Abstract
Sirtuin 1 (SIRT1) is a mammalian NAD+-dependent protein deacetylase that regulates cellular metabolism and inflammatory response. The organ-specific deletion of SIRT1 induces local inflammation and insulin resistance in dietary and genetic obesity. Macrophage-mediated inflammation contributes to insulin resistance and metabolic syndrome, however, the macrophage-specific SIRT1 function in the context of obesity is largely unknown. C57/BL6 wild type (WT) or myeloid-specific SIRT1 knockout (KO) mice were fed a high-fat diet (HFD) or normal diet (ND) for 12 weeks. Metabolic parameters and markers of hepatic steatosis and inflammation in liver were compared in WT and KO mice. SIRT1 deletion enhanced HFD-induced changes on body and liver weight gain, and increased glucose and insulin resistance. In liver, SIRT1 deletion increased the acetylation, and enhanced HFD-induced nuclear translocation of nuclear factor kappa B (NF-κB), hepatic inflammation and macrophage infiltration. HFD-fed KO mice showed severe hepatic steatosis by activating lipogenic pathway through sterol regulatory element-binding protein 1 (SREBP-1), and hepatic fibrogenesis, as indicated by induction of connective tissue growth factor (CTGF), alpha-smooth muscle actin (α-SMA), and collagen secretion. Myeloid-specific deletion of SIRT1 stimulates obesity-induced inflammation and increases the risk of hepatic fibrosis. Targeted induction of macrophage SIRT1 may be a good therapy for alleviating inflammation-associated metabolic syndrome.
Keywords: Hepatic steatosis, High-fat diet, Nuclear factor kappa B, Sirtuin 1
INTRODUCTION
Sirtuin 1 (SIRT1) is an NAD+-dependent deacetylase that regulates energy metabolism and stress response [1,2]. SIRT1 was shown to deacetylate nuclear factor kappa B (NF-κB) and regulate inflammatory and apoptotic responses [3]. The activated NF-κB promotes expression of inflammatory cytokines, such as tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6), and inhibits insulin signaling by insulin receptor substrate (IRS) serine phosphorylation or proteasomal degradation [4,5].
SIRT1 is ubiquitously expressed in various mammalian tissues such as brain, kidney, muscle, liver, and adipose tissue [6]. Its organ-specific functions include regulation of gluconeogenesis and fatty acid β oxidation in the liver, stimulation of insulin secretion in the pancreas, and regulation of energy homeostasis in the brain [7,8,9,10]. SIRT1 function has been extensively studied in tissue-specific knockdown or knockout mouse models, among which hepatic function of myeloid SIRT1 has been particularly interested in the pathology of obesity and diabetes.
Obesity is associated with a chronic low-grade inflammation, which contributes to diabetes, fatty liver disease, and other metabolic disorders. Inflammatory cytokines released by adipose tissue macrophages induce insulin resistance and the activated macrophages are infiltrated in various tissues, leading to systemic inflammation [11,12]. Hepatic macrophages are a major source of pro-inflammatory cytokines and reactive oxygen species and hepatocyte-specific SIRT1 knockout results in hepatic inflammation and steatosis [9]. The expression of SIRT1 was reduced in non-alcoholic fatty liver disease (NAFLD) induced by HFD-fed rats [13], and SIRT1 activators improved insulin sensitivity and reduced adipose tissue inflammatory responses in obese Zucker diabetic fatty rats [14]. In dietary- and genetically-induced obesity, a chronic insulin-resistant state of being hyperglycemic, hyperlipidemic, hypoadiponectinemic, and hyperleptinemic, may also contribute to systemic inflammation mediated by macrophage SIRT1 [15,16].
In this study, we investigated the role of macrophage SIRT1 on obesity-induced metabolic and inflammatory changes. We examined the effect of myeloid cell-specific SIRT1 deletion on insulin resistance, inflammation, hepatic steatosis and fibrogenesis, to provide a therapeutic potential of modulating SIRT1 expression for treating obesity-induced metabolic syndrome.
METHODS
Animals and genotyping
SIRT1flox/flox mice (B6;129- SIRT1tm1Ygu/J) and LysM-Cre mice (B6;129P2-lyz2tm1(cre)Ifo/J) were received from Dr. Lee SI (Gyenongsnag National Univeristy School of Medicine). The SIRT1flox/flox and homozygous LysM-Cre mice were crossed to obtain myeloid cell-specific SIRT1 KO mice (Supplementary Fig. 1A) [17]. SIRT1flox/flox; LysM-Cre+/+ (SIRT1 KO, n=20) and SIRT1flox/flox; LysM-Cre-/- (SIRT1 wild type [WT, n=20]) mice older than 6 weeks were fed ad libitum either a standard laboratory chow diet (ND) or a high-fat diet (HFD) (D12079B; Research Diets, New Brunswick, NJ, USA) for 12 weeks. The experiments were performed in accordance with the National Institutes of Health Guidelines on the Use of Laboratory Animals (GNU-130306-M0021).
Primers used for genotyping are listed. Floxed Sirt1 primers are Forward-5' GGT TGA CTT AGG TCT TGT CTG 3' and Reverse-5' CGT CCC TTG TAA TGT TTC CC 3'. Cre+/+ primers are WT- 5' TTA CAG TCG GCC AGG CTG AC 3' and Common-5' CTT GGG CTG CCA GAA TTT CTC 3'. Cre-/- primers are Mutant-5' CCC AGA AAT GC AGA TTA CG 3' and Common-5' CTT GGG CTG CCA GAA TTT CTC 3'.
Glucose tolerance test (GTT) and insulin tolerance test (ITT)
For the GTT, mice received D-glucose (2 g/kg, Sigma-Aldrich, St. Louis, MO, USA) by intraperitoneal injection after an overnight fast (16 h), and blood samples were collected before and 30, 60, 90, and 120 minutes after the injection. For the ITT, mice were given intraperitoneal injections of insulin (0.75 U/kg, Humulin-R; Eli Lilly, Indianapolis, IN, USA), and blood samples were collected before and 15, 30, 45, and 60 minutes after the injection. Blood glucose was measured using an Accu-Chek glucometer (Roche Diagnostics GmbH, Mannheim, Germany).
