Significance
Ribosome biogenesis requires a number of assembly factors. However, the exact mechanism of this process remains largely unknown. We could successfully reconstitute the 50S subunit from the 45S precursor by a 2′-O-methylation of U2552 (Um2552) mediated by rRNA methyltransferase RlmE, in the presence of the wash fraction from crude ribosomes. To our knowledge, this experiment is the first demonstration of enzymatic formation of a ribosomal subunit in vitro from its precursor via the action of an assembly factor. Mechanistic studies revealed that RlmE-mediated Um2552 formation promotes interdomain interactions of 23S rRNA, in concert with the incorporation of L36, triggering late steps of 50S subunit assembly. RlmE and Um2552 are conserved in other organisms, including human, indicating the functional importance of this process in ribosome biogenesis.
Keywords: ribosome assembly, post-transcriptional modification, rRNA methyltransferase, RlmE, L36
Abstract
Ribosome biogenesis requires multiple assembly factors. In Escherichia coli, deletion of RlmE, the methyltransferase responsible for the 2′-O-methyluridine modification at position 2552 (Um2552) in helix 92 of the 23S rRNA, results in slow growth and accumulation of the 45S particle. We demonstrate that the 45S particle that accumulates in ΔrlmE is a genuine precursor that can be assembled into the 50S subunit. Indeed, 50S formation from the 45S precursor could be promoted by RlmE-mediated Um2552 formation in vitro. Ribosomal protein L36 (encoded by rpmJ) was completely absent from the 45S precursor in ΔrlmE, and we observed a strong genetic interaction between rlmE and rpmJ. Structural probing of 23S rRNA and high-salt stripping of 45S components revealed that RlmE-mediated methylation promotes interdomain interactions via the association between helices 92 and 71, stabilized by the single 2′-O-methylation of Um2552, in concert with the incorporation of L36, triggering late steps of 50S subunit assembly.
Ribosomes are essential ribonucleoprotein complexes that translate the genetic information encoded by mRNA into protein. Intact bacterial ribosomes, which have a sedimentation coefficient of 70S, consist of large (50S) and small (30S) subunits. Each subunit can be reconstituted in vitro from its individual component ribosomal RNAs and proteins (1–5), indicating that these components contain all of the information necessary to automatically assemble into functional subunits. Because the intermediate particles observed in an in vitro reconstitution of the subunits are similar to those observed in vivo, assembly maps that describe the order of ribosomal protein binding in vitro are useful tools for understanding ribosome biogenesis in the cell (6). However, assembly is slower in vitro than in vivo, and nonphysiological conditions, such as high-salt concentration and high temperature, are required for in vitro assembly.
In the cell, ribosome assembly is coordinated with the transcription and processing of large precursor rRNAs encoded by the rrn operon (3, 7). Once transcription starts, nascent transcripts rapidly form local secondary and tertiary structures that serve as binding sites for the primary ribosomal proteins, thereby initiating ribosome assembly. However, hierarchical assembly starting at the 5′-terminal domains of the 16S and 23S rRNAs is not essential for ribosome formation in the cell (8) because nonribosomal factors, termed “assembly factors,” facilitate rapid and efficient assembly of ribosomal particles. Ribosome biogenesis is an energy-consuming process in which a number of assembly factors, including RNA helicases, GTPases, protein chaperones, and rRNA-modifying enzymes, support efficient assembly of each subunit (6, 9, 10). Individual mutation or deletion of several assembly factors causes accumulation of immature 50S subunits that sediment at 40S or 45S. For example, the RNA helicase SrmB participates in the early stage of 50S assembly (11, 12); deletion of srmB results in accumulation of a 40S particle lacking L13, one of the primary binders that initiates 50S assembly in vitro (13). Deletion of the RNA helicase DeaD also results in accumulation of a 40S particle; however, none of the primary binders are missing from this particle, indicating that DeaD acts later than SrmB during 50S assembly (9). The 40S particle that accumulates in ΔdeaD is a genuine precursor of 50S, rather than a dead-end particle (14). DbpA, another RNA helicase, is also involved in 50S assembly. Overexpression of the R331A active-site mutant of DbpA in Escherichia coli results in accumulation of a particle that sediments at 45S at low Mg2+ concentration, but increases in density and comigrates with the 50S subunit at high Mg2+ concentration (15), suggesting that the 45S particle is structurally flexible and can undergo conformational changes upon binding of Mg2+. A similar 45S particle that undergoes Mg2+-dependent alteration in density appears in cells expressing the G80E/D85N mutant of ObgE (CgtAE) (16), an essential GTPase involved in the late steps of 50S assembly (10).
