Abstract
Phylogeography contributes to our knowledge of regional biotas by integrating spatial and genetic information. In New Zealand, comprising two main islands and hundreds of smaller ones, phylogeography has transformed the way we view our biology and allowed comparison with other parts of the world. Here we review studies on New Zealand terrestrial and freshwater invertebrates. We find little evidence of congruence among studies of different taxa; instead there are signatures of partitioning in many different regions and expansion in different directions. A number of studies have revealed unusually high genetic distances within putative species, and in those where other data confirm this taxonomy, the revealed phylogeographic structure contrasts with northern hemisphere continental systems. Some taxa show a signature indicative of Pliocene tectonic events encompassing land extension and mountain building, whereas others are consistent with range expansion following the last glacial maximum (LGM) of the Pleistocene. There is some indication that montane taxa are more partitioned than lowland ones, but this observation is obscured by a broad range of patterns within the sample of lowland/forest taxa. We note that several geophysical processes make similar phylogeographic predictions for the same landscape, rendering confirmation of the drivers of partitioning difficult. Future multi-gene analyses where applied to testable alternative hypotheses may help resolve further the rich evolutionary history of New Zealand's invertebrates.
Keywords: range expansion, endemicity, pliocene, pleistocene, insect, species
1. Introduction
Phylogeography uses the geographic distribution of genetic variants to interpret the role of historical processes in the development of biological distributions. As originally circumscribed, phylogeography deals primarily with the structuring of populations within species [1]. This focus distinguishes it from phylogenetics and the use of species-level phylogenies to infer biogeography [2–5]. However, many of the pitfalls of biogeographic interpretation from species trees apply also to intraspecific phylogeography—there being a natural evolutionary continuum underpinning population genetics, intraspecific phylogeography, interspecific phylogeography and species phylogeny [6].
In its simplest form, two types of information can be gleaned from phylogeographic studies: first, the spatial distribution of genetic variation (e.g., whether the same DNA sequence for a gene is found in individuals from several locations) and second, the extent of differentiation between genetic variants (e.g., the proportion of nucleotide differences among DNA sequences for a given gene). Approaches founded in population genetic theory and latterly statistical phylogeography using coalescent modeling [7,8] allow description and consideration of the amount of variation present and how that variation is partitioned. Thus phylogeographic traits scale from shallow to deep in terms of divergence among sequence variants, with either intense partitioning (heterogeneous) or thorough mixing (homogenous) in terms of spatial distribution (Table 1). Many intermediary permutations are possible.
Table 1.
Information from phylogeography involves two main parameters: the degree of difference among genetic variants (branch length) and the way this diversity is distributed in space.
| Spatial Distribution | High Diversity | Low Diversity |
|---|---|---|
| Homogenous | deeper coalescence larger population higher gene flow associated with persistent range |
shallower coalescence smaller population higher gene flow associated with recent range expansion |
| Heterogenous | deeper coalescence larger metapopulation reduced gene flow spatial partitioning from older event(s) |
shallower coalescence smaller metapopulation reduced gene flow spatial partitioning from more recent event(s) |
Phylogeographic analysis uses sampling in current time, but has the goal of making inferences about past populations. As with all phylogenetically-based methods, what results is a hypothesis, and the fit of the hypothesis to some other (prior) information is usually used as the basis for “testing” between alternative historic scenarios. An obvious limitation is that the method may not be able to discriminate among alternative historic processes that predict the same or similar distribution of genetic diversity. Even with the advantage of molecular clock calibration, there remains uncertainty about which extrinsic factors have influenced gene flow, because: (a) drivers may be contiguous or even coincide in time; and (b) divergence time estimates are imprecise for many reasons at the scale that is relevant in phylogeography [9–11].
1.1. The New Zealand Phylogeographic Context and the Development of the Fauna
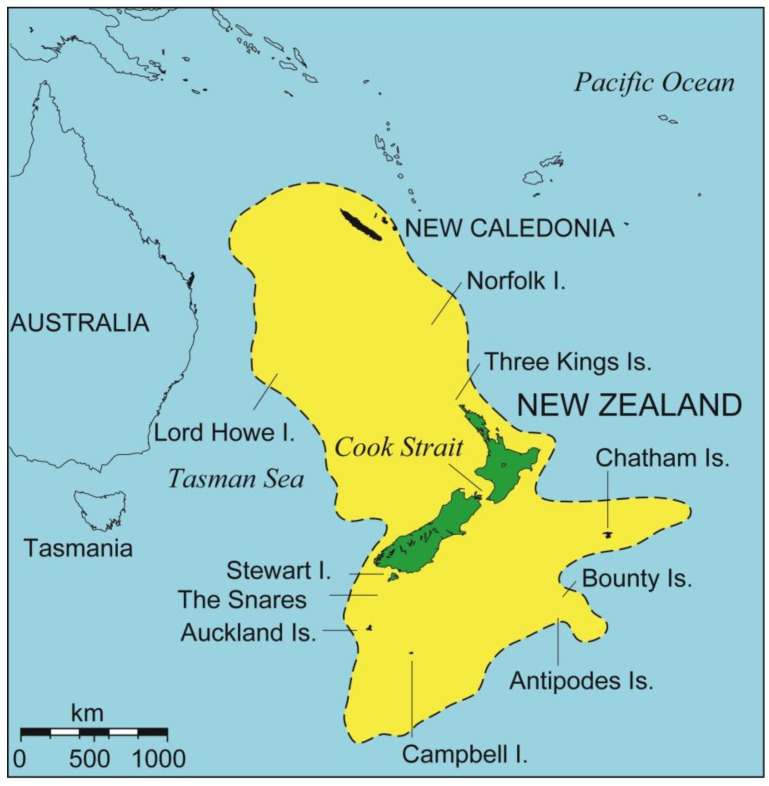
New Zealand is an archipelago of nearly 270,000 km2 situated in the south-western Pacific Ocean [12] (Figure 1). There are two main islands separated by a narrow seaway, Cook Strait, with a much greater distance of ocean to other significant land areas. Australia is a minimum of 1500 km to the west and the island of New Caledonia is 1500 km north. There are a number of small island groups within New Zealand waters including Chatham (east), Three Kings (north) and subantarctic (south).
Figure 1.
New Zealand's place in the Pacific. The approximate position of the largely submerged continental crust of Zealandia is indicated in yellow.
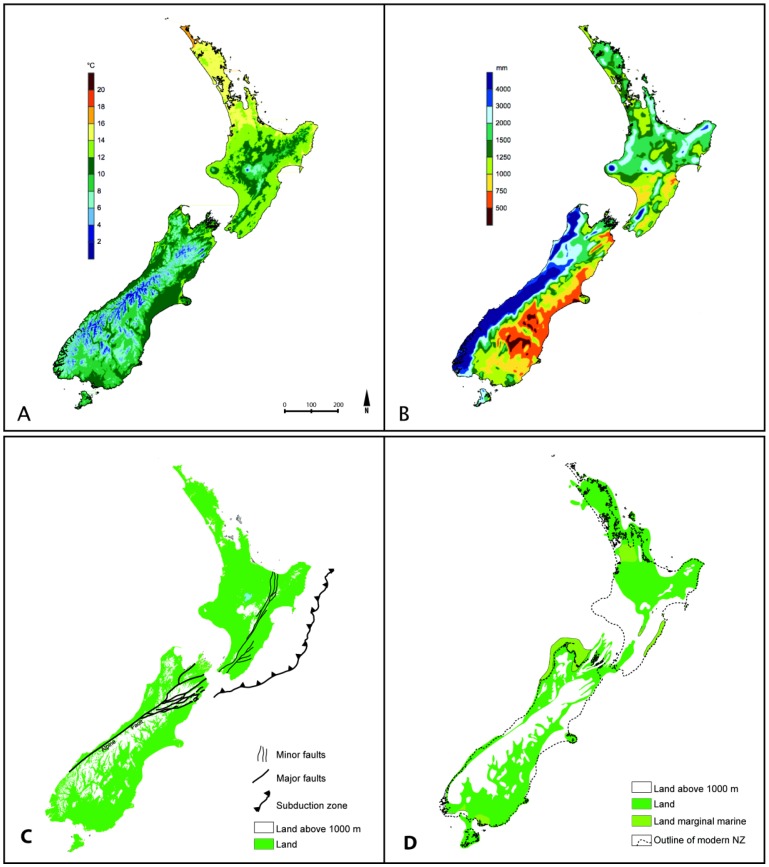
Uncertainty about the history of the New Zealand landscape (neither certainly continental nor oceanic [13]), and the presence of some peculiar elements in the fauna, have led biologists to view the New Zealand biota as extraordinary, even “the nearest approach to life on another planet” [14]. However, ecologically and morphologically distinct species are also a common feature [15] of islands much younger than the widely-assumed antiquity of New Zealand (e.g., 70 million years [16]). While plate tectonics and the breakup of Gondwana underpin the early physical development of the region, recent analyses have emphasized the onset of continental plate boundary activity at the start of the Miocene (∼25 Ma), in formation of New Zealand [17–19]. Most if not all of the pre-existing continent of Zealandia, which is an order of magnitude larger than New Zealand, is today below the sea [21,22] (Figure 1). A more youthful perspective on New Zealand geology has enabled biological thinking to move away from a focus on ancient isolation and toward evolutionary dynamism [5,22–27]. Even since Pliocene time (∼5 Ma), New Zealand has experienced extensive mountain building (Southern Alps), land extension (southern North Island), volcanism (Taupo Volcanic Zone), and climate/land changes associated with Pleistocene glaciations [20,28] (Figure 2).
Figure 2.
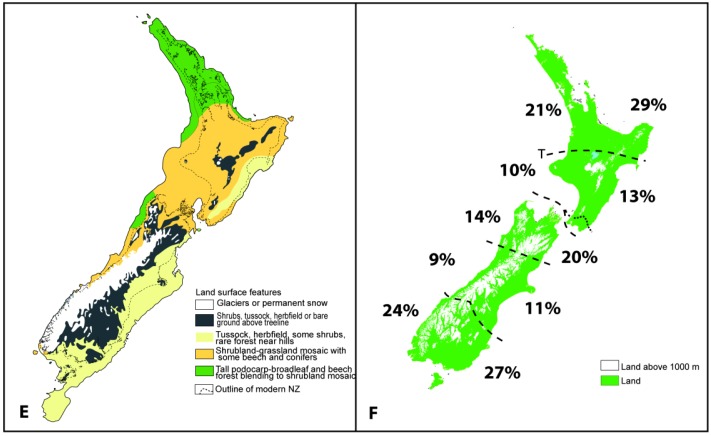
Geophysical and biogeographic features of New Zealand past and present. Environmental heterogeneity: A, mean annual temperature, B, mean annual rainfall, C, elevation. Temporal changes: D, Pliocene palaeogeography at 3 Ma, E, Pleistocene LGM, may yield uneven distribution of biodiversity (F). F, regional insect endemicity in a sample of 1724 species, % of species in a region that are endemic to that region (left), % of all 596 regional endemics that are endemic to a particular region (right). Thus phylogeographic (population) structure is a product of current and past environmental structure. Climate maps (A, B), courtesy of NIWA [91]. Palaeogeographic reconstructions (D, E) based on [92] and [93] respectively. Regional insect endemicity (F) from analysis of data in Fauna of New Zealand series volumes (2,3,12,15, 16,20–21,23,25,27,30,3–36,39–50,53,54,57–59,62,63,65) containing suitable information.
