Abstract
We report here two novel HIV-1 recombinant forms (CRF01_AE/B) isolated from two HIV-positive male subjects infected through homosexual contact in Beijing, China. Recombination contributes substantially to the genetic diversity of HIV-1, and is likely to occur in populations in which multiple subtypes circulate. Molecular epidemiological studies showed that subtype B, CRF01_AE, and CRF07_BC are currently cocirculating in parallel among men who have sex with men (MSM) in China, providing the opportunity for the emergence of new recombinants. Phylogenetic analysis of near full-length genome (NFLG) sequences showed that the unique recombinant forms (URFs) were composed of gene regions from CRF01_AE and subtype B. The CRF01_AE region of the recombinants clustered together with a previously described cluster 4 lineage of CRF01_AE. The B regions of both the recombinants clustered within the B strains. The two recombinants were quite similar with six breakpoints in common. These data highlight the importance of continuous surveillance of the dynamic change of HIV-1 subtypes and new recombinants among the MSM population.
HIV-1 is characterized by its high degree of genetic diversity. Phylogenetic analyses of HIV-1 isolates derived from different geographic locations have classified HIV-1 strains into four major groups: M, O, N, and group P.1 HIV-1 group M is predominant in HIV-1 infections worldwide and can be further divided into nine subtypes (A–D, F–H, J, and K), five subsubtypes (A1–A3, F1, and F2), and numerous recombinant forms. To date, 72 circulating recombinant forms (CRFs) have been identified in the Los Alamos HIV database (www.hiv.lanl.gov). In addition, many unique recombinant forms (URFs) have developed in populations in which multiple HIV-1 strains cocirculate.1
Recombination substantially contributes to the overall genetic diversity of HIV-1. It has been reported that multiple HIV-1 genotypes, including subtype B, CRF01_AE, and CRF07_BC, are currently circulating in the men who have sex with men (MSM) population in China.2 HIV-positive MSM constituted 7.3% of the national total of newly reported HIV cases in 2005. This proportion increased to 32.5% in 2009 and stabilized at around 27% in 2011.3 The cocirculation and rapid transmission of multiple HIV-1 genotypes provide the opportunity for the generation of new recombination forms. Indeed, we identified and characterized two novel recombinants derived from subtype B and CRF01_AE, which were highly different from the previously reported CRF01_AE/B recombinant.4–8
Plasma samples were collected from two HIV-positive MSM from an IRB approved study conducted by China CDC. Both of them were infected with HIV-1 through MSM, and were confirmed as HIV-1 seropositive in May 2013 for BJMP3037B and in September 2013 for BJMP3194B, respectively. Participant BJMP3037B had more than 30 MSM sexual partners in his lifetime. Subject BJMP3194B had four male sexual contacts with two in the past 3 months. Subject BJMP3194B was reported to have used nitrite inhalants. The CD4+ T cell counts were 293 cells/μl and 1,312 cells/μl and the viral loads were 57,300 copies/ml and 29,700 copies/ml for BJMP3037B and BJMP3194B, respectively. They are not sexual partners. More detailed information concerning the two participants is given in Table 1.
Table 1.
Demographics, HIV-Related Behaviors and Disease Status of Two HIV-Infected Study Participants
| Variable | BJMP3037B | BJMP3194B |
|---|---|---|
| Age (year) | 28 | 32 |
| Education | Graduate student | College |
| Marital status | Single | Single |
| Predominant role in anal sex | Receptive | Insertive |
| Number of lifetime male sex partners | 30 | 4 |
| Number of male sexual partners in the past 3 months | 3 | 2 |
| Ever used illicit drugs | No | Yes |
| Viral loads (copies/ml) | 57,300 | 29,700 |
| CD4+ T cell count (cells/μl) | 293 | 1,312 |
Near full-length genome (NFLG) sequences (9.0 kb) were determined using a single-genome amplification method with two sets of primers designed for the determination of the 5′ and 3′ halves of the HIV-1 genome sequence. RNA was extracted from 280 μl of plasma by the column purification method (QIAamp Viral Mini Kit, Qiagen, Hilden, Germany). Extracted RNA was reversely transcribed into cDNA using Superscript III (Invitrogen, USA) with primers: CRF01_AE-FL1.5 (5′-CCTTGAGTGCTTAAAGTGGTGTGTGCCCGTCTGT-3′, HXB2: 538–571) and CRF01_AE-FL1.3 (5′-ACCACTTTA-AGCACTCAAGGCAAGCTTTATTG-3′, HXB2: 9644–9611) for the first round polymerase chain reaction (PCR) and CRF01_AE-FL2.5 (5′-AGTGGTGTGTGCCCGTCTGTGTTAGGACTC-3′, HXB2: 551–581) and CRF01_AE-FL2.3 (5′-TTAAGCACTCAAGGCAAGCTTTATTGAGGCTTA-3′, HXB2: 9636–9603) for the second round. Reaction profiles were the same as the reference.9 The PCR products were purified using a QIAquick Gel Extraction Kit (Qiagen) and sequenced by an ABI 3730XL sequencer using BigDye terminators (Applied Biosystems, Foster City, CA). The chromatogram data were cleaned and assembled using Sequencher v.4.10.1 (Gene Codes, Ann Arbor, MI). The sequences were blasted with all the sequences obtained in the laboratory to confirm previous results and to avoid possible cross-contamination.
