Significance
The circadian clock, an endogenous time-keeping mechanism common to most species, allows organisms to coordinate biological processes with specific times of day. In plants, the role of the clock extends to almost every aspect of growth and development, including responses to biotic and abiotic stresses. The core molecular components and circuits of the clock have been well studied in the model organism Arabidopsis thaliana; however, how this mechanism connects to clock-controlled outputs remains poorly understood. Here, we performed a genome-wide characterization of the direct targets of a key clock component in Arabidopsis. Our results emphasize the broad role of the plant clock in regulating multiple biological functions and provide direct links between the oscillator and clock-regulated outputs.
Keywords: genome-wide, circadian clock, clock-controlled outputs, transcriptional regulation
Abstract
The circadian clock in Arabidopsis exerts a critical role in timing multiple biological processes and stress responses through the regulation of up to 80% of the transcriptome. As a key component of the clock, the Myb-like transcription factor CIRCADIAN CLOCK ASSOCIATED1 (CCA1) is able to initiate and set the phase of clock-controlled rhythms and has been shown to regulate gene expression by binding directly to the evening element (EE) motif found in target gene promoters. However, the precise molecular mechanisms underlying clock regulation of the rhythmic transcriptome, specifically how clock components connect to clock output pathways, is poorly understood. In this study, using ChIP followed by deep sequencing of CCA1 in constant light (LL) and diel (LD) conditions, more than 1,000 genomic regions occupied by CCA1 were identified. CCA1 targets are enriched for a myriad of biological processes and stress responses, providing direct links to clock-controlled pathways and suggesting that CCA1 plays an important role in regulating a large subset of the rhythmic transcriptome. Although many of these target genes are evening expressed and contain the EE motif, a significant subset is morning phased and enriched for previously unrecognized motifs associated with CCA1 function. Furthermore, this work revealed several CCA1 targets that do not cycle in either LL or LD conditions. Together, our results emphasize an expanded role for the clock in regulating a diverse category of genes and key pathways in Arabidopsis and provide a comprehensive resource for future functional studies.
In anticipation of the daily changes in light and temperature, the plant’s internal circadian clock regulates a large portion of the transcriptome, nearly 80% in rice, poplar, and Arabidopsis (1, 2). The clock provides temporal coordination of multiple biological functions to ensure optimal efficiency. For example, before dawn, photosynthetic transcripts accumulate in anticipation of the expected sunlight. Even when shifted to constant light (LL) and temperature, in the absence of day/night cues, transcripts associated with photosynthesis are still up-regulated before the subjective day (1–4). Not only are light- and energy-related activities restricted to particular times, but other aspects of plant growth, including water use, hormone activity, UVB response, and low-temperature response, show this gating effect (5–9). This temporal partitioning of biological responses ultimately provides an optimal integration of the plant’s organismal functions with the environment.
The circadian clock in plants involves a posttranscriptional component and a well-studied transcriptional–translation feedback regulation between the Myb-like transcription factors CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) and a member of the PSEUDO RESPONSE REGULATOR (PRR) family, TIMING OF CAB2 EXPRESSION 1 (TOC1) (10–16). Together these three components contribute to the rhythmic expression of several other circadian regulators and output genes (17–19). CCA1, along with LHY and 11 other members of REVEILLE (RVE) proteins, belongs to a subfamily of MYB-domain–containing transcription factors. Primarily, CCA1 functions as a transcriptional repressor that binds to the evening element (EE), a motif enriched in promoters of genes with peak expression in the evening (3, 13, 20–22). CCA1 mRNA accumulates around dawn and thus represses the expression of its evening-phased targets (13, 18). However, as CCA1 protein levels decrease later in the day, this repression is relaxed, and the expression of CCA1 targets can accumulate, peaking in the evening (13). Although considered a bona fide transcriptional repressor, CCA1 also can activate the expression of the clock genes PSEUDO-RESPONSE REGULATOR 9 (PRR9) and PRR7 by binding directly to their promoters (23). CCA1 is considered a master regulator of clock function, and its misexpression causes severe clock phenotypes that result in altered plant growth and stress responses in Arabidopsis (11, 22, 24, 25).
To gain a better understanding of the role that the circadian clock plays in the control of these multiple biological processes through CCA1, either directly or through indirect regulation of downstream factors, we performed ChIP sequencing (ChIP-Seq) analyses to comprehensively identify the regions of CCA1 occupancy in the Arabidopsis genome. Surprisingly, in addition to CCA1 occupancy at EE-containing, evening-expressed, clock-regulated genes, we also observed occupancy near many genes with peak expression in the morning and in proximity to genes that do not cycle in LL conditions. We further investigated the role of CCA1 in diel (light:dark, LD) conditions and found additional CCA1-occupied targets. Together, these connections between CCA1 and a wide range of circadian output targets indicate a further role for the Arabidopsis circadian clock and provide previously unidentified, direct connections between the molecular oscillator and circadian-regulated outputs.
Results
CCA1-Occupied Regions Are Enriched for the EE and Evening-Expressed Genes in LL.
To identify direct CCA1 targets, we performed ChIP-Seq analysis of plants expressing CCA1-GFP driven by the endogenous CCA1 promoter (CCA1p::CCA1-GFP) (26). Plants were grown in 12-h:12-h LD cycles and then were shifted to LL and harvested at zeitgeber time (ZT) 26 and ZT38 (2 and 14 hrs after subjective dawn on the second day in LL, respectively). For the ZT26 time point, we performed three independent ChIP-Seq experiments, which were highly correlated (Fig. S1A). Few reads were obtained from the ZT38 time point, as expected, because the CCA1 protein driven by its own promoter should be present in very low levels in the evening (11). Therefore we did not perform any additional replicates for ZT38, which is shown here only as a comparison and which was not included in the quantitative analysis presented below.
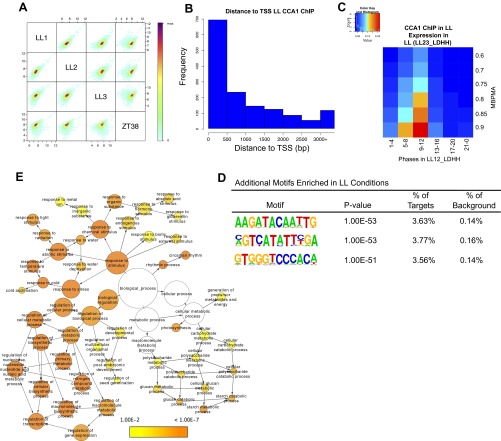
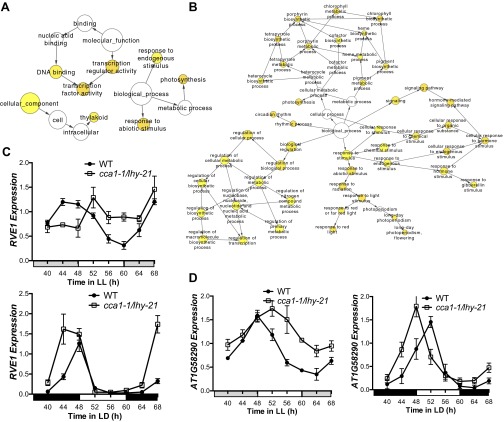
Fig. S1.
(A) Correlation of sequence tags between each of three CCA1 ChIP-Seq experiments performed at ZT26 in LL and one ChIP-Seq experiment performed at ZT38 in LL. R2 values of correlation between the three ZT26 replicates are LL1–LL2, 0.93; LL2–LL3, 0.77; and LL1–LL3, 0.82. (B) Histogram showing distances between the identified peaks of CCA1 occupancy in LL and the TSS of the nearest gene (bin size = 0–500 bp; range, 0–3,000 bp). (C) Phase enrichment of cycling CCA1-occupied targets. The expression datasets include transcripts that display >0.5 MBPMA in LL (LL23_LDHH). Genes were grouped by phase and MBPMA score (strength of cycling) into 36 groups. The ratio of CCA1 occupancy to the number of genes in each group is indicated by the intensity of the heat map (red and blue colors indicate maximum and minimum ratios of putative targets in the bin). (D) Additional motifs enriched in CCA1-occupied targets in LL. (E) Functional enrichment analysis with full GO biological process category of the 1,100 CCA1 targets identified in LL with a peak within 1 kb of the TSS. Circle size is proportional to gene numbers, and the color of each circle represents the enrichment P value for the GO term label on that circle, with orange representing highest enrichment and yellow the lowest enrichment above the cutoff (FDR corrected 0.01). Distance between nodes was arranged manually to optimize readability. The graph was generated using BINGO software (63).