As in most parts of the world, the New Zealand fauna is dominated by invertebrates and in particular insects (∼60% of 34,636 recorded New Zealand Animalia are arthropods [29]). This situation is emphasized by deficiencies in the extant native biota of several vertebrate groups (no snakes, no terrestrial mammals) [26,30,31], although among invertebrates too, a number of groups are underrepresented (e.g., within Orthoptera there are no native Gryllacrididae, and few Gryllidae and Tettigoniidae). As would be expected from its location, New Zealand fauna has its closest affinity with animals in other southern hemisphere lands. This southern distribution pattern is often referred to as “Gondwanan”, but this term can be confusing as it confounds current distribution pattern with a particular process that might have created it (namely plate tectonic breakup of Gondwana) [23]. It is increasingly recognized that the presence of “Gondwanan” lineages in New Zealand provides only limited information about the biogeographic process in development of the biota; vicariant and dispersal hypotheses often yield similar distribution patterns [23,32]. To what degree presence, absence, diversity of lineages and ecological traits are reliable indicators of the timing and extent of past occupation, arrival and extinction, is unclear and often difficult to ascertain [27]. What is more readily achieved is an understanding of the way current diversification has developed within extant lineages. In the last decade or so, a rapid expansion in the number of taxa that have been subject to some type of phylogeographic analysis has transformed our knowledge (Tables 2 and 3; see also [26]). While initial studies were largely descriptive of spatial patterns inferred from the partitioning of genetic data, they resulted in the development of hypotheses about past evolutionary processes in New Zealand invertebrate biology. They have also informed our understanding of biodiversity, systematics, taxonomy and conservation status, and enabled empirical comparison with biota in other parts of the world.
Table 2.
Summary of New Zealand invertebrate phylogeography/phylogenetic biogeography within genera, highlighting inferred geophysical process that have left signatures in the distribution and depth of genetic diversity. -Last glacial maximum (LGM) ending 20 k years ago.
| Time | Event/Process | Invertebrate taxa | Pattern/Evidence |
|---|---|---|---|
| recent | Taupo volcanics | Acanthophlebia mayfly[33], Orthopsyche caddisfly[34] | Low diversity |
|
|
|||
| >2,000 | Hemideina tree weta[35] | Chromosome contact zone | |
|
|
|||
| years | Argosarchus stick insect[36], Peripatoides peripatus[37] | High diversity | |
|
| |||
| LGM | Glaciation of central South Island (beech gap) | Brachynopus fungus beetle[38], six stonefly genera[39], freshwater snail Potamopyrgus[40] | Extirpation during a recent glaciation |
|
|
|||
|
Agyrtodes fungus beetle[41] cicada M. campbelli [42,43] |
West coast forest refugia Colonization of South Island during glaciation |
||
|
|
|||
| LGM | North and South Islands connected (no Cook Strait) | Hemideina tree weta[44,45,46], peripatus[37], Paranephrops koura[47], amphipods[48], Lyperobus huttoni weevils[49], Celatoblatta vulgaris cockroaches[49], Pachyrhamma edwardsii cave weta[50], Epistranus fungus beetles[51] | Similar or identical haplotypes straddling Cook Strait |
|
|
|||
| Wainuia snails[52], Kikihia cicadas[53], earthworms[54], Hemiandrus ground weta[55], Talitropsis cave weta[23,56], Pristoderus fungus beetles[51] | Phylogeographic gaps at Cook Strait | ||
|
|
|||
| LGM | South and Stewart Islands connected | freshwater isopods[57], Kikihia cicada[58], koura[47] | Similar or identical haplotypes straddling Foveaux Strait |
|
|
|||
| LGM | Expanded alpine species | Wiseana hepialid moths[58], Brachaspis grasshoppers[59], some Kikihia[60] and Maoricicada cicadas[43] | Speciation inferred to be within the Pleistocene |
|
|
|||
| LGM | Glacial refugia | Hemideina tree weta[61], Clitarchus stick insects[62,63], Acanthophlebia mayflies[33] | Higher diversity in Northland |
|
|
|||
| Paranephrops koura[47], Hemideina tree weta[45], mite harvestmen[64], Hemiandrus ground weta[55], Potamopyrgus freshwater snails[40], Epistranus fungus beetles[51], Agyrtodes[41] | Higher diversity in Nelson region | ||
|
|
|||
| Prodontria scarabaeid beetles[65], Powelliphanta snails[66], Peripatoides peripatus[67], Maoricicada cicadas[42,43], Niveaphasma stick insect[68] | Deep lineages in Southland | ||
|
| |||
| Pliocene | Southern Alp formation | scree weta D. Connectens[69], mite harvestman[64], Neocicindela[70] | Intraspecific structure |
|
|
|||
| Peripatoides peripatus[49], Lyperobius weevils[49], Celatoblatta cockroaches[49,56], Mecodema beetles[49], Neocicindela[70] | Interspecific structure | ||
|
|
|||
| Pliocene | Southern Alp formation | hepialid moths[58], Lycosid spiders[71], Kikihia cicadas[72,73], Sigara water boatmen[74], Sigaus grasshoppers[75], Celatoblatta cockroaches[76], Mecodema beetles[48,77], Lyperobius weevils[49], peripatus[49], 14 species of Maoricicada[78,79] | Radiations attributed to Pliocene uplift |
|
|
|||
| flightless beetles Prodontria[65], Deinacrida weta[80] | Multiple origins of alpine adaptation | ||
|
|
|||
| Paranephrops koura[47], Sigara water boatmen[74], Brachynopus fungus beetles[38], Kikihia cicadas[60], Celatoblatta cockroaches[49,56,76] | Splits across the alps | ||
|
|
|||
| Agyrtodes fungus beetle[41] | Connections across the alps | ||
|
|
|||
| Pliocene | Northland archipelago | Hemideina tree weta[61], Placostylus snails[81,82], Clitarchus stick insects[83], Amborhytida snails[84] | Structure concordant with islands |
|
|
|||
| Paryphanta snails[84], corophiid amphipods[85] | East-west split (not island concordant) | ||
|
|
|||
| Three Kings Is | Placostylus snails[86], rhytidid snails[84], Pseudoclitarchus stick insects[87], Epistranus fungus beetles[51] | Divergence from NZ consistent with island age | |
|
|
|||
| Campbell Island | freshwater isopods[57] | Divergence from NZ consistent with island age | |
|
|
|||
| 4 MYA | Chatham Island | stag beetles[56,77], cave weta[23,56,77], cockroaches[56,76,77], spiders[71], damselflies[88], cicadas[72,73,89], freshwater isopods[57], amphipods[48] | Divergence from NZ consistent with age of Chatham Islands |
|
| |||
| Miocene | East Coast Islands | Hemideina tree weta (H. trewicki)[44,45,46], Peripatoides peripatus (P. morgani)[37], Kikihia cicadas[87] | Divergent clades in Hawkes Bay |
|
|
|||
| Miocene | Banks Peninsula volcanic Is | Hemideina tree weta (H.ricta)[90] | Pleistocene landbridge lead to endemic taxa |
Table 3.
Intraspecific phylogeographic studies of New Zealand invertebrates. Pairwise distances calculated for taxa marked with * using HKY (Hasegawa, Kishino & Yano) from Genbank accessions using Geneious Pro v5.3.4. [112]. Numbers: Indiv.- individuals, Haps.- haplotypes. Regions: N-North Island, S-South Island, C-Chatham Islands. IBD- isolation by distance inferred from correlation between geographic and genetic distance (in most studies a Mantel test was used).
| Taxa | Mtdna Sequence | Nuclear | Number of: | Region | Maximum Intraspecific Distance | Cryptic Species Likely |
I B D |
Patterns | Reference | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||
| Species | Indiv. | Locations | Haps | Environment | NI | SI | CI | ||||||||||
| stick insects | Acanthoxyla | COI & COII | ITS sequence | 7 | 33 | 11 | 14 | forest | N | S | 0.022 | observed | asexual | no | recent expansion | [103,113] | |
| stick insect | Argosarchus horridus | COI & COII | sexual/asexual | 1 | 90 | 49 | 46 | forest | N | S | C | 0.033 | Tamura-Nei 1993 | no | out of north | [36] | |
