Significance
Amphibians are among the most threatened animal groups. Population declines and extinctions have been linked, in part, to emerging infectious diseases. One such emerging disease has been attributed to Perkinsea-like protists causing mass mortality events in the United States. Using molecular methods, we evaluated the diversity of Perkinsea parasites in livers sampled from a wide taxonomic collection of tadpoles from six countries across three continents. We discovered a previously unidentified phylogenetically distinct infectious agent of tadpole livers present in a broad range of frogs from both tropical and temperate sites and across all sampled continents. These data demonstrate the high prevalence and global distribution of this infectious protist.
Keywords: frog decline, emerging disease, parasite, alveolates, molecular diversity
Abstract
The decline of amphibian populations, particularly frogs, is often cited as an example in support of the claim that Earth is undergoing its sixth mass extinction event. Amphibians seem to be particularly sensitive to emerging diseases (e.g., fungal and viral pathogens), yet the diversity and geographic distribution of infectious agents are only starting to be investigated. Recent work has linked a previously undescribed protist with mass-mortality events in the United States, in which infected frog tadpoles have an abnormally enlarged yellowish liver filled with protist cells of a presumed parasite. Phylogenetic analyses revealed that this infectious agent was affiliated with the Perkinsea: a parasitic group within the alveolates exemplified by Perkinsus sp., a “marine” protist responsible for mass-mortality events in commercial shellfish populations. Using small subunit (SSU) ribosomal DNA (rDNA) sequencing, we developed a targeted PCR protocol for preferentially sampling a clade of the Perkinsea. We tested this protocol on freshwater environmental DNA, revealing a wide diversity of Perkinsea lineages in these environments. Then, we used the same protocol to test for Perkinsea-like lineages in livers of 182 tadpoles from multiple families of frogs. We identified a distinct Perkinsea clade, encompassing a low level of SSU rDNA variation different from the lineage previously associated with tadpole mass-mortality events. Members of this clade were present in 38 tadpoles sampled from 14 distinct genera/phylogroups, from five countries across three continents. These data provide, to our knowledge, the first evidence that Perkinsea-like protists infect tadpoles across a wide taxonomic range of frogs in tropical and temperate environments, including oceanic islands.
It is widely recognized that amphibians are among the most threatened animal groups: for example, in 2008, 32% of species were listed as “threatened or extinct” and 42% were listed as in decline (www.iucnredlist.org/initiatives/amphibians/analysis; accessed October 29, 2014) (1, 2). The main causes of amphibian decline have been identified as habitat loss, environmental change, and the introduction of nonnative species (e.g., refs. 3–6). Emerging infectious diseases have also been shown to play a key role in many amphibian declines: for example, the chytrid fungal pathogen Batrachochytrium dendrobatidis has caused mass mortality events (MMEs) in Australia, in Europe, and across the Americas (e.g., refs. 7–9). MMEs have also been associated with infection by Ranavirus in, for example, the United Kingdom (UK), United States (US), and Canada (10, 11). Recent work has linked local MMEs in the United States with the infection of larval frogs (tadpoles) of the genera Lithobates and Acris by a protist (SI Appendix, Fig. S1) (12, 13). In 2006, histological examinations of tadpole tissues revealed the presence of thousands of small spherical cells preferentially infecting livers of tadpoles of the Southern Leopard Frog (Lithobates sphenocephalus, formerly Rana sphenocephala) sampled from an MME in Georgia (United States) (12). Small subunit (SSU) ribosomal DNA (rDNA) PCR and direct-amplicon sequencing, combined with phylogenetic tree reconstruction, showed that a lineage of protists closely related to Perkinsus, a parasite of marine bivalves (14), was the likely infectious agent (12).
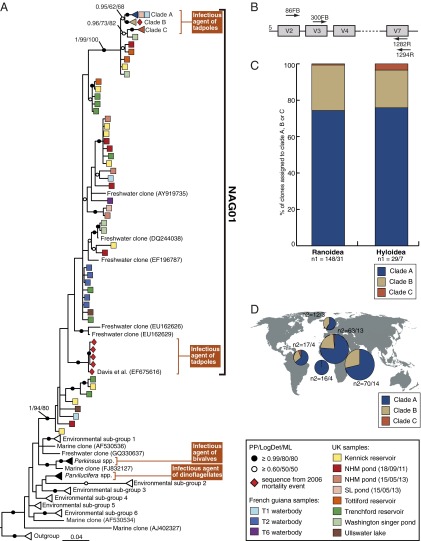
Perkinsea alveolates were first described as being affiliated with the Apicomplexa (14, 15), which includes important human pathogens such as Toxoplasma gondii and Plasmodium spp. (the causative agents of malaria). Phylogenetic analysis has shown that Perkinsea are a deeply divergent sister-group of dinoflagellate alveolates (16). Only three representative groups of Perkinsea were previously described morphologically and taxonomically: Perkinsus spp., parasites of marine bivalves (e.g., oysters and clams), Parvilucifera spp., parasites of dinoflagellates, and Rastrimonas subtilis (previously Cryptophagus subtilis), parasites of cryptophyte algae (17–20). However, environmental sequence analysis has considerably expanded the known diversity of Perkinsea-like organisms (21–25). The putative causal agent of the L. sphenocephalus MME lies within a clade (monophyletic group) of this environmental sequence diversity (Fig. 1A) (12) named here, for convenience, Novel Alveolate Group 01 (NAG01).
Fig. 1.
(A) Phylogenetic tree of Perkinsea SSU rDNA sequences focusing on the NAG01 group that includes two separate phylogenetic groups recovered from tadpole liver tissue samples. The phylogeny is estimated from a masked alignment consisting of 292 taxa and 776 characters. Bayesian posterior probability (6,000 samples from 2,000,000 MCMCMC generations), LogDet distance bootstrap (1,000 pseudoreplicates), and maximum likelihood bootstrap (1,000 pseudoreplicates) values are added to each node using the following convention: support values are summarized by black circles when all are equal to or greater than 0.9/80%/80%, and white circles when the topology support is less but equal to or greater than 0.6/50%/50%. Five sequences of Amoebophrya sp. were used as outgroup. Each square represents one environmental operational taxonomic unit (OTU), and the provenance of the OTUs is indicated by colored boxes (see key for the detail of sample provenance) (SI Appendix, Table S2 provides more details on the environments sampled). A red diamond indicates the individual clone sequences from the L. sphenocephalus 2006 mass mortality event in Georgia (United States) (12). A subset of the published environmental sequences have been reduced to representative triangles (see SI Appendix, Table S10 for detail of each environmental clade). (B) Representation of the V4 hyper-variable region of the template SSU rDNA and the relative position of the different primers used in this study (not to scale). (C) Histogram representing the percentage of clones per clade A, B, and C within each host superfamily from infected tadpoles. n1 represents the number of total clones sequenced per host superfamily/number of infected tadpoles per host superfamily. (D) Geographical distribution of clade A, B, and C. Pie charts represent the proportion of clones per clade A, B, and C in each of the five geographical locations where NAG01 was detected (United Kingdom, French Guiana, São Tomé, Cameroon, and Tanzania). n2 represents the number of clones sequenced per geographical location/number of infected tadpoles per geographical location.