To identify high-confidence sites of CCA1 occupancy, the three independent biological replicates were combined using the HOMER peak-calling software and ENCODE’s irreproducible discovery rate (IDR) analysis (details are given in Materials and Methods) (27–30). We identified 1,433 peaks that were associated with the nearest transcription start site (TSS) of 1,305 unique nuclear-encoded putative CCA1 direct targets (Dataset S1). As expected for a transcriptional regulator, many of these peaks were located in the promoter region (Fig. 1A). Even for peaks not classified as predominantly in the promoter region, the majority (68%) were within 1,000 bp of a TSS (Fig. S1B). Additionally, many peaks were identified as associated with the transcription termination site (TTS); this association was unexpected, because previously CCA1 has been observed to interact in the promoter region (20, 24, 31). However, closer examination of these sites revealed that many were between two gene loci oriented in the same direction, and although the peak is within close proximity (<1 kb) to the TSS of the downstream gene, it is closer to the TTS of the upstream gene, reflecting this assignment.
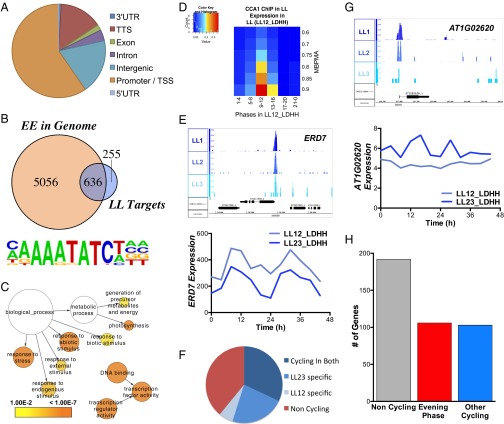
Fig. 1.
(A) Genomic distribution of CCA1-occupied sites in LL. Promoter/TSS is defined as the region from −1,000 to +1 from the TSS. (B) The Venn diagram shows the overlap of the presence of an EE in a region 1-kb upstream of genes closest to CCA1-occupied peaks relative to the genome. (C) Functional enrichment analysis against GO Slim of the 1,100 putative CCA1 targets identified in LL with a peak within 1 kb of the TSS. The sizes of circles are based on the gene numbers. The color of each circle represents the enrichment P value for the GO term label on that circle, with orange representing highest enrichment and yellow the lowest enrichment above the cutoff [false-discovery rate (FDR) corrected 0.01]. The distance between nodes was arranged manually to optimize readability. The graph was generated using BINGO software (63). (D) Phase enrichment of cycling CCA1-occupied targets. The expression datasets include transcripts that display >0.5 MBPMA in constant light [LL12_LDHH, entrained in 12-h:12-h LD cycles at constant temperature (HH) and then subjected to LL; data from the Kay laboratory] (3). Genes were grouped by phase and MBPMA score (strength of cycling) into 36 groups. The ratio of CCA1 occupancy to the number of genes in each group is indicated by the intensity of the heat map (red and blue colors indicate the maximum and minimum ratios of putative targets in the bin, respectively. (E) Example of an EE-containing, evening-phased, and cycling CCA1 target, EARLY-RESPONSIVE TO DEHYDRATION 7 (ERD7, AT2G17840). (Upper) Normalized tag counts by location in the genome for each of the three replicate ChIP-Seq experiments in LL. Genome annotation shows the ERD7 gene visualized using IGB (55). (Lower) ERD7 shows rhythmic expression in two of the previously published time series experiments in constant conditions, LL12_LDHH and LL23_LDHH [entrained in 12-h:12-h LD cycles at constant temperature (HH) and then subjected to LL; data are from the Millar laboratory (4)]. (F) The putative CCA1 targets were grouped based on their rhythmic expression in previous studies. Genes were classified as noncycling in either of the time series experiments performed in LL, as cycling in both experiments, or as cycling in only one experiment or the other. (G) Example of a CCA1 target, Ras-related small GTP-binding family protein (AT1G02620), which does not cycle in either published LL time series. (H) CCA1 targets, selected as having strong peaks of CCA1 occupancy within 1 kb of the TSS, were examined for their phase of peak expression in two published LL time series. Here we classified evening genes as those with peak phase of expression between ZT10 and ZT16.
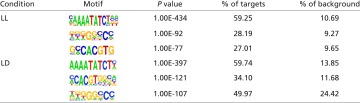
Of the high-confidence peaks, 62% contained the sequence matching the canonical EE (Fig. 1B). Several other motifs were enriched in the CCA1-occupied peaks, including the previously identified protein box (PBX) element (GGGCCCA) and G-box (CACGTG) and other motifs not previously reported to be enriched in circadian-regulated genes (Table 1 and Fig. S1D) (1, 21, 32). The 891 genes with a high-scoring CCA1 peak within 1 kb of the TSS were enriched for Gene Ontology (GO) terms describing responses to both external and internal stimuli and photosynthesis (Fig. 1C and Fig. S1E). Consistent with the known role of CCA1 in regulating responses to abiotic and biotic stresses, we observe enrichment in genes with GO terms associated with response to both biotic and abiotic stresses, including response to low temperature and light (Fig. S1E) (8, 9, 25, 33–35). Putative CCA1 targets also are enriched for genes involved in hormone response, including gibberellic acid and abscisic acid, as is consistent with the known role of the circadian clock in the regulation of daily cycles of hormone sensitivity (22, 36).
Table 1.
Motifs enriched in LL and LD conditions
 |
The majority of the transcripts associated with sites enriched for CCA1 occupancy showed strong cycling expression patterns with an evening phase (Fig. 1D). Specifically, the putative targets with a high-scoring peak of CCA1 occupancy within 1 kb of the TSS are enriched for a peak phase of expression between ZT9 and ZT12, based on published time series expression analyses performed in LL (Fig. 1D and Fig. S1C) (3, 21). These potential CCA1 targets also are strongly cycling, as indicated by enrichment with a model-based, pattern-matching algorithm (MBPMA) >0.8 in the existing processed data (Fig. 1 D and E and Fig. S1C) (1). Surprisingly, when we analyzed the transcripts associated with CCA1-occupied sites in the LL time series datasets, we observed a large portion of putative CCA1 targets (37%) that did not show a rhythmic expression pattern in either LL time series (Fig. 1 F and G). Although the two published datasets (LL12_LDHH and LL23_LDHH) are both in LL conditions, we observe that some transcripts cycle in one and not in the other, even when the cycling transcripts have different phases in the two datasets. This observation is consistent with previous reports that laboratory-specific conditions might contribute to noticeable differences in cycling calls between similarly derived datasets (1, 21). More than 50% of the peaks associated with these noncycling targets contained the AAATATCT sequence of the EE, suggesting that many clock-controlled genes probably are not rhythmic in LL. It is possible that the stress caused by the artificial conditions of light at night when plants are not expecting it may mask the cycling expression pattern of some genes (37). To determine if any of our putative CCA1 targets might be clock-regulated but masked for cycling expression in LL, we examined the expression of these noncycling CCA1 targets for their rhythmic expression in two LD time courses (LDHH_SM and LDHH_ST) (38, 39). Using the MBPMA cutoff of 0.8 as the threshold for cycling, about 50% of these potential targets were cycling in one of these two LD arrays, many with an evening phase of expression (Fig. 1H) (1). This finding suggests that many potential direct targets of the Arabidopsis circadian clock do not cycle in LL conditions and that the estimate of circadian-controlled genes obtained from experiments in LL conditions may underestimate the extensive role of the clock in plants.
Additional CCA1-Occupied Sites Are Observed in LD Conditions.