| stick insect | Clitarchus hookeri | COI & COII | sexual/asexual | 1 | 83/170 | 30/105 | 62/99 | forest | N | S | 0.030 | HKY* | no | yes | out of north | [62, 63] | |
| stick insect | Niveaphasma annulata | COI | EF1-alpha sequence | 1 | 97 | 66 | 48 | subalpine/forest | S | 0.044 | HKY+ Γ+I | no | yes | glaciation causing isolation | [68] | ||
| grasshopper | Brachaspis nivalis | COI | 2 | 26 | 13 | 22 | subalpine/open | S | 0.106 | K2P | yes | regional | [59, 114] | ||||
| grasshopper | Sigaus australis | COI & 12S | 1 | 130 | 22 | 32 | subalpine | S | 0.083 | uncorrected | yes | regional | [75, 114] | ||||
| grasshopper | Sigaus piliferus | COI | 1 | 51 | 13 | 31 | montane | N | 0.064 | K2P | yes | regional | [114] | ||||
| weta (cave) | Talitropsis sedilloti | COI & COII | 1 | 56 | 43 | 35 | forest | N | S | 0.031 | uncorrected | no | no | out of south | [23, 56, 77] | ||
| weta (giant) | Denacrida connectens | COI | allozymes | 1 | 78 | 24 | 40 | subalpine | S | 0.130 | GTR+Γ+I | no | regional | [69, 115, 116] | |||
| weta (ground) | Hemiandrus maculifrons | COI | morphology | 2 | 41 | 24 | 41 | forest | N | S | 0.120 | uncorrected | yes | yes | out of south | [55, 117] | |
| weta (ground) | Hemiandrus pallitarsis | COI | drumming & morphology | 1 | 88 | 18 | 55 | forest | N | 0.089 | TVM+ Γ+I | no | no | out of south | [55, 118] | ||
| weta (stone) | Hemideina maori | COII | 1 | 27 | 10 | 13 | subalpine | S | 0.055 | Tamura-Nei 1993 | no | regional | [90] | ||||
| weta (tree) | Hemideina crassidens | COI | allozymes &chromosomes | 1 | 12 | 12 | 12 | forest | N | S | 0.127 | HKY+I | no | out of Nelson | [44, 46] | ||
| weta (tree) | Hemideina thoracica | COI &12S | allozymes & chromosomes | 1 | 191 | 49 | 60 | forest | N | 0.095 | uncorrected | no | no | out of north | [35, 109] | ||
| beetle | Agyrtodes labralis | COI | 1 | 187 | 79 | 97 | forest | S | 0.056 | HKY* | expansion across alps west to east | [41] | |||||
| beetle | Brachynopus scutellaris | COI | 1 | 113 | 34 | 73 | forest | S | 0.060 | HKY* | yes | north/south glaciation gap | [38] | ||||
| beetle | Epistranus lawsoni | COI | 1 | 168 | 78 | 116 | forest | N | S | 0.246 | uncorrected | yes | yes | spatially & temporally continuous | [51] | ||
| beetle | Hisparonia hystrix | COI | 1 | 105 | 39 | 47 | forest | S | 0.018 | HKY* | no | yes | north/south glaciation gap | [38] | |||
| beetle | Pristoderus bakewelli | COI | 1 | 88 | 53 | 77 | forest | N | S | C | 0.149 | uncorrected | yes | no | spatially & temporally continuous | [51] | |
| cicada | Kikihia subalpina | COI & COII | song | 1 | 114 | 79 | 58 | montane | N | S | 0.035 | HKY+I | no | North I vs South I/out of Nelson | [59] | ||
| cicada | Kikihia muta | COI & COII | song & colour | 1 | 162 | 88 | 107 | open | N | S | C | 0.067 | uncorrected | yes | regional | [89] | |
| cicada | Maoricicada campbelli | COI & A6-A8 | song | 1 | 212 | 91 | 95 | open | N | S | 0.066 | HKY+I | no | out of north and southern refuge | [42, 43] | ||
| stonefly | Zelandoperla decorata | COI | H3 sequence | 1 | 144 | 63 | 45 | aquatic | S | 0.024 | HKY | no | recent expansion/gene flow | [105] | |||
| stonefly | Zelandoperla fenestrata | COI | H3 sequence | 1 | 186 | 81 | 71 | aquatic | S | 0.091 | Tamura-Nei 1993 | no | regional | [105] | |||
| mayfly | Acanthophlebia cruentata | Cytb | 1 | 186 | 19 | 34 | aquatic | N | 0.037 | HKY* | no | yes | out of Northland, Taupo extinction | [33] | |||
| caddisfly | Orthopsyche fimbriata | COI | 1 | 157 | 16 | 23 | aquatic | N | 0.047 | HKY* | no | yes | Taupo extinction? | [34] | |||
| damselfly | Xanthocnemis zealandica | COI | allozymes | 1 | 27 | 15 | 13 | aquatic | N | S | C | 0.012 | uncorrected | no | recent expansion/dispersal | [88] | |
| waterboatman | Sigara potamius | COI | 1 | 35 | 28 | 24 | aquatic | S | 0.071 | HKY* | east vs west/regional | [74] | |||||
| amphipods | Paracalliope fluviatilis | COI | allozymes | 1 | 54 | 14 | 17 | aquatic | N | S | 0.260 | uncorrected | yes | regional, out of south? | [107] | ||
| isopod | Austridotea lacustris | COI | ITS | 1 | 24 | 12 | 19 | aquatic | S | C | 0.113 | HKY | dispersal to Chathams | [57] | |||
| mite havestmen | Aoraki denticulata | COI | morphology | 1 | 119 | 17 | 84 | forest | S | 0.192 | uncorrected | yes | out of Nelson? | [64] | |||
| centipede | Craterostigmus crabilli | COI & 16S | 18S & 28S sequence | 2 | 14 | 9 | 13 | forest | N | S | 0.325 | HKY* | yes | out of Nelson | [104] | ||
| peripatus | Peripatoides sympatrica | COI | allozymes | 1 | 41 | 14 | 16 | forest | N | 0.027 | GTR+Γ+I | no | little pattern | [37, 108] | |||
| peripatus | Peripatoides n. sp. | COI | 1 | 47 | 21 | 18 | forest | S | 0.110 | K2P | yes | regional | [67] | ||||
| koura | Paranephrops planifrons | COI | 1 | 62 | 43 | 62 | aquatic | N | S | 0.135 | GTR+Γ+I | yes | yes | out of Nelson northwards | [47] | ||
| koura | Paranephrops zealandicus | COI | 1 | 43 | 33 | 39 | aquatic | N | S | 0.227 | GTR+Γ+I | yes | yes | regional/out of Nelson souththwards | [47] | ||
| snail | Potamopyrgus antipodarum | CytB | Microsats. for ploidy | 1 | 638 | 20 | 45 | aquatic | N | S | 0.037 | GTR+I | no | yes | out of north | [40] | |
1.2. Predicting the Past
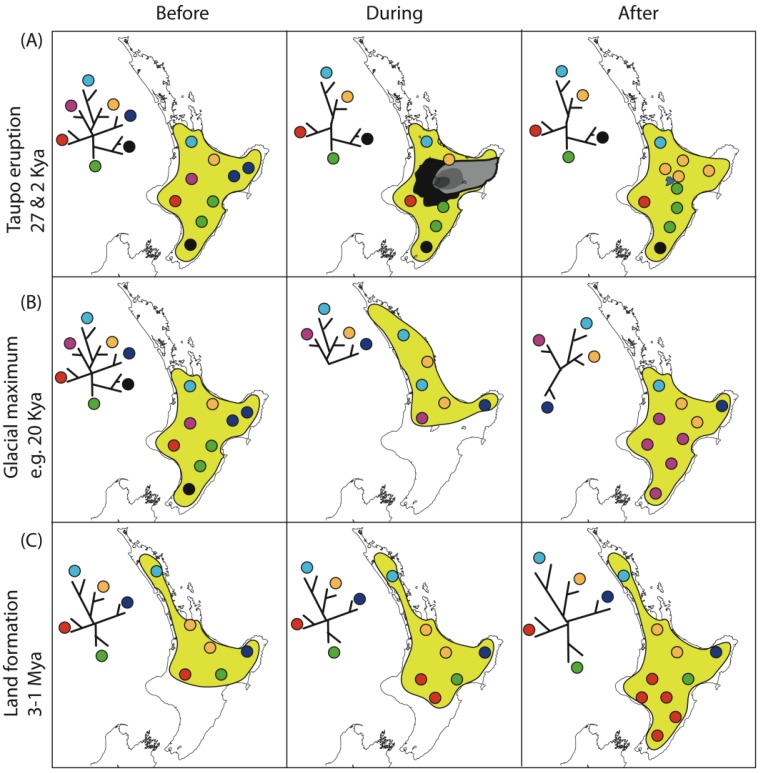
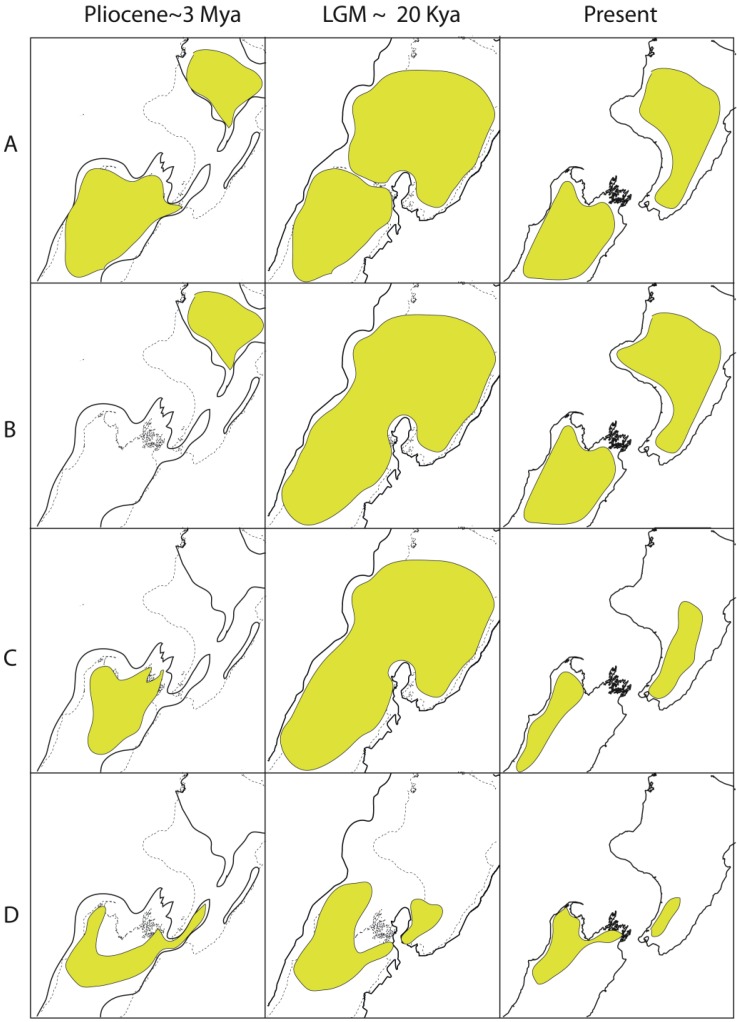
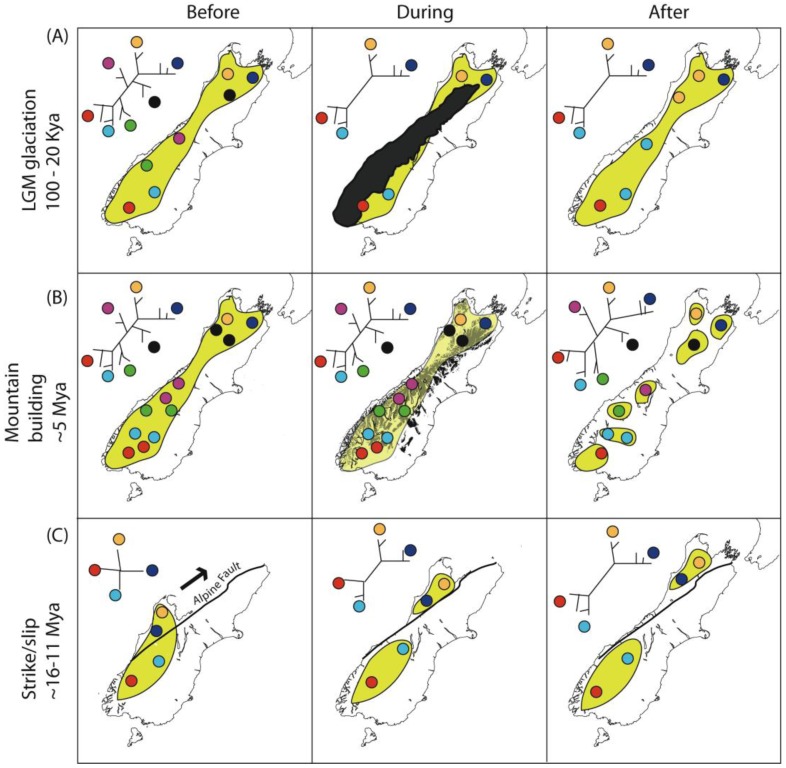
Spatial variation in climate, topography and vegetation generates fairly steep north-south, and in some places east-west gradients (Figure 2). Spatial variation intersects with temporal variation in these features, as changes in land area have been considerable in New Zealand during and since Pliocene time (Figure 2) [92,94]. Together, these processes have resulted in a stark pattern of regional diversity. This diversity, more usually expressed in terms of regional endemicity, was first notably documented in plants [95], but is also evident among insect groups (Figure 2E). More recent geophysical changes have probably erased large amounts of older biodiversity and population structure [92]. For example, the effect of volcanic activity in the central North Island Taupo Volcanic Zone less than 2000 years ago may have obscured population structure of forest species that had accumulated since the last glacial maximum (LGM ∼20,000 years ago), and a larger volcanic eruption in the same place 27,000 years ago may have overwritten the population structure that accumulated in North Island during the previous Pleistocene interglacials (Figure 3A). Not only are more recent geological and climatic events likely to obscure earlier patterns, but different events can sometimes leave similar spatial patterns of genetic diversity (Figures 3–5). For example, the effects of southward extension of forest habitat after the LGM (Figure 3B) could result in similar distribution of diversity as earlier southward extension of land in North Island (Figure 3C). Similarly, in South Island, low genetic diversity in a central swathe resulting from extinction of lineages yields a pattern similar to vicariance models involving glaciation or alpine fault movement (Figure 4) [49,96]. Where alternative process generate similar or identical tree hypotheses, comparison of tree topology will not distinguish them, but branch lengths can in principle exclude alternatives. Such an approach is, however, dependent on the availability of suitable molecular clock calibrations, sufficient precision for the time frame of interest, and consistency among genes and lineages. All these attributes are problematic for most phylogeographic studies, but emerging genome scale data, and coalescent model based approaches promise to enhance the sensitivity of phylogeographic testing [7,97,98].