To determine the subtype, phylogenetic trees were constructed using the neighbor-joining method of MEGA v.6.06 (www.megasoftware.net/). The reference sequences for classifying HIV-1 subtypes were obtained from the Los Alamos HIV databases (www.hiv.lanl.gov). SimPlot, version 3.5.1, was used to evaluate recombination breakpoints in BJMP3037B and BJMP3194B.10 To confirm the recombinant structure of the two strains, the sequences were submitted to the recombination identification program (RIP) (www.hiv.lanl.gov/content/sequence/RIP/RIP.html).
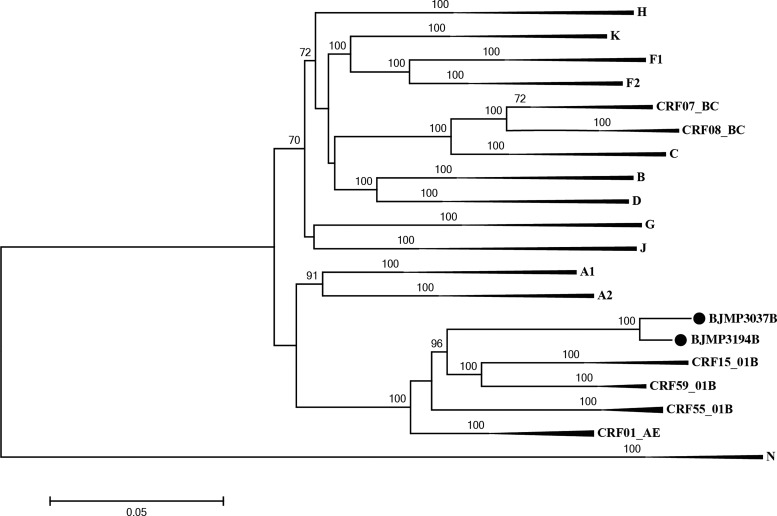
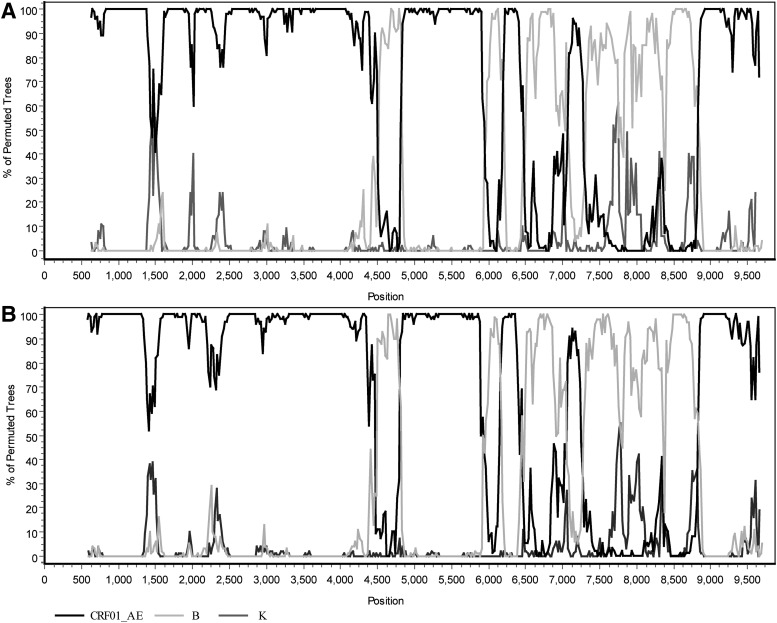
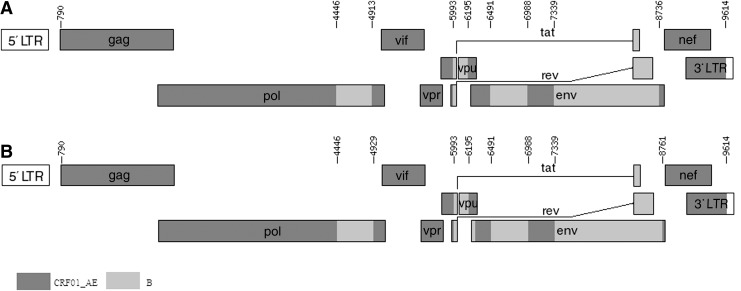
In the phylogenetic trees constructed with full genome reference sequences (Fig. 1), the two URFs clustered with CRF15_01B and CRF59_01B reference sequences with a high bootstrap value (96%). The result of subgenomic phylogenetic analyses revealed a CRF01_AE parental backbone with four subtype B fragments inserted in the pol, vpu, and env regions (Figs. 2 and 3). There is a very small region derived from CRF01_AE in the env gene; it appears as a “blip” in the bootscanning near position 8,200 with a 300 base window. To demonstrate the genomic graphic structure, the Map-Draw Tool available at the Los Alamos HIV sequence database (www.hiv.lanl.gov/content/sequence/DRAW_CRF/recom_mapper.html) was used to generate the genomic map of the two new HIV URFs (Fig. 3).