Measurement of serum metabolic parameters
Mice were intramuscularly anesthetized with Zoletil (5 mg/kg; Virbac Laboratories, Carros, France), after which blood samples were extracted transcardially and allowed to clot for 2 h at room temperature. After centrifugation, the serum samples were stored at -80℃ until analysis. Serum aspartate aminotransferase (AST), alanine aminotransferase (ALT), free fatty acid (FFA), total cholesterol, and triglyceride (TG) levels were determined by enzymatic colorimetric assays from Green Cross Reference Laboratory (Yongin-si, South Korea). Serum insulin, leptin, and adiponectin concentrations (n=7 mice per each group) were measured using mouse insulin (Shibayagi Co., Gunma, Japan), leptin (Crystal Chem Inc., IL, USA), and adiponectin (Shibayagi) ELISA kits, respectively.
Hepatic TG colorimetric assay
Frozen liver was freshly homogenized and centrifuged and the supernatants were used to determine TG levels. TG concentration (n=7 mice per each group) was measured using a TG colorimetric assay kit (Cayman Chemical Company, Ann Arbor, MI, USA).
Tissue preparation
Mice (n=4 mice per each group) were anesthetized with Zoletil and perfused transcardially with heparinized saline followed by 4% paraformaldehyde. Liver was processed for paraffin embedding and cut into 5-µm thick sections.
Oil Red O, Sirius red and Masson Trichrome staining
To determine hepatic lipid accumulation, frozen liver sections (5-µm thick) were stained with 0.5% Oil Red O (Sigma-Aldrich) for 10 min, washed, and counterstained with Mayer's hematoxylin (Sigma-Aldrich) for 45 sec. Data for Oil Red O staining were presented as the mean percentage of stained area to a total hepatic region in 10 fields from each liver section. Quantitative analysis was performed using analySIS-FIVE program (Olympus Soft Imaging System, Münster, Germany). To examine collagen deposition and hepatic fibrosis, the deparaffinized liver sections were stained with Sirius red (Sigma-Aldrich) and Masson trichrome (Sigma-Aldrich). The sections were visualized under a BX51 light microscope (Olympus, Tokyo, Japan).
Sircol collagen assay
The Sircol collagen assay is a quantitative dye-binding method for the analysis of acid and pepsin-soluble collagens. The collagen concentration from frozen liver tissues was determined by using a Sircol assay kit (Bioclor Ltd., Northern Ireland, UK).
Immunohistochemistry
The deparaffinized sections of liver was placed in 0.3% H2O2 for 10 minutes, washed, and incubated in blocking serum for 20 min. Sections were incubated in primary antibodies (Supplementary Table 1) at 4℃ overnight and with a secondary biotinylated antibody for 1 h at room temperature. After washing, the sections were incubated in avidin-biotin-peroxidase complex solution (Vector Laboratories, Burlingame, CA, USA) and developed with 0.05% diaminobenzidine (Sigma-Aldrich) containing 0.05% H2O2. The sections were then dehydrated in graded alcohols, cleared in xylene, and mounted under a coverslip with Permount (Sigma-Aldrich). The CD68-positive immunostained cells were counted from three sections (n=3~4 per each group) by observers blinded to the treatment conditions using 200x objectives. The sections were visualized under a BX51 light microscope (Olympus) and the quantitative analysis was performed using analySIS-FIVE program (Olympus Soft Imaging System).
Western blot analysis
For protein extraction (n=6 mice per each group), frozen liver tissues were used. For cytosolic and nuclear fraction preparation, livers were chopped in ice-cold lysis buffer (10 mM HEPES-KOH [pH7.9], 1.5 mM MgCl2, 10 mM KCl, protease inhibitors), homogenized, and centrifuged for 1 min at 12,000 rpm. Then, the supernatant was collected as a cytosol fraction and nuclear pellet was resuspended in high-salt extraction buffer (20 mM HEPES-KOH [pH7.9], 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 25% glycerol, protease inhibitors, 0.5 mM DTT), incubated on ice for 20 min, and centrifuged for 10 min at 12,000 rpm. Then, the supernatant was collected as a nuclear fraction. For total protein preparation, frozen liver and epididymal pad fats were homogenized in ice-cold lysis buffer (15 mM HEPES [pH 7.9], 0.25 M sucrose, 60 mM KCl, 10 mM NaCl, 1 mM ethylene glycol tetraacetic acid, 1 mM phenylmethylsulfonyl fluoride, and 2 mM NaF). Samples were probed with primary antibodies (Supplementary Table 1) and protein bands were detected using enhanced chemiluminescence substrates (Pierce, Rockford, IL, USA). The Multi-Gauge image analysis program (version 3.0; Fujifilm, Tokyo, Japan) was used for densitometry analysis.
Statistical analysis
Differences between groups were determined by one-way ANOVA, followed by post hoc analysis by using the Bonferroni test. Results were presented as mean±standard error of the mean (SEM), and p<0.05 was considered significant.
RESULTS
Effects of SIRT1 deletion on body weight and metabolic changes in HFD-fed mice
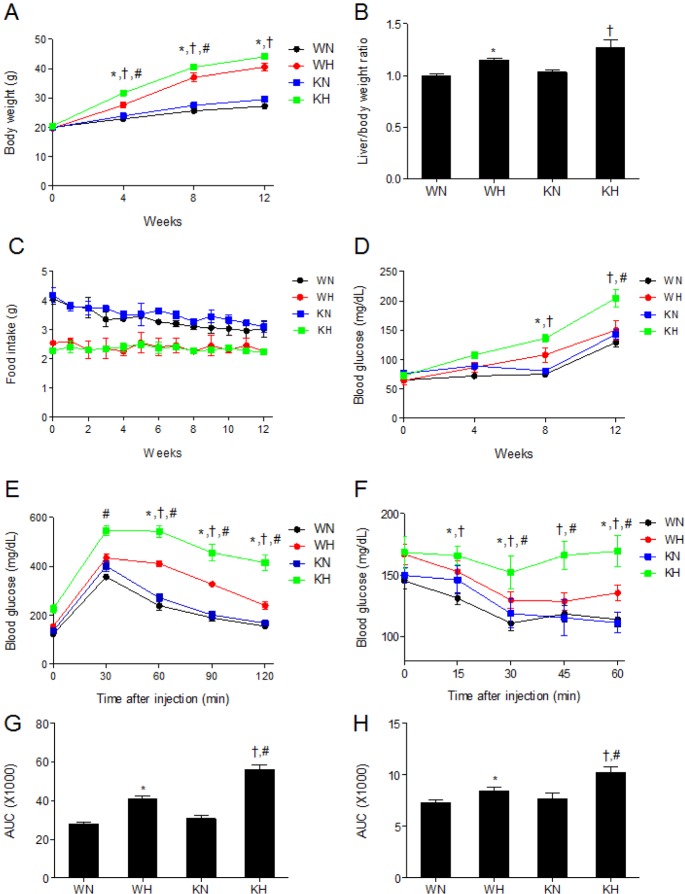
We first examined the effect of SIRT1 deletion on HFD-induced changes in metabolic parameters. Body weights were significantly increased in HFD-fed WT (WH) and HFD-fed KO (KH) mice compared with ND-fed WT (WN) and ND-fed KO (KN) mice, respectively (at weeks 4, 8, and 12); additionally, KH mice had significantly elevated weight gain compared with WH mice (at weeks 4 and 8) (Fig. 1A). HFD also significantly increased liver/body weight ratio in both KO and WT mice, the fold-changes in ratio induced by HFD were greater in KO mice than in WT mice (Fig. 1B). Although food intake was lower in HFD-fed mice than in ND-fed mice, SIRT1 deletion did not significantly affect food intake in either diet (Fig. 1C).