Figure 3.
Phylogeographic outcomes of different geophysical events in North Island New Zealand may be similar. (A) Taupo volcanic; (B) LGM forest range; (C) land emergence since 2 million years ago. Yellow area indicates range of hypothetical taxon. (A) Black and grey indicate area affected by pyroclastic flow and ash deposits from Taupo eruption. Existing diversity, which may or may not be partitioned in space is extinguished close to centre and subsequently replaced by range expansion. This is expected to result in reduced diversity around the centre; (B) Climate cooling during glacial events resulted in retraction of forest northwards, and formation of potential refugium. Subsequent expansion of habitat is expected to result in lower diversity in south compared to north through leading-edge re-colonization; (C) A near identical phylogeographic pattern is expected to result from land formation which resulted in southward extension of North Island, but branch lengths may be greater than B and might be associated with taxonomic subdivision. Sequential events in the same region might yield a wide number of permutations in different taxa reflecting ecological or stochastic processes.
Figure 5.
Cook Strait connections. A wide range of scenarios for South Island-North Island phylogeography are plausible. Yellow area indicates range of hypothetical taxon. (A) Population might be allopatric on older islands having moved between by oversea dispersal, before coming parapatric during LGM and so remaining specific to different islands; (B) Taxon might exist on one island only, expanding its range during LGM and then being partitioned as sea-level rises; (C) Taxon might initially be restricted to alpine environment on one island and colonizing new alpine environment when able during extension of lowered alpine habitat in LGM; (D) Taxon range in former South Island might result in occupation of southern North Island as it forms before LGM.
Figure 4.
South Island (A) Habitat partitioning by glaciation, (B) Formation of alps, (C) Alpine fault displacement. On a long narrow island a widely distributed taxon is likely to develop a pattern of isolation by distance, even without any habitat heterogeneity. Geophysical processes may influence the gene genealogy among populations and species that evolve. Yellow area indicates range of hypothetical taxon. (A) Glaciation (black area) might cause extinction of some populations (and their genetic lineages), and partition residual populations in the north and south. Subsequent retraction of glaciers could allow expansion of forest taxa through leading-edge colonization; (B) Formation of alps (black areas) might yield habitat heterogeneity and reduce gene flow among populations leading to formation of allopatric species; (C) Alpine fault displacement (alpine fault line in black) might sunder adjacent populations enabling their independent evolution over time, resulting in similar phylogeographic structure resulting from lineage extinction A.
1.3. Pattern and Process
Identifying the pattern of genetic structuring among populations (or other samples) is just a first step in phylogeography, as the main motivation of such work is to inform on the nature and timing of extrinsic processes that have driven population structuring. Despite a traditional focus in New Zealand biogeography on ancient processes (e.g., Gondwanan breakup, land reduction during the Oligocene), most if not all species-level diversity is much younger and coincides with the geophysically active period during the Plio-Pleistocene (Figure 2). As phylogeography deals with contemporary samples, we can be more confident about the influence of events that occurred recently than that of earlier events, because the former will tend to obscure the signal from the latter.
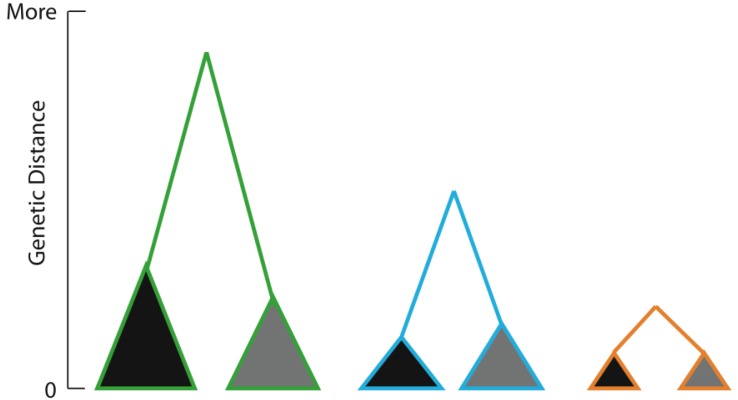
A general expectation in biogeography and phylogeography is that organisms occupying the same spatial range are likely to have been subject to the same evolutionary drivers and this should be evident in their similar phylogeographic signatures [1,99–101]. It has, however, often transpired that regional phylogeographic structure is only broadly concordant and that different types of plants and animals respond in different ways [26]. One complication is that different taxon groups may sample different (spatially overlapping) episodes in history. In addition, ecological traits (e.g., fecundity, dispersal behaviour) are likely to influence population structuring. Thus, similar patterns of spatial partitioning within different species may reflect distinct temporal episodes that have operated across the same geographic space. Only comparison of genetic distance data as an estimator of time (with caution) might enable their distinction (Figure 6).
Figure 6.
Three hypothetical taxa (green, blue, gold), each with populations in the same two areas (grey, black). Coalescent depth may differ among them, but this might be due to population size, gene/taxon specific mutation rate, lineage sorting effects or timing of historical event. Distinguishing between events requires clear statements of assumptions made in dating of nodes, to allow testing among alternative drivers of population partitioning if circularity is to be avoided [7].
1.4. Sampling for Phylogeography
The scope of studies of New Zealand invertebrates that involve some spatial and genetic information is enormous, ranging from multispecies to single species, widely sampled to narrowly sampled, with shallow genetic diversity to variation at the informative limits of cytochrome oxidase I (COI) sequencing (Table 4).
Table 4.
Exemplars of New Zealand invertebrate phylogeography studies matching hypothetical extremes in terms of depth of diversity and patchiness of diversity. See Tables 2 and 3 for additional detail.
| Spatial Distribution | High diversity | Low diversity |
|---|---|---|
| Homogenous | Beetle- Epistranus lawsoni Spiders- Anoteropsis species |
Stick insect- Acanthoxyla Damselfly - Xanthocnemis zealandica stonefly - Zelandoperla decorata |
| Heterogenous | Regional Cicada- Maoricicada campbelli Weta- Deinacrida connectens Koura- Paranephrops |
Regional Cicada- Kikihia subalpina beetle- Hisparonia hystrix Graded Stick insect- Clitarchus hookeri |
More than 50 studies deal with species-level variation, usually with relatively small samples of individuals from several species (Table 2), and have been used to infer processes ranging in age from 2000 years ago until the Miocene. For almost all the patterns observed there are invertebrate examples that show contrasting patterns, revealing the complexity of New Zealand's geological history and the ecological variety sampled. We include 37 population-level phylogeographic studies of invertebrates, of which 28 studies are of insects (Table 3). These latter papers report mtDNA sequence data from individuals of (putatively) single species sampled at multiple locations. More than half these studies also include some data representing the nuclear genome including morphology, song, allozymes, cytogenetics and DNA sequences of nuclear genes. Nuclear gene sequences can provide additional gene genealogies to inform phylogeographic analysis [102], and this has contributed to some New Zealand studies [39,60,68,79,104–106]. However, nuclear markers of some form are also essential for identification of cryptic species, or conversely, confirmation that samples are from conspecifics. For example, inclusion of allozyme data provided confidence that cryptic species were sampled in the amphipod Paracalliope fluviatilis [107] and morphologically-conservative peripatus Peripatoides novaezealandiae [108]. In contrast, chromosome and allozyme analyses of the tree weta Hemideina thoracica confirmed that cryptic species were not present [61,109] and challenged the status of two species of Prodontria [65]. High mtDNA diversity within the fungus beetles Pristoderus bakewelli and Epistranus lawsoni [51], and koura (freshwater crayfish) Paranephrops zealandicus [47] await further analysis to determine whether reproductive isolation is likely. The mitochondrial genome is haploid and maternally inherited and therefore it cannot be used to delimit species boundaries. Without such confirmation, spurious inferences about high intraspecific mtDNA diversity or the existence of cryptic species are likely, and this undermines the power of data to test among alternative hypotheses about phylogeographic process, timing and outcome (Figures 3 and 4).
Taxon selection in phylogeographic studies tends to be biased toward widespread species, because these are perceived as having the potential to provide data about large-scale phenomena. Widespread species are more likely to be chosen because their ranges will more often encompass geophysical features of interest. These species by their very nature, however, are more likely to have high gene flow and connectedness [110], and may not be representative of a tendency in other lineages to evolve reproductive isolation mechanisms or morphologically distinct units. In contrast, lineages that have speciated extensively (e.g., stoneflies [39]; cockroaches [49,76]; cicada [60,72,73,79]; peripatus [37,49,67,108] tend not be used for the same kind of study. Interspecific studies might provide useful insights into historical processes in evolution where species are primarily allopatric or parapatric [9,49,60]. Expansion of species' ranges into sympatry will tend to obscure biogeographic patterns and conceal the historical drivers of lineage formation [108], but where interactions are limited it remains possible to estimate and test evolutionary scenarios [70] (Table 2).
2. Refugia and Expansion
Variation in the level of mtDNA diversity may by used to infer population expansion from an area of high diversity towards one of low diversity [111]. Half of the New Zealand intraspecific phylogeographic studies show a genetic pattern from which range expansion is inferred (18 of 37; Table 3). Source areas of range expansion (putative refugia), usually related to post-glacial climate change, that have been inferred from phylogeography include northern North Island, southern South Island and northern South Island (references in Table 3).
We note from an inventory of regional New Zealand insect endemicity (Figure 2F) that diversity is not homogenous or even graded across latitude. Instead, areas of relatively high endemicity exist in the northern North Island, northern South Island and southern South Island, similar to that identified in plants [95]. Although there is some correspondence between areas of high endemicity and location of phylogeographic refugia, the relationship between these different data has yet to be explained. Probably they relate to instances of speciation, range expansion and extinction in quite different time frames.