In this study, we developed SSU rDNA primers that preferentially target the NAG01 group. We used these PCR primers for targeted screening of NAG01 diversity in both tropical and temperate freshwater environments, demonstrating the efficacy of this PCR protocol and expanding the diversity of Perkinsea-like sequences sampled from freshwater environments. Using the same protocol, we also detected NAG01 from livers of a wide taxonomic diversity of tadpoles from five countries across three continents. Within our study sample, we found evidence of infection by lineages different from the Perkinsea-like L. sphenocephalus pathogen identified by Davis et al. (12), suggesting that multiple Perkinsea lineages infect tadpoles.
Results and Discussion
Development of a Targeted PCR Assay for Perkinsea-Like Infection of Frog Tissue.
Culture-independent environmental DNA methods can be useful for detecting microbial lineages from the environment, including from the tissues of plants and animals (e.g., refs. 26–28). To investigate the prevalence and diversity of Perkinsea-like parasites infecting tadpoles, we designed two independent sets of PCR primers targeting ∼800 base pairs (bp) of the small subunit (SSU) rRNA-encoding gene, including the V4 variable region (29) (see Fig. 1B and SI Appendix, Table S1 for primers used in this study). The primers were designed to amplify the rDNA sequences of the wider NAG01 group (Fig. 1A), including the previously sampled SSU rDNA sequence of the Perkinsea-like infectious agent of the Southern Leopard Frog (12). To investigate the specificity of these two pairs of primers, we performed PCR on 10 environmental DNA samples from three tropical (French Guiana) and seven temperate (United Kingdom) planktonic freshwater samples (SI Appendix, Table S2). A total of 248 clones were sampled from the UK temperate water masses and 60 clones from the French Guiana water masses (SI Appendix, Table S2). Among these 308 clones, sequences from 240 clustered together into 46 nonidentical sample-specific NAG01 sequences (SI Appendix, Table S2 and S3) that were included in the phylogenetic analysis (Fig. 1A). The remaining 68 clones were non-NAG01 sequences encompassing a mixed assemblage of Fungi, Cryptophyta, and nematode sequences. Based on these results, our PCR protocol was judged as adequate for preferentially targeted environmental clone library analyses of NAG01, including the previously identified Perkinsea-like L. sphenocephalus infectious agent and seven environmental DNA sequences present in the GenBank database that were recovered from freshwater planktonic samples (30–32) (SI Appendix, Table S4).
Investigating the Global Prevalence of NAG01 Infections in Tadpoles.
Using the same PCR protocol used to detect NAG01 from freshwater environmental samples, we screened for NAG01 sequences in DNA extractions from liver samples dissected from 182 ethanol-preserved tadpoles. We sampled tadpoles from French Guiana (80 individuals from 8 sampling localities), Cameroon (37 from 14), Tanzania (15 from 1), the island of São Tomé (4 from 1), the United Kingdom (40 from 5), and the Czech Republic (6 from 3), of which 38 (21%) were PCR-positive for NAG01. See SI Appendix, Table S5 for more details, including the following: GPS location, sampling date, and description of environment where the tadpoles were found. For each positive sample, the PCR was repeated three times, and the amplicons were pooled. Each PCR product was checked on a 1% agarose gel for the presence of a single band of ∼800 bp. The PCR product was then purified, cloned, and sequenced. We sequenced six clones using M13F primers from each PCR-positive liver sample, resulting in 228 sequences. Based on preliminary sequence analysis of the forward reads, all NAG01 SSU sequences that were unique were double-strand sequenced. This sequencing effort encompassed a minimum of four clones per tadpole liver DNA clone library even though most libraries included fewer nonidentical clones (SI Appendix, Table S6).
Phylogenetic analysis demonstrated that all 177 sample-specific, unique NAG01 sequences recovered from tadpole livers form three discrete strongly supported and closely related clades (labeled clades A, B, and C in Fig. 1A). Clades A, B, and C do not correspond directly with host taxon or geographic origin (Figs. 1 C and D and 2), a similar result to that described for Perkinsus sp. (33) parasites of marine bivalves. Interestingly, 21 of the 38 (55%) NAG01-positive tadpole liver samples yielded sequences from both clade A and B (Fig. 2), suggesting that these livers harbored representatives from different nonclonal strains/species or that there is intranuclear SSU rDNA variation within NAG01 genomes sampled. Indeed, there is less than 3% nucleotide variation among clade A, B, and C sequences (SI Appendix, Fig. S2), consistent with intranuclear variation such as that observed in SSU-5.8S-LSU paralogues and pseudogenes. For example, variant copies of the rRNA gene are known to occur in alveolates (34, 35) with SSU rRNA gene paralogues, with 11% difference (36) transcribed at different stages of Plasmodium spp. life cycle (37).
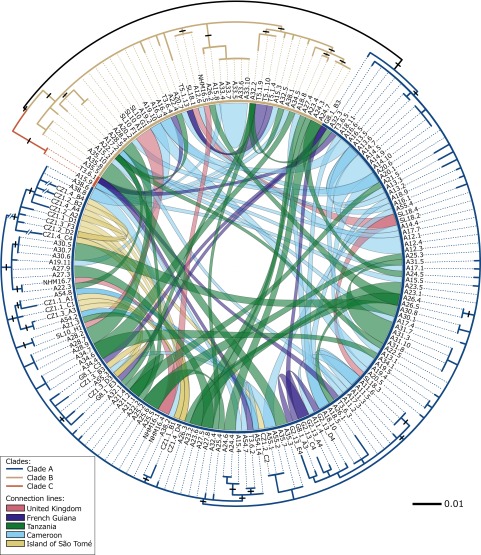
Fig. 2.
A maximum likelihood phylogenetic circle tree (inverted) showing clades A, B, and C and demonstrating the provenance of NAG01 phylotypes detected in tadpole liver tissue. The RAxML phylogeny is estimated from a masked alignment consisting of 177 NAG01 sequences and 806 characters from infected tadpoles. Branches proportionally shortened by 1/2 are labeled with a double-slashed line. Each clone sequence detected from the same liver sample is connected across the central circle. The colors of the connected lines were defined by the geographical location of tadpoles sampled (see the key). Black lines on the branches mark sequence variation confirmed across multiple samples and therefore cannot be the product of PCR error during clone library construction.
