Abstract
This study examined developmental changes and sexual dimorphisms in hypothalamic microRNAs, and whether gestational exposures to environmental endocrine-disrupting chemicals (EDCs) altered their expression patterns. Pregnant rat dams were treated on gestational days 16 and 18 with vehicle, estradiol benzoate, or a mixture of polychlorinated biphenyls. Male and female offspring were euthanized on postnatal days (P) 15, 30, 45, or 90, and microRNA and mRNA targets were quantified in the medial preoptic nucleus (MPN) and ventromedial nucleus (VMN) of the hypothalamus. MicroRNAs showed robust developmental changes in both regions, and were sexually dimorphic in the MPN, but not VMN. Importantly, microRNAs in females were up-regulated by EDCs at P30, and down-regulated in males at P90. Few changes in mRNAs were found. Thus, hypothalamic microRNAs are sensitive to prenatal EDC treatment in a sex-, developmental age-, and brain region-specific manner.
Keywords: Endocrine-disrupting chemicals (EDCs), Polychlorinated biphenyls (PCBs), MicroRNA, Hypothalamus, Medial Preoptic Nucleus (MPN), Ventromedial Nucleus (VMN)
Introduction
During the period of brain sexual differentiation in gestation and early postnatal life, gonadal hormones organize the development of brain structures that govern sex-typical physiology and behavior (1, 2). Exposure to exogenous hormones or endocrine-disrupting chemicals (EDCs) during this life stage results in structural and functional neurobiological changes (3). The underlying molecular pathways for these effects are varied, and can involve gene and protein expression, apoptosis, neurogenesis, and molecular epigenetic mechanisms such as DNA methylation and histone modifications (4–8).
A recently identified player in brain sexual differentiation are microRNAs, a family of small regulatory noncoding RNAs that bind to the 3′-untranslated region of a target mRNA, causing mRNA translational repression and/or degradation (9). The expression of some microRNAs is hormone-sensitive, and microRNAs, in turn, influence the expression of genes involved in mediating hormone responses (10, 11). Individual microRNAs are expressed in the nervous system in a region- and development-specific manner (12, 13). Some, such as mir-124a and mir-9 are important in neurodevelopment (14, 15), and are highly expressed in the hypothalamus (e.g. mir-7, mir-132, mir-219) (16–18). Furthermore, a number of hypothalamic microRNAs are expressed in a sexually-dimorphic manner during development (e.g., lin28/let-7 family) (19). Although research on links between prenatal hormones on microRNA expression on the brain is limited, work on prenatal or maternal stress demonstrates effects on expression of a subset of these and other microRNAs (20, 21).
The effects of prenatal EDCs on developmental expression of microRNAs have not been studied in the hypothalamus, but their effects have been shown in other tissues including mouse Sertoli cells (22), whole brains, and livers (23); rat penile shafts (24) and hippocampal cultures (25); and human breast carcinoma (26, 27) and placental cell lines (28). In the current study, we addressed whether a class of prenatal EDCs affect the expression of microRNAs during brain sexual differentiation following prenatal exposure of rats to A1221, a mixture of polychlorinated biphenyls (PCBs) that has previously been shown to perturb expression of sexually dimorphic genes in the brain and cause reproductive and behavioral phenotypic changes in adulthood (4, 5, 29–32). We also assayed several mRNA targets of the microRNAs. Work was conducted on two sexually-dimorphic, hormone-sensitive hypothalamic regions involved in reproductive physiology and behavior (33–38), the medial preoptic nucleus (MPN) and the ventromedial nucleus (VMN). These regions function as complementary regulatory hubs for motivated behaviors such as social interactions and sex, feeding, aggression, and fear (33–38), and are important potential but under-studied targets of endocrine disruption.
Materials and Methods
Animals and Treatments
All protocols were performed in accordance with the guidelines from the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee at the University of Texas at Austin. Brain regions were collected from rats used for a published study on effects of EDCs on gene expression in other hypothalamic regions, and detailed husbandry is provided in that report (31). In brief, Sprague-Dawley rat dams and sires (Harlan, Houston, TX) were purchased, and the dams fed low phytoestrogen Harlan Teklad 2019 Global Diet ad libidum for at least 2 weeks prior to mating. The first day of successful pregnancy was termed embryonic day (E) 0. At the beginning of the third trimester, corresponding to the onset of the period of hypothalamic sexual differentiation, on E16 and E18, the dams were injected with one of three treatments: 1 mg/kg A1221 (AccuStandard, New Haven, Connecticut, number C221N, i.p.): 50 μg/kg estradiol benzoate (EB; Sigma number E8515, s.c.); the vehicle [100% dimethylsulfoxide (DMSO) Sigma number D4540; Sigma, St. Louis, Missouri] was alternated i.p. and s.c.; all in 0.1 ml volume. No effect of route was found on the outcomes examined and therefore the two DMSO routes (s.c. and i.p. injections) groups were combined for the analysis. The dosage, route, and timing of exposure were based on published work showing effects on reproductive function and gene expression in exposed rats (4, 5, 29–32). Average age of vaginal opening in females was 34 days, with no treatment effects (31). Preputial separation in males occurred on P42.1 (DMSO), P42.5 (EB), and P42.9 (A1221), with the DMSO and A1221 groups significantly different (p < 0.05) from one another (31).
Brain tissue collection and storage
On P15, P30, P45, and P90, one male and one female littermate were euthanized 2–3 hours before lights out by rapid decapitation. There were a total of 10 litters per treatment, with 30 litters total. The final sample size was n=10 animals per treatment per age. Trunk blood was collected, allowed to clot, and centrifuged to collect the serum for hormone assays. Body and organ weights were also measured at euthanasia. The brains were dissected and cut into 1 mm coronal sections using a rat brain matrix, and bilateral micropunches of MPN and VMN were collected using a Palkovits punch (0.98 mm in diameter) (30). Post-pubertal females were monitored daily by vaginal smears and euthanized on proestrus.
RNA isolation, preparation, and real-time PCR
Total RNA was isolated from frozen MPN and VMN punches of individual male and female rats using a mirVana microRNA isolation kit according to the manufacturer’s protocols (catalog no. AM1560, Life Technologies, Carlsbad, CA). All RNA samples were analyzed for quantity by Nanodrop spectrophotometry and run on the Bioanalyzer 2100 using the Agilent RNA 6000 Nano Kit (catalog no. 5067–1511, Agilent Technologies, Santa Clara, CA) to assess RNA purity and integrity. Only RNA samples with the RIN of 8 or higher were used.
Total RNA (200 ng total for mRNA, 10 ng for microRNA) was used to generate cDNA. Taqman MicroRNA Reverse Transcription kit (catalog no. 4366596) with Taqman RT primers (catalog no. 4440886, Life Technologies) and high-capacity cDNA reverse transcription kit with RNase inhibitor (catalog no. 4374966, Life Technologies) were used for microRNA and mRNA, respectively, according to the manufacturer’s recommended protocols.
For microRNA analysis, we selected eight specific microRNA assays based on evidence for their expression in the hypothalamus (let-7a, let-7b, mir-124a, mir-132, mir-145, mir-219, mir-7, mir-9) (16, 39, 40). Due to the small amount of starting material, only biological replicates were used for analysis. Technical replicates were run prior to the experiment to validate each assay and intra-assay CV was determined to be <2%. For these and other assays, a no-reverse transcription control was run to confirm the absence of genomic DNA contamination, and a positive control was run on each plate to control for inter-plate variability. Inter-plate variability was <4%.
The choice of mRNAs was based on a combination of in silico predictions from the results of the Comir analysis of microRNA results, together with a review of the literature on neuroendocrine functions controlled by these genes (4, 5, 30, 31, 41). We conducted real-time PCR analysis using Taqman primer and probe sets for subsets of 8 genes (Ar, Clock, Lepr, Lin28b, Ppara, Grin2a, Igf1r, Ar, Pgr) based on microRNA results by sex and brain region. All samples were run in triplicate to account for pipetting errors. No sample had a CV > 2%, however, those replicates that were > 1.5 SD from the mean of the individual were omitted from the analysis, with up to 2 samples per gene removed.