Some rRNA-modifying enzymes play critical roles in ribosome assembly. RlmE (RrmJ, FtsJ) is an S-adenosylmethionine (AdoMet)-dependent methyltransferase that catalyzes 2′-O-methylation of uridine at position 2552 (Um2552) in the A-loop (Helix 92) of 23S rRNA (Fig. 1 A and B) (17). RlmE is highly conserved in all domains of life, as well as in organelles, indicating the functional importance of this enzyme. Um2552 is situated at 5′ adjacent to G2553 (Fig. 1B), which is an essential base that anchors the CCA terminus of A-site tRNA in the 50S subunit (18). The absence of a 2′-O-methyl group at Um2552 in the rlmE deletion strain of E. coli (ΔrlmE) decreases the rate of programmed frameshifting and read-through of stop codons (19), indicating that Um2552 is required for optimization of translational accuracy and thus allows natural recoding events.
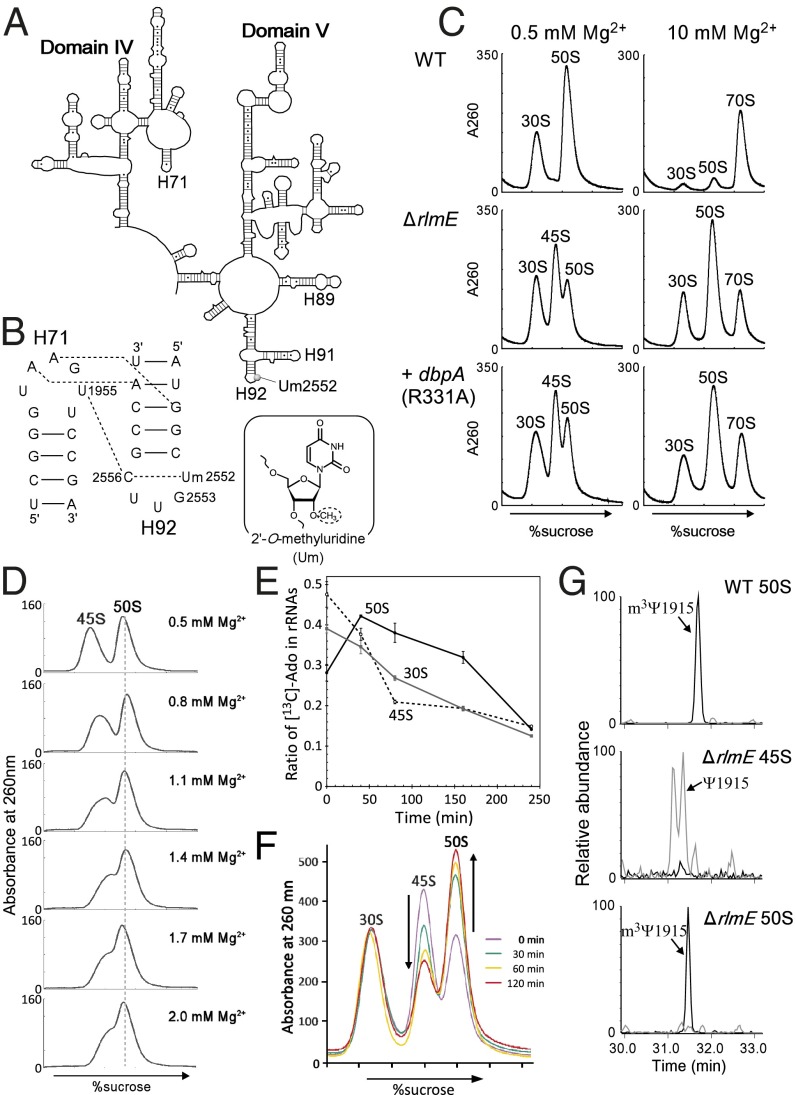
Fig. 1.