It is possible that the NAG01 DNA detected could have arisen from contamination from the environment and/or gastrointestinal tissue in the liver samples. However, this possibility was judged unlikely because all of the NAG01 SSU sequences detected from tadpole liver sequences grouped into one discrete phylogenetic subgroup whereas the primers used were capable of detecting a wide diversity of Perkinsea-like sequences from environmental DNA samples (Fig. 1A). To further test for cases of environmental contamination, DNA was extracted from tail samples (muscle and fin) taken from the same 182 tadpoles as a control to identify possible sources of nontissue-specific PCR detection or environmental contamination of NAG01 sequences. Although infections of an unknown alveolate-like parasite have been identified in the muscles of adult frogs (38), the tadpoles sampled here showed no evidence of disease progression so this experimental approach was judged as an adequate control to identify cases of environmental and wider animal tissue contamination. All controls were negative for the two primer-paired NAG01-specific PCR protocols, suggesting that detection of NAG01 was not an artifact of environmental contamination but instead a tissue-specific signal consistent with infection of a Perkinsea-like protist associated with the liver of these tadpoles. Interestingly, these liver-derived NAG01 sequences are closely related to some of the sequences recovered from filtered plankton environmental DNA sample sequences, suggesting that this protist group is found both associated with tadpoles and either as free living stage or associated with additional microbial hosts. Indeed, experimental manipulations have shown that infection of putative members of the NAG01 clade likely occur through ingestion of spores and/or zoospores from the watercolumn (39).
Diversity of Perkinsea Parasites Recovered from Liver Tissues from United States Mass Mortality Event.
Sequences generated from the tadpole livers sampled here are from a different NAG01 Perkinsea lineage from that detected in liver tissues of L. sphenocephalus tadpoles sampled from an MME in Georgia (United States) in 2006 (Fig. 1A). Only formalin-fixed, paraffin-embedded liver samples were available from this event (12). We successfully performed two DNA extractions from a historical sample. The standard NAG01 primers failed to generate an amplicon from both DNA samples, most likely because the template DNA was highly fragmented. Consequently, we targeted a shorter template using the 300F-B forward primer in combination with a eukaryotic general reverse primer 600R (SI Appendix, Table S1), and we amplified 414 bp of the SSU rDNA. PCR reactions were repeated three times for both DNA samples, and amplicons were pooled and cloned separately for the two DNA samples. In total, across the two DNA samples, we sequenced 45 clones in the forward and reverse direction. Phylogenetic analysis revealed that 44 clones represented four unique sequences that are closely related to the published sequence of the L. sphenocephalus infectious agent (EF675616) and one unique sequence closely related to NAG01 clade B (Fig. 1A).
Barcode Sequencing Reveals a Wide Host–Taxon Diversity and Biogeography for NAG01.
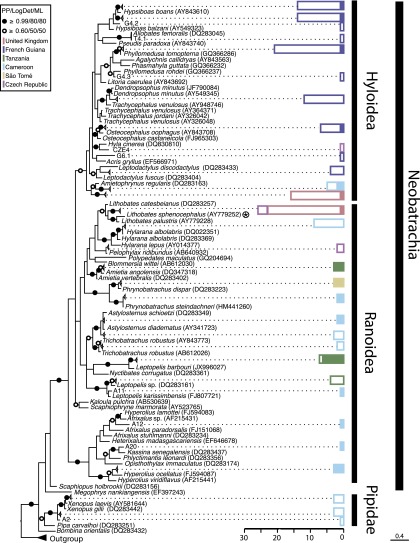
Precise taxonomic identification of tadpoles using morphological characters can be difficult, especially in geographic regions with high species diversity. We amplified a mitochondrial 16S rDNA barcode, shown to be effective for higher taxonomic assignment of amphibians (40, 41), for all 182 sampled tadpoles. The majority (n = 175) of the tadpoles sampled are members of the Neobatrachia (which comprises >95% of extant frogs) whereas the other 7 are tadpoles of the Pipidae: i.e., Xenopus (Fig. 3). We note that, although the pipid tadpoles sampled all tested negative, increased sampling is required to more confidently determine the presence/prevalence of this protist in this group of frogs.
Fig. 3.
Bayesian 16S rDNA phylogenetic tree of tadpole diversity sampled in this study, with histograms showing prevalence of NAG01 detection. The phylogeny is inferred from a masked alignment consisting of 247 taxa and 440 characters. Bayesian posterior probability (6,000 samples from 2,000,000 MCMCMC generations), LogDet distance bootstrap (1,000 replicates), and maximum likelihood bootstrap (1,000 replicates). Support values are summarized by black circles when all are equal to or greater than 0.9/80%/80%, and a white circle when topology support is weaker but all values are equal to or greater than 0.6/50%/50%. Sequences of Ambystoma sp. and Pleurodeles sp. (salamanders) were used as outgroup. Some frog species with multiple nonidentical 16S rDNA sequences recorded in GenBank are retained. The color-coded histogram represents the number of NAG01-negative tadpole samples (uncolored bars) and the number of NAG01-positive samples (colored bars). Each color corresponds to the tadpole’s country of origin as detailed in the key. The superfamily and suborder of the tadpoles tested is indicated on the histogram. The circled star indicates the host species described by Davis et al. (12) during the 2006 mortality event.
NAG01 DNA was detected in tadpoles of two of the largest groups of neobatrachians, with 6% of sampled Hyloidea (n = 102) infected and 42% of Ranoidea (n = 73) infected (Fig. 3). In separate studies, a Perkinsea-like parasite linked to local mortality events was detected using histology in six species of Ranoidea (L. sphenocephalus, Lithobates capito, Lithobates sevosus, Lithobates catesbeianus, Lithobates heckscheri, and Lithobates sylvaticus) and one species of Hyloidea (Acris gryllus) (www.nwhc.usgs.gov/publications/quarterly_reports; accessed October 29, 2014) (SI Appendix, Fig. S1). Similar dissection-based approaches have also shown related infectious agents present in a wide diversity of ranids, as well as A. gryllus, Hyla femoralis, Hyla gratiosa, Pseudacris ornata, and Gastrophryne carolinensis (39). In the absence of molecular data, it is not clear whether these infections were of protists from NAG01 clades A, B, or C reported here, the L. sphenocephalus parasite detected by Davis et al. (12), or a lineage not yet sampled for DNA analysis. However, the NAG01 sequences from tadpole livers detected in this study were recovered in five of the six countries sampled, including both tropical and temperate environments and an oceanic island. Mean prevalence estimate per country and 95% confidence intervals (CIs) using the Jeffreys method were as follows: 3% (four of 80, 95% CI of 0.4–34%) in French Guiana (eight sampling events); 55.2% (13 of 37, 11–78% CI) in Cameroon (14 sampling events); 93.3% (14 of 15, 72–99% CI) in Tanzania (one sampling event); 100% (four of four, 73–100% CI) on the Island of São Tomé (one sampling event); and 9% (three of 40, 1–40% CI) in the United Kingdom (five sampling events). Taken together, these data suggest a high prevalence and broad spatial distribution of infection by a specific subclade of NAG01 Perkinsea-like protists in neobatrachian tadpole populations specifically of the superfamily Ranoidea. Information connecting the putative phylogeny inferred taxonomy of the host tadpole to the presence of the NAG01 sequence type is given in SI Appendix, Table S5 to supplement the results shown in Fig. 3.