Real-time PCR for microRNA and mRNA analysis was carried out on an ABI ViiA7 using Applied Biosystems TaqMan Universal PCR Master Mix (catalog no. 4324018, Life Technologies) and using the following run parameters: 95°C for 10 minutes, 50 cycles of 95°C for 15 seconds, and 60°C for 1 minute.
In silico analysis of combinatorial microRNA activity
To determine the potential mRNA targets of the microRNAs that were affected by sex, treatment, or age, we used the prediction program ComiR, chosen for its combinatorial approach in gene target identification, to generate a list of mRNA targets for each region and sex (43, 44). The selected genes were verified for the number of binding sites of the predicted microRNAs using Targetscan. The bioinformatic analysis was done across age in four groups: female MPN, male MPN, female VMN, and male VMN. The microRNA groups used for ComiR analysis were mir-145 and mir-7 in female MPN; mir-132, mir-219, mir-9, mir-145, let-7a, and mir-124a in male MPN; let-7a, mir-124a, and mir-219 in female VMN; mir-124a in male VMN. DAVID was used to provide biological interpretation of the large gene lists generated by ComiR and annotate them by gene families and function (45, 46). Putative targets were chosen for further analysis based on their known role in neuronal function and their algorithm score, as determined by the computer modeling programs (43–46).
Hormone Assays
Serum testosterone and estradiol levels in the same rats used herein have previously been measured, and assay characteristics and results were published previously (31). For the current study, hormone concentrations were used for correlation analysis with microRNA and mRNA levels in the MPN and VMN in bionetwork analyses.
Statistics
All raw CT microRNA and mRNA data were normalized within sex to the median of the DMSO P15 group in an R statistical package for qPCR analysis which utilized a generalized linear mixed model with Poisson-lognormal errors and a Bayesian Marco Chain Monte Carlo sampling scheme (47). All data were normally distributed and homoscedastic. The statistical analysis was done using the multiple comparisons analysis of variance (ANOVA), which addresses the false discovery rates of multiple comparisons, to compare each endpoint (genes and microRNAs) using sex, age, and treatment as independent variables. The R package was run in naïve form, without specifying any control genes, and the statistical significance was set at P < 0.05. Post hoc analyses included t-test for sex effect and Tukey HSD for treatment effect and interactions. MicroRNA expression data were graphed as fold change in expression using DMSO female P15 group as control group for females and males. For gene expression data, since different genes were analyzed in females and males in the MPN and VMN, DMSO P15 group within sex was used as reference group. All data shown are mean ± SEM.
Bionetwork analysis
To examine possible relationships among microRNAs, genes, and serum estradiol and testosterone levels (the latter from a companion paper already published on these rats) (31) throughout development, the data were analyzed using a bootstrap technique, as previously reported (30, 31).
Results
Effects of sex, age, and EDC treatment on microRNA expression in the MPN
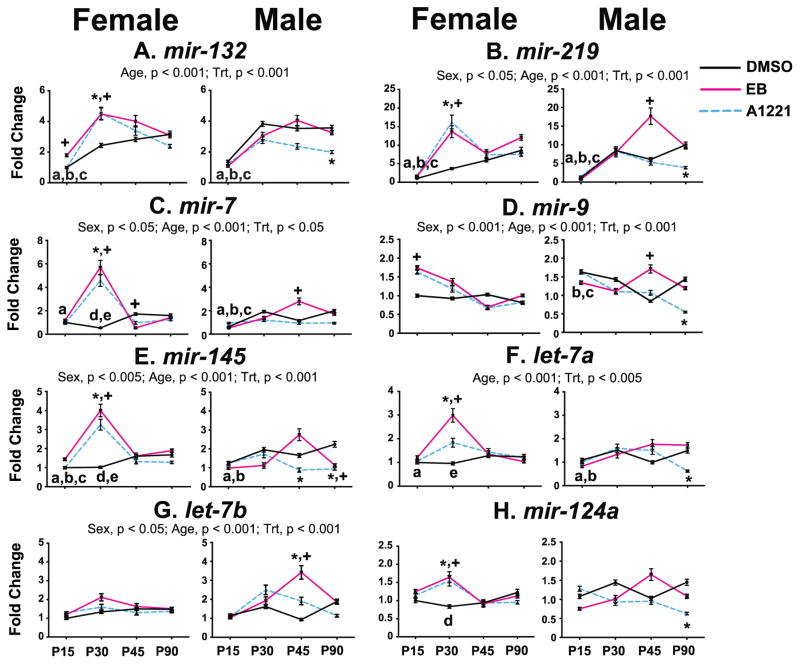
Effects of sex, age, treatment, and their interactions, on microRNA expression in the MPN were analyzed (Figure 1), and detailed statistical results and p-values are presented in Supplemental Table 1. In the MPN, five microRNAs showed significant sex effects, of which three were higher in females than males [mir-219, mir-7 (both, p < 0.05), mir-145 (p < 0.005)], and two were higher in males than females [mir-9 (p < 0.001) and let-7b (p < 0.05)]. Significant age effects were found for six microRNAs, all of which increased with advancing age [mir-132, mir-219, mir-7, mir-145, let-7a, let-7b (all p < 0.005)]. Mir-9 expression decreased significantly with age (p < 0.005).
Figure 1.
Developmental profiles of the 8 microRNAs in the MPN of female and male rats are shown. Main effects of age, sex, and treatment are indicated above the panels. Post-hoc analyses of significant age effects are indicated as: a, P15 vs P30; b, P15 vs P45; c, P15 vs P90; d, P30 vs P45; e, P30 vs P90 (all p < 0.05). Post-hoc analyses for significant age x treatment interactions are indicated as *p < 0.05, A1221 vs. DMSO at the same age; +p < 0.05, EB vs. DMSO at the same age. Abbreviation: Trt, Treatment.
The effects of prenatal treatment in the MPN were age- and sex-specific. A1221 and EB females had significantly increased expression of six microRNAs and this was specific to one age, P30 [mir-219, mir-132, mir-7, mir-145, let-7a, and mir-124a (all p < 0.005)]. Expression of mir-132 and mir-9 was increased by EB at P15 (both p < 0.005), and mir-7 was decreased at P45 in the EB females. In the males, A1221 animals had decreased expression of 6 microRNAs, specifically at P90 [mir-132, mir-219, mir-9, mir-145, let-7a, and mir-124a (all p < 0.005)]. Mir-145 was decreased in A1221 males (p < 0.005) and let-7b was increased (p < 0.005) at P45. EB males had decreased expression of mir-145 at P90 (p < 0.005). EB males had increased expression of mir-219, mir-7, mir-9, and let-7b (all p < 0.005) at P45.