Fig. 1. SIRT1 deletion aggravates HFD-induced weight gain and insulin resistance. (A) Body weights, (B) liver/body weight ratio, (C) food intake, (D) fasting blood glucose levels, (E) GTT, (F) ITT, (G) area under curve (AUC) for GTT and (H) AUC for ITT of WT and KO mice fed ND or HFD. Data are presented as mean±SEM. *p<0.05 for WH mice versus WN mice. †p<0.05 for KH mice versus KN mice. #p<0.05 for KH mice versus WH mice. WN, ND-fed WT mice; WH, HFD-fed WT mice; KN, ND-fed KO mice; KH, HFD-fed KO mice.
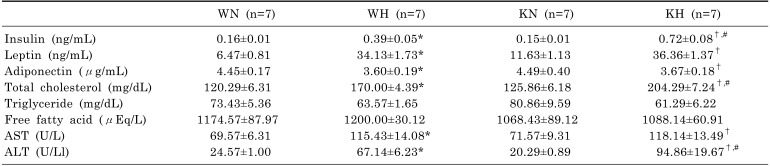
Results of measurement in serum metabolic hormones, lipids, and hepatic enzymes (Table 1) showed that HFD induced hyperinsulinemia, hyperleptinemia, hypoadiponectinemia, and hypercholesterolemia in both WT and KO mice compared to ND mice. Insulin and total cholesterol levels were significantly higher in KH mice than in WH mice. Serum TG and FFA levels were not significantly different among the groups. AST and ALT levels were significantly increased in HFD-fed mice compared with ND-fed mice; KH mice had significantly higher ALT level than WH mice.
Table 1. Serum metabolic parameters in WT and SIRT1 KO mice fed with ND or HFD.
*p<0.05. vs. WN mice, †p<0.05. vs. KN mice, #p<0.05. vs. WH mice.
Effects of SIRT1 deletion on insulin resistance in HFD-fed mice
We found that HFD significantly increased serum insulin levels in KO mice compared with WT mice (Table 1). Fasting blood glucose levels were significantly higher in KH mice than in KN mice (at weeks 8 and 12) or WH mice (at weeks 12) (Fig. 1D). Further, KH mice had impaired blood glucose regulation following D-glucose injection, and exhibited significantly elevated blood glucose levels compared with WH mice at all measurement intervals of the GTT (Fig. 1E and G). The hypoglycemic response to insulin was also impaired in KH mice, as they exhibited significantly elevated blood glucose levels compared with WH mice at all measurement intervals of the ITT (Fig. 1F and H).
Effects of SIRT1 deletion on NF-κB acetylation and inflammation in HFD-fed mice
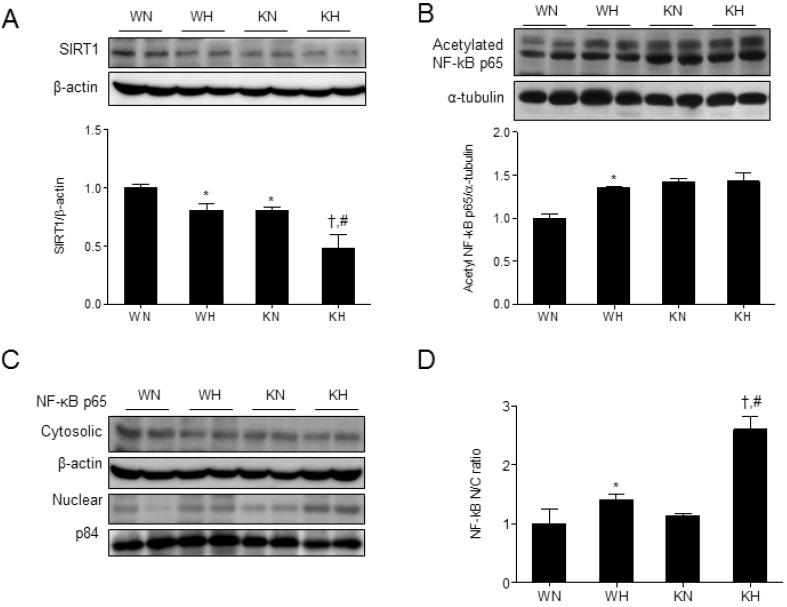
SIRT1 is known to suppress inflammatory responses by deacetylating NF-κB p65 [17] and thus, we investigated the correlation between hepatic SIRT1 and acetylated NF-κB expression. SIRT1 expression was decreased in KO mice compared with WT mice due to SIRT1 silencing in myeloid-derived liver macrophages (Kupffer cells), and the level was further reduced by HFD (Fig. 2A). As previously reported [17], we confirmed the reduced SIRT1 expression in liver and epididymal fat of KO mice in which myeloid-derived cells, Kupffer cells and peritoneal macrophages are enriched, respectively (Supplementary Fig. 1B). Among sirtuin family, SIRT1 and SIRT3 are closely related proteins as belonging to class I sirtuins [18]. However, the expression of mitochondrial SIRT3 in liver of both WT and KO was not affected by myeloid SIRT1 deletion (Supplementary Fig. 2).
Fig. 2. SIRT1 deletion increases NF-κB acetylation and induces NF-κB nuclear translocation in HFD-fed mice. Western blots of SIRT1 (A) and acetylated NF-κB p65 (B) in liver homogenates from WT and KO mice fed ND or HFD. (C, D) Western blots of cytosolic and nuclear NF-κB and the nuclear/cytosolic (N/C) ratio of NF-κB levels. Band intensity was normalized to β-actin or α-tubulin. Data are presented as mean±SEM. *p<0.05 for WH mice versus WN mice. †p<0.05 for KH mice versus KN mice. #p<0.05 for KH mice versus WH mice. WN, ND-fed WT mice; WH, HFD-fed WT mice; KN, ND-fed KO mice; KH, HFD-fed KO mice.