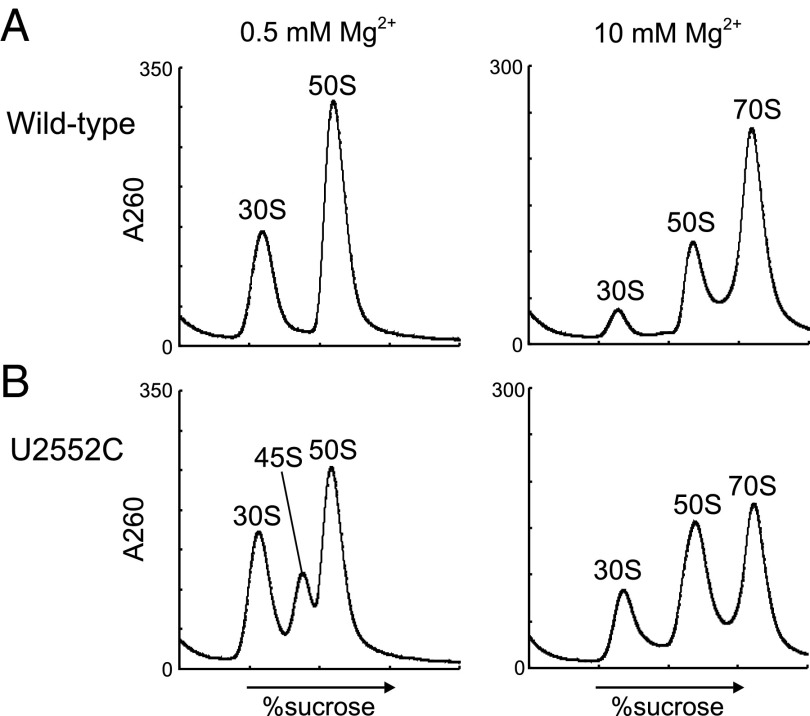
The 45S particle is an authentic assembly intermediate. (A) Secondary structure of E. coli 23S rRNA domains IV and V. (B, Left) interactions between helices 92 and 71, indicated by dotted lines (24). (Right) The chemical structure of 2′-O-methyluridine. (C) Sucrose density gradient (SDG) profiles of ribosomes from WT (Top), ΔrlmE (Middle), and DbpA R331A-overexpressing (Bottom) strains, performed at low Mg2+ concentration (Left) and high Mg2+ concentration (Right). All strains were cultured at 22 °C. Throughout the figures, the y axis unit for SDG analyses is milli-absorbance units at 260 nm. (D) SDG analyses of the 45S particle at various Mg2+ concentrations (0.5–2.0 mM). Purified 45S particle and the 50S subunit were mixed and subjected to SDG centrifugation at the indicated Mg2+ concentrations. (E) Monitoring ratio of 13C-labled Ado in rRNAs from 30S, 45S, and 50S in E. coli ΔrlmE strain. E. coli ΔrlmE strain labeled with [13C] Ado was transferred to LB medium with nonlabeled Ado and cultured for several hours; the 13C-to-12C ratio of Ado in rRNAs from 30S, 45S, and 50S was monitored by mass spectrometry. Averaged values of three independent experiments with SD values are shown. (F) Ribosome profiles in ΔrlmE after inhibition of transcription. Aliquots of cultures were taken at various time points, after addition of rifampicin, and then analyzed by SDG at 0.5 mM Mg2+. SDG profiles at 0 min (violet), 30 min (green), 60 min (yellow), and 120 min (red) are overlaid. (G) Absence of 70S-specific m3Ψ1915 modification in 45S particle. Mass-spectrometric analysis of RNase T1-digested 23S rRNAs obtained from WT 50S (Top), ΔrlmE 45S (Middle), and ΔrlmE 50S (Bottom). Mass chromatograms produced by integration of multiple-charged negative ions of m3Ψ1915-containing fragment (ΨAACm3ΨAΨAACGp; m/z 1774.24, 1182.49, 886.62, and 709.09; black lines) and unmethylated fragment (ΨAACΨAΨAACGp; m/z 1767.23, 1177.82, 883.11, and 706.29; gray lines) are shown.