NAG01 Perkinsea-Like Protist and Disease.
All our tadpole field samples were preserved in ethanol, and, as such, we could not attempt to purify NAG01 clade A, B, or C into culture. Infection by Perkinsea-like parasites identified using histological and/or dissection microscopy techniques have been reported as visible from Gosner life cycle stages 24–42 (39, 42). In some cases, the disease phenotype of tadpoles infected by Perkinsea-like parasites has previously been described as bloated, lethargic, and showing cutaneous hemorrhages (12, 13, 42, 43) although we note that these symptoms are not diagnostic for Perkinsea disease of tadpoles, because other diseases can cause similar pathologies. The majority of the 38 infected tadpoles analyzed in this study were sampled from early Gosner stages, with the majority being of, or close to, Gosner stage 25, with one at Gosner stage 42 (SI Appendix, Table S5). The infected tadpoles showed no gross morphological symptoms of disease. A more precise definition of the disease in the cases of Perkinsea infection is infiltration of the liver and other visceral organs by large numbers of Perkinsea-like organisms (12, 38). Indications of tissue level disease in samples showing molecular evidence for the presence of NAG01 were sought through histology of liver with H&E staining of 5-μm sections from representative samples (two from French Guiana, three from Cameroon, and three from Tanzania; for examples, see SI Appendix, Fig. S3). These data showed no identifiable tissue damage consistent with disease and no cells attributable to the NAG01 microbes detected using molecular methods, demonstrating that these infectious agents are either a small cellular form and/or badly preserved in histological sections, or, alternatively, that the population of NAG01 is very low, suggesting a low infection intensity in these tadpoles. Thus, we cannot confirm that the infectious protists identified here are pathogenic, either because (i) the tadpoles were all sampled in an early phase of disease progression and/or the infectious protists are currently dormant, (ii) NAG01 clades A, B, or C detected in this study and that branch in a different part of the phylogenetic tree to previously reported disease-causing Perkinsea of tadpoles (Fig. 1A and ref. 12) have a limited or absent disease pathology, and/or (iii) disease is caused only in association with other infectious pathogens such as Ranavirus (43) or other forms of host stress (44). Other parasitic Perkinsea, such as Perkinsus sensu stricto (parasites of bivalves), are also widely geographically distributed, but infection and catastrophic host population MMEs are localized, with pathogenicity related in part to abiotic factors (33, 45–47).
These data demonstrate that the NAG01 protists detected in tadpole livers in this study (i) are not a ubiquitous agent or contaminant but instead are liver-associated, (ii) represent a closely related phylogenetically distinct subgroup within NAG01, (iii) lie within the phylum Perkinsea, for which all known taxa are potential parasites (e.g., refs. 18, 48, and 49), and (iv) are prevalent in a range of tadpole developmental stages (Gosner stages 25–42) (SI Appendix, Table S5). Although frog MMEs associated with a Perkinsea-like parasite have been recorded only in the United States, our results demonstrate that a greater diversity of Perkinsea-like protist infections of tadpoles are widespread. There are increasing efforts to monitor the health of wild amphibian populations (50–52), with justifiable focus on fungal chytrid pathogens and Ranavirus, both of which have been identified as causing disease in adult frogs (6–8, 10). Further studies on the etiology of tadpole (and other larval amphibian) infections are necessary to understand the impact of these protists on amphibian populations and to inform conservation planning.
Materials and Methods
Developing Group-Specific Primers.
Based on a multiple sequence alignment of available Perkinsea SSU rDNA sequences (SI Appendix, Tables S4 and S7) assembled using ARB (53), sets of “NAG01-specific primers” were designed to recover a central portion of the SSU rRNA-encoding gene, including the variable V4 region (Fig. 1B and SI Appendix, Table S1). Specificity of the PCR primers was checked first in silico by submitting sequences to the National Center for Biotechnology Information (NCBI) nonredundant (nr) DNA database (using Primer-BLAST–May 2014) and SILVA (TestProbe search–May 2014) databases (54) and, second, in situ using an environmental DNA clone library approach.
Environmental DNA Sampling of Freshwater Environments.
Water samples from the surface of the watercolumn were collected from multiple freshwater environments in the United Kingdom and French Guiana (details of each sample are given in SI Appendix, Table S2). For the sampling in French Guiana, water samples were prefiltered through 10-μm polycarbonate filters (Merck Millipore), and the filtrate was then serially filtered through 5-μm and then 2-μm polycarbonate filters to collect size-specific subsections of the microbial community. This process was conducted until each filter became saturated (the volume of water filtered for each sample is reported in SI Appendix, Table S2). Saturated filters were then submerged in RNAlifeguard (MoBio) and then stored at −20 °C for 2 weeks before being transported back to the United Kingdom at ambient temperatures and then finally stored at −80 °C. The UK samples were processed in a similar manner but were serially passed through 20-μm and then 2-μm or 0.2-μm filters and transformed directly to storage at −80 °C. DNA extraction was performed using the PowerWater DNA Isolation Kit (MoBio) using the protocol recommended by the manufacturer. Details of filtration size for positive detection of NAG01 are given in SI Appendix, Table S2 and indicate the presence of this group in both the 0.2- to 20-μm and 2- to 5-μm size fraction samples.
Sampling of Tadpole Tissue for NAG01 Molecular Screening.
A total of 182 ethanol-preserved tadpoles were examined: Details of the phylogenetic diversity sampled are given in Fig. 3, and the highest BLAST hit for each sample in the NCBI nr database (accessed October 2014) is given in SI Appendix, Table S8. SI Appendix, Table S5 provides details of the environmental provenance of the tadpoles. Tadpoles preserved whole in ethanol were dissected using sterile tools. A piece of liver of each tadpole was removed (taking care not to pierce the gut) and placed in a fresh tube of ethanol. Concurrently, a similarly sized piece of tail was excised and placed in a separate tube of ethanol. Total DNA was extracted from all tadpole tissue samples using the Blood and Tissue DNeasy extraction kit (Qiagen) following the manufacturer’s protocol, with an overnight lysis and incubation step.