Accordingly, the acetylated NF-κB p65 levels were increased in KO mice and the level was comparable to that in WH mice (Fig. 2B). Acetylated NF-κB is known to translocate to the nucleus, where it regulates transcriptional activity. To determine whether HFD induces the nuclear localization of NF-κB, nuclear and cytosolic fractions were separated and NF-κB levels were analyzed in each fraction. Nuclear NF-κB was increased by HFD in both KO and WT mice (Fig. 2C), and the nuclear-to-cytosolic (N/C) ratio was significantly increased in KH mice compared to WH mice (Fig. 2D).
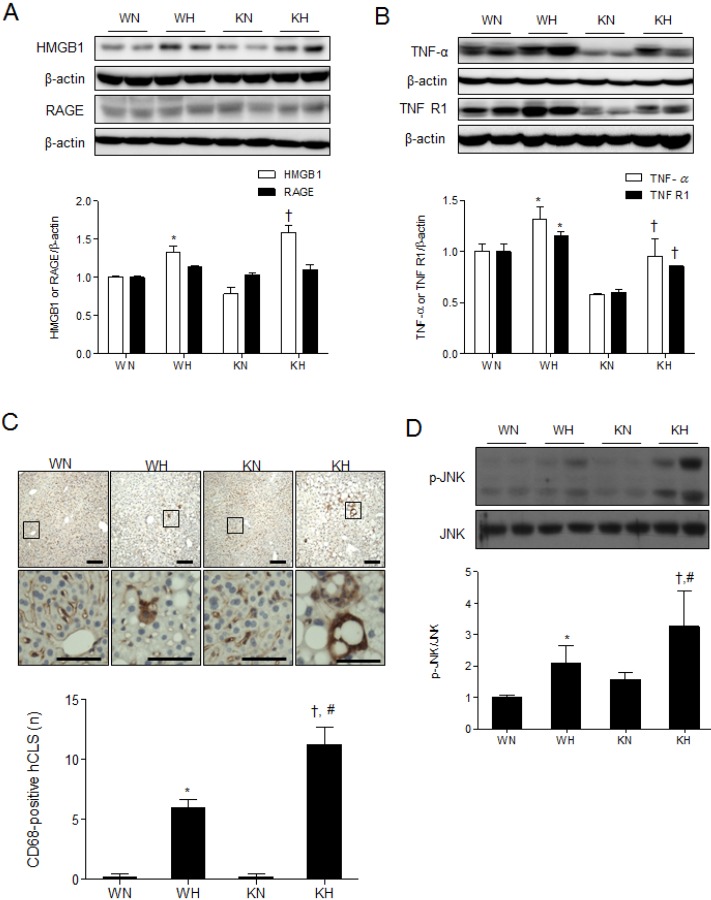
We then examined whether NF-κB activation by SIRT1 deletion aggravates hepatic inflammation induced by HFD. TNF-α signaling contributes to hepatic steatosis and fibrosis by activating Kupffer cells and hepatic stellate cells (HSCs) and reducing apoptosis of activated HSCs [18]. Western blot analysis showed that HFD increased high mobility group protein B1 (HMGB1), TNF-α, and TNF receptor 1 (TNFR1) expression over ND in both WT and KO mice (Fig. 3A and B). Although TNF-α and TNFR1 expression was lower in KO mice than WT mice, the fold-changes in expression induced by HFD were greater in KO mice than in WT mice.
Fig. 3. SIRT1 deletion aggravates HFD-induced hepatic inflammation and JNK activation. (A, B) Western blots of HMGB1, RAGE, TNF-α, and TNFR1 in liver homogenates from WT and KO mice fed an ND or HFD. (C) Immunohistochemistry using a CD68 antibody for detection of activated macrophages in liver sections. The areas in black squares in the top panels were magnified on the bottom. The CD68-positive cells were quantified. Scale bar=100 µm (high magnification, 50 µm). (D) Western blots of p-JNK/JNK in liver homogenates from WT and KO mice fed ND or HFD. Data are presented as mean±SEM. *p<0.05 for WH mice versus WN mice. †p<0.05 for KH mice versus KN mice. #p<0.05 for KH mice versus WH mice. WN, ND-fed WT mice; WH, HFD-fed WT mice; KN, ND-fed KO mice; KH, HFD-fed KO mice.
We showed that HFD increased hepatic inflammation as indicated by induction of CD68-positive mature macrophages in both KO and WT mice; the effect was much greater in KO mice compared to WT mice (Fig. 3C). We also observed HFD-induced hepatic crown-like structures (hCLS) which are macrophage aggregates surrounding the fatty hepatocytes and positively correlated with hepatic inflammation [19]. The number of hCLS, positive for CD68 staining, was significantly increased in KH mice compared to WH mice (Fig. 3C). The results suggest that HFD increased TNF-α-mediated hepatic inflammation and macrophage activation, and SIRT1 deletion aggravated the phenotype.
The c-Jun N-terminal kinase (JNK) activity in macrophages is a feature of obesity-induced insulin resistance and inflammation [20]. Thus, we examined the effect of SIRT1 deletion on JNK activity in HFD-fed mice and found that phosphorylated JNK was dramatically increased by HFD compared to ND; the effect was much more severe in KH mice than in WH mice (Fig. 3D).
Effects of SIRT1 deletion on hepatic steatosis in HFD-fed mice
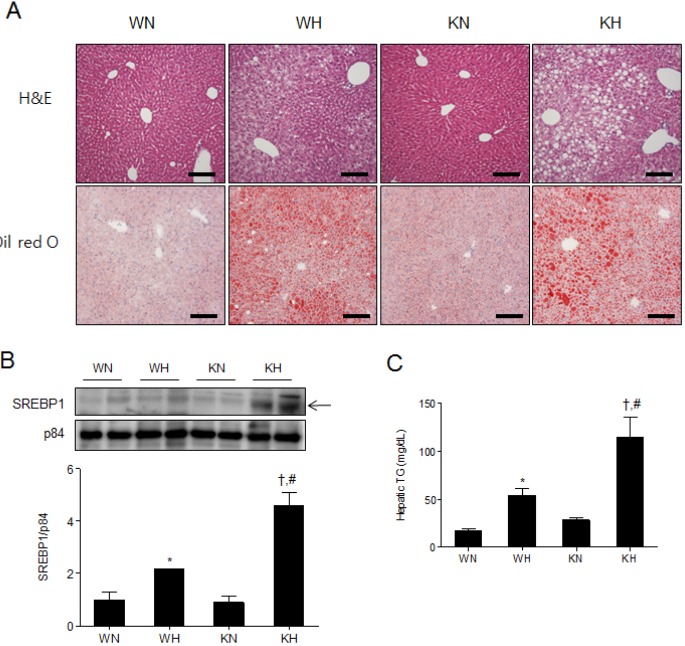
Hepatic steatosis is characterized by accumulation of TG vacuoles and hepatomegaly, which is an early stage of liver damage and could further develop into hepatic fibrosis and cirrhosis [21]. To assess the phenotype of hepatic steatosis, we performed hematoxylin and eosin (H&E) and Oil Red O staining (Fig. 4A). We found that WH and KH mice exhibited enlarged cytoplasmic lipid droplets compared to WN and KN mice, and lipid droplet sizes were larger in KH mice than in WH mice (Fig. 4A). Oil Red O-positive areas (%) in WN and WH mice were 7.71±0.58 and 32.85±1.27, respectively; the positive areas (%) in KN and KH mice were 15.01±0.72 and 40.19±2.13, respectively. The results demonstrate that KN mice showed a mild hepatic steatosis phenotype that was aggravated by HFD.