Deletion of rlmE resulted in a growth defect and accumulation of a 45S particle, especially at low temperature (Fig. 1C) (20). The particle was similar to that observed in E. coli cells overexpressing mutants of DbpA (Fig. 1C) (15) or ObgE (16). It remains unknown, however, whether this 45S particle is able to form an active 50S subunit (21). The cold-sensitive phenotype, along with accumulation of the 45S particle, was alleviated by overexpression of the obgE or engA GTPase (22), indicating that RlmE is an assembly factor that acts at a late step of 50S formation. However, the mechanism by which RlmE-mediated Um2552 formation facilitates late 50S assembly remains unknown.
In this study, we demonstrate that the 45S particle that accumulates in ΔrlmE is a genuine precursor of the 50S subunit. Furthermore, we partially recapitulate the formation of 50S from 45S in the presence of recombinant RlmE and AdoMet in vitro. Genetic and biochemical studies suggested that the Um2552 formation by RlmE facilitates interdomain interactions between H92 and H71, in concert with the incorporation of L36 and other ribosomal proteins, leading to correct placement of the central protuberance and other domains during late steps of 50S assembly.
Results
The 45S Particle Is an Assembly Intermediate of the 50S Subunit.
In sucrose density gradient (SDG) centrifugation analysis (Fig. 1C), the 45S particle in ΔrlmE cultured at low temperature (22 °C) sedimented between 30S and 50S at a low Mg2+ concentration (0.5 mM), as previously reported (20), but increased its sedimentation coefficient to comigrate with the 50S subunit at a high Mg2+ concentration (10 mM); thus, the 45S particle is unable to associate with the 30S subunit to form the 70S ribosome, even at 10 mM Mg2+. As noted above, a similar particle that also exhibits Mg2+-dependent alteration of density appears in cells overexpressing the R331A mutant of DbpA (Fig. 1C) (15). To characterize the physical properties of the 45S particle of ΔrlmE, we used SDG centrifugation to analyze the 45S particle together with the 50S subunit at various Mg2+ concentrations (Fig. 1D). The particle gradually increased its density in response to the increasing Mg2+ concentration and almost overlapped with the 50S subunit at 2.0 mM Mg2+, indicating that the 45S particle is structurally flexible and can alter its conformation consecutively in response to the Mg2+ concentration. The result clearly ruled out the possibility that the precursor has two distinct structural conformers bearing 45S and 50S.
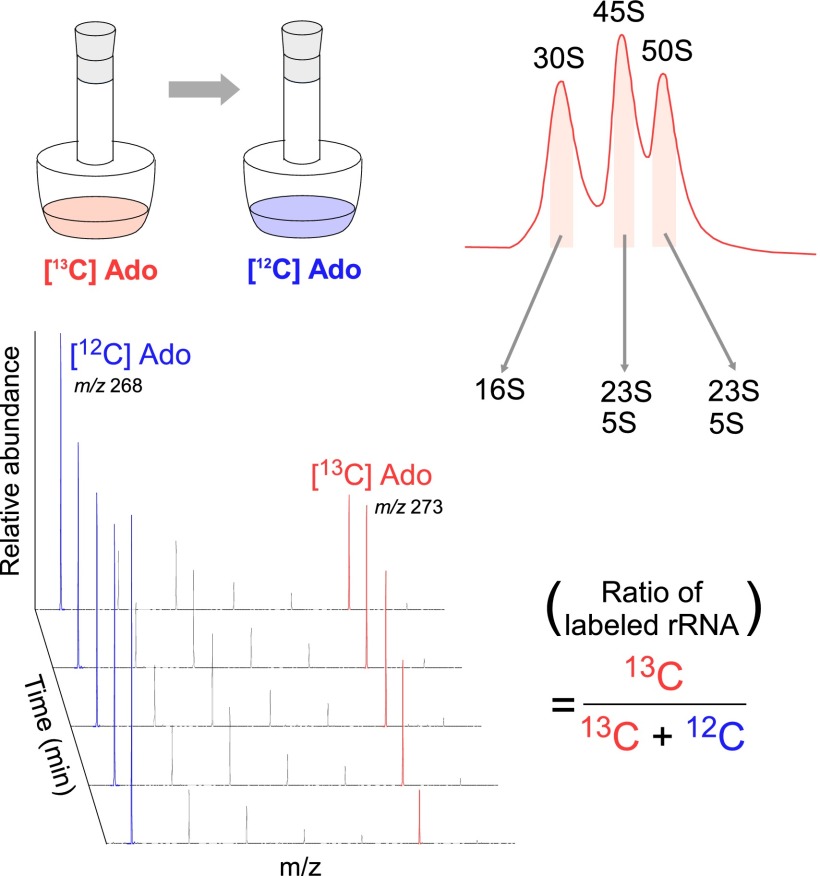