Effects of sex, age, and EDC treatment on microRNA expression in the VMN
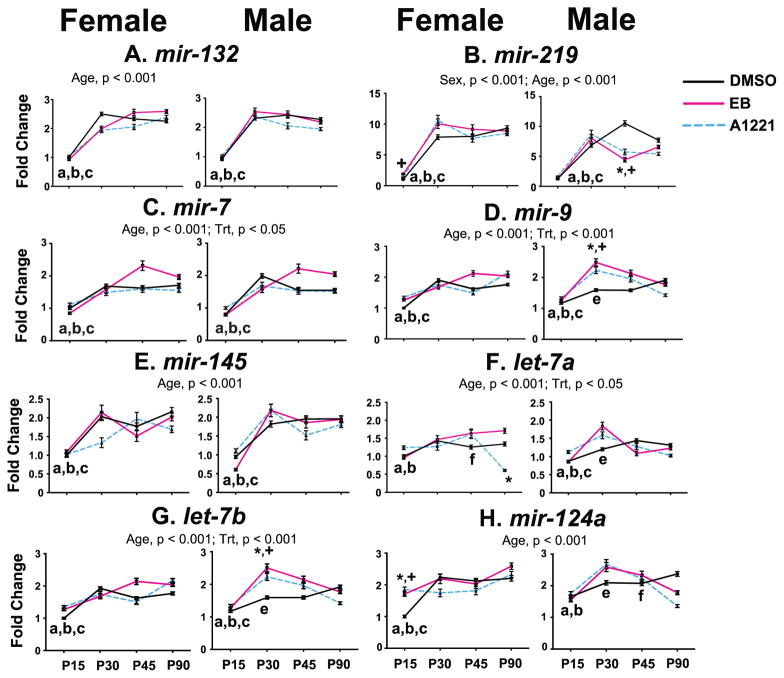
In the VMN, main effects of sex, age, treatment, and their interactions on microRNA expression were examined (Figure 2; detailed statistics and p-values in Supplemental Table 1). Only one sex difference was found, for mir-219 (p < 0.005), with expression higher in the females compared to males. All eight microRNAs increased expression with advancing age in the VMN (p < 0.005).
Figure 2.
Developmental profiles of the 8 microRNAs in the VMN of female and male rats are shown. Main effects of age, sex, and treatment are indicated above the panels. Post-hoc analyses for significant age effects are indicated as: a, P15 vs P30; b, P15 vs P45; c, P15 vs P90; e, P30 vs P90; f, P45 vs P90 (all p < 0.05). Post-hoc analyses for significant age x treatment interactions are indicated as *p < 0.05, A1221 vs. DMSO at the same age; +p < 0.05, EB vs. DMSO at the same age. Abbreviations: Trt, Treatment.
The effects of prenatal treatment in the VMN were age- and sex-specific. In the A1221 females, mir-124a expression was increased at P15 (p < 0.005), and let-7a expression was decreased at P90 (p < 0.005). EB females had increased expression at P15 of mir-219 (p < 0.0005) and mir-124a (p < 0.005). In the males, A1221 and EB rats had increased expression at P30 of mir-9 and let-7b (both p < 0.005), and decreased expression of mir-219 at P45 (p < 0.0005). A1221 males also had decreased expression of mir-124a at P90 (p < 0.005).
In silico analysis of combinatorial microRNA activity
In silico analysis was conducted separately for the female MPN, female VMN, male MPN, and male VMN, to identify mRNA targets. The microRNA groupings were: mir-132, mir-219, mir-9, mir-145, let-7a, and mir-124a in male MPN; mir-145 and mir-7 in female MPN; let-7a, mir-124a, and mir-219 in female VMN; mir-124a in male VMN. To provide an example, the DAVID results in the males in the MPN are discussed. DAVID analysis of the combined male MPN target list from ComiR yielded 158 clusters, with 25 significantly enriched (score >1). Annotation clusters 1 and 2 (enrichment scores of 2.5) had the gene clusters that belonged to the nuclear-hormone receptor, ligand/DNA-binding receptors families. The pathways included post-transcriptional silencing by small RNAs, nuclear receptor transcription pathway and nuclear receptors among the top six which highlighted the hormone regulatory function of chosen microRNA. The genes chosen for qPCR in each group are shown in Table 1.
Table 1.
mRNA qPCR assays in the MPN and VMN.
List of mRNAs selected for real-time PCR assays in the MPN and VMN of male and female rats. Numbers of microRNA binding sites for each mRNA are indicated in parentheses.
| Gene ID | Gene name | Tissue | Predicted Targeting microRNA (# of binding sites) |
|---|---|---|---|
| Igf1r | Insulin-like growth factor 1 receptor | Female MPN | mir-145 (2); mir-7 (3) |
| Grin2a | Glutamate receptor, ionotropic, N-methyl-D-aspartate 2A | Female MPN | mir-7 (4); mir-145 (3) |
| Ar | Androgen receptor | Male MPN Male VMN |
mir-124a (1) |
| Pgr | Progesterone receptor | Male MPN | mir-9 (2); let-7a (2); mir-124 (1) |
| Ppara | Peroxisome proliferator-activated receptor alpha | Female VMN Male MPN |
mir-124a (2); mir-9 (1); let-7a (1); mir-219 (1) |
| Lin28b | Lin-28 homolog B | Female VMN | let7a (4) |
| Clock | Clock gene | Female VMN Male VMN |
mir-124a (1) |
| Lepr | Leptin receptor | Female VMN | mir-219 (1) |
Effects of age and EDC treatment on mRNA expression in the MPN
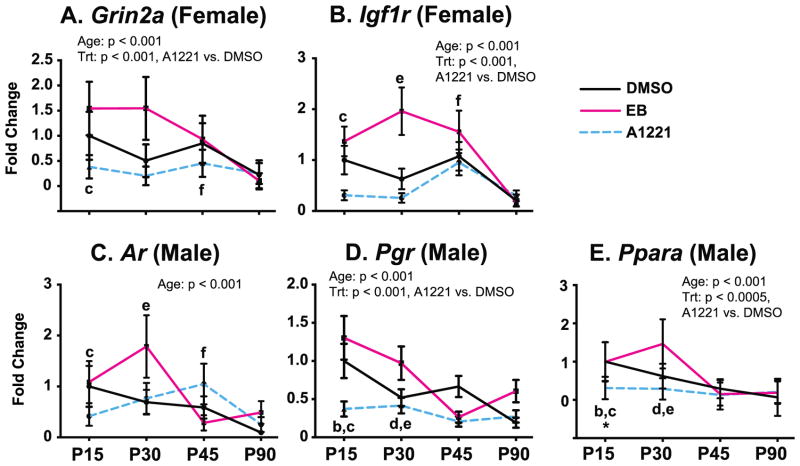
Because gene targets were different in males and females, analyses were conducted separately by sex and region. In the female MPN, gene expression decreased with increasing developmental age for Grin2a and Igf1r (p < 0.005 for both, Figure 3A, 3B; detailed statistics and p-values in Supplemental Table 2). A main effect of treatment was also found for both genes, with decreased expression of Grin2a and Igf1r in the A1221-treated females compared to DMSO females (p < 0.005). There were no treatment by age interactions in females. In the male MPN (Figure 3C, 3D, 3E), expression of Ar, Pgr, and Ppara decreased with developmental age (all p < 0.005). A main effect of treatment was found for Pgr and Ppara, with decreased expression in A1221 compared to DMSO males (both p < 0.005). A treatment by age interaction for A1221 was also found for Ppara at P15, which was lower in A1221 compared to DMSO vehicle males (p < 0.005).
Figure 3.
Developmental profiles of mRNAs in the MPN that were significantly affected by age, treatment, or had age x treatment interactions, in female (A, B) and male (C, D, E) rats. Main effects of age and treatment are indicated above the panels. Post-hoc analyses for age are indicated as b, P15 vs P45; c, P15 vs P90; d, P30 vs P45; e, P30 vs P90; f, P45 vs P90 (all p < 0.05). A significant age x treatment interaction was found only for male Ppara expression at P15 (*, p < 0.05 A1221 vs. DMSO). Abbreviations: Trt, Treatment.