Fig. 4. SIRT1 deletion aggravates hepatic steatosis in HFD-fed mice. (A) Histological analysis of hepatic fat accumulation by H&E and Oil Red O staining. Scale bar=100 µm. (B) Western blots of nuclear SREBP1 in liver homogenates from WT and KO mice fed an ND or HFD. Band intensity was normalized to nuclear p84 protein. (C) Hepatic TG levels in supernatant fractions of liver homogenates. Data are presented as mean±SEM. *p<0.05 for WH mice versus WN mice. †p<0.05 for KH mice versus KN mice. #p<0.05 for KH mice versus WH mice. WN, ND-fed WT mice; WH, HFD-fed WT mice; KN, ND-fed KO mice; KH, HFD-fed KO mice.
We examined the expression of SREBP1 which is a lipogenic gene, being transcriptionally activated in hepatic steatosis. HFD increased mature SREBP1 in the nuclear fractions of both WT and KO mice, and KH mice displayed higher levels of mature SREBP1 than WH mic (Fig. 4B). In addition, we found that the elevated hepatic TG in KH mice compared to WH mice (Fig. 4C). Taken together, the results suggest that SIRT1 deletion favors hepatic lipogenesis and aggravates hepatic steatosis under HFD.
Effects of SIRT1 deletion on hepatic fibrogenesis in HFD-fed mice
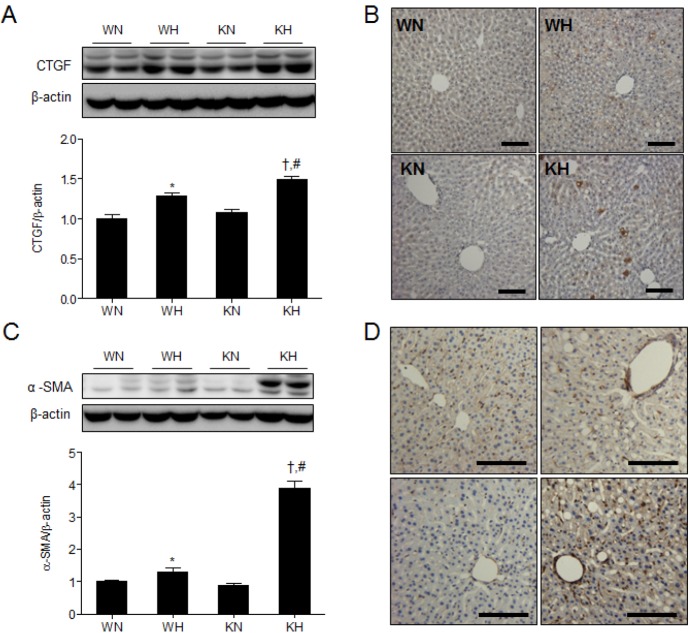
We examined hepatic expression of connective tissue growth factor (CTGF) and alpha-smooth muscle actin (α-SMA) which are indicative of hepatic fibrogenesis (Fig. 5). CTGF and α-SMA are overexpressed in the patients with severe hepatic fibrosis [22,23]. Compared with WH or KN mice, KH mice showed a significant increase in hepatic CTGF expression and CTGF-positive immuno-stained cells (Fig. 5A and B). In addition, KH mice showed a significant increase in hepatic α-SMA expression and α-SMA -positive immuno-stained cells (Fig. 5C and D).
Fig. 5. SIRT1 deletion increases hepatic CTGF and α-SMA expression in HFD-fed mice. (A) Western blots of CTGF in liver homogenates from WT and KO mice fed ND or HFD. (B) Immunohistochemistry for CTGF detection in liver sections. (C) Western blots of α-SMA in liver homogenates from WT and KO mice fed ND or HFD. (D) Immunohistochemistry for α-SMA detection in liver sections. Band intensity was normalized to β-actin. Data are presented as mean±SEM. *p<0.05 for WH mice versus WN mice. †p<0.05 for KH mice versus KN mice. #p<0.05 for KH mice versus WH mice. WN, ND-fed WT mice; WH, HFD-fed WT mice; KN, ND-fed KO mice; KH, HFD-fed KO mice. Scale bar=100 µm.