With only 37 independent intraspecific phylogeographic studies of invertebrates in New Zealand we find few statistically supported patterns although trends are revealing. Forest (n = 17) and aquatic (n = 11) species show all manner of phylogeographic patterns (Table 3). In contrast, species sampled from subalpine, montane and open habitats (n = 9) show a high proportion with intense regional differentiation; only two such species showing genetic evidence of “expansion”. This is consistent with the idea that topographical heterogeneity facilitates partitioning. Winged species (n = 12) are not more prevalent among the examples of range expansion as might be expected of animals capable of flight, though they do contribute relatively few studies (17%) with maximum intraspecific pairwise distances above 0.10. Species with high intraspecific pairwise distances come from all environments, but are slightly more common in forest species (41%). In six of the seven forest species where high intraspecific pairwise distances were encountered this may be partially explained by a high likelihood of cryptic species being sampled in contiguous/homogenous habitat. Conversely, a relative paucity of high intraspecific genetic distances among taxa in heterogenous environments (e.g., alpine) may be because allopatric population partitioning corresponds more frequently with species taxonomy. Fewer than half the studies tested for isolation-by-distance (IBD), but of those that did, 11 showed a positive relationship between genetic and geographic distance, and five did not. Interestingly, lack of IBD occurs in species with both small COI divergences (e.g., Acanthoxyla; 0.022 uncorrected) and large (e.g., P. bakewell; 0.149 uncorrected).
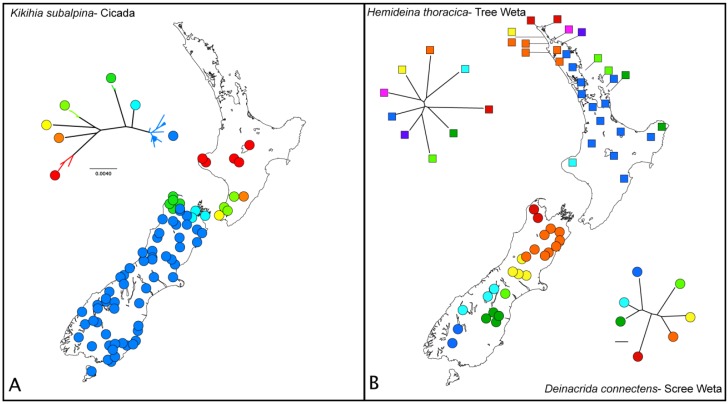
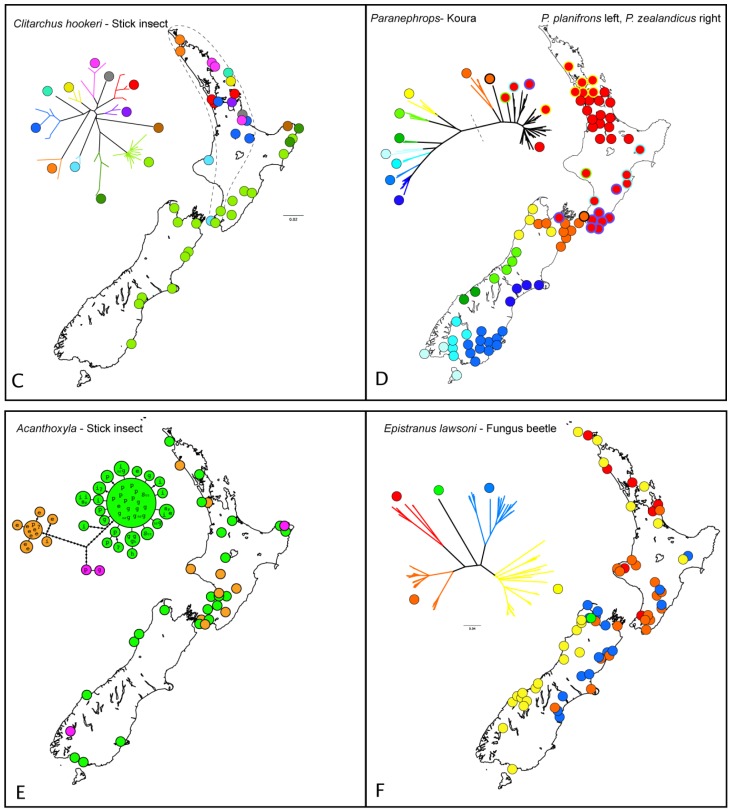
Patterns such as “out-of-north” almost certainly result from more than one process. For example, cicada, snail, caddisfly, mayfly and stick insects (Kikihia subalpina, Potamopyrgus antipodarum, Orthopsyche fimbriata, Acanthophlebia cruentata, Clitarchus hookeri, Argosarchus horridus) might have expanded their ranges southward in a similar and relatively recent time-frame as maximum mtDNA distances within each are similar and relatively small (3–4.7%; Table 3, Figure 7). A common inference in such cases is that these taxa were influenced by Pleistocene climatic cycling (see Figure 2). However, it is unlikely (assuming population size and rate of molecular evolution are similar- Figure 6) that mite-harvestman (Aoraki denticulata) and tree weta (Hemideina crassidens and H. thoracica) were expanding southwards at the same time, as they show quite different intraspecific pairwise mtDNA distances (19.2, 12.7 and 9.5% respectively). A refugium (at some undefined time in the past) in the Nelson region (northern South Island) is possible for centipedes (mtDNA genetic distances up to 32%), koura (13.5%; Figure 7), mite-harvestman (19.2%), tree weta (12.7%), and fungus beetles (24.6%). As the processes involved are likely to be different (based on the range of genetic and taxonomic/ecological diversity sampled) it can be misleading to identify similar patterns. Furthermore, most New Zealand phylogeographic studies find a number of patterns and inferred processes [70] within a single taxon (Table 3).
Figure 7.
Exemplars of phylogeographic patterns revealed in species of New Zealand invertebrates. In each case mtDNA lineages are color coded and mapped, with cooler colors (blue, green) to south and warmer ones (reds) to north. Trees for A, C and F inferred using Neighbor-Joining of HKY distances in Geneious Pro v5.3.4 with mtDNA COI sequences download from GenBank; tree topology did not differ significantly from that originally reported. (A) Cicada Kikihia subalpina [59]; (B) Tree weta Hemideina thoracica only major mtDNA lineages for each species are indicated here [61], scree weta Deinacrida connectens [37]; (C) Within the stick insect Clitarchus hookeri [62], lineages associated with sexual populations are multicolored within dashed line whereas green populations are parthenogenetic; (D) The two New Zealand koura or freshwater crayfish Paranephrops are partitioned north and south, tree redrawn as above [47]; (E) Maximum parsimony network of the stick insect genus Acanthoxyla comprises numerous parthenogenetic morphospecies: e A. inermis, P A. prasina, i A. intermedia, g A. geisovii, Sp A. speciosa, Stt A. suteri, nrg A. nr geisovii (Myers et al. subm). Refer to Table 2 for data details; (F) The fungus beetle Epistranus lawsoni is likely to include cryptic species, tree redrawn as above [51]
3. Gaps and Regional
From the 37 putative population-level studies we can conclude that many invertebrates were apparently unaffected by the Pleistocene LGM, as they have widespread, high genetic diversity (e.g., fungus beetles Epistranus lawsoni and Pristoderus bakewelli, ground weta Hemiandrus maculifrons and H. pallitarsis, scree weta Deinacrida connectens, mite harvestman Aoraki denticulata, stick insect Niveaphasma annulata, stonefly Zelandoperla fenestrata, waterboatman Sigara potamius) (Table 3). For example, we see regional diversity and widespread sympatry of divergent haplotype lineages in the forest (fungus beetle Figure 7F). There is clearly differentiated regional diversity in alpine (weta Figure 7B), aquatic (stonefly) and open grass/scrub taxa (cicada Figure 7A). Species that are likely to have extended their ranges during cold glacial cycles, such as alpine, sub-alpine and open- habitat species are well sampled in the New Zealand phylogeographic literature (Table 2). These taxa show regional variation, distinguishing populations that, although currently isolated, could have been connected at lower altitudes during colder times. Alpine environments are thought to have first appeared about five million years ago when fault movement started the formation of the Southern Alps. Evidence of multiple origins of alpine adaptation comes from studies of weta [80] and flightless scarabaeid beetles [65]. The formation of the alpine zone resulted in species radiations (e.g., spiders, moths, cicada, cockroaches, grasshoppers) and the origin of intraspecific diversity (e.g., scree weta, cicada). In contrast, a few insects with low diversity and little geographic structure might be the product of recent population expansion post LGM, such as the lineage of parthenogenetic stick insects Acanthoxyla that have arisen via hybridisation (Figure 7E), and the damselfly Xanthocnemis zealandica.
The most obvious gap in terrestrial habitat in mainland New Zealand is Cook Strait, a narrow seaway (minimum 25 km) between North Island and South Island [92]. This feature corresponds approximately with one margin of an area of relatively low endemicity in southern North Island among plants and insects (Figure 2F) [95,119]. This current division of North and South Islands is no older than 500 ky [120], and was almost certainly bridged during the LGM when sea level dropped (Figure 2E) [121]. Although two main islands have probably existed at least since the late Miocene, the position and extent of seaways between them has changed over time with a general southward migration of a wave of uplift and subsidence [92]. Eight phylogeographic studies of invertebrate species reveal individuals with similar or identical haplotypes either side of Cook Strait, while six show evidence of a phylogeographic break there (Table 2). While sharing of similar haplotypes across the Strait must indicate recent gene flow (either by over-sea dispersal or LGM landbridge), partitioning could be the result of anything from recent lineage sorting via small populations (a lower level of gene flow) to clade formation on islands older than Cook Strait with their current close proximity resulting from land emergence in southern North Island (see Figures 3 and 5). Numerous genealogical histories are consistent with the formation of Cook Strait, but few if any may have originated as a result of it. Range expansion during LGM, or species ranges that were spanning old North Island and South Island, or species ranges with a connection to southern tip of North Island during Pliocene, all make subtly different genealogical predictions, the relative probability of which might be estimated in future using multigene phylogeography (Figure 5).
4. Dispersal Ability
With their flighted and flightless forms, insects display a wide range of dispersal capability and behaviour. Flightlessness evolves quickly and repeatedly, even showing polymorphism within species [122], presenting opportunities to examine its effect on phylogeographic structure on an otherwise closely related genetic background. It is often assumed that capacity for dispersal is readily inferred from presence of wings, but many factors might influence dispersal ability [56,77]. A comparison of phylogeographic structure among macropterous, micropterous and apterous forms of the stonefly Zelandoperla fenestrata, found all three to be quite highly structured, consistent with the field observation that the winged form is rarely, if ever, seen in flight [105]. In contrast, the stronger flying Z. decorata showed much less structure. In a cicada, lineages are finely subdivided, despite the cicada being widespread and apparently a good flier [89]. Studies of insect phylogeography encompassing New Zealand and its offshore islands are revealing in this respect. In particular, data for Chatham Islands fauna provide compelling evidence that dispersal can take many forms and is not necessarily linked to presence of wings [23]. Successful migrants to the Chatham Islands [56] include damsel flies [88], isopod [57], cicada [89], wingless beetles and crickets [77], beetles [51] and worms [54].
5. Partitioning and Pruning
A hypothesized effect of glaciation in South Island is vicariance (division) of widespread species into isolated populations [39]. Glaciation tends to sunder populations allowing their independent evolution (vicariance), but may also prune out intervening diversity non-randomly. The net result is phylogenetic trees with long internal branches that correspond to widely spaced and often disjunct distributions in the modern fauna (Figure 4), and such patterns are commonly reported in intra and interspecific phylogeographic studies of taxa with north-south disjunct distributions in South Island [39,49] (Table 2). Vicariance may have little to do with lineage formation in such cases, and pruning will tend to push back the inferred age of origin of surviving lineages if this is not taken into account [5]. Pleistocene glaciation consisted of a series of some 20 potential isolation/pruning episodes, and it is possible that deeper splits in phylogenetic trees were related to Pliocene diversification during mountain building [69].