Effects of age and EDC treatment on mRNA expression in the VMN
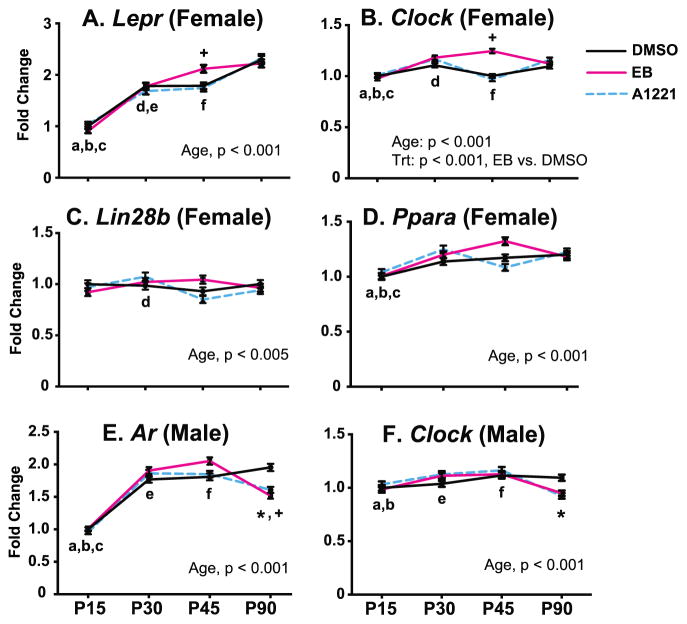
Age and treatment effects on mRNA gene expression were found in the VMN (Figure 4; detailed statistics and p-values in Supplemental Table 2). In the females (Figure 4A, 4B, 4C, 4D), significant developmental increases were found for Lepr, Clock, and Ppara (all p < 0.005). A main effect of EB treatment was found, with increased gene expression of Clock in the female VMN (p < 0.001). There were also treatment by age interactions in the female VMN for prenatal EB treatment, with increased expression at P45 of Lepr and Clock (both p < 0.005). In the males (Figure 4E, 4F), Ar expression increased with age (p < 0.005) and Clock (p < 0.005) decreased with age. Treatment by age interactions of prenatal A1221 and EB treatments were also found. Specifically, at P90, A1221 and EB males had decreased expression of Ar (p < 0.0005), and Clock expression was decreased in A1221 males (p < 0.05).
Figure 4.
Developmental profiles of mRNAs in the VMN that were significantly affected by age, treatment, or had age x treatment interactions, in female (A – D) and male (E, F) rats. Main effects are indicated below the panels. Post-hoc analyses for age are indicated as: a, P15 vs P30; b, P15 vs P45; c, P15 vs P90; d, P30 vs P45; e, P30 vs P90; f, P45 vs P90 (all p < 0.05). Age x treatment interaction were found for Lepr and Clock at P45 in females, and for Ar and Clock at P90 in males (*p < 0.05, A1221 vs. DMSO at the same age; +p < 0.05, EB vs. DMSO at the same age). Abbreviations: Trt, Treatment.
Bionetwork analysis of microRNAs, mRNAs, and hormones
Bionetwork analysis was conducted using Pearson’s correlation coefficients to enable investigation into relationships among microRNAs, genes, and hormones, and to determine whether there were inverse relationships between microRNAs and their predicted mRNA targets. There were no treatment effects on testosterone and estradiol serum hormone concentrations in males and females at P30 and P45. In the males on P30, testosterone concentrations (ng/ml, for the DMSO, EB, and A1221 groups, respectively) were 0.03, 0.02, and 0.05, and on P45, they were 0.20, 0.20, and 0.34. In the females at P30, testosterone was undetectable in the DMSO and A1221 groups, and was 0.11 ng/ml in EB females. On P45, testosterone concentrations were 0.02, 0.03, 0.03 ng/ml for DMSO, EB, and A1221, respectively. For serum estradiol in males, concentrations (pg/ml for the DMSO, EB, and A1221 groups, respectively) on P30 were 8.50, 8.25, and 9.96, and on P45, they were 7.20, 7.84, and 8.77. In the females, estradiol concentrations on P30 were 7.09, 7.72, and 7.06, and on P45, they were 21.94, 20.36, and 28.78.
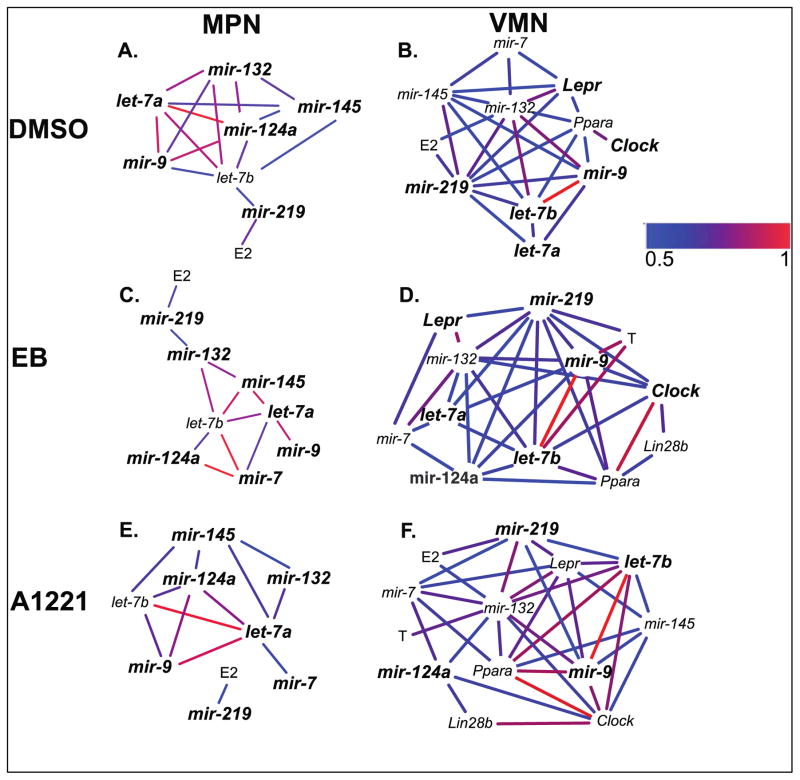
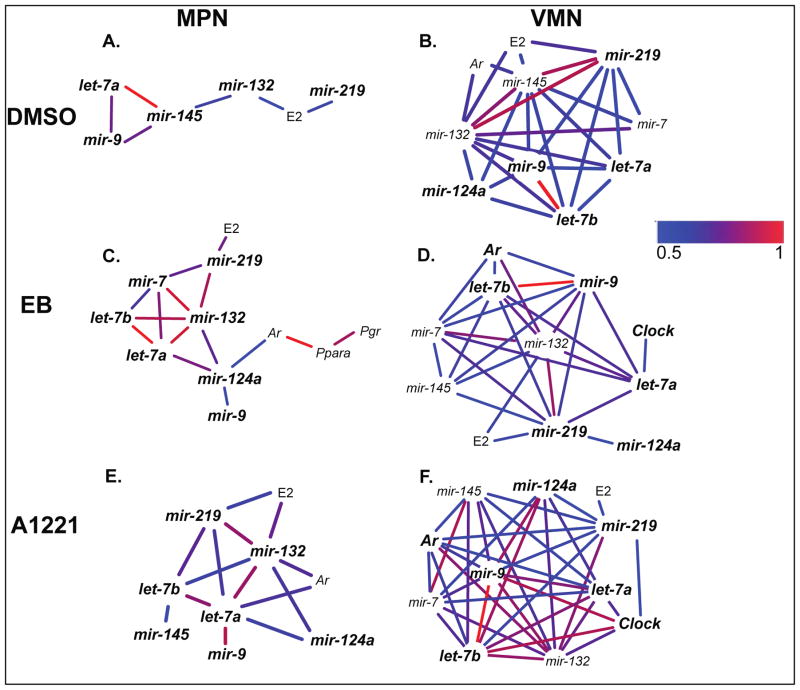
Only positive correlations were detected, with the correlation strength ranging from 0.5 to 1 (Figures 5, 6). The networks were examined within each region, and the numbers of identical (same as vehicle) and novel (different from vehicle) correlations were identified in A1221 and EB networks. In the females, there were few differences in correlations with treatment in either brain region (Figure 5). For example, in the female MPN, mir-132 and mir-145, mir-124a and let-7b, and mir-9 and let-7a, were significantly correlated in all three treatment groups. In the female VMN, mir-132 and mir-219, mir-132 and mir-7, and others, showed similar correlations across the different treatment networks. By contrast, males showed more differences between networks with treatment (Figure 6). In the MPN, the A1221 and EB males had correlations not seen in the DMSO males, such as for mir-124a, mir-7, let-7b, Ar, Ppara, and Pgr. The male VMN networks did not differ substantially by treatment. Interestingly, serum estradiol concentrations were positively correlated with mir-219 in both the male MPN and VMN of all treatment groups, and estradiol and mir-132 were correlated in four of these six groups (Figure 6). In females (Figure 5) estradiol was also correlated with mir-219 in the three treatment groups in the MPN, and in the DMSO and A1221 VMN.