SSU rDNA Clone Libraries from Environmental DNA and Tadpole Liver DNA.
Environmental DNA and tadpole liver and tadpole tail DNA extractions were used as a source template to construct NAG01-specific SSU rDNA gene clone libraries. Two sets of primers were used, including two NAG01-specific forward primers paired with two general eukaryotic reverse primers (Fig. 1B). For every PCR, we included a negative control (distilled H2O). All PCR amplification reactions were performed in 25 µL of total volume containing 8 ng of DNA and PCR MasterMix (Promega). Cycling reactions were as follows: 2 min at 95 °C, followed by 35 cycles of 30 s at 95 °C, 30 s at 55 °C, and 120 s at 72 °C, with an additional 10-min extension at 72 °C. For each template, three independent PCR reactions were performed, mixed together, purified using the Wizard SV Gel and PCR Clean-Up System kit (Promega), and cloned using the Strataclone PCR cloning kit (Stratagene) according to the manufacturer’s instructions. Clones were blue/white screened, and a subset was selected for PCR using M13F and M13R primers, which flank the vector insertion site. Clones showing the presence of an insert of the correct size were sequenced using M13F primers. For each environmental clone library, a minimum of 20 clones was sequenced. All of the tadpole tail samples were PCR-negative. For each tadpole liver sample with a positive PCR result, we sequenced six clones using the M13F primer. All M13F NAG01 SSU sequences that were unique in one or more position were double-strand sequenced.
Mitochondrial Encoded 16S SSU rRNA Gene Barcoding of Tadpoles.
Tadpole liver DNA extracts were subject to PCR using the 16Sar-F and 16Sar-R primers (40), which amplify an ∼600-bp region of the 16S SSU rRNA-encoding gene. PCR amplification reactions were performed in 50 µL of total volume containing 8 ng of DNA and PCR MasterMix (Promega). Cycling reactions were as follows: 2 min at 95 °C, followed by 25 cycles of 30 s at 95 °C, 30 s at 55 °C, and 120 s at 72 °C, with an additional 10-min extension at 72 °C. PCR products were checked on 1% agarose gel and purified using the Wizard SV Gel and PCR Clean-up System kit (Promega).
DNA Extraction and SSU rDNA Clone Library Construction from Formalin-Fixed, Paraffin-Embedded Liver Tissue from the United States Southern Leopard Frog MMEs.
Frozen and/or ethanol-preserved tadpole tissue from the 2006 mortality event (12) was unavailable due to theft of copper wiring from the freezers of the M.J.Y. laboratory, resulting in loss of these samples. Thus, only formalin-fixed and paraffin-embedded tissues used for microscopy sections were available for molecular analysis. Two liver sections were excised from the paraffin block using sterile scalpel blades. Each section was incubated with xylene at 50 °C for 3 min until the paraffin had dissolved. After this process, the liver section still constituted a compact tissue aggregate. The xylene solution was removed by pipetting, and the tissue sample was washed twice using pure ethanol and dried for 15 min at 37 °C. DNA was then extracted using the DNeasy Blood and Tissue kit (Qiagen) protocol with an overnight incubation in 100 μL of lysis buffer at 55 °C. The DNA extractions were conducted twice on the two different formalin-fixed, paraffin-embedded liver samples.
The DNA extractions were checked using a 2100 Bio-analyzer (Agilent Technologies) demonstrating highly fragmented template DNA (similar to ancient DNA samples) with an average fragment size of ∼160 bp. We thus amended our PCR protocol to target a shorter amplicon using the forward NAG01-specific 300F-B primer with the eukaryotic general reverse primer 600R (see SI Appendix, Table S1 for details). The PCR amplification reaction was performed in 25 µL of total volume of PCR MasterMix (Promega) again with an additional negative control reaction (distilled H2O). Cycling reactions were as follows: 2 min at 95 °C, followed by 35 cycles of 30 s at 95 °C, 30 s at 59 °C, and 120 s at 72 °C, with an additional 10-min extension at 72 °C. For each clone library, three independent PCRs were completed, mixed together, and cloned. Two clone libraries were constructed using the Strataclone cloning kit (Stratagene) according to the manufacturer’s instructions. Forty-five independent clones with insertions of appropriate size were selected and sequenced in both directions using M13F and M13R primers.
Sequencing and Assembly.
All sequencing was performed externally by Beckman Coulter Genomics. The final sets of sequences were trimmed to regions of high sequencing quality; vector sequences were removed, sequence reads were assembled into a contiguous sequence using Sequencher (Genecodes), and ambiguous sites were corrected.
Multiple Sequence Alignment and Phylogenetic Analysis.
Our NAG01 clone library sequencing resulted in 177 sample-specific unique clones from the tadpole liver samples (SI Appendix, Table S6), 46 sample-specific unique clones from the environmental DNA samples, and five sample-specific unique clones from formalin-fixed, paraffin-embedded liver samples (see SI Appendix, Table S3 for details). These sequences were assembled into a multiple sequence alignment with 59 Perkinsea-like sequences and Amoebophrya sp. SSU rDNA outgroup sequences (HQ658161, HM483395, HM483394, AY208894, and AF472555) recovered from the NCBI nr database (accessed May 2014; see SI Appendix, Tables S4 and S7). The sequences were aligned using MUSCLE (55), available via the graphical multiple sequence alignment viewer Seaview v4.2.12 (56), using default settings. The alignment was then checked and masked manually in Seaview, resulting in a data matrix of 292 sequences and 776 alignment positions. We note that the alignment included some partial database sequences to best sample the diversity of sequences sampled previously. However, Perkinsea-like sequences with the major central portion of the sequence absent were excluded from the final analysis (e.g., EUY162621, EUY162622, and EUY162623). This data matrix contained 487 variable alignment positions (excluding alignment positions with gaps) and 571 parsimony informative sites (including gaps).
All Perkinsea SSU rDNA sequences generated as a part of this study have been deposited in GenBank (see SI Appendix, Tables S3 and S6 for details). The Perkinsea alignment is available in the Seaview (56) Mase format with the alignment mask information retained and is available at doi 10.5281/zenodo.12712.
The 182 tadpole 16S SSU rDNA sequences were aligned with a collection of frog 16S SSU rDNA sequences (SI Appendix, Table S9) using the same approach described for the NAG01 sequences and using MUSCLE (55) via Seaview v4.2.12 (56). All tadpole 16S SSU rDNA sequences generated in this study were BLASTn searched against the NCBI nr database (accessed May 2014), and the most similarly named frog sequence based on sequence similarity was noted (SI Appendix, Table S8). Preliminary phylogenies were compared with published trees (57), and additional frog 16S SSU rDNA sequences representing intermediate branches (arbitrarily selected) were added to the phylogeny. Sequences of Caudata (salamanders) were chosen as outgroup for the phylogeny: Pleurodeles waltl (DQ283445), Pleurodeles nebulosus (DQ092266), Ambystoma mexicanum (EF107170), and Ambystoma tigrinum (DQ283407). The alignment was then checked and masked manually in Seaview, resulting in a data matrix of 247 sequences and 440 alignment positions with 225 variable alignment positions (excluding alignment positions with gaps) and with 232 parsimony informative sites (including sites with gaps in the masked data matrix).
