The new tRNA naming convention echoes the systematic naming adopted for microRNAs in miRBase40. In brief, each unique mature tRNA transcript is named by isotype and codon (i.e. isodecoder), numbered in ascending order (e.g., tRNA-Ala-AGC-1, tRNA-Ala-AGC-2, etc.), from most "canonical" to least canonical (canonical is objectively defined by the bit score given to each tRNA by tRNAscan-SE using the default general tRNA model19). As with microRNAs, there are often multiple genome loci encoding identical mature tRNAs, so a secondary index number is assigned to denote specific tRNA gene loci (i.e., tRNA-Ala-AGC-1-1, tRNA-Ala-AGC-1-2, tRNA-Ala-AGC-1-3 describe different gene loci, but produce identical mature tRNA transcripts). Thus, labels for mature tRNA transcripts include only the first index number, which refers to the specific unique tRNA (e.g., tRNA-Ala-AGC-2), whereas labels for tRNA genes also include a second index, which refers to the locus number (i.e., tRNA-Ala-AGC-2-1). The new naming convention has been applied to all tRNAs in the Genomic tRNA Database20, and has been adopted by the HUGO Gene Nomenclature Committee, and by RNAcentral41. For convenience in cross-referencing, Tables S1 and S2 also include legacy labels from the genomic tRNA database, where tRNA genes were labeled by chromosome number and order of occurrence20. By this new naming convention, we count 414 possible unique mature tRNAs in the GRCh37/hg19 assembly of the human genome (not including the 10 tRNA predictions with undetermined anticodons).
Correspondence to modifications annotated in Modomics
Predicted mature tRNA sequences were compared to those from the Modomics database (downloaded January 2015) to annotate modifications. tRNAs were labeled with annotated modifications from Modomics when these contained matching anticodons and the sequence of originating (un-modified) bases in Modomics matched those of the genomically encoded tRNAs with three or fewer nucleotide mismatches. tRNAs that did not match Modomics tRNA sequences using these criteria were labeled as “not documented.”
Code availability
The software pipeline developed for this study includes components for trimming of raw sequencing reads, merging of paired-end reads, read mapping of small RNAs (including pre-tRNAs & mature tRNAs), abundance estimation, and differential expression analysis (current version available at http://lowelab.ucsc.edu/software/).
Supplementary Material
Acknowledgements
We thank Patricia Chan for her assistance with technical edits and final figure improvements. This work was supported by the National Institutes of Health NHGRI grant 5R01HG006753 to T.M.L. E.Q. and E.M.P. were also supported by NIH grant GM052347 to E.M.P.
Footnotes
Accession codes
FASTQ files for all sequencing runs are deposited in the NCBI Sequence Read Archive under project number SRP056032.
Author contributions
E.M.P. & T.M.L. conceived project; A.E.C., E.Q., A.D.H., E.H.R., E.M.P. & T.M.L. designed and performed research; A.E.C. & A.D.H. contributed new analytic tools; A.E.C., E.M.P & T.M.L. wrote the paper.
Statement of competing financial interests
All authors are named on a provisional patent application describing this method, filed by the University of California.
References
- 1.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haussecker D, et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole C, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanada T, et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature. 2013;495:474–480. doi: 10.1038/nature11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maute RL, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mei Y, et al. tRNA binds to cytochrome c and inhibits caspase activation. Mol Cell. 2010;37:668–678. doi: 10.1016/j.molcel.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Couvillion MT, Bounova G, Purdom E, Speed TP, Collins K. A Tetrahymena Piwi bound to mature tRNA 3' fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol Cell. 2012;48:509–520. doi: 10.1016/j.molcel.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selitsky SR, et al. Small tRNA-derived RNAs are increased and more abundant than microRNAs in chronic hepatitis B and C. Scientific reports. 2015;5:7675. doi: 10.1038/srep07675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motorin Y, Muller S, Behm-Ansmant I, Branlant C. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol. 2007;425:21–53. doi: 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed] [Google Scholar]
- 11.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Studte P, et al. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Molecular microbiology. 2008;69:1266–1277. doi: 10.1111/j.1365-2958.2008.06358.x. [DOI] [PubMed] [Google Scholar]
- 13.Schaefer M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanco S, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–2039. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc Natl Acad Sci U S A. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aas PA, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 17.Machnicka MA, et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falnes PO. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:6260–6267. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopper AK. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics. 2013;194:43–67. doi: 10.1534/genetics.112.147470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson J, et al. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishikura K, De Robertis EM. RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in the spliced transfer RNA. J Mol Biol. 1981;145:405–420. doi: 10.1016/0022-2836(81)90212-6. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki T, Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014;42:7346–7357. doi: 10.1093/nar/gku390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlile TM, et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz S, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 28.Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009;37:e12. doi: 10.1093/nar/gkn954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Squires JE, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yarham JW, Elson JL, Blakely EL, McFarland R, Taylor RW. Mitochondrial tRNA mutations and disease. Wiley Interdiscip Rev RNA. 2010;1:304–324. doi: 10.1002/wrna.27. [DOI] [PubMed] [Google Scholar]
- 31.Ryvkin P, et al. HAMR: high-throughput annotation of modified ribonucleotides. RNA. 2013;19:1684–1692. doi: 10.1261/rna.036806.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Findeiss S, Langenberger D, Stadler PF, Hoffmann S. Traces of post-transcriptional RNA modifications in deep sequencing data. Biol Chem. 2011;392:305–313. doi: 10.1515/BC.2011.043. [DOI] [PubMed] [Google Scholar]
- 33.Ebhardt HA, et al. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res. 2009;37:2461–2470. doi: 10.1093/nar/gkp093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawaji H, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishimura R, et al. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 19.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D'Silva S, Haider SJ, Phizicky EM. A domain of the actin binding protein Abp140 is the yeast methyltransferase responsible for 3-methylcytidine modification in the tRNA anti-codon loop. RNA. 2011;17:1100–1110. doi: 10.1261/rna.2652611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quartley E, et al. Heterologous expression of L. major proteins in S. cerevisiae: a test of solubility, purity, and gene recoding. Journal of structural and functional genomics. 2009;10:233–247. doi: 10.1007/s10969-009-9068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The, R.C. RNAcentral: an international database of ncRNA sequences. Nucleic Acids Res. 2015;43:D123–D129. doi: 10.1093/nar/gku991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.