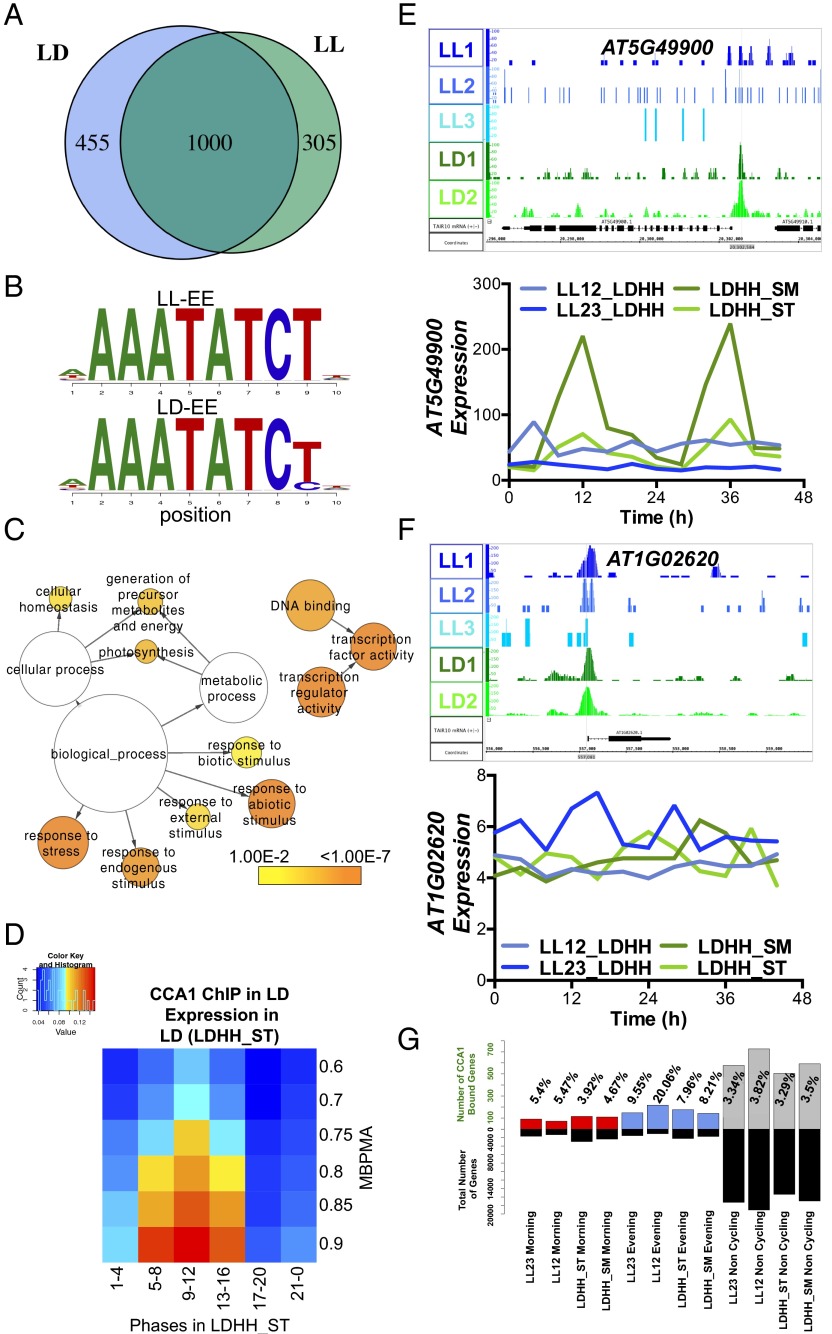
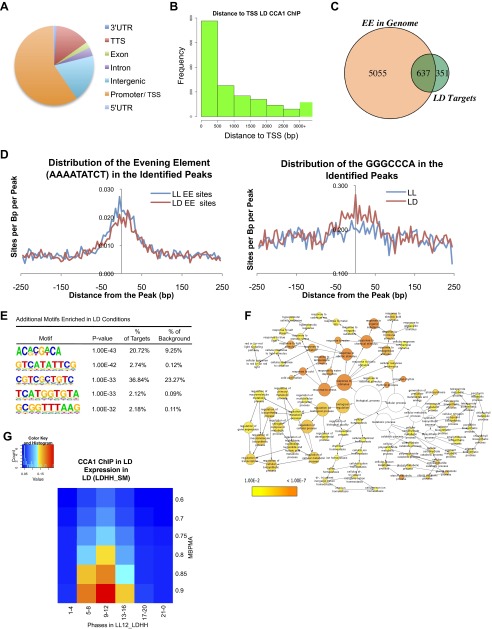
To determine if additional CCA1 targets might be identified and to examine the role of CCA1 transcriptional regulation in LD conditions, we performed two independent ChIP-Seq experiments in LD. As in the LL analysis, we focused on high-confidence sites using IDR analysis and peak calls by HOMER (27–30). We identified 1,607 top-scoring peaks and selected a stringent list of 1,455 putative CCA1 target genes (Dataset S2). To examine the degree of overlap between CCA1-occupied sites in LL and LD, we systematically compared the CCA1 targets from LL and LD. Of the 1,455 occupied sites observed in LD conditions, 1,000 overlapped with the 1,305 sites observed in LL (Fig. 2A). Although the majority of peaks identified in LL also were observed in LD, the overlap between peaks identified in LL and LD was not complete (Fig. 2A), perhaps because of a lack of complete coverage with only two and three biological replicates or because of specific binding environments in LL versus LD (29). Again, the identified peaks were enriched for promoter regions, and most (>80%) occurred within 1 kb of a TSS (Fig. S2 A and B). The top motif in both ChIP-Seq datasets was the EE, and, as in LL, the EE was highly enriched in the CCA1-occupied regions, along with the PBX element (GGGCCCA) and G-box (CACGTG) (Fig. 2B, Fig. S2C, and Table 1). In fact, for some promoters, more than one previously identified motif associated with circadian rhythms was identified, suggesting multiple regulatory inputs for the promoters of those genes. Although the EE was located at the peak center in both LL and LD, the position of the PBX element was more centered in the peaks identified in LD than in the peaks identified in LL (Fig. S2D). In addition to these motifs, we also observed several other enriched motifs identified only in the CCA1 ChIP performed in LD (Fig. S2E).
Fig. 2.
(A) Comparison of CCA1-occupied peaks in LL and LD. The Venn diagram shows the number of targets that are LD-specific (455) shaded in blue, the targets that are LL-specific (305) shaded in green, and the overlap between LD and LL targets (1,000 targets). (B) Alignment of the EE motif identified in LL and LD targets. (C) Functional enrichment analysis against GO Slim of the 1,231 CCA1 targets in LD with a peak within 1 kb of the TSS. The sizes of circles are based on the gene numbers. The color of each circle represents the enrichment P value for the GO term label on that circle, with orange representing highest enrichment and yellow the lowest enrichment above the cutoff (FDR corrected 0.01). (D) Phase enrichment of cycling CCA1-occupied targets. The expression datasets include transcripts that display >0.5 MBPMA in LD conditions [LDHH_ST, entrained in 12-h:12-h LD cycles and constant temperature (HH), data are from the Stitt laboratory (ST)] (39). Genes were grouped by phase and MBPMA score. The ratio of CCA1 occupancy to the number of genes in each group is indicated by intensity of the heat map (red and blue colors indicate maximum and minimum ratios of putative targets in the bin, respectively). (E) Example of a CCA1-occupied target in LD visualized using IGB (55). β-Glucosidase, GBA2 type family protein (AT5G49900), shows a rhythmic expression pattern in two of the previously published LD expression experiments (green) but not in LL (blue). (F) An example of a noncycling CCA1-occupied target in LD and LL, AT1G02620, visualized using IGB (55). Genome annotation for target gene structures is shown below. (G) Percentage of the CCA1-occupied targets in LD for each category of expression in each of the four time series datasets. The upper bars represent the percent of CCA1 targets that match the category: red bars represent morning-phased (ZT22–ZT4) genes, blue bars represent evening-phased (ZT10– ZT16) genes, and gray bars represent noncycling genes (MBPMA <0.80). The lower black bars show the total number of genes matching the respective category in that particular dataset.
Fig. S2.
(A) Genomic distribution of CCA1-occupied sites in LD conditions. Promoter/TSS is defined as the region from −1,000 to +1 from the TSS. (B) Histogram showing distances between the identified peaks of CCA1 occupancy in LD and the TSS of the nearest gene (bin size = 0–500 bp; range, 0–3,000 bp). (C) Overlap of EE occurrence in the region 1-kb upstream of genes closest to CCA1-occupied peaks relative to the genome in LD. (D) Comparison between the distribution of the EE motif (AAAATATCT) and the PBX motif (GGGCCCA) in the CCA1-occupied peaks in LL and LD. (E) Additional motifs enriched in CCA1-occupied targets in LD conditions. (F) Functional enrichment analysis with full GO biological process category of the 1,231 CCA1 targets identified in LD with a peak within 1 kb of the TSS. Circle size is proportional to gene numbers, and the color of each circle represents the enrichment P value for the GO term label on that circle, with orange representing highest enrichment and yellow the lowest enrichment above the cutoff (FDR corrected 0.01). The distance between nodes was arranged manually to optimize readability. The graph was generated using BINGO software (63). (G) Phase enrichment of cycling CCA1-occupied targets. The expression datasets include transcripts that display >0.5 MBPMA in LD conditions [LDHH_SM, entrained in 12-h:12-h LD cycles and constant temperature (HH); data are from the Smith laboratory (SM)] (38). Genes were grouped by phase and MBPMA score. The ratio of CCA1 occupancy to the number of genes in each group is indicated by intensity of the heat map (red and blue colors indicate the maximum and minimum ratios of putative targets in the bin, respectively).
Similar to the putative CCA1 targets in LL, functional analysis of the genes associated with LD peaks included categories for transcriptional regulation, photosynthesis, and responses to stresses (Fig. 2C and Fig. S2F). In addition, the GO analysis of the genes associated with CCA1 peaks in LD conditions also revealed enrichment for genes associated with response to osmotic stress and salinity and with response to metal ions, including cadmium. A link between cadmium and the circadian clock has been observed previously in Arabidopsis and other organisms (40–42).
Comparison of the CCA1-occupied targets in LD conditions with published expression datasets representing two LD time courses again showed enrichment for genes with high MBPMA scores, highlighting groups with robust rhythms and evening-phased expression (Fig. 2D and Fig. S2G). CCA1-occupied sites in LD also are associated with many noncycling genes in LL (Fig. 2 E and F). Many of these targets also were not considered as cycling in any of the four expression time series datasets in LD and LL (Fig. 2G). Specifically, 30% of these high-confidence CCA1 peaks did not appear to be cycling (MBPMA <0.8) in any of the 12-h:12-h conditions examined (1). However, the sequence in the region associated with the peaks of many (33, 40%) of these targets contains a perfectly matched EE (AAATATCT).