FIG. 1.
Neighbor-joining phylogenetic tree of BJMP3037B and BJMP3194B. The tree was constructed using MEGA 6.06. A solid circle (●) marks the indicated sequences. The stability of each node was confirmed by bootstrapping with 1,000 replicates and only significant bootstrap values ≥70% are shown at the corresponding nodes. The scale bar represents 5% genetic distance.
FIG. 2.
Bootscan analysis of the near full-length nucleotide sequences of BJMP3037B (A) and BJMP3194B (B). Bootscan analysis was performed using CRF01_AE, B, and subtype K reference sequences. The bootscan window was 300 bp with a step size of 10 bp using SimPlot 3.5.1 software. The x-axis is the nucleotide position in the HIV genomic sequence; the y-axis indicates the percentage supporting the clustering with reference sequences.
FIG. 3.
Genetic map of BJMP3037B (A) and BJMP3194B (B). Map-draw tool is available at the Los Alamos HIV sequence database (www.hiv.lanl.gov/content/sequence/DRAW_CRF/recom_mapper.html).
The phylogenetic trees of the individual gene regions confirmed the breakpoints of the two NFLG sequences as follows (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/aid): I (790–4445 nt) CRF01_AE, II (4446 –4912 nt) B, III (4913–5992 nt) CRF01_AE, IV (5993–6194 nt) B, V (6195–6490 nt) CRF01_AE, VI (6491–6987 nt) B, VII (6988–7398 nt) CRF01_AE, VIII (7339–8735 nt) B, IX (8736–9614 nt) CRF01_AE (BJMP3037B); I (790–4445 nt) CRF01_AE, II (4446–4928 nt) B, III (4929–5992 nt) CRF01_AE, IV (5993–6279 nt) B, V (6280–6490 nt) CRF01_AE, VI (6491–6987 nt) B, VII (6988–7398 nt) CRF01_AE, VIII (7339–8760 nt) B, IX (8761–9614 nt) CRF01_AE (BJMP3194B). Bootscan analysis of the two NFLG sequences revealed that they shared six identical breakpoints (nt 4446, nt 4921±8, nt 5993, nt 6491, nt 6988, and nt 7339), using HXB2 as a reference, as shown in the genomic map (Fig. 3). As MSM without any known epidemiological links, it is noteworthy that they are infected with a high degree of consistency in the breakpoints.
We analyzed the parental origin of all the fragments of the two NFLGs. The CRF01_AE regions for both URFs were from the CRF01_AE cluster 4 lineage11 (Supplementary Fig. S1), which is circulating primarily among Beijing MSM. The subtype B regions clustered within the B strains. Subtype B and CRF01_AE have been circulating in China since the 1990s, predominantly among MSM for subtype B and among heterosexuals for CRF01_AE.12,13 Previously, CRF01_AE was found among MSM in 200514 and then experienced a rapid increase, from 3.7% to 30.2% in 2006 and 2008,2,15 and finally replacing subtype B as the most predominant strain in the MSM population after 2008.16 Overall, cocirculation of CRF01_AE and subtype B among MSM is likely to facilitate the generation of different unique recombinant forms of CRF01_AE and subtype B, as reported here in two novel URFs.
We have previously described two URF NFLG sequences composed of CRF01_AE/CRF07_BC second-generation recombinants among Beijing MSM.17 As the cocirculation of multiple HIV-1 subtypes continues among the MSM population, it is likely that new recombinants of HIV-1 will continue to emerge. Since HIV infections are increasing alarmingly within the MSM risk group in China, monitoring the diversity may provide a means of following the developing epidemic in China.