All frog SSU rDNA sequences generated as apart of this study have been deposited in GenBank (see SI Appendix, Table S5 for details). The amphibian alignment is available in the Seaview (56) Mase format with the mask information retained and is available at doi 10.5281/zenodo.12712.
The best-fitting nucleotide substitution model for each alignment was determined using the information criterion and likelihood ratio tests implemented in Modelgenerator v0.85 (58). For Perkinsea and amphibian alignments, GTR+Γ and GTR+I+Γ models were selected, respectively. The α parameters for the Γ distributions were 0.38 and 0.30, respectively, with eight discrete rate categories whereas the I parameter for the amphibian alignment was 0.28. These parameters, where possible, were input into a Bayesian analysis using MrBayes v3.1.2 (59): i.e., lset, nst = 6 rates = gamma (or invgamma in the case of the amphibian alignment), and, in both cases, we included the covarion parameter search. Two independent runs of four Metropolis-coupled (MC) Markov chain Monte Carlo (MCMC) chains (with a heat parameter of 2) were run for 2,000,000 generations. Trees were sampled every 250 generations. In both analyses, the MCMC searches had converged within the first 25% of the generations sampled; as such, the first quarter of the search results were discarded (as the burnin). Convergence between the runs and burn-in were assessed using Tracer v1.6 (tree.bio.ed.ac.uk/software/tracer). The consensus topologies and posterior probabilities of each node were then calculated from the remaining sampled trees.
Support for the Bayesian tree topology was evaluated using two bootstrap methods and the Bayesian posterior probabilities from the MrBayes runs. Bootstrap support values were estimated using (i) RAxML v8.0.3 (60) with 1,000 pseudoreplicates and (ii) LogDet distance analysis with 1,000 pseudoreplicates using a BioNJ search method (available through Seaview v4.2.1). This second bootstrap analysis was conducted for comparison because it uses a method that minimizes artifacts arising from biases in base composition across the alignment (61).
To further investigate the phylogenetic diversity of the NAG01 sequences recovered from the tadpole livers, we resampled the alignment mask specifically focusing on sequences recovered from clades A, B, and C with the aim of maximizing unambiguously aligned sites. This analysis excluded all environmental sequences and retained only tadpole-associated Perkinsea sequences from clades A, B, and C. This new alignment resulted in a data matrix of 177 sequences and 806 alignment positions. This data matrix was analyzed with Modelgenerator, selecting the GTR + Γ (α parameter of 1.18) using the standard Akaike information criterion. The phylogeny was then estimated using RAxML v8.0.3 (60) using the GTR + Γ substitution model. For this analysis, we aimed to calculate a single tree that best displays the phylogenetic relationships of the NAG01 sequences sampled from tadpole livers. This alignment encompassed little sequence variation, and tree searches did not result in a consensus tree with consistently high/moderate bootstrap support values. This phylogenetic analysis therefore primarily serves to demonstrate the distribution of different SSU types across the different tissue samples and not to present a resolved phylogeny. To display codetection of NAG01 clades A, B, and C from specific liver samples, we used the Circos tool. Fig. 2 was created using Inkscape (https://inkscape.org/en/), Circos (62), and the Interactive Tree of Life (IToL) (itol.embl.de) (63). Connections highlighting phylogenetically disparate clones from the same frog were plotted and color-coded by geographic sampling location (see key in Fig. 2). This plot was then combined with an inverted circular tree generated by IToL using Inkscape.
Histology of Representative Tadpole Tissue Samples.
For eight samples, half of the liver that was not used for DNA extraction was stored in 100% ethanol at 4 °C. Each liver was then embedded in paraffin wax (Sigma-Aldrich) and cut into serial sections 5 um thick using a Shadon tissue processor (Thermo Electron Corporation). Sections were collected onto glass slides and stained with hematoxylin and eosin staining methods (Thermo Fisher). The sections of each slide were mounted using Histomount (National Diagnostics). Sections were examined by light microscopy (Microscope Olympus IX73) for the presence of putative parasites, and digital images were obtained using the Infinity 3 camera (Lumenera Corporation).
Supplementary Material
Acknowledgments
D.J.G., T.M.D.-B., and M.W. thank Marcel Kouete, Solo Ndeme, Gespo Alobwede, and many local people, especially in Mamfe Division. For access to Tanzanian material, we thank Michele Menegon and Simon Loader. We thank Hendrik Müller for tadpole expertise and David Blackburn for assistance in identifying Xenopus life cycle stages. We thank Caroline Ware of the Natural History Museum Wildlife Garden for assistance with sampling and Anke Lange of Biosciences at the University of Exeter for assistance in histological analysis. For permits and approvals for fieldwork in French Guiana, G.B.B.-S., D.J.G., and M.W. thank Myrian Virevaire (Directions de l'Environnement de l'Aménagement et du Logement) and Le Comité Scientifique Régional du Patrimoine Naturel. Additionally, for assisting our fieldwork, we thank Elodie Courtoise, Antoine Fouquet, Philippe Gaucher, Fausto Starace and family, and Jeannot and Odette (Camp Patawa). F.M. is a joint Natural History Museum, London and University College London PhD student. This research was funded primarily by the United Kingdom Department for Environment, Food and Rural Affairs, through the Systematics and Taxonomy scheme run by the Linnean Society of London and the Systematics Association UK (awarded to T.A.R.), and by fellowship awards (to A.C.). A.C. is supported by Marie Curie Intra-European Fellowship Grant FP7-PEOPLE-2011-IEF-299815 PARAFROGS and European Molecular Biology Organization (EMBO) Long-Term Fellowship ATL-1069-2011. T.A.R. is an EMBO Young Investigator and is supported by research grants from the Gordon and Betty Moore Foundation (Grant GBMF3307), the Natural Environment Research Council, the Leverhume Trust, the Biotechnology and Biological Sciences Research Council, the Canadian Institute for Advanced Research, and the Royal Society. Cameroon fieldwork was permitted by the Ministry of Forestry & Wildlife (no. 5032) funded by the US Fish & Wildlife’s Wildlife Without Borders–Amphibians in Decline scheme, the Royal Geographical Society, and the Zoological Society of London’s Evolutionarily Distinct and Globally Endangered Fellowship scheme. M.J. was supported by Czech Science Foundation Grant P506/10/2330 and the Institute of Parasitology, Biology Centre, Academy of Sciences of the Czech Republic (institutional support RVO: 60077344).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database. For a list of accession numbers, see SI Appendix.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500163112/-/DCSupplemental.