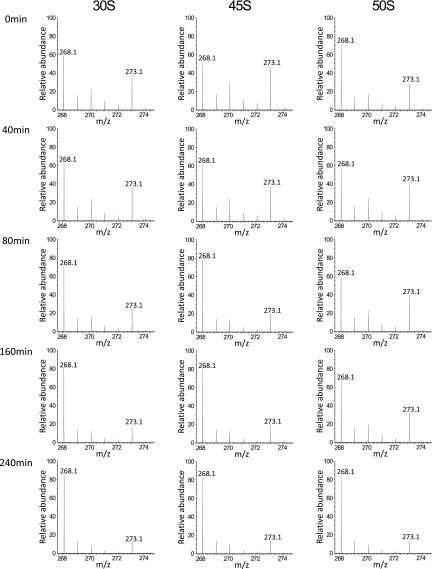
The 45S particle of the ΔrlmE strain has been speculated to be a dead end or breakdown product, rather than a bona fide precursor, of the 50S subunit (21). To investigate whether the 45S particle in ΔrlmE is an authentic precursor of the 50S subunit, we cultured the E. coli ΔrlmE strain in the presence of [13C] adenosine (Ado), to label rRNAs with 13C-Ado, and then cultured the cells for several hours in LB medium with nonlabeled Ado. We then used mass spectrometry to monitor the 13C-to-12C ratio of Ado in rRNAs from 30S, 45S, and 50S (Figs. S1 and S2). As shown in Fig. 1E, the proportion of 13C-labeled 16S rRNA in 30S decreased moderately over time at a constant speed in normal medium because newly synthesized 16S rRNA composed of 12C-Ado constantly replaced 13C-labeled 16S rRNA. On the other hand, 13C-labeled rRNAs in 50S clearly increased during the first 40 min after cultivation in normal medium, followed by a moderate decrease, as newly synthesized rRNA replaced 13C-labeled rRNA. Consequently, the proportion of 13C-labeled rRNAs in 45S plunged during the first 80 min. If the 45S particle is not a precursor for 50S, 13C-rRNA in 50S should not increase in normal medium in this experiment. Moreover, we monitored ribosome profiles in ΔrlmE over a time course after blocking transcription with rifampicin (Fig. 1F). In SDG analysis performed at 0.5 mM Mg2+, the level of 45S particle gradually decreased, and about half of the particle was converted to 50S within 120 min. In SDG analysis performed at 10 mM Mg2+, the 70S peaks increased over time whereas the 50S peak decreased proportionally (Fig. S3). Taken together, these results clearly demonstrate that the 45S particle is not a dead-end product; instead, at least half of this 45S particle is a genuine precursor that can be assembled into a 50S subunit, which, in turn, can productively associate with the 30S subunit to form the 70S ribosome.
Fig. S1.
Schematic depiction of monitoring biogenesis of ribosomal subunits using stable isotope labeling of rRNAs. E. coli ΔrlmE/Δrna strain was cultured in a medium containing [13C] Ado and then transferred to a medium containing nonlabeled Ado and cultured for several hours to chase the 13C-to-12C ratio of Ado in rRNAs from 30S, 45S, and 50S by mass spectrometry.
Fig. S2.
Mass spectrometric analysis of stable isotope-labeled adenosines from ribosomal subunits. Mass spectra for proton-adduct of [12C]Ado (m/z 268) and [13C] Ado (m/z 273) from rRNAs of 30S, 45S, and 50S at different period after chasing with nonlabeled Ado.
Fig. S3.