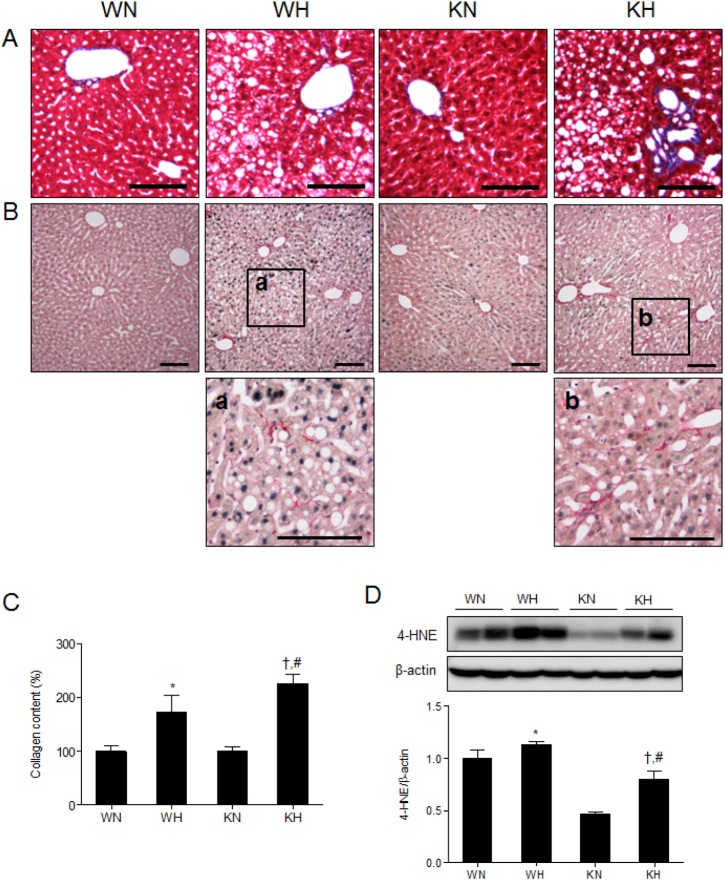
To determine hepatic collagen accumulation, we performed Masson Trichrome and Sirius red staining (Fig. 6). We found that collagen deposition around portal triad were larger in KH mice compared to WH mice (Fig. 6A and B). In addition, KH mice showed a greater accumulation of hepatic collagen than WH mice, indicating that SIRT1 deletion may precipitate the development of hepatic fibrosis (Fig. 6C). The expression of hepatic 4-hydroxynonenal (4-HNE), a marker of lipid peroxidation [24,25] is associated with hepatic fibrogenesis [26]. Although KN mice has lower 4-HNE levels than WN mice, the fold-induction induced by HFD was greater than WN mice (Fig. 6D), suggesting the fibrogenic potential of SIRT1 deletion. Taken together, the results suggest that SIRT1 deletion contributes to hepatic fibrogenesis under HFD.
Fig. 6. SIRT1 deletion induces hepatic fibrogenesis in HFD-fed mice. Histological analysis of hepatic collagen deposition by Masson trichrome (A) and Sirius red staining (B). Scale bar=100 µm. (C) Hepatic collagen levels measured in liver extracts using a Sircol collagen assay kit. (D) Western blots of 4-HNE in liver homogenates from WT and KO mice fed ND or HFD. Band intensity was normalized to β-actin. Data are presented as mean±SEM. *p<0.05 for WH mice versus WN mice. †p<0.05 for KH mice versus KN mice. #p<0.05 for KH mice versus WH mice. WN, ND-fed WT mice; WH, HFD-fed WT mice; KN, ND-fed KO mice; KH, HFD-fed KO mice.
DISCUSSION
We previously showed that a 24-week of HFD significantly induces microglial activation and reduces cholinergic neurotransmission in the hypothalamus of myeloid-specific SIRT1 KO mice [27]. However, neither hypothalamic inflammation nor food intake response was changed after a 12-week of HFD from our previous observation. SIRT1 deletion may affect insulin-responsive organs such as liver early, and later impairs hypothalamic function. In this study, we showed that myeloid-specific SIRT1 deletion aggravated the HFD-induced weight gain, insulin resistance, and hepatic inflammation, and further showed hepatic fibrogenesis. SIRT1 deletion induced a significant increase of NF-κB activity and inflammatory protein expression, and severe macrophage infiltration in HFD-fed mice. Previous studies identified SIRT1 as an anti-inflammatory factor [4,14,27,28], and showed that modest overexpression of SIRT1 suppressed the inflammation [29], whereas conditional SIRT1 deletion increased inflammatory response in a mouse model of arthritis [29,30]. Consistently, we showed that myeloid-specific SIRT1 deletion aggravated HFD-induced inflammation in liver.
SIRT1 heterozygote mice fed a HFD are shown to be more obese and insulin-resistant and typically develop hepatomegaly [31]. Consistently, we showed that myeloid-specific SIRT1 KO mice were more vulnerable to HFD-induced changes, as they gained more body and liver weight and more insulin-resistant compared to WH mice. In our study, KH mice showed severe lipid accumulation and inflammation in liver, as previously reported in HFD-fed SIRT1 heterozygote mice [31,32] and liver-specific SIRT1 KO mice [9]. In addition, we found that KH mice showed a significant induction of 4-HNE, a marker of lipid peroxidation, compared to WH mice. A recent study showed that the treatment of antioxidant resveratrol or N-acetyl-L-cysteine (NAC) improves insulin sensitivity in liver-specific SIRT1 KO mice [25], suggesting that HFD-induced oxidative stress is involved in SIRT1-mediated regulation.
In the present study, we showed an increase of hepatic TG and lipid accumulation and induction of mature SREBP1 in KH mice, and suggest that SIRT1-mediated lipid metabolism is a potential therapeutic target for treating fatty liver disease. CTGF, a potent HSC-activating cytokine, is associated with HSC activation and progression and shown to be overexpressed in patients with fatty liver disease [33]. Hyperglycemia and hyperinsulinemia contribute to the progression of fibrosis in patients with hepatic steatohepatitis or ob/ob mice through the up-regulation of CTGF [22,34]. In addition, we observed a significant increase of hCLS in HFD-fed KO mice; hCLS precedes the development of collagen deposition and is often located close to fibrogenic lesions [19]. Thus, our findings of increased CTGF expression, the hCLS and collagen levels in KH mice indicate that myeloid deletion of SIRT1 has a fibrogenic effect and is critically involved in the progression from simple steatosis to non-alcoholic steatohepatitis (NASH).
Obesity is associated with a chronic low-grade inflammation, and particularly, insulin resistance is closely linked to the activation of inflammatory cytokines, TNF-α and IL-6, that in turn promote insulin resistance by inhibiting insulin receptor substrate-1 (IRS-1) [35,36]. Consistently, we showed that TNF-α and TNFR1 expression and HMGB1 secretion were significantly increased by HFD. Although NF-κB p65 acetylation was increased in KN mice compared to WN mice due to the reduced deacetylase enzyme activity, the NF-κB nuclear translocation was not significantly induced in KN mice (Fig. 2B, D); it may explain that the basal levels of TNF-α and TNFR1 were reduced in KN mice. However, upon HFD challenge, the fold-changes in TNF-α and TNFR1 expression were greater in KO mice than in WT mice. A previous report showed that macrophage-derived TNF-α contributes to hepatic steatosis and inflammation in diet-induced obesity [37]. We speculate that the myeloid-specific SIRT1 deletion may also contribute to the reduced production of TNF-α and suppressed immune responses in KN mice. In this study, we showed that HFD stimulated the nuclear translocation of acetylated NF-κB and induced TNF-α-mediated signaling in KO mice; the effect was severe compared with WT mice. Other signaling factors linking SIRT1, NF-κB and TNF-α/TNFR1 in the pathway may be addressed in future study.