Sequence Data
The NFLG sequences of BJMP3037B and BJMP3194B have been deposited in GenBank with accession numbers KP418805 and KP418806 respectively.
Supplementary Material
Acknowledgments
This study was supported by the Chinese National Natural Science Foundation of China (81261120393, 81471962), the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award R01AI094562, and the Chinese National Science and Technology Major Projects for Infectious Diseases Control and Prevention (2012ZX1001-002). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Plantier JC, Leoz M, Dickerson JE, et al. : A new human immunodeficiency virus derived from gorillas. Nat Med 2009;15:871–872 [DOI] [PubMed] [Google Scholar]
- 2.Wang W, Jiang S, Li S, et al. : Identification of subtype B, multiple circulating recombinant forms and unique recombinants of HIV type 1 in an MSM cohort in China. AIDS Res Hum Retroviruses 2008;24:1245–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Ministry of Health of the People's Republic of China: 2011. Update on the HIV/AIDS epidemic in China. www.moh.gov.cn
- 4.Tovanabutra S, Watanaveeradej V, Viputtikul K, et al. : A new circulating recombinant form, CRF15_01B, reinforces the linkage between IDU and heterosexual epidemics in Thailand. AIDS Res Hum Retroviruses 2003;19:561–567 [DOI] [PubMed] [Google Scholar]
- 5.Tee KK, Li XJ, Nohtomi K, et al. : Identification of a novel circulating recombinant form (CRF33_01B) disseminating widely among various risk populations in Kuala Lumpur, Malaysia. J Acquir Immune Defic Syndr 2006;43:523–529 [DOI] [PubMed] [Google Scholar]
- 6.Tovanabutra S, Kijak GH, Beyrer C, et al. : Identification of CRF34_01B, a second circulating recombinant form unrelated to and more complex than CRF15_01B, among injecting drug users in northern Thailand. AIDS Res Hum Retroviruses 2007;23:829–833 [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Tee KK, Liao H, et al. : Identification of a novel second generation circulating recombinant form (CRF48_01B) in Malaysia: A descendant of the previously identified CRF33_01B. J Acquir Immune Defic Syndr 2010;54:129–136 [DOI] [PubMed] [Google Scholar]
- 8.Wang W, Meng Z, Zhou M, et al. : Near full-length sequence analysis of two new HIV type 1 unique (CRF01_AE/B) recombinant forms among men who have sex with men in China. AIDS Res Hum Retroviruses 2012;28(4):411–417 [DOI] [PubMed] [Google Scholar]
- 9.Rousseau CM, Birditt BA, McKay AR, et al. : Large-scale amplification, cloning and sequencing of near full-length HIV-1 subtype C genomes. J Virol Methods 2006;136(1–2):118–125 [DOI] [PubMed] [Google Scholar]
- 10.Lole KS, Bollinger RC, Paranjape RS, et al. : Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 1999;73(1):152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y, He X, Hsi JH, et al. : The rapidly expanding CRF01_AE epidemic in China is driven by multiple lineages of HIV-1 viruses introduced in the 1990s. AIDS 2013;27(11):1793–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Lu L, Ba L, et al. : Dominance of HIV-1 subtype CRF01_AE in sexually acquired cases leads to a new epidemic in Yunnan province of China. PLoS Med 2006;3:e443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao J, Zhang F, He Z, et al. : Subtype and sequence analysis of the C2-V3 region of env gene among HIV-1 infected homosexual men in Beijing. Chin J STD/AIDS Prev Contr 2002;8:131–133 [Google Scholar]
- 14.Zhang X, Li S, Li X, et al. : Characterization of HIV-1 subtypes and viral antiretroviral drug resistance in men who have sex with men in Beijing, China. AIDS 2007;21(Suppl 8):S59–65 [DOI] [PubMed] [Google Scholar]
- 15.Guo H, Wei JF, Yang H, et al. : Rapidly increasing prevalence of HIV and syphilis and HIV-1 subtype characterization among men who have sex with men in Jiangsu, China. Sex Transm Dis 2009;36:120–125 [DOI] [PubMed] [Google Scholar]
- 16.Wang W, Xub J, Jiang S, et al. : The dynamic face of HIV-1 subtypes among men who have sex with men in Beijing, China. Curr HIV Res 2011;9:136–139 [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Wei H, Feng Y, et al. : Genomic characterization of two novel HIV-1 second-generation recombinant forms among men who have sex with men in Beijing, China. AIDS Res Hum Retroviruses. 2015;31(3):342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.