References
- 1.Wake DB, Vredenburg VT. Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc Natl Acad Sci USA. 2008;105(Suppl 1):11466–11473. doi: 10.1073/pnas.0801921105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monastersky R. Biodiversity: Life—A status report. Nature. 2014;516(7530):158–161. doi: 10.1038/516158a. [DOI] [PubMed] [Google Scholar]
- 3.Rohr JR, Raffel TR. Linking global climate and temperature variability to widespread amphibian declines putatively caused by disease. Proc Natl Acad Sci USA. 2010;107(18):8269–8274. doi: 10.1073/pnas.0912883107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson PTJ, et al. Aquatic eutrophication promotes pathogenic infection in amphibians. Proc Natl Acad Sci USA. 2007;104(40):15781–15786. doi: 10.1073/pnas.0707763104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins JP, Storfer A. Global amphibian declines: Sorting the hypotheses. Divers Distrib. 2003;9(2):89–98. [Google Scholar]
- 6.Pounds JA, et al. Widespread amphibian extinctions from epidemic disease driven by global warming. Nature. 2006;439(7073):161–167. doi: 10.1038/nature04246. [DOI] [PubMed] [Google Scholar]
- 7.Crawford AJ, Lips KR, Bermingham E. Epidemic disease decimates amphibian abundance, species diversity, and evolutionary history in the highlands of central Panama. Proc Natl Acad Sci USA. 2010;107(31):13777–13782. doi: 10.1073/pnas.0914115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lips KR, et al. Emerging infectious disease and the loss of biodiversity in a Neotropical amphibian community. Proc Natl Acad Sci USA. 2006;103(9):3165–3170. doi: 10.1073/pnas.0506889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher MC, Garner TW, Walker SF. Global emergence of Batrachochytrium dendrobatidis and amphibian chytridiomycosis in space, time, and host. Annu Rev Microbiol. 2009;63:291–310. doi: 10.1146/annurev.micro.091208.073435. [DOI] [PubMed] [Google Scholar]
- 10.Daszak P, et al. Emerging infectious diseases and amphibian population declines. Emerg Infect Dis. 1999;5(6):735–748. doi: 10.3201/eid0506.990601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duffus ALJ, Pauli BD, Wozney K, Brunetti CR, Berrill M. Frog virus 3-like infections in aquatic amphibian communities. J Wildl Dis. 2008;44(1):109–120. doi: 10.7589/0090-3558-44.1.109. [DOI] [PubMed] [Google Scholar]
- 12.Davis AK, Yabsley MJ, Keel MK, Maerz JC. Discovery of a novel alveolate pathogen affecting Southern Leopard frogs in Georgia: Description of the disease and host effects. EcoHealth. 2007;4:310–317. [Google Scholar]
- 13.Green DE, Feldman SH, Wimsatt J. Emergence of a Perkinsus-like agent in anuran liver during die-offs of local populations: PCR detection and phylogenetic characterization. Proc Am Assoc Zoo Vet. 2003;2003:120–121. [Google Scholar]
- 14.Azevedo C. Fine structure of Perkinsus atlanticus n. sp. (Apicomplexa, Perkinsea) parasite of the clam Ruditapes decussatus from Portugal. J Parasitol. 1989;75(4):627–635. [PubMed] [Google Scholar]
- 15.Lester RJG, Davis GHG. A new Perkinsus species (Apicomplexa, Perkinsea) from the abalone, Haliotis ruber. J Invertebr Pathol. 1981;37:181–187. [Google Scholar]
- 16.Bachvaroff TR, et al. Dinoflagellate phylogeny revisited: Using ribosomal proteins to resolve deep branching dinoflagellate clades. Mol Phylogenet Evol. 2014;70(0):314–322. doi: 10.1016/j.ympev.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brugerolle G. Apicomplexan parasite Cryptophagus renamed Rastrimonas gen. nov. Eur J Protistol. 2003;39:101. [Google Scholar]
- 18.Brugerolle G. Cryptophagus subtilis: A new parasite of cryptophytes affiliated with the Perkinsozoa lineage. Eur J Protistol. 2002;37:379–390. [Google Scholar]
- 19.Mackin JG. Histopathology of infection of Crassostrea virginica (Gmelin) by Dermocystidium marinum Mackin, Owen and Collier. Bull Mar Sci. 1951;1(1):72–87. [Google Scholar]
- 20.Park MG, Yih W, Coats DW. Parasites and phytoplankton, with special emphasis on dinoflagellate infections. J Eukaryot Microbiol. 2004;51(2):145–155. doi: 10.1111/j.1550-7408.2004.tb00539.x. [DOI] [PubMed] [Google Scholar]
- 21.Lefranc M, Thénot A, Lepère C, Debroas D. Genetic diversity of small eukaryotes in lakes differing by their trophic status. Appl Environ Microbiol. 2005;71(10):5935–5942. doi: 10.1128/AEM.71.10.5935-5942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lepère C, Domaizon I, Debroas D. Unexpected importance of potential parasites in the composition of the freshwater small-eukaryote community. Appl Environ Microbiol. 2008;74(10):2940–2949. doi: 10.1128/AEM.01156-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangot JF, Lepère C, Bouvier C, Debroas D, Domaizon I. Community structure and dynamics of small eukaryotes targeted by new oligonucleotide probes: New insight into the lacustrine microbial food web. Appl Environ Microbiol. 2009;75(19):6373–6381. doi: 10.1128/AEM.00607-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bråte J, et al. Freshwater Perkinsea and marine-freshwater colonizations revealed by pyrosequencing and phylogeny of environmental rDNA. ISME J. 2010;4(9):1144–1153. doi: 10.1038/ismej.2010.39. [DOI] [PubMed] [Google Scholar]
- 25.Chambouvet A, et al. Diverse molecular signatures for ribosomally ‘active’ Perkinsea in marine sediments. BMC Microbiol. 2014;14(1):110. doi: 10.1186/1471-2180-14-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fredricks DN, Relman DA. Sequence-based identification of microbial pathogens: A reconsideration of Koch’s postulates. Clin Microbiol Rev. 1996;9(1):18–33. doi: 10.1128/cmr.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Relman DA, Schmidt TM, MacDermott RP, Falkow S. Identification of the uncultured bacillus of Whipple’s disease. N Engl J Med. 1992;327(5):293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 28.Anderson BE, Dawson JE, Jones DC, Wilson KH. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J Clin Microbiol. 1991;29(12):2838–2842. doi: 10.1128/jcm.29.12.2838-2842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wuyts J, et al. Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA. Nucleic Acids Res. 2000;28(23):4698–4708. doi: 10.1093/nar/28.23.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards TA, Vepritskiy AA, Gouliamova DE, Nierzwicki-Bauer SA. The molecular diversity of freshwater picoeukaryotes from an oligotrophic lake reveals diverse, distinctive and globally dispersed lineages. Environ Microbiol. 2005;7(9):1413–1425. doi: 10.1111/j.1462-2920.2005.00828.x. [DOI] [PubMed] [Google Scholar]