Figure 5.
Cytoscape analysis of microRNAs, genes, and hormones for the three treatment groups in the females, collapsed across development, in the MPN (A–C) and VMN (D–F). Abbreviations: E2, estradiol, T, testosterone.
Figure 6.
Cytoscape analysis of microRNAs, genes, and hormones for the three treatment groups in the males, collapsed across development, in the MPN (A–C) and VMN (D–F). Abbreviations: E2, estradiol, T, testosterone.
Discussion
In the current study we profiled the expression of eight microRNAs in the developing MPN and VMN of the hypothalamus, and determined effects of gestational exposures to PCBs on microRNA expression. We further related changes in microRNA expression to changes in target gene mRNA expression changes in the same animals. Our major findings were that many more of the selected microRNAs were sexually dimorphic, and affected by prenatal EDC treatment, in the MPN than the VMN. Importantly, EDC-treated females showed up-regulation of microRNAs at P30, in the midst of pubertal development, whereas microRNAs in males were affected (down-regulated) at P90, in adulthood. Relatively few EDC effects were found on the mRNA targets of these microRNAs, the implications for which are discussed below. As a whole, the results add to knowledge about EDC effects on microRNA expression, and provide new information about sex differences and developmental change in the hypothalamus.
MicroRNA expression in the MPN changes with postnatal development and is sexually dimorphic
The most common finding of our study was a developmental increase in microRNA expression as animals progressed from the juvenile through the pubertal period and into adulthood. In the MPN, six microRNAs (mir-132, mir-219, mir-7, mir-145, let-7a, and let-7b) exhibited this pattern. The lin28/let-7 family has previously been investigated in the preoptic area, for which the expression of let-7a, let-7b, mir-132, and mir-145 increased with postnatal development. Changes in their expression were suggested to be involved in the mechanisms permitting or leading to puberty onset (19), as Lin28 overexpression in mice resulted in delayed puberty and increased body size (48). Another study showed that seven of the eight microRNAs of the let-7 family were highly expressed in hypothalamic arcuate and paraventricular nuclei of adult rats (49). Our finding that let-7a, let-7b, mir-132 and mir-145 expression increase with age in the MPN is consistent with those reports (19) and adds greater regional specificity by our focus on the MPN, which is a smaller sub-region of the entire preoptic area.
To our knowledge, this is the first report on developmental changes in mir-219 in the brain. Together with mir-132, mir-219 is thought to be involved in the regulation of the biological clock (40), with recent reports implicating a role in early development (50), oligodendrocyte differentiation (51), and modulation of NMDA receptor-mediated effects (18). Interestingly, the mir-219 and mir-132 expression patterns were quite similar across development, and most of our bionetwork analyses showed a significant positive correlation between these two microRNAs. Furthermore, these two microRNAs also frequently correlated with serum estradiol concentrations, especially in males. A previous study comparing mir-219 and mir-132 in the fetal, adult, and diseased hippocampus, showed similar expression profiles (52), and NMDAR activation downregulated mir-219 and mir-132 expression in the adult dentate gyrus in vivo (53). Therefore, we speculate that in the MPN, and possibly other regions, mir-132 and mir-219 are involved in a common signaling pathway.
A developmental decrease in mir-9 was observed for both sexes. Mir-9 is involved in regulating neurogenesis and maturational events at earlier, fetal stages of brain development (54–56). For example, mir-9 expression was highest in the fetal hippocampus and much lower in adult tissue (52). Our results are consistent with that finding.
Several microRNAs in the MPN had sexually dimorphic expression. Females had higher expression of mir-219, mir-7, and mir-145 than males, an effect driven by differences at P30. Although we do not know what this may represent, females mature earlier than males and are farther along in pubertal development than males at this age. There were also sexual dimorphisms in expression of mir-9 and let-7b, with males having higher levels of expression than females at P45, an age when males have just begun to attain adult reproductive function and where serum testosterone concentrations are at their peak (57). We suggest that changes in sexually dimorphic microRNAs may be involved in, or reflect, pubertal changes in the MPN, but further work is needed to get at causal relationships.
MicroRNA expression in the MPN increased by A1221 and EB in P30 females, and decreased by A1221 in P90 males
In the females, prenatal A1221 and EB treated rats had increased expression of mir-219, mir-132, mir-7, mir-145, let-7a, and mir-124a at P30. Similarities between A1221 and EB are consistent with A1221’s ability to act via estrogenic mechanisms (58). Effects of estrogens on neural microRNA expression have primarily been investigated in adults (10). In the aging female brain, for example, estradiol treatment differentially altered microRNA levels in an age- and brain region-dependent manner (10). We are unaware of any studies that examined the effect of prenatal estradiol on microRNA expression in the hypothalamus; however adult exposure studies in zebrafish and rodents (10, 59), in addition to in vitro reports on effects of estradiol on microRNA expression in breast cancer cell lines [reviewed in (11)], underscore the sensitivity of microRNA expression to estradiol treatment.
In the male MPN, expression of all eight microRNAs examined was decreased in prenatally treated A1221 males on P90, an effect not mimicked by EB. This suggests that the mechanism for this A1221 effect in males likely involves a pathway other than through estrogen receptors. A reason for the sex difference may be the already greater exposure of prenatal male than female brains to estradiol (60, 61), such that the addition of low-levels of exogenous EB had little influence in males compared to females. A1221 is weakly estrogenic but also has been shown to be anti-androgenic at low doses (62, 63) and the decrease in microRNA expression observed in A1221 males may be caused by an antagonistic effect on the androgen receptor, something that could potentially be discerned using an anti-androgenic positive control in future studies.
The finding that prenatal EDC treatment effects are manifested at only a subset of developmental ages is consistent with our previously published study on these same rats, in which we measured mRNA expression in the anteroventral periventricular nucleus (AVPV) and arcuate nucleus (ARC) of the hypothalamus (31). Results from that study showed individual postnatal gene expression profiles that were often affected at one or two ages, but not all ages, in prenatally exposed individuals. In other words, the age of analysis of EDC effects is a key determinant of the outcome. This is not surprising, as the profiles of different genes and proteins in the brain undergo dynamic change throughout postnatal development and may continue to change through the life cycle, up to and including aging (64).
MicroRNA expression in the VMN changes with postnatal development but have few sex differences or EDC effects
In the VMN, all of the microRNAs studied increased their expression with age in both sexes. Our results for mir-132, mir-7, mir-145, let-7a, and let-7b are consistent with recent studies on microRNA expression in the medial basal hypothalamus (19). Mir-9, on the contrary, decreased during postnatal development in the medial basal hypothalamus (MBH) (19), a result that may be explained by our VMN punch being a small sub-region within the entire MBH in which gene expression levels differ from those in neighboring regions.