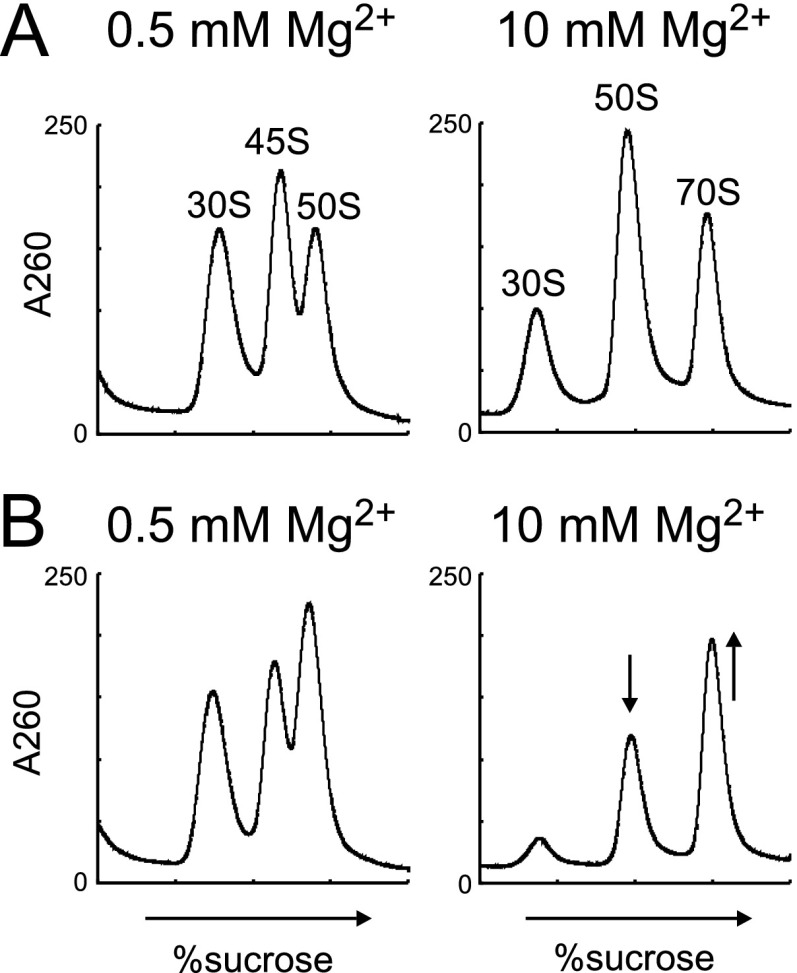
Ribosome profiles in ΔrlmE after inhibition of transcription. Aliquots of cultures were taken at 0 min (A) and 60 min (B), after addition of rifampicin, and then analyzed by SDG at 0.5 mM (Left) and 10 mM (Right) Mg2+ concentrations.
The RlmH methyltransferase, which is responsible for 3-methylpseudouridine (m3Ψ) formation at position 1915 (m3Ψ) in 23S rRNA, employs 70S ribosome as a substrate (23). Therefore, if the 45S particle of ΔrlmE were a breakdown product of mature 50S subunits, m3Ψ1915 would be present in 23S rRNA of the 45S particle. We analyzed the posttranscriptional modification of 23S rRNAs obtained from the 45S particle and 50S subunit of ΔrlmE cells. The extracted 23S rRNA was digested by RNase T1 and analyzed by capillary liquid chromatography (LC)/nanoelectrospray ionization mass spectrometry (MS). In 50S subunits from both the WT and ΔrlmE, only the m3Ψ1915-containing fragment (ΨAAC m3ΨAΨAACGp) was detected, and very little unmethylated fragment was observed (Fig. 1G). By contrast, in the 45S particle of ΔrlmE, a large fraction of the corresponding fragment was unmethylated (Fig. 1G). Because m3Ψ1915 is the sole methylation site in this fragment, we can conclude that position 1915 is unmethylated in the 45S particle. It is reasonable to attribute the small fraction of methylated fragments to contaminating 50S subunits. Thus, the 45S particle is a genuine precursor, rather than a breakdown product, of the mature 50S subunit. Therefore, we will refer to this particle as the “45S precursor.”
Enzymatic Formation of the 50S Subunit from the 45S Precursor by RlmE-Mediated Um2552 Methylation.