- 31.Lefèvre E, et al. Unveiling fungal zooflagellates as members of freshwater picoeukaryotes: Evidence from a molecular diversity study in a deep meromictic lake. Environ Microbiol. 2007;9(1):61–71. doi: 10.1111/j.1462-2920.2006.01111.x. [DOI] [PubMed] [Google Scholar]
- 32.Lefèvre E, Roussel B, Amblard C, Sime-Ngando T. The molecular diversity of freshwater picoeukaryotes reveals high occurrence of putative parasitoids in the plankton. PLoS One. 2008;3(6):e2324. doi: 10.1371/journal.pone.0002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villalba A, Reece KS, Ordás MC, Casas SM, Figueras A. Perkinsosis in molluscs: A review. Aquat Living Resour. 2004;17:411–432. [Google Scholar]
- 34.Stern RF, et al. Evaluating the ribosomal internal transcribed spacer (ITS) as a candidate dinoflagellate barcode marker. PLoS One. 2012;7(8):e42780. doi: 10.1371/journal.pone.0042780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galluzzi L, et al. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a Dinoflagellate) Appl Environ Microbiol. 2004;70(2):1199–1206. doi: 10.1128/AEM.70.2.1199-1206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCutchan TF, et al. Primary sequences of two small subunit ribosomal RNA genes from Plasmodium falciparum. Mol Biochem Parasitol. 1988;28(1):63–68. doi: 10.1016/0166-6851(88)90181-8. [DOI] [PubMed] [Google Scholar]
- 37.Nishimoto Y, et al. Evolution and phylogeny of the heterogeneous cytosolic SSU rRNA genes in the genus Plasmodium. Mol Phylogenet Evol. 2008;47(1):45–53. doi: 10.1016/j.ympev.2008.01.031. [DOI] [PubMed] [Google Scholar]
- 38.Jones MEB, Armién AG, Rothermel BB, Pessier AP. Granulomatous myositis associated with a novel alveolate pathogen in an adult southern leopard frog (Lithobates sphenocephalus) Dis Aquat Organ. 2012;102(2):163–167. doi: 10.3354/dao02539. [DOI] [PubMed] [Google Scholar]
- 39.Cook JO. 2008. Transmission and Occurence of Dermomycoides sp. in Rana sevosa and Other Ranids in the North Central Gulf of Mexico States. Master's thesis (University of Southern Mississippi)
- 40.Palumbi SR, et al. 1991. The Simple Fool’s Guide to PCR: A Collection of PCR Protocols (University of Hawaii, Honolulu), Version 2.
- 41.Vences M, Thomas M, van der Meijden A, Chiari Y, Vieites DR. Comparative performance of the 16S rRNA gene in DNA barcoding of amphibians. Front Zool. 2005;2(1):5. doi: 10.1186/1742-9994-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gahl MK. 2007. Spatial and temporal patterns of amphibian disease in Acadia National Park wetlands. PhD dissertation (The University of Maine, Orono, Maine)
- 43.Landsberg JH, et al. Co-infection by alveolate parasites and frog virus 3-like ranavirus during an amphibian larval mortality event in Florida, USA. Dis Aquat Organ. 2013;105(2):89–99. doi: 10.3354/dao02625. [DOI] [PubMed] [Google Scholar]
- 44.Rollins-Smith LA. Metamorphosis and the amphibian immune system. Immunol Rev. 1998;166(1):221–230. doi: 10.1111/j.1600-065x.1998.tb01265.x. [DOI] [PubMed] [Google Scholar]
- 45.Casas SM, La Peyre JF, Reece KS, Azevedo C, Villalba A. Continuous in vitro culture of the carpet shell clam Tapes decussatus protozoan parasite Perkinsus atlanticus. Dis Aquat Organ. 2002;52(3):217–231. doi: 10.3354/dao052217. [DOI] [PubMed] [Google Scholar]
- 46.Chu FLE, La Peyre JF. Perkinsus marinus susceptibility and defense-related activities in Eastern oysters Crassostrea virginica: Temperature effects. Dis Aquat Organ. 1993;16:223–234. [Google Scholar]
- 47.Cook T, Folli M, Klinck J, Ford S, Miller J. The relationship between increasing sea surface temperature and the northward spread of Perkinsus marinus (Dermo) disease epizootics in oysters. Estuar Coast Shelf Sci. 1998;40:587–597. [Google Scholar]
- 48.Andrews JD. Epizootiology of the disease caused by the oyster pathogen Perkinsus marinus and its effects on the oyster industry. Am Fish Soc Spec Publ. 1988;18:47–63. [Google Scholar]
- 49.Garcés E, Alacid E, Bravo I, Fraga S, Figueroa RI. Parvilucifera sinerae (Alveolata, Myzozoa) is a generalist parasitoid of dinoflagellates. Protist. 2013;164(2):245–260. doi: 10.1016/j.protis.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 50.Young BE, et al. Population declines and priorities for amphibian conservation in Latin America. Conserv Biol. 2001;15:1213–1223. [Google Scholar]
- 51.Buckley J, Beebee TJC, Schmidt BR. Monitoring amphibian declines: Population trends of an endangered species over 20 years in Britain. Anim Conserv. 2014;17:27–34. [Google Scholar]
- 52.Duffus AL. Chytrid blinders: What other disease risks to amphibians are we missing? EcoHealth. 2009;6(3):335–339. doi: 10.1007/s10393-009-0271-8. [DOI] [PubMed] [Google Scholar]
- 53.Ludwig W, et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004;32(4):1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pruesse E, et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35(21):7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12(6):543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 57.Frost DR, et al. 2006. The Amphibian Tree of Life (American Museum of Natural History, New York), Bulletin No. 297, pp 1–291.
- 58.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McInerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 60.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lockhart PJ, Steel MA, Hendy MD, Penny D. Recovering evolutionary trees under a more realistic model of sequence evolution. Mol Biol Evol. 1994;11(4):605–612. doi: 10.1093/oxfordjournals.molbev.a040136. [DOI] [PubMed] [Google Scholar]
- 62.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Letunic I, Bork P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23(1):127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.