There were relatively few sex differences or effects of EDCs on microRNA expression in the VMN. In the females, there were modest changes in let-7a, mir-124a, and mir-219 in response to the EDCs that were developmental age-specific. In the male VMN, similarly to the male MPN, A1221 decreased the expression of mir-124a. To our knowledge, there are no other EDC studies examining microRNA expression in the VMN.
mRNA expression of targets of selected microRNAs
We examined the expression levels of mRNAs that were selected based on the microRNA results together with the literature. In the MPN, few EDC effects were found, but there were several changes with age. In the female MPN, Igf1r and Grin2a decreased expression with advancing developmental age. Consistent with that result, we previously reported that Igf1r mRNA decreased postnatally from P1 through P60 in the whole preoptic area (POA) of developing male and female rats (65). However that same study showed a small developmental age-related increase in Grin2a, with disparate results from the current study presumably due to differences in the dissection size from the previous (whole POA) and current (MPN) work. In our males, expression of Ar, Pgr, and Ppara decreased across development. These results were surprising, as the expression of Ar in the MPN increased with age in male hamsters during puberty (66), and in our earlier rat studies, Ar as well as Pgr expression underwent significant postnatal developmental increases in the whole POA (65,67). We are unaware of any reports on Ppara expression in the developing MPN.
Results in the VMN also indicated few gene expression changes due to treatment, but there were several age effects. In females, an age-related increase in expression of Lepr, Ppara, and Clock was detected. These genes have not been previously examined in the postnatal developing VMN, to our knowledge. In the male VMN, Ar increased with age, consistent with other work in whole MBH (65), and Clock expression decreased with age. As the only mRNA measured in the VMN of both sexes, Clock expression had opposite developmental patterns (increase in females, decrease in males).
Interestingly, for all of the mRNA results in both regions, we did not see a predicted inverse relationship with the microRNA expression. In fact, our network analysis revealed only positive correlations among mRNAs, microRNAs, and hormones. The lack of such a finding is attributable to a number of possibilities. First, we were only able to measure a small number of microRNAs and their mRNA targets. Other microRNAs were not measured, and these molecules their combinations, are likely to have a stronger association with mRNAs, and vice versa, than the subset measured herein. Second, there are other molecular mechanisms for the regulation of gene expression that include DNA methylation, histone modifications, transcription factors, and post-transcriptional processes, that were not explored in the current study, but which contribute to the absolute expression of any gene. Finally, we have not looked at protein expression, and thus changes in microRNA expression might affect protein translation. Moreover, the temporal relationship between changes in microRNA and subsequently mRNA may not enable detection of a change in a microRNA leading to a mRNA change. During the time period between the ages analyzed (15 or 45 days) there may not be a linear continuum in expression of genes; in fact, there may be ups and downs in expression between the ages examined. To get at these relationships we would need to follow animals at much shorter, possibly day-to-day intervals.
Conclusions and implications
Several global conclusions can be drawn for the microRNAs and mRNAs measured herein. First, while both the MPN and VMN are sexually dimorphic in expression of various genes and proteins (68,69), of the microRNAs measured here, expression was sexually dimorphic in the MPN, but not the VMN. Second, the PCB mixture A1221 affected microRNA expression in the MPN, and to a lesser degree, in the VMN, in a region-, sex-, and age-specific manner. This finding indicates the importance of analyzing endpoints in both sexes and at multiple ages during postnatal development. Third, few treatment changes in the target mRNA expression were found, contrary to the microRNA results. It is also notable that the changes in mRNA with age and treatment were far smaller than those seen for microRNAs, underscoring that different levels of molecular analysis have individual developmental profiles and sensitivity to EDCs.
This research also has implications for developmental sex differences in the brain. Sexual dimorphisms in microRNA expression have become an area of investigation for neurobiological disorders with biases in prevalence and nature, such as autism, schizophrenia, and stroke (70–72). Furthermore, an increase in EDC exposures is beginning to be linked to the rise in these multifactorial disorders (73–75). While it is premature to draw any strong inferences from these correlations, further research on connections among EDCs, microRNAs, and neurobehavioral outcomes is warranted.
Supplementary Material
Supplemental Table 1. Statistical results of sex, age, and treatment and their interactions on miRNA expression in the MPN and VMN.
Supplemental Table 2. Statistical results of mRNA analysis in the MPN and VMN.
Highlights.
MicroRNA expression in the MPN undergoes robust developmental change in a sexually dimorphic manner.
Prenatal PCB exposures increase microRNA expression in the MPN of females at P30, and decrease it in males at P90.
MicroRNA expression in the VMN increases during postnatal development, but has few sex or prenatal EDC effects
Targeted mRNA expression did not have a clear relationship to the microRNA changes, suggesting other epigenetic mechanisms contribute to the mRNA expression patterns.
Acknowledgments
We thank Lauren Wagner and Jayden Chen for technical assistance in running the microRNA assays, Dr. Dean Kirson for generously allowing us to use his Matlab script for bionetworks analysis, and Saazina Afsah for help with running the script. We thank Dr. Stephen Topper for running the statistical analyses and for critically reading and commenting upon the manuscript.
Footnotes
Disclosure statement: Funding was provided by NIH grants RO1 ES020662 and RO1 ES023254 to ACG, and F31 AG034813 to DMW. The authors have nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wallen K. The Organizational Hypothesis: Reflections on the 50th anniversary of the publication of Phoenix, Goy, Gerall, and Young (1959) Horm Behav. 2009;55:561–565. doi: 10.1016/j.yhbeh.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 2.Phoenix CH, Goy RW, Gerall AA, Young WC. Organizing action of prenatally administered testosterone propionate on the tissues mediating mating behavior in the female guinea pig. Endocrinology. 1959;65:369–382. doi: 10.1210/endo-65-3-369. [DOI] [PubMed] [Google Scholar]