CCA1 Occupies Promoters of Morning-Phased Transcripts.
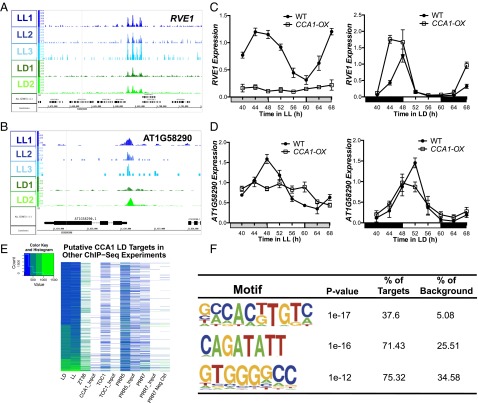
Our analysis of the CCA1 ChIP-Seq data in LL and LD revealed a potentially larger role for CCA1 in the regulation of morning-expressed genes (Fig. 2G). CCA1 previously has been implicated in the control of specific morning-expressed genes, so perhaps the large number of targets with a morning peak of expression should not have been surprising (9, 23). Overall, these morning-expressed CCA1-occupied targets are enriched for a range of biological processes and functions (Fig. S3 A and B). From our top-scoring targets from the LD CCA1 ChIP, we considered those that are morning expressed and have a peak of CCA1 binding within 1 kb of the TSS. Of the CCA1 targets identified in LD that showed strong rhythmic expression in at least one of the four LL or LD time courses (923 transcripts), about 40% showed a peak of expression in the morning (defined as ZT22– ZT4). A similar percent was observed for the CCA1-associated transcripts identified in LL. We selected targets that were in the top-scoring CCA1 peak list in both LL and LD, that were strongly cycling with a peak of expression around dawn in all four time courses, and that contained an EE in the 1-kb upstream region upstream of the TSS.
Fig. S3.
(A) Functional enrichment analysis against GO Slim of the morning-expressed CCA1 targets identified in LD with a peak within 1 kb of the TSS. (B) Functional enrichment analysis with full GO biological process category of the morning-phased CCA1-occupied targets identified in LD. The circle size is proportional to gene numbers, and the color of each circle represents the enrichment P value for the GO term label on that circle, with orange representing the highest enrichment and yellow representing the lowest enrichment above the cutoff (FDR corrected 0.01). The distance between nodes was arranged manually to optimize readability. The graph was generated using BINGO software (63). (C and D) qRT-PCR of RVE1 (C) and HEMA1 (D) transcript levels in wild-type (Col-0) and cca1-1/lhy-21 plants grown in LL after 10 d entrainment in 12-h:12-h LD cycles or collected after growth only in LD.
The genes associated with the top two peak scores that met these criteria were the CCA1 homolog RVE1 and HEMA1, a gene involved in chlorophyll biosynthesis. For both genes, the CCA1 ChIP peak is centered around the EE in the 1-kb upstream region in all LL and LD ChIP experiments (Fig. 3 A and B). In plants grown in LL, overexpression of CCA1 (CCA1-OX) and/or loss of CCA1 expression in combination with the close CCA1 homolog LHY (cca1-1/lhy-21) disrupts the rhythmic expression of these two transcripts, indicating that they indeed are regulated by CCA1 (Fig. 3 C and D and Fig. S3 C and D). In LD conditions, CCA1-OX plants still show a sharp peak in RVE1 expression in the morning, but the peak is slightly wider with an advanced phase (Fig. 3C) in the CCA1-OX plants compared to WT. For HEMA1, the peak of expression also occurs around dawn in CCA1-OX plants (Fig. 3D). These observations suggest that although the expression of both RVE1 and HEMA1 is severely affected by CCA1 overexpression in LL, CCA1 overexpression has a less significant effect in LD, reflecting the complicated nature of the circadian clock. Furthermore, although the effects of CCA1 overexpression on RVE1 and HEMA1 are similar in LL, the differences in effects of CCA1 overexpression on these two targets in LD conditions highlight possible feedback interactions between light signals and other clock components even on direct CCA1 targets. For example, RVE1 is a direct target of CCA1 in our analysis but also has been identified as a direct target of PRR5 and PRR7 (43, 44). This potential for combinatorial regulation by CCA1 and other circadian regulators appears to be abundant. For example, comparison of the CCA1 targets in LD with TOC1, PRR5, and PRR7 ChIP-Seq targets showed that the 500-bp regions surrounding the promoters of many of the CCA1 targets are occupied by these other clock genes as well (Fig. 3E) (43, 44).
Fig. 3.
(A and B) Examples of CCA1-occupied regions in the morning-expressed genes RVE1 (AT5G17300) (A) and HEMA1 (AT1G58290) (B) in LL and LD visualized using IGB (55). Normalized tag counts by location in the genome are shown for each of the three replicate LL ChIP experiments (blue) and for each of the two replicate LD ChIP experiments (green). Genome annotation for RVE1 or HEMA1 gene structures is shown below. (C and D) Real-time qRT-PCR of RVE1 (C) and HEMA1 (D) transcript levels in wild-type (Col-0) and CCA1-OX plants grown in LL after 10 d entrainment in 12-h:12-h LD cycles or only in LD. mRNA levels were normalized to IPP2 and PP2A expression (mean values ± SD, n = 2, two independent experiments. (E) Heatmap comparing the number of reads in this study and published ChIP-Seq experiments for other clock genes: TOC1 (AT5G61380), PRR5 (AT5G24470), and PRR7 (AT5G02810) that map to the 500-bp region surrounding the peaks identified in LD (15, 43, 44). Read counts are normalized to 10 million reads. Normalization factors are LD, 7.48; LL, 3.44 ZT38, 439.75; CCA1_Input, 0.73; TOC1, 6.43; TOC1_Input, 2.24; PRR5, 1.38; PRR5_Input, 5.66; PRR7, 5.10; PRR7_Input, 0.64; and PRR7_Negative control, 2.04. Each row represents a peak in LD, and the number of reads in that peak area in the corresponding ChIP-Seq experiments for each column is indicated from low (blue) to high (green). Read totals less ≤80 are mapped as white. (F) Motifs significantly enriched in CCA1-occupied targets with a morning phase of expression.
Motif analysis of these putative CCA1-regulated morning transcripts indicated that the EE was not enriched above background in this set of targets. Instead, we observed three significant motifs, the first of which shows some similarity to the G-box and E-box motifs, suggesting that CCA1 might interact with basic helix-loop-helix transcription factors to regulate some of these morning-expressed genes (Fig. 3F).
Expression of CCA1 Target Genes Is Disrupted by Overexpression of CCA1.
To determine if CCA1 occupancy plays a functional role in the regulation of the predicted targets, we analyzed target gene mRNA levels by quantitative RT-PCR (qRT-PCR) in wild-type and CCA1-OX plants. As proof of concept, we first validated genes previously identified as direct targets of CCA1. For example, the promoters of the core clock gene TOC1, a recently identified clock component JUMONJI DOMAIN PROTEIN 5 (JMJD5), and three genes involved in reactive oxygen species homeostasis and oxidative stress responses, COLD-REGULATED GENE 27 (COR27), CATALASE 3 (CAT3), and the zinc-finger protein, ZAT12, were all occupied by CCA1 in both LD and LL (Fig. 4A and Fig. S4 A–C) (13, 24, 31, 45). We further examined previously uncharacterized CCA1 targets that contain an EE and are evening expressed. Among others, we observe misregulation of RING MEMBRANE-ANCHOR 1 (RMA1), an uncharacterized protein with possible E3 ubiquitin ligase activity, and DREB2C, a member of the DREB subfamily A-2 of the ERF/AP2 transcription factor family implicated in drought stress responses (Fig. 4 B–E) (46–49). Interestingly, in addition to DREB2C, 39 other transcription factors identified as CCA1 targets in this study recently have been shown to interact with the CCA1 promoter in a yeast one-hybrid assay (Dataset S3) (50). Of the 30,000 accessible genomic loci (or the actual 41,671 loci), our list of CCA1 targets is enriched for the transcription factors that show reciprocal regulation based in the yeast one-hybrid assay (hypergeometric P value <0.01). Finally, we found that the expression of noncycling CCA1 target genes, such as AT1G02620 (a Ras-related small GTP-binding family protein) (shown in Figs. 1 and 2), and ABCG2 (ATP-binding cassette G2, AT2G37360), is altered in CCA1-OX plants in LL (Fig. 4 F and G and Fig. S4D). These results indicate that a functional relationship likely exists between CCA1 and noncycling transcripts and imply a broader role for the clock in the regulation of plant biological functions.
Fig. 4.