6. Conclusions
The current data indicate that all manner of phylogeographic patterns are identifiable among New Zealand invertebrate taxa (Table 4), sustaining inferences of diverse geophysical events in their evolution. Although it is well established that phylogenies are gene trees, the phylogeographic and systematics literature shows continual lapses into treating them as taxon trees. Similarly, the associated inference of particular historic processes in genetic partitioning that are in fact only weakly consistent with the data are common-place. The emergence and availability of next generation sequencing will be a boon to phylogeography: it will become commonplace to use multiple nuclear markers and differences in these gene histories will continually remind us of their fickle reflection of the taxon trees, of which they are but a fragment. Equally, there are challenges, not least in how we use these clouds of gene trees to estimate phylogeographic history [123–125].
Even with the best multigene data to infer phylogeographic patterns, it will remain difficult to distinguish among some events that might be drivers of population partitioning and speciation in New Zealand. The geophysical events that have shaped New Zealand often overlap in their temporal influence, so future phylogeographic studies should concentrate on alternative hypotheses that can be distinguished, such as Pliocene versus Pleistocene effects of habitat availability.
In the meantime, we have to remember that inferences about history are susceptible to the general problems of biogeographic testing and the difficulty of assigning confidence [5]. It is essential that data-rich phylogeography, bolstered by emerging sophisticated analytical tools, is not allowed to descend to the domain of story-telling that has marred so much biogeographic ‘analysis’ of this region in the past.
Acknowledgements
Thanks to Lesley van Essen for compiling the taxonomic inventory using Fauna of New Zealand series [126], and Andrew Tait at NIWA (National Institute of Water and Atmospheric Research) for providing climate maps. Thanks also for the support and enthusiasm of the Phoenix Lab. Evolutionary Ecology and Genetics group: http://www.massey.ac.nz/∼strewick/.
References
- 1.Avise J.C. Phylogeography. The History and Formation of Species. Harvard University Press; Cambridge, MA, USA: 2000. [Google Scholar]
- 2.Cracraft J. Species diversity, biogeography, and the evolution of biotas. Amer. Zoologist. 1994;34:33–47. [Google Scholar]
- 3.Arbogast B.S, Kenagy G.J. Comparative phylogeography as an integrative approach to historical biogeography. J. Biogeogr. 2001;28:819–825. [Google Scholar]
- 4.Funk V.A. Foundations of biogeography. In: Lomolino M.V., Sax D.F., Brown J.H., editors. Revolutions in Historical Biogeography. The University of Chicago Press; Chicago, IL: 2004. pp. 647–657. [Google Scholar]
- 5.Crisp M.D., Trewick S.A., Cook L.G. Hypothesis testing in biogeography. Trends Ecol. Evol. 2011;26:66–72. doi: 10.1016/j.tree.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Page R.D.M. Maps between trees and cladistic analysis of historical associations among genes, organisms, and areas. Syst. Biol. 1994;43:58–77. [Google Scholar]
- 7.Knowles L.L. Statistical phylogeography. Annu. Rev. Ecol. Evol. Syst. 2009;40:593–612. [Google Scholar]
- 8.Knowles L.L., Maddison W.P. Statistical phylogeography. Mol. Ecol. 2002;11:2623–35. doi: 10.1046/j.1365-294x.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- 9.Ho S.W., Phillips M.J., Cooper A., Drummond A.J. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. [DOI] [PubMed] [Google Scholar]
- 10.Penny D. Relativity for molecular clocks. Nature. 2005;436:183–184. doi: 10.1038/436183a. [DOI] [PubMed] [Google Scholar]
- 11.Waters J.M., Rowe D.L., Burridge C.P., Wallis G.P. Gene trees versus species trees: reassessing life-history evolution in a freshwater fish radiation. Syst. Biol. 2010;59:504–517. doi: 10.1093/sysbio/syq031. [DOI] [PubMed] [Google Scholar]
- 12.Neall V.E., Trewick S.A. The age and origin of the pacific islands: A geological overview. Philos. T. Roy. Soc. B. 2008;363:3293–3308. doi: 10.1098/rstb.2008.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorton I. Island Colonization- the Origin and Development of Island Communities. Cambridge University Press; Cambridge, UK: 2007. [Google Scholar]
- 14.Diamond J. New Zealand As An Archipelago: An International Perspective in Ecological Restoration of New Zealand's Islands. Department of Conservation; Auckland, New Zealand: 1990. [Google Scholar]
- 15.Quammen D. The Song of the Dodo: Island Biogeography in an Age of Extinctions. Touchstone Press; New York, NY, USA: 1997. [Google Scholar]
- 16.Whittaker R.J., Fernández-Palacios J.M. Island Biogeography: Ecology, Evolution, and Conservation. 2nd ed. Oxford University Press; Oxford, UK: 2007. [Google Scholar]
- 17.Waters J.M., Craw D. Goodbye gondwana? New Zealand biogeography, geology, and the problem of circularity. Syst. Biol. 2006;55:351–356. doi: 10.1080/10635150600681659. [DOI] [PubMed] [Google Scholar]
- 18.Trewick S.A., Paterson A.M., Campbell H.J. Hello New Zealand. J. Biogeogr. 2007;34:1–6. [Google Scholar]
- 19.Didham R.K. New Zealand: ‘fly-paper of the Pacific?’. The Weta. 2005;29:1–5. [Google Scholar]
- 20.Campbell H., Hutching G. In Search of Ancient New Zealand. Penguin; Auckland, New Zealand: 2007. [Google Scholar]
- 21.Landis C.A., Campbell H.J., Begg J.G., Mildenhall D.C., Paterson A.M., Trewick S.A. The waipounamu erosion surface: Questioning the antiquity of the new zealand land surface and terrestrial fauna and flora. Geol. Mag. 2008;145:173–197. [Google Scholar]
- 22.Winkworth R.C., Wagstaff S.J., Glenny D., Lockhart P.J. Evolution of the New Zealand mountain flora: origins, diversification and dispersal. Org. Divers. Evol. 2005;5:237–247. [Google Scholar]
- 23.Goldberg J, Trewick S.A., Paterson A.M. Evolution of New Zealand's terrestrial fauna: a review of molecular evidence. Philos. T. Roy. Soc. B. 2008;363:3319–3334. doi: 10.1098/rstb.2008.0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDowall R.M. Process and pattern in the biogeography of New Zealand—A global microcosm. J. Biogeogr. 2008;35:197–212. [Google Scholar]
- 25.Waters J.M. Driven by the west wind drift? A synthesis of southern temperate marine biogeography, with new directions for dispersalism. J. Biogeogr. 2008;35:417–427. [Google Scholar]
- 26.Wallis G.P., Trewick S.A. New Zealand phylogeography: evolution on a small continent. Mol. Ecol. 2009;18:3548–3580. doi: 10.1111/j.1365-294X.2009.04294.x. [DOI] [PubMed] [Google Scholar]
- 27.Trewick S.A., Gibb G.C. Vicars, Tramps and the assembly of New Zealand avifauna: A review of molecular phylogenetic evidence. Ibis. 2010;152:226–253. [Google Scholar]
- 28.Graham I.J. A Continent on the Move: New Zealand Geosciences in the 21st Century. Geological Society of New Zealand; Wellington, NZ: 2008. [Google Scholar]
- 29.Gordon D.P. The New Zealand Inventory of Biodiversity. Volume 2: Kingdom Animalia Chaetognatha, Ecdysozoa, Ichnofossils. Canterbury University Press; Christchurch, NZ: 2010. [Google Scholar]
- 30.Gibbs G. Ghosts of Gondwana: The History of Life in New Zealand. Craig Potton Publishing; Nelson, NZ: 2006. [Google Scholar]
- 31.Trewick S.A., Morgan-Richards M. New Zealand biology. In: Gillespie R.G., Clague D.A., editors. Encyclopedia of Islands. University of California Press; Berkeley, CA, USA: 2009. pp. 665–673. [Google Scholar]
- 32.Waters J.M., Dijkstra L.H., Wallis G.P. Biogeography of a southern hemisphere freshwater fish: how important is marine dispersal. Mol. Ecol. 2000;9:1815–1821. doi: 10.1046/j.1365-294x.2000.01082.x. [DOI] [PubMed] [Google Scholar]
- 33.Smith P.J., McVeagh S.M., Collier K.J. Genetic diversity and historical population structure in the new zealand mayfly Acanthophlebia cruentata. Freshwater Biol. 2006;51:12–24. [Google Scholar]
- 34.Smith P.J., McVeagh S.M., Collier K.J. Population-genetic structure in the New Zealand caddisfly orthopsyche fimbriata revealed with mitochondrial DNA. New Zeal. J. Mar. Fresh. Res. 2006;40:141–148. [Google Scholar]
- 35.Morgan-Richards M., Trewick S.A., Wallis G.P. Characterization of a hybrid zone between two chromosomal races of the weta hemideina thoracica following a geologically recent volcanic eruption. Heredity. 2000;85:586–592. doi: 10.1046/j.1365-2540.2000.00796.x. [DOI] [PubMed] [Google Scholar]
- 36.Buckley T.R., Marske K.A., Attanayake D. Identifying glacial refugia in a geographic parthenogen using palaeoclimate modelling and phylogeography: the New Zealand stick insect argosarchus horridus (white) Mol. Ecol. 2009;18:4650–4663. doi: 10.1111/j.1365-294X.2009.04396.x. [DOI] [PubMed] [Google Scholar]
- 37.Trewick S.A. Mitochondrial DNA sequences support allozyme evidence for cryptic radiation of new zealand peripatoides (onychophora) Mol. Ecol. 2000;9:269–281. doi: 10.1046/j.1365-294x.2000.00873.x. [DOI] [PubMed] [Google Scholar]
- 38.Leschen R.A.B., Buckley T.R., Harman H.M., Shulmeister J. Determining the origin and age of the westland beech (nothofagus) gap, New Zealand, using fungus beetle genetics. Mol. Ecol. 2008;17:1256–1276. doi: 10.1111/j.1365-294X.2007.03630.x. [DOI] [PubMed] [Google Scholar]
- 39.McCulloch G.A., Wallis G.P., Waters J.M. onset of glaciation drove simultaneous vicariant isolation of alpine insects in New Zealand. Evolution. 2010;64:2033–2043. doi: 10.1111/j.1558-5646.2010.00980.x. [DOI] [PubMed] [Google Scholar]
- 40.Neiman M., Lively C.M. Pleistocene glaciation is implicated in the phylogeographical structure of potamopyrgus antipodarum, a New Zealand snail. Mol. Ecol. 2004;13:3085–3098. doi: 10.1111/j.1365-294X.2004.02292.x. [DOI] [PubMed] [Google Scholar]