In the present study, JNK activity, measured by a p-JNK level, was significantly increased in KH mice. The activated JNK promotes serine phosphorylation of IRS-1, which down-regulates insulin signaling causing insulin resistance [38]. Consistently, selective JNK deficiency in macrophages was shown to be insulin-sensitive in HFD-fed mice, and the protection was associated with reduced macrophage infiltration and polarization [20]. These studies demonstrate that JNK is required for obesity-induced insulin resistance and inflammation and SIRT1 in macrophages may be an important regulator of JNK activity.
We demonstrate that SIRT1 deficiency in macrophages aggravated HFD-induced inflammation, insulin resistance, and fatty liver disease. Activating SIRT1 signaling may reduce obesity-induced inflammation and metabolic syndrome, and importantly, targeted induction of macrophage SIRT1 may be a good therapy for alleviating systemic levels of obesity-induced inflammation.
ACKNOWLEDGMENTS
This study was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A111436) and the Basic Science Research Program through the National Research Foundation (NRF) of Korea (No. 2014R1A2A1A11049588). Myeloid-specific SIRT1 KO mice were a gift from Dr. Lee SI (Gyeongsang National University School of Medicine). The authors declare that they have no competing interests.
ABBREVIATIONS
- SIRT1
sirtuin 1
- NAD
nicotinamide adenine dinucleotide
- NF-κB
nuclear factor kappa B
- TNF-α
tumor necrosis factor-α
- IL-6
interleukin-6
- IRS-1
insulin receptor substrate-1
- NAFLD
non-alcoholic fatty liver disease
- GTT
glucose tolerance test
- ITT
insulin tolerance test
- AST
aspartate aminotransferase
- ALT
alanine aminotransferase
- FFA
free fatty acids
- TG
triglyceride
- HMGB1
high mobility group protein B1
- RAGE
receptor for advanced glycation end-products
- TNFR1
TNF receptor 1
- SREBP1
sterol regulatory element-binding protein 1
- ACC
acetyl CoA carboxylase
- CTGF
connective tissue growth factor
- α-SMA
alpha-smooth muscle actin
- 4-HNE
4-hydroxynonenal
- HSC
hepatic stellate cells
- hCLS
hepatic crown-like structures
- JNK
c-Jun N-terminal kinase
- KN
normal-diet fed knockout mice
- KH
high-fat diet-fed knockout mice
- WN
normal-diet fed wild type mice
- WH
high-fat diet-fed wild type mice
SUPPLEMENTARY MATERIALS
Supplementary data including one figure can be found with this article online at http://pdf.medrang.co.kr/paper/pdf/Kjpp/Kjpp019-05-08-s001.pdf.
List of primary antibodies
Myeloid-specific SIRT1 deletion decreases SIRT1 expression (A) PCR genotyping results from tail DNA. (B) Western blots of SIRT1 in liver and epididymal fat homogenates from WT and KO mice. Band intensity was normalized to β-actin. Data are presented as mean±SEM. *p<0.05 for WT versus KO mice.
SIRT3 expression in KO mice. Western blots of SIRT3 in liver mitochondria from WT and KO mice. Band intensity was normalized to voltage-dependent anion channel 1(VDAC1). Data are presented as mean±SEM. *p<0.05 for WT versus KO mice.
References
- 1.Li X. SIRT1 and energy metabolism. Acta Biochim Biophys Sin (Shanghai) 2013;45:51–60. doi: 10.1093/abbs/gms108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci U S A. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schug TT, Xu Q, Gao H, Peres-da-Silva A, Draper DW, Fessler MB, Purushotham A, Li X. Myeloid deletion of SIRT1 induces inflammatory signaling in response to environmental stress. Mol Cell Biol. 2010;30:4712–4721. doi: 10.1128/MCB.00657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 7.Anastasiou D, Krek W. SIRT1: linking adaptive cellular responses to aging-associated changes in organismal physiology. Physiology (Bethesda) 2006;21:404–410. doi: 10.1152/physiol.00031.2006. [DOI] [PubMed] [Google Scholar]
- 8.Cohen DE, Supinski AM, Bonkowski MS, Donmez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009;23:2812–2817. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chawla A. Control of macrophage activation and function by PPARs. Circ Res. 2010;106:1559–1569. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamei N, Tobe K, Suzuki R, Ohsugi M, Watanabe T, Kubota N, Ohtsuka-Kowatari N, Kumagai K, Sakamoto K, Kobayashi M, Yamauchi T, Ueki K, Oishi Y, Nishimura S, Manabe I, Hashimoto H, Ohnishi Y, Ogata H, Tokuyama K, Tsunoda M, Ide T, Murakami K, Nagai R, Kadowaki T. Overexpression of monocyte chemoattractant protein-1 in adipose tissues causes macrophage recruitment and insulin resistance. J Biol Chem. 2006;281:26602–26614. doi: 10.1074/jbc.M601284200. [DOI] [PubMed] [Google Scholar]
- 13.Deng XQ, Chen LL, Li NX. The expression of SIRT1 in nonalcoholic fatty liver disease induced by high-fat diet in rats. Liver Int. 2007;27:708–715. doi: 10.1111/j.1478-3231.2007.01497.x. [DOI] [PubMed] [Google Scholar]
- 14.Yoshizaki T, Schenk S, Imamura T, Babendure JL, Sonoda N, Bae EJ, Oh DY, Lu M, Milne JC, Westphal C, Bandyopadhyay G, Olefsky JM. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am J Physiol Endocrinol Metab. 2010;298:E419–E428. doi: 10.1152/ajpendo.00417.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 16.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnes PJ, Adcock IM, Ito K. Histone acetylation and deacetylation: importance in inflammatory lung diseases. Eur Respir J. 2005;25:552–563. doi: 10.1183/09031936.05.00117504. [DOI] [PubMed] [Google Scholar]