- 3.Diamanti-Kandarakis E, Bourguignon JP, Giudice LC, Hauser R, Prins GS, Soto AM, Zoeller RT, Gore AC. Endocrine-disrupting chemicals: an Endocrine Society scientific statement. Endocrine Rev. 2009;30:293–342. doi: 10.1210/er.2009-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dickerson SM, Cunningham SL, Gore AC. Prenatal PCBs disrupt early neuroendocrine development of the rat hypothalamus. Toxicol Appl Pharmacol. 2011;252:36–46. doi: 10.1016/j.taap.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickerson SM, Cunningham SL, Patisaul HB, Woller MJ, Gore AC. Endocrine disruption of brain sexual differentiation by developmental PCB exposure. Endocrinology. 2011;152:581–594. doi: 10.1210/en.2010-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komada M, Asai Y, Morii M, Matsuki M, Sato M, Nagao T. Maternal bisphenol A oral dosing relates to the acceleration of neurogenesis in the developing neocortex of mouse fetuses. Toxicology. 2012;295:31–38. doi: 10.1016/j.tox.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bredfeldt TG, Greathouse KL, Safe SH, Hung MC, Bedford MT, Walker CL. Xenoestrogen-induced regulation of EZH2 and histone methylation via estrogen receptor signaling to PI3K/AKT. Molecular Endocrinology. 2010;24:993–1006. doi: 10.1210/me.2009-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Rao YS, Mott NN, Wang Y, Chung WC, Pak TR. MicroRNAs in the aging female brain: a putative mechanism for age-specific estrogen effects. Endocrinology. 2013;154:2795–2806. doi: 10.1210/en.2013-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klinge CM. Estrogen Regulation of MicroRNA Expression. Current genomics. 2009;10:169–183. doi: 10.2174/138920209788185289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziats MN, Rennert OM. Identification of differentially expressed microRNAs across the developing human brain. Mol Psychiatry. 2014;19:848–852. doi: 10.1038/mp.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olsen L, Klausen M, Helboe L, Nielsen FC, Werge T. MicroRNAs show mutually exclusive expression patterns in the brain of adult male rats. PloS one. 2009;4:e7225. doi: 10.1371/journal.pone.0007225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan SL, Ohtsuka T, Gonzalez A, Kageyama R. MicroRNA9 regulates neural stem cell differentiation by controlling Hes1 expression dynamics in the developing brain. Genes to cells. 2012;17:952–961. doi: 10.1111/gtc.12009. [DOI] [PubMed] [Google Scholar]
- 16.Sakai A, Saitow F, Miyake N, Miyake K, Shimada T, Suzuki H. miR-7a alleviates the maintenance of neuropathic pain through regulation of neuronal excitability. Brain. 2013;136:2738–2750. doi: 10.1093/brain/awt191. [DOI] [PubMed] [Google Scholar]
- 17.Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, Willoughby D, Kenny PJ, Elsworth JD, Lawrence MS, Roth RH, Edbauer D, Kleiman RJ, Wahlestedt C. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci USA. 2012;109:3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kocerha J, Faghihi MA, Lopez-Toledano MA, Huang J, Ramsey AJ, Caron MG, Sales N, Willoughby D, Elmen J, Hansen HF, Orum H, Kauppinen S, Kenny PJ, Wahlestedt C. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc Natl Acad Sci USA. 2009;106:3507–3512. doi: 10.1073/pnas.0805854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sangiao-Alvarellos S, Manfredi-Lozano M, Ruiz-Pino F, Navarro VM, Sanchez-Garrido MA, Leon S, Dieguez C, Cordido F, Matagne V, Dissen GA, Ojeda SR, Pinilla L, Tena-Sempere M. Changes in hypothalamic expression of the Lin28/let-7 system and related microRNAs during postnatal maturation and after experimental manipulations of puberty. Endocrinology. 2013;154:942–955. doi: 10.1210/en.2012-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgan CP, Bale TL. Early prenatal stress epigenetically programs dysmasculinization in second-generation offspring via the paternal lineage. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31:11748–11755. doi: 10.1523/JNEUROSCI.1887-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zucchi FC, Yao Y, Ward ID, Ilnytskyy Y, Olson DM, Benzies K, Kovalchuk I, Kovalchuk O, Metz GA. Maternal stress induces epigenetic signatures of psychiatric and neurological diseases in the offspring. PloS one. 2013;8:e56967. doi: 10.1371/journal.pone.0056967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi JS, Oh JH, Park HJ, Choi MS, Park SM, Kang SJ, Oh MJ, Kim SJ, Hwang SY, Yoon S. miRNA regulation of cytotoxic effects in mouse Sertoli cells exposed to nonylphenol. Reprod Biol Endocrinol. 2011;9:126. doi: 10.1186/1477-7827-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B, Pan X. RDX induces aberrant expression of microRNAs in mouse brain and liver. Environ Health Perspect. 2009;117:231–240. doi: 10.1289/ehp.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovanecz I, Gelfand R, Masouminia M, Gharib S, Segura D, Vernet D, Rajfer J, Li DK, Kannan K, Gonzalez-Cadavid NF. Oral Bisphenol A (BPA) given to rats at moderate doses is associated with erectile dysfunction, cavernosal lipofibrosis and alterations of global gene transcription. Intl J Impotence Res. 2014;26:67–75. doi: 10.1038/ijir.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesiak A, Zhu M, Chen H, Appleyard SM, Impey S, Lein PJ, Wayman GA. The environmental neurotoxicant PCB 95 promotes synaptogenesis via ryanodine receptor-dependent miR132 upregulation. J Neurosci. 2014;34:717–725. doi: 10.1523/JNEUROSCI.2884-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tilghman SL, Bratton MR, Segar HC, Martin EC, Rhodes LV, Li M, McLachlan JA, Wiese TE, Nephew KP, Burow ME. Endocrine disruptor regulation of microRNA expression in breast carcinoma cells. PloS one. 2012;7:e32754. doi: 10.1371/journal.pone.0032754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng Y, Manavalan TT, Hu C, Medjakovic S, Jungbauer A, Klinge CM. Endocrine disruptors fludioxonil and fenhexamid stimulate miR-21 expression in breast cancer cells. Toxicological sciences: an official journal of the Society of Toxicology. 2013;131:71–83. doi: 10.1093/toxsci/kfs290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avissar-Whiting M, Veiga KR, Uhl KM, Maccani MA, Gagne LA, Moen EL, Marsit CJ. Bisphenol A exposure leads to specific microRNA alterations in placental cells. Reprod Toxicol. 2010;29:401–406. doi: 10.1016/j.reprotox.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinberg RM, Juenger TE, Gore AC. The effects of prenatal PCBs on adult female paced mating reproductive behaviors in rats. Horm Behav. 2007;51:364–372. doi: 10.1016/j.yhbeh.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker DM, Kermath BA, Woller MJ, Gore AC. Disruption of reproductive aging in female and male rats by gestational exposure to estrogenic endocrine disruptors. Endocrinology. 2013;154:2129–2143. doi: 10.1210/en.2012-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker DM, Goetz BM, Gore AC. Dynamic postnatal developmental and sex-specific neuroendocrine effects of prenatal polychlorinated biphenyls in rats. Molecular Endocrinology. 2014;28:99–115. doi: 10.1210/me.2013-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinberg RM, Walker DM, Juenger TE, Woller MJ, Gore AC. Effects of perinatal polychlorinated biphenyls on adult female rat reproduction: development, reproductive physiology, and second generational effects. Biol Reprod. 2008;78:1091–1101. doi: 10.1095/biolreprod.107.067249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathews D, Edwards DA. Involvement of the ventromedial and anterior hypothalamic nuclei in the hormonal induction of receptivity in the female rat. Physiol Behav. 1977;19:319–326. doi: 10.1016/0031-9384(77)90345-6. [DOI] [PubMed] [Google Scholar]
- 34.Clark AS, Pfeifle JK, Edwards DA. Ventromedial hypothalamic damage and sexual proceptivity in female rats. Physiol Behav. 1981;27:597–602. doi: 10.1016/0031-9384(81)90228-6. [DOI] [PubMed] [Google Scholar]
- 35.Malsbury CW, Kow LM, Pfaff DW. Effects of medial hypothalamic lesions on the lordosis response and other behaviors in remale golden hamsters. Physiol Behav. 1977;19:223–237. doi: 10.1016/0031-9384(77)90331-6. [DOI] [PubMed] [Google Scholar]
- 36.Hull EM, Dominguez JM. Getting his act together: roles of glutamate, nitric oxide, and dopamine in the medial preoptic area. Brain Res. 2006;1126:66–75. doi: 10.1016/j.brainres.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 37.Hoshina Y, Takeo T, Nakano K, Sato T, Sakuma Y. Axon-sparing lesion of the preoptic area enhances receptivity and diminishes proceptivity among components of female rat sexual behavior. Behav Brain Res. 1994;61:197–204. doi: 10.1016/0166-4328(94)90160-0. [DOI] [PubMed] [Google Scholar]
- 38.Kato A, Sakuma Y. Neuronal activity in female rat preoptic area associated with sexually motivated behavior. Brain Res. 2000;862:90–102. doi: 10.1016/s0006-8993(00)02076-x. [DOI] [PubMed] [Google Scholar]