- 41.Marske K.A., Leschen R.A.B., Barker G.M., Buckley T.R. Phylogeography and ecological niche modeling implicate coastal refugia and trans-alpine dispersal of a New Zealand fungus beetle. Mol. Ecol. 2009;18:5126–5142. doi: 10.1111/j.1365-294X.2009.04418.x. [DOI] [PubMed] [Google Scholar]
- 42.Hill K.B.R., Simon C., Marshall D.C., Chambers G.K. Surviving glacial ages within the biotic gap: phylogeography of the New Zealand cicada maoricicada campbelli. J. Biogeogr. 2009;36:675–692. [Google Scholar]
- 43.Buckley T.R., Simon C., Chambers G.K. Phylogeography of the New Zealand cicada maoricicada campbelli based on mitochondrial DNA sequences: ancient clades associated with cenozoic environmental change. Evolution. 2001;55:1395–1407. doi: 10.1111/j.0014-3820.2001.tb00661.x. [DOI] [PubMed] [Google Scholar]
- 44.Morgan-Richards M., Daugherty C.H., Gibbs G. Taxonomic status of tree weta from stephens island, Mt Holdsworth and Mt Arthur, based on allozyme variation. J. Roy. Soc. New Zeal. 1995;25:301–312. [Google Scholar]
- 45.Morgan-Richards M. Robertsonian translocations and b chromosomes in the wellington tree weta, hemideina crassidens (orthoptera: anostostomatidae) Hereditas. 2000;132:49–54. doi: 10.1111/j.1601-5223.2000.00049.x. [DOI] [PubMed] [Google Scholar]
- 46.Morgan-Richards M. Fission or fusion? mitochondrial dna phylogenetics of the chromosome races of hemideina crassidens (orthoptera: anostostomatidae) Cytogenet. Genome Res. 2002;96:217–222. doi: 10.1159/000063042. [DOI] [PubMed] [Google Scholar]
- 47.Apte S., Smith P.J., Wallis G.P. Mitochondrial phylogeography of New Zealand freshwater crayfishes, paranephrops spp. Mol. Ecol. 2007;16:1897–1908. doi: 10.1111/j.1365-294X.2007.03224.x. [DOI] [PubMed] [Google Scholar]
- 48.Stevens M.I., Hogg I.D. Population genetic structure of New Zealand's endemic corophiid amphipods: evidence for allopatric speciation. Biol. J. Linn. Soc. 2004;81:119–133. [Google Scholar]
- 49.Trewick S.A., Wallis G.P. Bridging the “Beech-Gap”: New Zealand invertebrate phylogeography implicates pleistocene glaciation and pliocene isolation. Evolution. 2001;55:2170–2180. doi: 10.1111/j.0014-3820.2001.tb00733.x. [DOI] [PubMed] [Google Scholar]
- 50.Cook L.D., Trewick S.A., Morgan-Richards M., Johns P. Status of the New Zealand cave weta (rhaphidophoridae) genera pachyrhamma, gymnoplectron and turbottoplectron. Invertebr. Syst. 2010;24:131–138. [Google Scholar]
- 51.Marske K.A., Leschen R.A.B., Buckley T.R. reconciling phylogeography and ecological niche models for new zealand beetles: looking beyond glacial refugia. Mol. Phylogenet. Evol. 2011;59:89–102. doi: 10.1016/j.ympev.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 52.Efford M., Howitt R., Gleeson D. phylogenetic relationships of wainuia (mollusca: pulmonata)-biogeography and conservation implications. J. Roy. Soc. New Zeal. 2002;32:445–456. [Google Scholar]
- 53.Marshall D.C., Hill K.B.R., Fontaine K.M., Buckley T.R., Simon C. Glacial refugia in a maritime temperate climate: cicada (kikihia subalpina) MtDNA phylogeography in New Zealand. Mol. Ecol. 2009;18:1995–2009. doi: 10.1111/j.1365-294x.2009.04155.x. [DOI] [PubMed] [Google Scholar]
- 54.Buckley T.R., James S., Allwood J., Bartlam S., Howitt R., Prada D. Phylogenetic analysis of new zealand earthworms (oligochaeta: megascolecidae) reveals ancient clades and cryptic taxonomic diversity. Mol. Phylogenet. Evol. 2011;58:85–96. doi: 10.1016/j.ympev.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 55.Pratt R.C., Morgan-Richards M., Trewick S.A. Diversification of New Zealand weta (orthoptera: ensifera: anostostomatidae) and their relationships in Australasia. Philos. T. Roy. Soc. B. 2008;363:3427–3437. doi: 10.1098/rstb.2008.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldberg J., Trewick S.A. Exploring Phylogeographic Congruence in a Continental Island System. Insects. 2011 doi: 10.3390/insects2030369. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McGaughran A., Hogg I., Stevens M.I., Chadderton W.L., Winterbourn M.J. Genetic divergence of three freshwater isopod species from southern New Zealand. J. Biogeogr. 2006;33:23–30. [Google Scholar]
- 58.Brown B., Emberson R.M., Paterson A.M. Phylogeny of “oxycanus” lineages of hepialid moths from new zealand inferred from sequence variation in the mtDNA COI and II regions. Mol. Phylogenet. Evol. 1999;13:463–473. doi: 10.1006/mpev.1999.0662. [DOI] [PubMed] [Google Scholar]
- 59.Trewick S.A. Identity of an endangered grasshopper (acrididae: brachaspis): taxonomy, molecules and conservation. Conserv. Genet. 2001;2:233–243. [Google Scholar]
- 60.Marshall D.C., Slon K., Cooley J.R, Hill K.B.R., Simon C. Steady plio-pleistocene diversification and a 2-million-year sympatry threshold in a New Zealand cicada. Mol. Phylogenet. Evol. 2008;48:1054–1066. doi: 10.1016/j.ympev.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 61.Morgan-Richards M., Trewick S.A., Wallis G.P. Chromosome races with pliocene origins: evidence from MtDNA. Heredity. 2001;86:303–312. doi: 10.1046/j.1365-2540.2001.00828.x. [DOI] [PubMed] [Google Scholar]
- 62.Morgan-Richards M., Trewick S.A., Stringer I.A.N. Geographic parthenogenesis and the common tea-tree stick insect of New Zealand. Mol. Ecol. 2010;19:1227–1238. doi: 10.1111/j.1365-294X.2010.04542.x. [DOI] [PubMed] [Google Scholar]
- 63.Buckley T.R., Marske K., Attanayake D. phylogeography and ecological niche modelling of the New Zealand stick insect clitarchus hookeri (white) support survival in multiple coastal refugia. J. Biogeogr. 2010;37:682–695. [Google Scholar]
- 64.Boyer S.L., Baker J.M., Giribet G. Deep genetic divergences in aoraki denticulata (arachnida, opiliones, cyphophthalmi): A widespread ‘mite harvestman’ defies DNA taxonomy. Mol. Ecol. 2007;16:4999–5016. doi: 10.1111/j.1365-294X.2007.03555.x. [DOI] [PubMed] [Google Scholar]
- 65.Emerson B.C., Wallis G.P. Phylogenetic relationships of the prodontria (coleoptera; scarabaeidae; subfamily melolonthinae), derived from sequence variation in the mitochondrial cytochrome oxidase II gene. Mol. Phylogenet. Evol. 1995;4:433–447. doi: 10.1006/mpev.1995.1040. [DOI] [PubMed] [Google Scholar]
- 66.Trewick S.A., Walker K.J., Jordan C.J. Taxonomic and conservation status of a newly discovered giant landsnail from mount augustus, New Zealand. Conserv. Genet. 2008;9:1563–1575. [Google Scholar]
- 67.Trewick S.A. Molecular diversity of dunedin peripatus (onychophora: peripatopsidae) New Zeal. J. Zool. 1999;26:381–393. [Google Scholar]
- 68.O'Neill S.B., Buckley T.R., Jewell T.R., Ritchie P.A. Phylogeographic history of the New Zealand stick insect niveaphasma annulata (phasmatodea) estimated from mitochondrial and nuclear loci. Mol. Phylogenet. Evol. 2009;53:523–536. doi: 10.1016/j.ympev.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 69.Trewick S.A., Wallis G.P., Morgan-Richards M. Phylogeographical pattern correlates with pliocene mountain building in the alpine scree weta (orthoptera, anostostomatidae) Mol. Ecol. 2000;9:657–666. doi: 10.1046/j.1365-294x.2000.00905.x. [DOI] [PubMed] [Google Scholar]
- 70.Pons J., Fujisawa T, Claridge E.M., Savill R.A., Barraclough T.G., Vogler A.P. Deep mtDNA subdivision within linnean species in an endemic radiation of tiger beetles from New Zealand (genus neocicindela) Mol. Phylogenet. Evol. 2011;59:251–262. doi: 10.1016/j.ympev.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 71.Vink C.J., Paterson A.M. Combined molecular and morphological phylogenetic analyses of the New Zealand wolf spider genus anoteropsis (araneae: lycosidae) Mol. Phylogenet. Evol. 2003;28:576–587. doi: 10.1016/s1055-7903(03)00219-7. [DOI] [PubMed] [Google Scholar]
- 72.Arensburger P., Buckley T.R., Simon C., Moulds M., Holsinger K.E. Biogeography and phylogeny of the New Zealand cicada genera (hemiptera: cicadidae) based on nuclear and mitochondrial DNA data. J. Biogeogr. 2004;31:557–569. [Google Scholar]
- 73.Arensburger P., Simon C., Holsinger K. Evolution and phylogeny of the New Zealand cicada genus kikihia dugdale (homoptera: auchenorrhyncha: cicadidae) with special reference to the origin of the kermadec and norfolk islands' species. J. Biogeogr. 2004;31:1769–1783. [Google Scholar]
- 74.Buckley T.R., Young E.C. A revision of the taxonomic status of sigara potamius and s. limnochares (hemiptera: corixidae), water boatmen of braided rivers in New Zealand. New Zeal. Entomol. 2008;31:47–57. [Google Scholar]
- 75.Trewick S.A. DNA barcoding is not enough: mismatch of taxonomy and genealogy in New Zealand grasshoppers (orthoptera: acrididae) Cladistics. 2008;23:1–15. [Google Scholar]
- 76.Chinn W.G., Gemmell N.J. Adaptive radiation within New Zealand endemic species of the cockroach genus celatoblatta johns (blattidae): a response to plio-pleistocene mountain building and climate change. Mol. Ecol. 2004;13:1507–1518. doi: 10.1111/j.1365-294X.2004.02160.x. [DOI] [PubMed] [Google Scholar]
- 77.Trewick S.A. Molecular evidence for dispersal rather than vicariance as the origin of flightless insect species on the Chatham Islands, New Zealand. J. Biogeogr. 2000;27:1189–1200. [Google Scholar]
- 78.Chambers G.K., Boon W.-M., Buckley T.R., Hitchmough R.A. Using molecular methods to understand the gondwanan affinities of the New Zealand biota: three case studies. Aust. J. Bot. 2001;49:377–387. [Google Scholar]
- 79.Buckley T.R., Simon C. Evolutionary radiation of the cicada genus maoricicada dugdale (hemiptera: cicadoidea) and the origins of the New Zealand alpine biota. Biol. J. Linn. Soc. 2007;91:419–435. [Google Scholar]
- 80.Trewick S.A., Morgan-Richards M. After the deluge: mitochondrial DNA indicates miocene radiation and pliocene adaptation of tree and giant weta (orthoptera: anostostomatidae) J. Biogeogr. 2005;32:295–309. [Google Scholar]