- 18.Tomita K, Tamiya G, Ando S, Ohsumi K, Chiyo T, Mizutani A, Kitamura N, Toda K, Kaneko T, Horie Y, Han JY, Kato S, Shimoda M, Oike Y, Tomizawa M, Makino S, Ohkura T, Saito H, Kumagai N, Nagata H, Ishii H, Hibi T. Tumour necrosis factor alpha signalling through activation of Kupffer cells plays an essential role in liver fibrosis of non-alcoholic steatohepatitis in mice. Gut. 2006;55:415–424. doi: 10.1136/gut.2005.071118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh M, Kato H, Suganami T, Konuma K, Marumoto Y, Terai S, Sakugawa H, Kanai S, Hamaguchi M, Fukaishi T, Aoe S, Akiyoshi K, Komohara Y, Takeya M9, Sakaida I, Ogawa Y. Hepatic crown-like structure: a unique histological feature in non-alcoholic steatohepatitis in mice and humans. PLoS One. 2013;8:e82163. doi: 10.1371/journal.pone.0082163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han MS, Jung DY, Morel C, Lakhani SA, Kim JK, Flavell RA, Davis RJ. JNK expression by macrophages promotes obesity-induced insulin resistance and inflammation. Science. 2013;339:218–222. doi: 10.1126/science.1227568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.den Boer M, Voshol PJ, Kuipers F, Havekes LM, Romijn JA. Hepatic steatosis: a mediator of the metabolic syndrome. Lessons from animal models. Arterioscler Thromb Vasc Biol. 2004;24:644–649. doi: 10.1161/01.ATV.0000116217.57583.6e. [DOI] [PubMed] [Google Scholar]
- 22.Paradis V, Perlemuter G, Bonvoust F, Dargere D, Parfait B, Vidaud M, Conti M, Huet S, Ba N, Buffet C, Bedossa P. High glucose and hyperinsulinemia stimulate connective tissue growth factor expression: a potential mechanism involved in progression to fibrosis in nonalcoholic steatohepatitis. Hepatology. 2001;34:738–744. doi: 10.1053/jhep.2001.28055. [DOI] [PubMed] [Google Scholar]
- 23.Iredale JP. Models of liver fibrosis: exploring the dynamic nature of inflammation and repair in a solid organ. J Clin Invest. 2007;117:539–548. doi: 10.1172/JCI30542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuda M, Shimomura I. Increased oxidative stress in obesity: implications for metabolic syndrome, diabetes, hypertension, dyslipidemia, atherosclerosis, and cancer. Obes Res Clin Pract. 2013;7:e330–e341. doi: 10.1016/j.orcp.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Wang RH, Kim HS, Xiao C, Xu X, Gavrilova O, Deng CX. Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. J Clin Invest. 2011;121:4477–4490. doi: 10.1172/JCI46243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svegliati Baroni G, D'Ambrosio L, Ferretti G, Casini A, Di Sario A, Salzano R, Ridolfi F, Saccomanno S, Jezequel AM, Benedetti A. Fibrogenic effect of oxidative stress on rat hepatic stellate cells. Hepatology. 1998;27:720–726. doi: 10.1002/hep.510270313. [DOI] [PubMed] [Google Scholar]
- 27.Rajendrasozhan S, Yang SR, Kinnula VL, Rahman I. SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;177:861–870. doi: 10.1164/rccm.200708-1269OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshizaki T, Milne JC, Imamura T, Schenk S, Sonoda N, Babendure JL, Lu JC, Smith JJ, Jirousek MR, Olefsky JM. SIRT1 exerts anti-inflammatory effects and improves insulin sensitivity in adipocytes. Mol Cell Biol. 2009;29:1363–1374. doi: 10.1128/MCB.00705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschöp MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hah YS, Cheon YH, Lim HS, Cho HY, Park BH, Ka SO, Lee YR, Jeong DW, Kim HO, Han MK, Lee SI. Myeloid deletion of SIRT1 aggravates serum transfer arthritis in mice via nuclear factor-κB activation. PLoS One. 2014;9:e87733. doi: 10.1371/journal.pone.0087733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purushotham A, Xu Q, Li X. Systemic SIRT1 insufficiency results in disruption of energy homeostasis and steroid hormone metabolism upon high-fat-diet feeding. FASEB J. 2012;26:656–667. doi: 10.1096/fj.11-195172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu F, Gao Z, Zhang J, Rivera CA, Yin J, Weng J, Ye J. Lack of SIRT1 (Mammalian Sirtuin 1) activity leads to liver steatosis in the SIRT1+/- mice: a role of lipid mobilization and inflammation. Endocrinology. 2010;151:2504–2514. doi: 10.1210/en.2009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams EJ, Gaça MD, Brigstock DR, Arthur MJ, Benyon RC. Increased expression of connective tissue growth factor in fibrotic human liver and in activated hepatic stellate cells. J Hepatol. 2000;32:754–761. doi: 10.1016/s0168-8278(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Jung J, Kim H, Heo RW, Yi CO, Lee JE, Jeon BT, Kim WH, Hahm JR, Roh GS. Exendin-4 improves nonalcoholic fatty liver disease by regulating glucose transporter 4 expression in ob/ob mice. Korean J Physiol Pharmacol. 2014;18:333–339. doi: 10.4196/kjpp.2014.18.4.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hotamisligil GS, Peraldi P, Budavari A, Ellis R, White MF, Spiegelman BM. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-alpha- and obesity-induced insulin resistance. Science. 1996;271:665–668. doi: 10.1126/science.271.5249.665. [DOI] [PubMed] [Google Scholar]
- 36.Kern PA, Ranganathan S, Li C, Wood L, Ranganathan G. Adipose tissue tumor necrosis factor and interleukin-6 expression in human obesity and insulin resistance. Am J Physiol Endocrinol Metab. 2001;280:E745–E751. doi: 10.1152/ajpendo.2001.280.5.E745. [DOI] [PubMed] [Google Scholar]
- 37.De Taeye BM, Novitskaya T, McGuinness OP, Gleaves L, Medda M, Covington JW, Vaughan DE. Macrophage TNF-alpha contributes to insulin resistance and hepatic steatosis in diet-induced obesity. Am J Physiol Endocrinol Metab. 2007;293:E713–E725. doi: 10.1152/ajpendo.00194.2007. [DOI] [PubMed] [Google Scholar]
- 38.Aguirre V, Uchida T, Yenush L, Davis R, White MF. The c-Jun NH(2)-terminal kinase promotes insulin resistance during association with insulin receptor substrate-1 and phosphorylation of Ser(307) J Biol Chem. 2000;275:9047–9054. doi: 10.1074/jbc.275.12.9047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primary antibodies
Myeloid-specific SIRT1 deletion decreases SIRT1 expression (A) PCR genotyping results from tail DNA. (B) Western blots of SIRT1 in liver and epididymal fat homogenates from WT and KO mice. Band intensity was normalized to β-actin. Data are presented as mean±SEM. *p<0.05 for WT versus KO mice.
SIRT3 expression in KO mice. Western blots of SIRT3 in liver mitochondria from WT and KO mice. Band intensity was normalized to voltage-dependent anion channel 1(VDAC1). Data are presented as mean±SEM. *p<0.05 for WT versus KO mice.