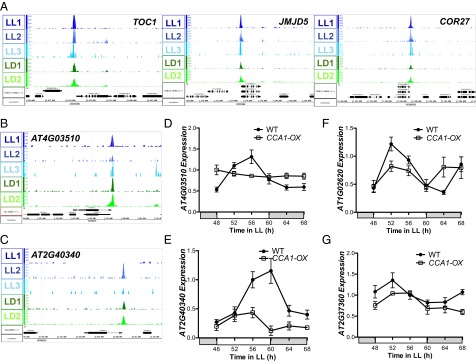
(A–C) CCA1-occupied peaks for the previously identified CCA1 targets TOC1 (AT5G61380), JMJD5 (AT3G20810), and COR27 (AT5G42900) (A) and uncharacterized CCA1 targets RMA1 (AT4G03510) (B) and DREB2C (AT2G40340) (C). Normalized tag counts by location in the genome are shown for each of the three replicate LL ChIP-Seq experiments (blue) and for each of the two replicate LD ChIP-Seq experiments (green). Genome annotation for each gene structure, visualized using IGB (55), is shown below. (D–G) qRT-PCR of wild-type and CCA1-OX plants grown in LL after 10-d entrainment in 12-h:12-h LD cycles for RMA1 (D), DREB2C (E), AT1G02620 (F), and ABCG2 (AT2G37360) (G). mRNA levels were normalized to IPP2 and PP2A expression (mean values ± SD, n = 2, two independent experiments).
Fig. S4.
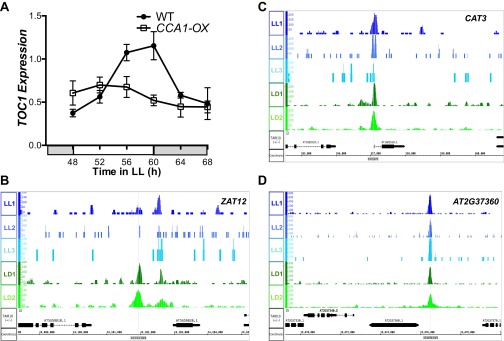
(A) qRT-PCR of TOC1 transcript levels in wild-type (Col-0) and CCA1-OX plants grown in LL after 10 d entrainment in 12-h:12-h LD cycles. mRNA levels were normalized to IPP2 and PP2A expression (mean values ± SD, n = 2, two independent experiments). (B–D) Normalized tag counts by location in the genome for CCA1 targets ZAT12 (AT5G59820) (B), CAT3 (AT1G20620) (C), and AT2G37360 (D) for each of the three replicate LL ChIP experiments (blue) and for each of the two replicate LD ChIP experiments (green). Genome annotation for the respective gene structures is shown below.
Our initial approach was to identify a set of high-confidence peaks that might be direct CCA1 targets to examine conservatively the potential role for this key circadian regulator. However, this stringent analysis misses some previously identified CCA1-regulated targets, such as PRR9, and therefore likely does not include many other real targets (Fig. S5A) (23). Our initial approach was to assign only the nearest TSS as a target of the peak, but we expanded our analysis to include all TSS within 1 kb of the peak, thus allowing more than one target per peak and the inclusion of bidirectional promoters (51, 52). This inclusive target list identified 2,327 TSS within 1 kb of the peaks in LL and 2,459 TSS in LD conditions (Dataset S4). These TSS correspond to 1,725 and 1,919 unique gene models that are within 1 kb of a high-confidence site of CCA1 occupancy in LL and LD, respectively. This inclusive target list identified NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED3 (LNK3) as a CCA1 target, and the severely altered LNK3 transcript level in CCA1-OX and cca1-1/lhy-21 plants in LL confirms that it is indeed regulated by CCA1 (Fig. S5 B–D) (53). We therefore prepared a more inclusive list of CCA1 targets and made the ChIP-Seq data available on the Integrated Genome Browser (IGB) Quickload site (bioviz.org/igb/index.html) (54) and through the AnnoJ browser (neomorph.salk.edu/Kay_CCA1.php) (55) to enable researchers to browse for potential CCA1 peaks near the promoters of their favorite gene (Dataset S4).
Fig. S5.
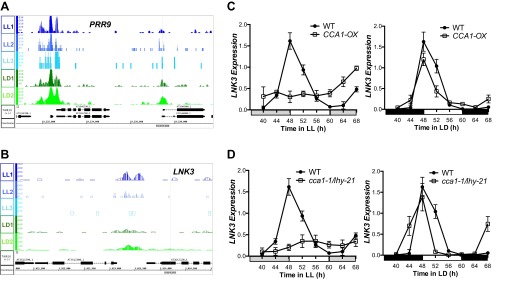
(A and B) Normalized tag counts by location in the genome for CCA1 targets PRR9 (AT2G46790) (A) and LNK3 (AT3G12320) (B) for each of the three replicate LL ChIP experiments (blue) and for each of the two replicate LD ChIP experiments (green). Genome annotation for the respective gene structures is shown below. (C and D) qRT-PCR of LNK3 transcript levels in wild-type (Col-0) (C and D), CCA1-OX (C), and cca1-1/lhy-21 (D) plants grown in LL after 10-d entrainment in 12-h:12-h LD cycles or collected in continuous LD. mRNA levels were normalized to IPP2 and PP2A expression (mean values ± SD, n = 2, two independent experiments).
Discussion
Our analysis and results from these CCA1 ChIP-Seq experiments show that the circadian clock in Arabidopsis plays a much greater role than previously established through examination of experiments in LL and LD. Additionally, we have identified a set of high-confidence sites of CCA1 occupancy that now can be examined for direct links between the circadian clock and known and previously unidentified circadian-regulated outputs. Based on conservative criteria, we identified more than 1,000 potential direct CCA1 targets from both LD and LL experiments that were enriched for a broad range of biological processes. The majority of the genes associated with the top-scoring peaks contained the known CCA1-binding site, the EE, and showed a rhythmic pattern with a peak of expression in the evening in at least one of four LL and LD microarray time series. Interestingly some of the putative CCA1 targets do not pass the cycling criteria in either of the time courses in LL. Therefore, these are CCA1 targets that previously have not been considered as circadian clock-regulated genes in the classic sense.
The ChIP-Seq analysis performed in this study was aligned to the Columbia-0 (Col-0) genome; however the CCA1:CCA1::GFP plants are in the Wassilewskija (Ws) background. We performed a parallel analysis on a genome backbone constructed by replacing the Columbia genome with WS SNPs from the 1,001 genomes project and observed a large increase in the number of reads that mapped to the genome and a relative increase in the number of identified peaks (Dataset S5) (56, 57). Although we focused here on the peaks that were consistent between Col-0 and Ws, future sequencing of the Ws genome or development of a proper reference may identify additional ecotype-specific peaks (58).
We identified CCA1 occupancy at promoters of genes that were noncycling and therefore likely were never included as clock-regulated transcripts in any expression datasets. It is possible that some of these targets cycle in specific tissues but are diluted in the expression datasets, which are based on whole seedlings. Performing a tissue-specific analysis similar to that recently described (59) could help distinguish true noncycling transcripts. Although LL has been the standard condition for identifying clock-controlled genes in plants, this criterion might overlook a subset of circadian-regulated targets, because not all direct targets of the clock necessarily exhibit rhythmic expression in LL. It is also possible that an underlying rhythmic transcription of these genes is masked by changes in mRNA stability (60, 61). In addition, we observe occupied sites specific for the LD conditions, suggesting a possible condition-specific role for CCA1 regulation of select targets. CCA1 occupancy at promoters of morning genes is enriched for motifs other than the EE, suggesting that CCA1 might regulate a large subset of its targets through interaction with other transcription factors. A yeast two-hybrid assay of CCA1 against the recently published Arabidopsis transcription factor library could help refine the mechanism of CCA1 regulation of target genes (50).
A comparison between our list of CCA1 targets and recently published CCA1 promoter interactors showed significant overlap. Interestingly, we observe peaks of CCA1 occupancy in the promoters of both transcription factors and their downstream targets as well as in the promoters of potential regulators of CCA1 expression, suggesting the possibility of both feedback and feed-forward loops between the clock and stress-response pathways. For example, DREB2C, a transcription factor up-regulated during heat stress, has a significant peak of CCA1 occupancy 88 bp upstream of the TSS centered directly above a canonical EE. In addition, CCA1 peaks also are observed at the DREB2C homologs C-repeat binding factor (CBF)1, -2, and -3 as previously reported, and, in fact, CBF1 also interacts with the CCA1 promoter (50). Many of the downstream targets of CBFs/DREB2C, including cold-responsive protein 6.6 (COR6.6/KIN2), COR27, and COR15B, are also occupied by CCA1, further highlighting the complexity and crosstalk between clock-controlled pathways (47).