- 81.Triggs S.J., Sherley G.H. Allozyme genetic diversity in placostylus land snails and implications for conservation. New Zeal. J. Zool. 1993;20:19–33. [Google Scholar]
- 82.Buckley T.R., Stringer I., Gleeson D., Howitt R., Attanayake D., Parrish R., Sherley G., Rohan M. A revision of the new zealand placostylus land snails using mitochondrial DNA and shell morphometric analyses, with implications for conservation. New Zeal. J. Zool. 2011;38:55–81. [Google Scholar]
- 83.Buckley T.R., Bradler S. Tepakiphasma ngatikuri, a new genus and species of stick insect (phasmatodea) from the far north of New Zealand. New Zeal. Entomol. 2010;33:118–126. [Google Scholar]
- 84.Spencer H.G., Brook F.J., Kennedy M. Phylogeography of kauri snails and their allies from northland, new zealand (mollusca: gastropoda: rhytididae: paryphantinae) Molec. Phylogenet. Evol. 2005;38:835–842. doi: 10.1016/j.ympev.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 85.Schnabel K.E., Hogg I.D., Chapman M.A. Population genetic structures of two New Zealand corophiid amphipods and the presence of morphologically cryptic species: implications for the conservation of diversity. New Zeal. J. Mar. Fresh. Res. 2000;34:637–644. [Google Scholar]
- 86.Ponder W.F., Colgan D.J., Gleeson D.M., Sherley G.H. Relationships of placostylus from lord howe island: an investigation using the mitochondrial cytochrome c oxidase 1 gene. Molluscan Res. 2003;23:159–178. [Google Scholar]
- 87.Trewick S.A., Morgan-Richards M., Collins L.J. Are you my mother? phylogenetic analysis reveals orphan hybrid stick insect genus is part of a monophyletic New Zealand clade. Molecular Phylogenetics and Evolution. 2008;48:799–808. doi: 10.1016/j.ympev.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 88.Nolan L., Hogg I.D., Sutherland D.L., Stevens M.I., Schnabel K.E. allozyme and mitochondrial dna variability within the new zealand damselfly genera xanthocnemis, austrolestes, and ischnura (odonata) New Zeal. J. Zool. 2007;34:371–380. [Google Scholar]
- 89.Marshall D.C., Hill K.B.R., Cooley J.R., Simon C. hybridization, mitochondrial dna phylogeography, and prediction of the early stages of reproductive isolation: Lessons from New Zealand cicadas (genus kikihia) Syst. Biol. 2011 doi: 10.1093/sysbio/syr017. [DOI] [PubMed] [Google Scholar]
- 90.King T.M., Kennedy M., Wallis G.P. Phylogeographic genetic analysis of the alpine weta, hemideina maori: evolution of a colour polymorphism and origins of a hybrid zone. J. Roy. Soc. New Zeal. 2003;33:715–729. [Google Scholar]
- 91.Wratt D.S., Tait A., Griffiths G., Espie P., Jessen M., Keys J., Ladd M., Lew D., Lowther W., Mitchell N., Morton J., Reid J., Reid S., Richardson A., Sansom J, Shankar U. Climate for crops: integrating climate data with information about soils and crop requirements to reduce risks in agricultural decision-making. Meteorol. Appl. 2006;13:305–315. [Google Scholar]
- 92.Trewick S.A., Bland K. Fire and slice: Palaeogeography for biogeography at New Zealand's North Island/South island juncture. J. Roy. Soc. New Zeal. 2011. in press. Available online: http://www.tandfonline.com/doi/abs/10.1080/03036758.2010.549493 (accessed on 16 June 2011)
- 93.Alloway B.V., Lowe D.J., Barrell D.J.A., Newnham R.M., Almond P.C., Augustinus P.C., Bertler N.A.N., Carter L., Litchfield N.J., McGlone M.S., Shulmeister J., Vandergoes M.J., Williams P.W. Towards a climate event stratigraphy for New Zealand over the past 30000 years (nz-intimate project) J. Quaternary Sci. 2007;22:9–35. [Google Scholar]
- 94.Suggate R.P., Stevens G.R., Te Punga M.T., editors. The geology of New Zealand. The Government Printer; Wellington, New Zealand: 1978. [Google Scholar]
- 95.Wardle P. Evolution and distribution of the New Zealand flora, as affected by quaternary climates. New Zeal. J. Bot. 1963;1:3–17. [Google Scholar]
- 96.Wallis G.P., Trewick S.A. Finding fault with vicariance: a critique of heads (1998) Syst Biol. 2001;50:602–609. [PubMed] [Google Scholar]
- 97.Hickerson M.J., Carstens B.C., Cavender-Bares J., Crandall K.A., Graham C.H., Johnson J.B., Rissler L., Victoriano P.F, Yoder A.D. phylogeography's past, present, and future: 10 years after avise, 2000. Mol. Phylogenet. Evol. 2010;54:291–301. doi: 10.1016/j.ympev.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 98.Templeton A.R. Coalescent-based, maximum likelihood inference in phylogeography. Mol Ecol. 2010;19:431–446. doi: 10.1111/j.1365-294X.2009.04514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Darwin C. On the Origin of Species by Means of Natural Selection. John Murray; London, UK: 1859. [Google Scholar]
- 100.Avise J.C. Molecular population structure and the biogeographic history of a regional fauna: a case history with lessons for conservation biology. Oikos. 1992;63:62–76. [Google Scholar]
- 101.Schneider C.J., Cunningham M., Moritz C. Comparative phylogeography and the history of endemic vertebrates in the wet tropics rainforests of Australia. Mol. Ecol. 1998;7:487–498. [Google Scholar]
- 102.Garrick G., Sunnucks P., Dyer R.J. Nuclear gene phylogeography using phase: Dealing with unresolved genotypes, lost alleles, and systematic bias in parameter estimation. BMC Evol. Biol. 2010;10:118. doi: 10.1186/1471-2148-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Morgan-Richards M., Trewick S.A. Hybrid origin of a parthenogenetic genus? Mol. Ecol. 2005;14:2133–2142. doi: 10.1111/j.1365-294X.2005.02575.x. [DOI] [PubMed] [Google Scholar]
- 104.Edgecombe G.D., Giribet G. A New Zealand species of the trans-tasman centipede order craterostigmomorpha (arthropoda: chilopoda) corroborated by molecular evidence. Invertebr. Syst. 2008;22:1–15. [Google Scholar]
- 105.McCulloch G.A., Wallis G.P., Waters J.M. Do Insects lose flight before they lose their wings? population genetic structure in subalpine stoneflies. Mol. Ecol. 2009;18:4073–4087. doi: 10.1111/j.1365-294X.2009.04337.x. [DOI] [PubMed] [Google Scholar]
- 106.Cranston P.S., Hardy N.B., Morse G.E., Puslednik L., McCluen S.R. When molecules and morphology concur: the ‘gondwanan’ midges (diptera: chironomidae) Syst. Entomol. 2010;35:636–648. [Google Scholar]
- 107.Hogg I.D., Stevens M.I., Schnabel K.E., Chapman M.A. Deeply divergent lineages of the widespread New Zealand amphipod paracalliope fluviatilis revealed using allozyme and mitochondrial DNA analysis. Freshwater Biol. 2006;51:236–248. [Google Scholar]
- 108.Trewick S.A. Sympatric cryptic species in new zealand onychophora. Biol. J. Linn. Soc. 1998;63:307–329. [Google Scholar]
- 109.Morgan-Richards M. Intraspecific karyotype variation is not concordant with allozyme variation in the auckland tree weta of New Zealand, hemideina thoracica (orthoptera: stenopelmatidae) Biol. J. Linn. Soc. 1997;60:423–442. [Google Scholar]
- 110.Slatkin M. Gene flow and the geographic structure of nnatural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- 111.Excoffier L., Foll M, Petit R.J. Genetic consequences of range expansions. Annu. Rev. Ecol. Evol. Syst. 2009;40:481–501. [Google Scholar]
- 112.Geneious v5.3 Available online: http://www.geneious.com/ (accessed on 20 June 2011)
- 113.Buckley T.R., Attanayake D., Park D., Ravindran S., Jewell T.R., Normark B.B. Investigating hybridization in the parthenogenetic New Zealand stick insect acanthoxyla (phasmatodea) using single-copy nuclear loci. Mol. Phylogenet. Evol. 2008;48:335–349. doi: 10.1016/j.ympev.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 114.Trewick S.A., Morris S. Diversity and Taxonomic Status of Some New Zealand Grasshoppers. Department of Conservation, Research & Development Series #290; Wellington, NZ: 2008. [Google Scholar]
- 115.Morgan-Richards M., Gibbs G. W. Colour, Allozyme karyotype variation show little concordance in the New zealand giant scree weta deinacrida connectens (orthoptera: stenopelmatidae) Hereditas. 1996;125:303–312. [Google Scholar]
- 116.Trewick S.A. Scree weta phylogeography: surviving glaciation and implications for pleistocene biogeography in New Zealand. N. Z. J. Zool. 2001;28:291–298. [Google Scholar]
- 117.Smith B.L.T. Unpublished BSc Honours Thesis. Massey University; Palmerston North, New Zealand: 2011. The Distribution, Phylogeography and Morphology of the New Zealand Ground Weta, Hemiandrus maculifrons. [Google Scholar]
- 118.Chappell E., Trewick SA., Morgan-Richards Shape and Sound Reveal Genetic Cohesion not Speciation in the New Zealand orthopteran, Hemiandrus pallitarsis, Despite High mtDNA Divergence. Biological Journal of the Linnean Society. 2011 in press. [Google Scholar]
- 119.Rogers G.M. The nature of the lower north island floristic Gap. N. Z. J. Bot. 1989;27:221–242. [Google Scholar]
- 120.Lewis K.B., Carter L., Davey F.J. The opening of cook strait: interglacial tidal scour and aligning basins at a subduction to transform plate edge. Mar. Geol. 1994;16:293–312. [Google Scholar]
- 121.Fleming C.A. The Geological History of New Zealand and its Life. Auckland University Press; Auckland, New Zealand: 1979. [Google Scholar]
- 122.Harrison R.G. Dispersal polymorphisms in insects. Annu. Rev. Ecol. Syst. 1980;11:95–118. [Google Scholar]
- 123.Edwards S.V. Is a new and general theory of molecular systematics emerging? Evolution. 2009;63:1–19. doi: 10.1111/j.1558-5646.2008.00549.x. [DOI] [PubMed] [Google Scholar]
- 124.Edwards S.V., Liu L., Pearl D.K. High-resolution species trees without concatenation. Proc. Natl. Acad. Sci. USA. 2007;104:5936–5941. doi: 10.1073/pnas.0607004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu L., Edwards S.V. Phylogenetic analysis in the anomaly zone. Syst. Biol. 2009;58:452–460. doi: 10.1093/sysbio/syp034. [DOI] [PubMed] [Google Scholar]
- 126.Crosby T. Fauna of New Zealand. Available online: http://www.landcareresearch.co.nz/research/biosystematics/invertebrates/faunaofnz/ (accessed on 15 June 2011)