- 39.Davis CJ, Clinton JM, Krueger JM. MicroRNA 138, let-7b, and 125a inhibitors differentially alter sleep and EEG delta-wave activity in rats. J Appl Physiol. 2012;113:1756–1762. doi: 10.1152/japplphysiol.00940.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casati L, Sendra R, Poletti A, Negri-Cesi P, Celotti F. Androgen receptor activation by polychlorinated biphenyls: epigenetic effects mediated by the histone demethylase Jarid1b. Epigenetics. 2013;8:1061–1068. doi: 10.4161/epi.25811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coronnello C, Hartmaier R, Arora A, Huleihel L, Pandit KV, Bais AS, Butterworth M, Kaminski N, Stormo GD, Oesterreich S, Benos PV. Novel modeling of combinatorial miRNA targeting identifies SNP with potential role in bone density. PLoS Comp Biol. 2012;8:e1002830. doi: 10.1371/journal.pcbi.1002830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coronnello C, Benos PV. ComiR: Combinatorial microRNA target prediction tool. Nucl Acids Res. 2013;41:W159–164. doi: 10.1093/nar/gkt379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucl Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 47.Matz MV, Wright RM, Scott JG. No control genes required: Bayesian analysis of qRT-PCR data. PloS one. 2013;8:e71448. doi: 10.1371/journal.pone.0071448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, Takeuchi A, Grasemann C, Rinn JL, Lopez MF, Hirschhorn JN, Palmert MR, Daley GQ. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nature Genet. 2010;42:626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amar L, Benoit C, Beaumont G, Vacher CM, Crepin D, Taouis M, Baroin-Tourancheau A. MicroRNA expression profiling of hypothalamic arcuate and paraventricular nuclei from single rats using Illumina sequencing technology. J Neurosci Meth. 2012;209:134–143. doi: 10.1016/j.jneumeth.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 50.Hudish LI, Blasky AJ, Appel B. miR-219 regulates neural precursor differentiation by direct inhibition of apical par polarity proteins. Developmental Cell. 2013;27:387–398. doi: 10.1016/j.devcel.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, Barres BA. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65:597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 53.Wibrand K, Panja D, Tiron A, Ofte ML, Skaftnesmo KO, Lee CS, Pena JT, Tuschl T, Bramham CR. Differential regulation of mature and precursor microRNA expression by NMDA and metabotropic glutamate receptor activation during LTP in the adult dentate gyrus in vivo. Eur J Neurosci. 2010;31:636–645. doi: 10.1111/j.1460-9568.2010.07112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Denli AM, Cao X, Gage FH. miR-9 and TLX: chasing tails in neural stem cells. Nature Struct Molec Biol. 2009;16:346–347. doi: 10.1038/nsmb0409-346. [DOI] [PubMed] [Google Scholar]
- 55.Kapsimali M, Kloosterman WP, de Bruijn E, Rosa F, Plasterk RH, Wilson SW. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8:R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zanato VF, Martins MP, Anselmo-Franci JA, Petenusci SO, Lamano-Carvalho TL. Sexual development of male Wistar rats. Brazilian J Med Biol Res. 1994;27:1273–1280. [PubMed] [Google Scholar]
- 58.Dickerson SM, Gore AC. Estrogenic environmental endocrine-disrupting chemical effects on reproductive neuroendocrine function and dysfunction across the life cycle. Rev Endo Metab Disorders. 2007;8:143–159. doi: 10.1007/s11154-007-9048-y. [DOI] [PubMed] [Google Scholar]
- 59.Cohen A, Shmoish M, Levi L, Cheruti U, Levavi-Sivan B, Lubzens E. Alterations in micro-ribonucleic acid expression profiles reveal a novel pathway for estrogen regulation. Endocrinology. 2008;149:1687–1696. doi: 10.1210/en.2007-0969. [DOI] [PubMed] [Google Scholar]
- 60.Forest MG. Differentiation and development of the male. Clinics Endocrinol Metab. 1975;4:569–596. doi: 10.1016/s0300-595x(75)80048-x. [DOI] [PubMed] [Google Scholar]
- 61.Roselli CE, Horton LE, Resko JA. Distribution and regulation of aromatase activity in the rat hypothalamus and limbic system. Endocrinology. 1985;117:2471–2477. doi: 10.1210/endo-117-6-2471. [DOI] [PubMed] [Google Scholar]
- 62.Bonefeld-Jorgensen EC, Andersen HR, Rasmussen TH, Vinggaard AM. Effect of highly bioaccumulated polychlorinated biphenyl congeners on estrogen and androgen receptor activity. Toxicology. 2001;158:141–153. doi: 10.1016/s0300-483x(00)00368-1. [DOI] [PubMed] [Google Scholar]
- 63.Portigal CL, Cowell SP, Fedoruk MN, Butler CM, Rennie PS, Nelson CC. Polychlorinated biphenyls interfere with androgen-induced transcriptional activation and hormone binding. Toxicol Appl Pharmacol. 2002;179:185–194. doi: 10.1006/taap.2002.9371. [DOI] [PubMed] [Google Scholar]
- 64.Kermath BA, Riha PD, Woller MJ, Wolfe A, Gore AC. Hypothalamic molecular changes underlying natural reproductive senescence in the female rat. Endocrinology. 2014;155:3597–3609. doi: 10.1210/en.2014-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walker DM, Kirson D, Perez LF, Gore AC. Molecular profiling of postnatal development of the hypothalamus in female and male rats. Biol Reprod. 2012;87:129. doi: 10.1095/biolreprod.112.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meek LR, Romeo RD, Novak CM, Sisk CL. Actions of testosterone in prepubertal and postpubertal male hamsters: dissociation of effects on reproductive behavior and brain androgen receptor immunoreactivity. Horm Behav. 1997;31:75–88. doi: 10.1006/hbeh.1997.1371. [DOI] [PubMed] [Google Scholar]
- 67.Walker DM, Juenger TE, Gore AC. Developmental profiles of neuroendocrine gene expression in the preoptic area of male rats. Endocrinology. 2009;150:2308–2316. doi: 10.1210/en.2008-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herbison AE, Spratt DP. Sexually dimorphic expression of calcitonin gene-related peptide (CGRP) mRNA in rat medial preoptic nucleus. Brain Res Mol Brain Res. 1995;34:143–148. doi: 10.1016/0169-328x(95)00144-h. [DOI] [PubMed] [Google Scholar]
- 69.Cao J, Patisaul HB. Sexually dimorphic expression of hypothalamic estrogen receptors alpha and beta and Kiss1 in neonatal male and female rats. J Comp Neurol. 2011;519:2954–2977. doi: 10.1002/cne.22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mundalil Vasu M, Anitha A, Thanseem I, Suzuki K, Yamada K, Takahashi T, Wakuda T, Iwata K, Tsujii M, Sugiyama T, Mori N. Serum microRNA profiles in children with autism. Molec Autism. 2014;5:40. doi: 10.1186/2040-2392-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guan F, Zhang B, Yan T, Li L, Liu F, Li T, Feng Z, Zhang B, Liu X, Li S. MIR137 gene and target gene CACNA1C of miR-137 contribute to schizophrenia susceptibility in Han Chinese. Schizophrenia Res. 2014;152:97–104. doi: 10.1016/j.schres.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 72.Selvamani A, Williams MH, Miranda RC, Sohrabji F. Circulating miRNA profiles provide a biomarker for severity of stroke outcomes associated with age and sex in a rat model. Clinical Science. 2014;127:77–89. doi: 10.1042/CS20130565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kalkbrenner AE, Schmidt RJ, Penlesky AC. Environmental chemical exposures and autism spectrum disorders: a review of the epidemiological evidence. Current Problems Ped Adolesc Health Care. 2014;44:277–318. doi: 10.1016/j.cppeds.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brown JS., Jr Effects of bisphenol-A and other endocrine disruptors compared with abnormalities of schizophrenia: an endocrine-disruption theory of schizophrenia. Schizophrenia Bull. 2009;35:256–278. doi: 10.1093/schbul/sbm147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Melzer D, Osborne NJ, Henley WE, Cipelli R, Young A, Money C, McCormack P, Luben R, Khaw KT, Wareham NJ, Galloway TS. Urinary bisphenol A concentration and risk of future coronary artery disease in apparently healthy men and women. Circulation. 2012;125:1482–1490. doi: 10.1161/CIRCULATIONAHA.111.069153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Statistical results of sex, age, and treatment and their interactions on miRNA expression in the MPN and VMN.
Supplemental Table 2. Statistical results of mRNA analysis in the MPN and VMN.