An often overlooked challenge of ChIP-Seq analysis is the assignment of target genes to identified peaks. The most common approaches, optimized for the mammalian genome through the ENCODE project, have limitations when implemented in other species. One typical approach, applied to the data presented here, is to assign the peak of occupancy for the transcription factor to the nearest TSS. Although in a large genome, such as human or mouse, this approach often provides an unequivocal assignment to a single gene, the peak assignment may be ambiguous in the smaller genome of Arabidopsis. For example, PRR9, a known target of CCA1, did not appear in our high-confidence target list in either LD or LL (23). Examination of the PRR9 genomic region revealed a significant peak centered less than 243 bp upstream of the PRR9 TSS. However, the locus AT2G46787 is located within this region upstream of PRR9, and the peak center is only 27 bp from the putative TSS of this region (Fig. S5A). Another example of a gene that would have been missed by this analysis is LNK3. A significant peak of CCA1 occupancy occurs 356 bp upstream of the LNK3 TSS. However, this peak is also 222 bp upstream of the TSS of AT3g12300 (Fig. S5B). In addition to relaxing the criteria for identifying transcripts associated with peaks of CCA1 occupancy, we also could relax our highly stringent peak-calling criteria to include additional peaks. Therefore, we have made the CCA1 ChIP-Seq data available through the Integrated Genome Browser's Quickload Site (bioviz.org/igb/index.html) (55) so that researchers can examine their regions of interest for peaks that did not meet the strict threshold used for considering peaks in our analysis (Dataset S4).
In summary, the data presented in this study can useful for direct functional characterization of specific target genes that link to key biological processes and can contribute significantly to our understanding of the underlying molecular mechanisms of clock-controlled pathways in Arabidopsis and possibly other plants.
Materials and Methods
Plant Materials.
Arabidopsis thaliana Columbia-0 (Col-0) or Wassilewskija (Ws) ecotypes were used unless otherwise indicated. The CCA1::GFP-CCA1 (Ws), CCA1-OX (Col-0), and cca1-1/lhy-21 (Col-0) lines were described previously (26, 62). Plants were grown on plates containing Murashige and Skoog (MS) medium (Sigma) [1.5% agar (wt/vol) (Sigma)] under 12-h:12-h LD cycles at 22 °C.
RNA Preparation and qRT-PCR.
Seeds were plated on MS plates supplemented with 3% (wt/vol) sucrose, stratified for 2–3 d at 4 °C, and grown in LD conditions at 22 °C for 10 d. For LL experiments, seedlings were transferred to LL for 2 or 3 d. Samples were collected every 4 h in both LD and LL conditions. Total RNA was isolated with the Qiagen RNeasy plant mini kit (Qiagen). cDNA was synthesized using 1 µg of total RNA and was reverse-transcribed with the iScript cDNA synthesis kit (Bio-Rad). Primers used in this study for mRNA expression were TOC1 (5′-AATAGTAATCCAGCGCAATTTTCTTC-3′; 5′-CTTCAATCTACTTTTCTTCGGTGCT-3′); RVE1 (5′-AGAACATGTGGGCTCAAAGAC-3′; 5′-GGTATTACAATCGGCTCTACTGAG-3′); AT1G58290 (5′-TCTCAGCATCGTGGAGTTAAAG-3′; 5′-GTTGACAAATCTCTGAAACTGGG-3′); AT4G03510 (5′-TGGTGATGGT TGGAGAAATGG-3′; 5′-GTCCCTGCGAGATTGTAAGTG-3′); AT1G02620 (5-′CACTGTCCCATTCCTCATCTTA-G3′; 5′-CACTTTGCCTTTACCAGTTGTG-3′); AT2G40340 (5′-CCGTCGGAGATTGTTGACAG-3′; 5′-GGAGGAGGCTTTCGGATTG-3′); AT2G37360 (5′-AAGTCCTTCAGAGAATCGCAC-3′; 5′-GAGGGAGATGTGTTGGTGAG-3′); and LNK3 (5′-CAAAGGCTTTCTATGGTGCTTC-3′; 5′-CCGACACTGTTATTGCTACTGG-3′). As a normalization control, we used isopentenyl-diphosphate delta-isomerase II (IPP2) (5′-GTATGAGTTGCTTCTGGAGCAAAG-3′; 5′-GAGGATGGCTGCAACAAGTGT-3′) and PP2A (5′-TAACGTGGCCAAAATGATGC-3′; 5′-GTTCTCCACAACCGATTGGT-3′). PCR conditions were 95 °C for 3 min followed by 40 cycles at 95 °C for 10 s, 55 °C for 15 s, and 72 °C for 15 s.
ChIP, Library Preparation, Deep Sequencing, and Peak Analysis.
Detailed descriptions of the CCA1 ChIP-Seq procedure and data analysis are provided in SI Materials and Methods. Raw fastq files, normalized bed files, and peak calls for the LL and LD ChIP-Seq experiments have been uploaded to NCBI Gene Expression Omnibus (GEO) and Sequence Read Archive under GEO series GSE70533. Normalized read counts were uploaded to IGB Quickload (bioviz.org/igb/index.html) (55) and the AnnoJ Browser (neomorph.salk.edu/Kay_CCA1.php) (54).
SI Materials and Methods
ChIP, Library Preparation, and Deep Sequencing.
Arabidopsis seedlings for CCA1::GFP-CCA1 were grown on MS plates for 12 d under 12-h:12-h LD cycles at 22 °C. Then seedlings were transferred to LL for 2 d or remained in LD for 2 d. Samples were collected and processed as follows: The detailed procedures of fixation and isolation of chromatin were performed as previously described (64). Fourteen-day-old Arabidopsis seedlings grown on MS plates were treated with 1% formaldehyde under a vacuum for 20 min at room temperature. The cross-linking reaction was stopped by adding glycine to a final concentration of 0.125 M. Seedlings were rinsed five times with water, frozen in liquid nitrogen, and ground to a fine powder. Arabidopsis nuclei were extracted, lysed in Nuclei Lysis Buffer [50 mM Tris⋅HCl (pH 8.0), 10 mM EDTA, 1% SDS, 1 mM PMSF, and Complete protease inhibitor mixture tablets (Roche)], and sonicated using a Biorupter sonicator (Diagenode) to shear the DNA to an average size of 100–300 bp. The chromatin solution was diluted 10-fold with a ChIP dilution buffer [16.7 mM Tris⋅HCl (pH 8.0), 167 mM NaCl, 1.1% Triton X-100, 1.2 mM EDTA]. Immunoprecipitation was performed using Dynabeads Protein G (Invitrogen). The beads were pretreated with either anti-GFP antibody (Invitrogen) or total rabbit IgG (mock control) (Jackson ImmunoResearch) and incubated with the chromatin solution for 1 h at 4 °C. Next the beads were washed at 4 °C, twice with low-salt buffer [20 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 0.2% SDS, 0.5% Triton X-100, 2 mM EDTA], once with high-salt buffer [20 mM Tris⋅HCl (pH 8.0), 500 mM NaCl, 0.2% SDS, 0.5% Triton X-100, 2 mM EDTA], and twice with Tris⋅EDTA. Immunocomplexes were eluted from the beads using elution buffer [50 mM Tris⋅HCl (pH 8.0), 10 mM EDTA, 1% SDS], and cross-linking was reversed by incubation at 65 °C overnight, followed by Proteinase-K treatment for 1 h. DNA was purified by phenol-chloroform extraction and recovered by ethanol precipitation in the presence of 20 μg glycogen. The DNA was resuspended in 60 μL of water. We performed three independent ChIP experiments (biological replicates) for LL samples at ZT26, two for the LD samples at an equivalent time in LD, and one for LL samples at ZT38.
Library preparation.
The immunoprecipitated DNA and input samples were first processed using the MinElute PCR Purification Kit (Qiagen) and were eluted twice in 21.25 µL EB (Qiagen) for a final volume of 34 µL. For samples LL3 and LD2, two immunoprecipitations were pooled; for the remaining samples, a single immunoprecipitation was used for each library preparation. Next, the samples were end repaired using the Paired-End DNA Sample Prep Kit (Illumina) according to the supplied protocol. The samples were incubated at 20 °C for 30 min, purified using the MinElute PCR Purification Kit (Qiagen), and eluted twice in 16 µL EB. ATP was added to the 3′ ends using Klenow (3′→5′ exo minus) (New England BioLabs) as follows: The 32-µL end-repaired DNA was mixed with 5 μL 10× Klenow buffer (NEBuffer 2; New England Biolabs), 10 μL 1 mM dATP, and 3 μL Klenow (3′→5′ exo minus) (5U/μL) and was incubated at 37 °C for 30 min. For the LL1 and ZT38 samples, the DNA was purified using the MinElute PCR Purification Kit (Qiagen) and eluted in 11.25 µL EB. The other samples were purified using Agencourt AMPure XP beads (Beckman Coulter). DA-tailing was performed using Klenow buffer (New England Biolabs), 1 mM dATP (New England Biolabs), and Klenow (New England Biolabs). To ligate adapters to DNA fragments, 10 µL adapters (diluted 1:50) were mixed with 11.25 µL DNA samples and 2.5 µL 10× DNA ligase buffer with ATP (Illumina) (samples LL1 and ZT38) or Quick Ligation Reaction Buffer and quick T4 DNA ligase (the remaining samples) and incubated at 16 °C overnight. After incubation, 25 µL of H2O was added for a final volume of 50 µL. LL1 and ZT38 samples were purified using the MinElute PCR Purification Kit (Qiagen) and were eluted twice in 15 µL EB. For all other samples, 3 uL USER enzyme mix (New England Biolabs) was added after ligation, and samples were incubated at 37 °C for 30 min. The samples were purified using Agencourt AMPure XP beads (Beckman Coulter), and the purification was repeated twice. DNA from samples LL1 and ZT38 was separated and excised from a 2% agarose gel. Bands in the range of 150–300 bp were excised with a new razor blade, purified using the MinElute Gel Extraction Kit (Qiagen), and eluted in 23 µL of EB. For all other samples, size selection was performed using AMPure XP beads (Beckman Coulter) as follows: 80 µL of resuspended AMPure XP beads (Beckman Coulter) were added to 100 µL DNA solution, vortexed, incubated for 5 min, and separated on a magnetic stand for 5 min. Supernatant was transferred to a new tube, and the beads were discarded. To the supernatant 1.2× of resuspended AMPure XP beads (Beckman Coulter) were added, vortexed, allowed to incubate for 5 min, and separated on a magnetic stand. The supernatant was removed and discarded. The beads were washed twice with 200 µL of freshly prepared 80% ethanol, incubated for 30 s, and then removed. Beads were allowed to air dry on the magnetic stand for 5–10 min. This bead-based size selection was performed twice. The first time the DNA was eluted in 100 µL water; the second time the DNA was eluted in 23 µL water. The DNA library was PCR amplified using Phusion High-Fidelity PCR Master mix with HF buffer (New England Biolabs). Indexed primers for Illumina (New England Biolabs) were used to enable multiplexing. DNA (23 µL) was mixed with 1 µL PCR primer 1.1, 1 µL PCR primer 2.1, and 25 µL Phusion DNA polymerase and was amplified using the following PCR protocol: 30 s at 98 °C; 15 cycles of 10 s at 98 °C, 30 s at 65 °C, and 30 s at 72 °C; final extension of 5 min at 72 °C. Samples were purified using the MinElute PCR Purification Kit (Qiagen) and were eluted twice in 10 µL EB. The amplified and purified library was diluted to 1.7 ng/µL. We used the BioAnalyzer DNA high-sensitivity kit (Agilent; www.agilent.com/home) and Fluorometer Qubit (Invitrogen; www.lifetechnologies.com/) to ensure appropriate DNA size and the quality of the library preparation.
Deep sequencing of samples.
Single-end ChIP and input libraries were sequenced up to 50 cycles using the Illumina GAIIx according to the manufacturer’s instructions. Image analysis and base calling were performed with the standard Illumina pipeline. Illumina sequencing-by-synthesis was performed.
ChIP-Seq Analysis.
Initial quality-control analysis was performed using FastQScreen and FastQC (64, 65). This analysis showed that the number of reads in the ZT38 sample was less than adequate for comparison (66). Additionally, at each step in preprocessing, samples were monitored for quality control and improvement by FastQC. Adaptors and barcodes were trimmed and duplicate sequences were removed using TagDust v1.12, Trimmomatic v0.33, and PRINSEQ v0.20.4, sequentially (67–70). Once processed, the reads were aligned using Bowtie, retaining only the best mapping position. We aligned the reads to both the Columbia-0 reference (TAIR9) and a sequence for Ws that we made by replacing the Col-0 sequence with the SNP variants available from the 1,001 genomes project (56, 57). We observed an increase in mapped reads when mapping to Ws; however, annotation for downstream analysis was confounded in the Ws alignment; therefore the results presented here are entirely from the Col-0 mapping. To estimate the number of uniquely mapped reads we used samtools high-quality mapping filter (samtools view –bq1) (Dataset S5) (71). We used HOMER software v4.7 for peak calling and applied the IDR to identify significant peaks (27–29). To apply IDR to HOMER-identified ChIP-Seq peaks, we used the method described by Karmel (30). In summary, we created tag directories for each individual sample, allowing only one tag per base pair to remove any remaining duplicate reads (-tbp 1) and the combined replicates each treatment. We next made peak calls with a very low threshold as required for IDR (findPeaks LL1 -P 0.1 -LP 0.1 -poisson 0.1 -fragLength 100 -style factor -i Input) on the individual samples, combined replicates, individual pseudoreplicates, and combined pseudoreplicates. We then applied the HOMER-IDR program to format the data for the IDR R package to determine the IDR threshold and identify the top peaks above that threshold (29, 30). This output defined our list of top-scoring peaks in LL and LD.
Annotating ChIP Peaks.
Identified peaks were mapped to the genome in two ways. First we used HOMER’s annotatepeaks.pl script (27). To identify not only the nearest TSS but all TSS within 1 kb of the peak, we converted the peak file to an R Genomic Ranges object and extended the peak by 1,000 bp on both sides of the 41,671 annotated loci in Arabidopsis (72). The overlap with TSS from TxDb.Athaliana.BioMart.plantsmart25 were identified using findOverlaps, with the parameters type= any, select= all, and ignore.strand =TRUE. Motifs were identified using HOMER2 v4.7 findMotifsGenome.pl and RGADEM v2.16.0 (27, 73, 74). WebLogos were generated using WebLogo v2.8.2 (75). GO enrichment was performed using BINGO in Cytoscape v3.1.1 using a hypergeometric test with a significance cutoff of 0.01, Benjamini and Hochberg FDR correction (63, 76). Nodes were rearranged manually for optimal position. Enrichments were calculated using the phyper function in R v3.2.0 (77). Heatmaps of peak enrichment were made by first dividing the expression data for all transcripts with an MBPMA >0.5 into bins based on the phase of peak expression and MBPMA score (1). The Z-score of the number of genes that were putative targets of CCA1 in each bin was calculated by comparing the actual number of putative CCA1 targets in each bin with number of putative CCA1 targets in a random sample of the same number of transcripts in the bin. Permutations were performed 10,000 times, and the mean of the putative CCA1 targets and SD for the population was calculated. Data in heatmaps are plotted as a ratio of the putative CCA1 targets in each bin to the total number of genes in the bin. Heatmaps were plotted using heatmap.2 from the gplots package (78). Peak comparisons between LD peaks and published ChIP-Seq data were made by downloading the original fastq files and processing the data as described above to the point of alignment with the genome (15, 43, 44). Tag directories were made for each ChIP using HOMER makeTagDirectory (27). For PRR7 the two PRR7-HA–tagged immunoprecipitations were combined, the two inputs were combined, and the two negative controls were combined. For PRR5 and TOC1 only one replicate was available. The tag counts were normalized to 10 million reads, and the number of counts overlapping the LD peaks was determined for a 500-bp window using HOMER annotatePeaks.pl. Regions with 80 or fewer reads were set to 0. Samples were binned and the displayed using heatmap.2 (78). Bins were set as 0 (white) and then from 80:100, 101:300, 301:500, 501:1,000, and 1,001:1,500 (ranging from light blue to dark green).
Accession Numbers.
The Arabidopsis Information Resource (TAIR10) accession numbers for the gene loci referenced in this manuscript are ABCG2, AT2G37360; β-glucosidase, GBA2 type family protein, AT5G49900; CAT3, AT1G20620; CBF1, AT4G24590; CBF2, AT4G24570; CBF3, AT4G24580; CCA1, AT2G46830; COR6.6/KIN2, AT3G60520; COR15B, AT2G42530; COR27, AT5G42900; DREB2C, AT2G40340; ERD7, AT2G17840; IPP2, AT3G02780; HEMA1, AT1G58290; JMJD5, AT3G20810; LHY, AT1G01060; LNK3, AT3G12320; PP2A, AT1G69960; PRR5, AT5G24470; PRR7, AT5G02810; PRR9, AT2G46790; Ras-related small GTP-binding family protein, AT1G02620; RMA1, AT4G03510; RVE1, AT5G17300; TOC1, AT5G61380; and ZAT12, AT5G59820.
Supplementary Material
Acknowledgments
We thank Sabrina Sanchez for critical reading of the manuscript, members of the S.A.K. laboratory for helpful discussions, Ryan Sartor for assistance and guidance with final formatting, and Ann Loraine and Huaming Chen for assistance uploading the data to IGB and AnnoJ, respectively. The research reported in this publication was supported by Ruth L. Kirschstein National Research Service Award F32GM090375 (to D.H.N.), National Science Foundation Postdoctoral Research Fellowship in Biology Award 0906055 (to C.J.D.), National Institutes of General Medicine of the NIH Awards R01GM056006, R01GM067837, and RC2GM092412 (to S.A.K.), National Science Foundation Award MCB-1024999 (to J.R.E.), and Howard Hughes Medical Institute (HHMI) and Gordon and Betty Moore Foundation (GBMF) Award GBMF3034 (to J.R.E.). J.R.E. is an investigator of the HHMI-GBMF.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database and Sequence Read Archive series GSE70533.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513609112/-/DCSupplemental.
References
- 1.Michael TP, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008;4(2):e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filichkin SA, et al. Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS One. 2011;6(6):e16907. doi: 10.1371/journal.pone.0016907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harmer SL, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290(5499):2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 4.Edwards KD, et al. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell. 2006;18(3):639–650. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carbonell-Bejerano P, et al. Circadian oscillatory transcriptional programs in grapevine ripening fruits. BMC Plant Biol. 2014;14(1):78. doi: 10.1186/1471-2229-14-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Covington MF, Harmer SL. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007;5(8):e222. doi: 10.1371/journal.pbio.0050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fehér B, et al. Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana. Plant J. 2011;67(1):37–48. doi: 10.1111/j.1365-313X.2011.04573.x. [DOI] [PubMed] [Google Scholar]
- 8.Fowler SG, Cook D, Thomashow MF. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005;137(3):961–968. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong MA, Farré EM, Thomashow MF. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc Natl Acad Sci USA. 2011;108(17):7241–7246. doi: 10.1073/pnas.1103741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaffer R, et al. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93(7):1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z-Y, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93(7):1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 12.Strayer C, et al. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289(5480):768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- 13.Alabadí D, et al. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293(5531):880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 14.Gendron JM, et al. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA. 2012;109(8):3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang W, et al. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336(6077):75–79. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- 16.Pokhilko A, et al. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol. 2012;8:574. doi: 10.1038/msb.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruneda-Paz JL, Kay SA. An expanding universe of circadian networks in higher plants. Trends Plant Sci. 2010;15(5):259–265. doi: 10.1016/j.tplants.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagel DH, Kay SA. Complexity in the wiring and regulation of plant circadian networks. Curr Biol. 2012;22(16):R648–R657. doi: 10.1016/j.cub.2012.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClung CR. Wheels within wheels: New transcriptional feedback loops in the Arabidopsis circadian clock. F1000Prime Rep. 2014;6:2. doi: 10.12703/P6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmer SL, Kay SA. Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell. 2005;17(7):1926–1940. doi: 10.1105/tpc.105.033035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9(8):R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60(1):357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 23.Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol. 2005;15(1):47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- 24.Lai AG, et al. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc Natl Acad Sci USA. 2012;109(42):17129–17134. doi: 10.1073/pnas.1209148109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodspeed D, Chehab EW, Min-Venditti A, Braam J, Covington MF. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc Natl Acad Sci USA. 2012;109(12):4674–4677. doi: 10.1073/pnas.1116368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323(5920):1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, Brown JB, Huang H, Bickel PJ. Measuring reproducibility of high-throughput experiments. Ann Appl Stat. 2011;5(3):1752–1779. [Google Scholar]
- 29.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22(9):1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karmel A (2014) homer-idr first-pass. Available at dx.doi.org/. Accessed June 27, 2015.
- 31.Jones MA, et al. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc Natl Acad Sci USA. 2010;107(50):21623–21628. doi: 10.1073/pnas.1014204108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michael TP, et al. A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 2008;6(9):e225. doi: 10.1371/journal.pbio.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang W, et al. Timing of plant immune responses by a central circadian regulator. Nature. 2011;470(7332):110–114. doi: 10.1038/nature09766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang C, et al. Crosstalk between the circadian clock and innate immunity in Arabidopsis. PLoS Pathog. 2013;9(6):e1003370. doi: 10.1371/journal.ppat.1003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou M, et al. Redox rhythm reinforces the circadian clock to gate immune response. Nature. 2015;523:472–476. doi: 10.1038/nature14449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penfield S, Hall A. A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell. 2009;21(6):1722–1732. doi: 10.1105/tpc.108.064022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velez-Ramirez AI, van Ieperen W, Vreugdenhil D, Millenaar FF. Plants under continuous light. Trends Plant Sci. 2011;16(6):310–318. doi: 10.1016/j.tplants.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Smith SM, et al. Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 2004;136(1):2687–2699. doi: 10.1104/pp.104.044347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bläsing OE, et al. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell. 2005;17(12):3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hinrichsen RD, Tran JR. A circadian clock regulates sensitivity to cadmium in Paramecium tetraurelia. Cell Biol Toxicol. 2010;26(4):379–389. doi: 10.1007/s10565-010-9150-x. [DOI] [PubMed] [Google Scholar]
- 41.Maistri S, DalCorso G, Vicentini V, Furini A. Cadmium affects the expression of ELF4, a circadian clock gene in Arabidopsis. Environ Exp Bot. 2011;72(2):115–122. [Google Scholar]
- 42.Jiménez-Ortega V, Cano Barquilla P, Fernández-Mateos P, Cardinali DP, Esquifino AI. Cadmium as an endocrine disruptor: Correlation with anterior pituitary redox and circadian clock mechanisms and prevention by melatonin. Free Radic Biol Med. 2012;53(12):2287–2297. doi: 10.1016/j.freeradbiomed.2012.10.533. [DOI] [PubMed] [Google Scholar]
- 43.Nakamichi N, et al. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci USA. 2012;109(42):17123–17128. doi: 10.1073/pnas.1205156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu T, Carlsson J, Takeuchi T, Newton L, Farré EM. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 2013;76(1):101–114. doi: 10.1111/tpj.12276. [DOI] [PubMed] [Google Scholar]
- 45.Lu SX, et al. The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 2011;155(2):906–915. doi: 10.1104/pp.110.167015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuda N, Suzuki T, Tanaka K, Nakano A. Rma1, a novel type of RING finger protein conserved from Arabidopsis to human, is a membrane-bound ubiquitin ligase. J Cell Sci. 2001;114(Pt 10):1949–1957. doi: 10.1242/jcs.114.10.1949. [DOI] [PubMed] [Google Scholar]
- 47.Lim CJ, et al. Over-expression of the Arabidopsis DRE/CRT-binding transcription factor DREB2C enhances thermotolerance. Biochem Biophys Res Commun. 2007;362(2):431–436. doi: 10.1016/j.bbrc.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 48.Chen H, et al. Arabidopsis DREB2C functions as a transcriptional activator of HsfA3 during the heat stress response. Biochem Biophys Res Commun. 2010;401(2):238–244. doi: 10.1016/j.bbrc.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 49.Hwang JE, et al. Overexpression of Arabidopsis dehydration- responsive element-binding protein 2C confers tolerance to oxidative stress. Mol Cells. 2012;33(2):135–140. doi: 10.1007/s10059-012-2188-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pruneda-Paz JL, et al. A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell Reports. 2014;8(2):622–632. doi: 10.1016/j.celrep.2014.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Q, et al. Searching for bidirectional promoters in Arabidopsis thaliana. BMC Bioinformatics. 2009;10(Suppl 1):S29. doi: 10.1186/1471-2105-10-S1-S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kourmpetli S, et al. Bidirectional promoters in seed development and related hormone/stress responses. BMC Plant Biol. 2013;13:187. doi: 10.1186/1471-2229-13-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rugnone ML, et al. LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc Natl Acad Sci USA. 2013;110(29):12120–12125. doi: 10.1073/pnas.1302170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133(3):523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25(20):2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gan X, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477(7365):419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmitz RJ, et al. Patterns of population epigenomic diversity. Nature. 2013;495(7440):193–198. doi: 10.1038/nature11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneeberger K, et al. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc Natl Acad Sci USA. 2011;108(25):10249–10254. doi: 10.1073/pnas.1107739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Endo M, Shimizu H, Nohales MA, Araki T, Kay SA. Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature. 2014;515(7527):419–422. doi: 10.1038/nature13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagel DH, Pruneda-Paz JL, Kay SA. FBH1 affects warm temperature responses in the Arabidopsis circadian clock. Proc Natl Acad Sci USA. 2014;111(40):14595–14600. doi: 10.1073/pnas.1416666111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 64. Andrews S Babraham Bioinformatics FastQ Screen. Available at www.bioinformatics.babraham.ac.uk/projects/fastq_screen/. Accessed June 27, 2015.
- 65. Andrews S (2013) FastQC: A quality control tool for high throughput sequence data. Available at www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed June 27, 2015.
- 66.Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318(5848):261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lassmann T, Hayashizaki Y, Daub CO. TagDust–a program to eliminate artifacts from next generation sequencing data. Bioinformatics. 2009;25(21):2839–2840. doi: 10.1093/bioinformatics/btp527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lassmann T. TagDust2: A generic method to extract reads from sequencing data. BMC Bioinformatics. 2015;16:24. doi: 10.1186/s12859-015-0454-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAM tools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lawrence M, et al. Software for computing and annotating genomic ranges. PLOS Comput Biol. 2013;9(8):e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Carlson M TxDb.Athaliana.BioMart.plantsmart25: Annotation package for TxDb object(s). Available at www.bioconductor.org/packages/release/data/annotation/html/TxDb.Athaliana.BioMart.plantsmart25.html. Accessed June 20, 2015.
- 74.Droit A, Gottardo R, Roberston G, Li L. 2014. rGADEM: De novo motif discovery. Available at www.bioconductor.org/packages/release/bioc/html/rGADEM.html. Accessed June 25, 2015.
- 75.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. RDC Team (2010) R: A Language and Environment for Statistical Computing (The R Project for Statistical Computing, Vienna) Available at www.R-project.org. Accessed June 25, 2015.
- 78.Warnes GR, et al. 2015 gplots: Various R Programming Tools for Plotting Data Available at cran.r-project.org/web/packages/gplots/index.html. Accessed June 25, 2015.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.