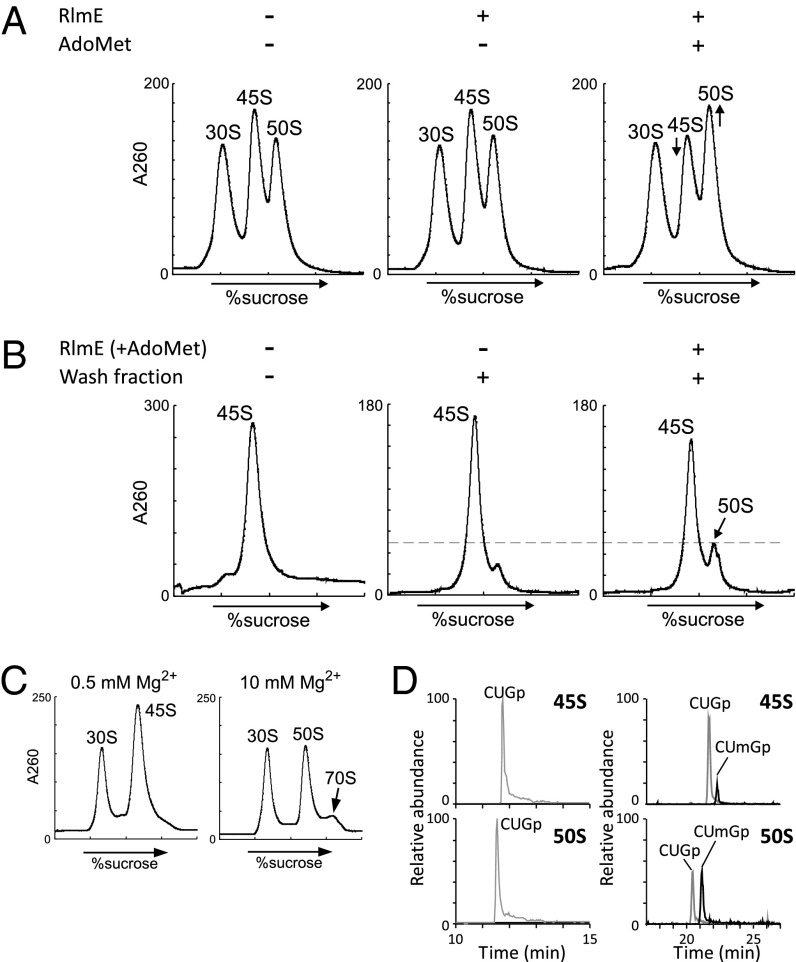
To investigate whether RlmE-mediated 2′-O-methylation of U2552 facilitates 50S assembly, we tried to generate the 50S subunit from the 45S precursor in vitro in the presence of recombinant RlmE and AdoMet. Initially, we used crude ribosomes obtained from ΔrlmE as a substrate for this experiment because many components required for 50S assembly, including missing ribosome proteins and assembly factors, are likely to be present in the crude ribosome fraction. The crude ribosomes were incubated with recombinant RlmE, in the presence or absence of AdoMet, and subjected to SDG analysis at 0.5 mM Mg2+ (Fig. 2A). The 50S peak slightly increased, concomitant with a decrease in the 45S peak, only in the presence of both RlmE and AdoMet, whereas the ribosome profile changed very little in the absence of AdoMet. In total, about 20% of the 45S precursor was converted to 50S subunits. Under these conditions, we also observed an increase in the 70S peak relative to the 50S and 30S peaks in SDG analysis at 10 mM Mg2+ (Fig. S4A). Thus, we proved that the newly synthesized 50S could productively associate with the 30S subunit to form the 70S ribosome. We next analyzed the 2′-O-methylation status of U2552 in 23S rRNA from both 45S and 50S particles after reconstitution. LC/MS analysis detected the RNA trinucleotide C2551–G2553 in 23S rRNA from 45S and 50S particles (Fig. S4B). Without incubation with RlmE and AdoMet, only unmethylated trimer (CUGp) was detected in both 45S and 50S. After incubation with RlmE and AdoMet, however, the methylated trimer (CUmGp) was clearly detected along with the unmethylated trimer (CUGp) (Fig. S4B). Approximately 50% of 23S rRNA in the 45S was methylated whereas 70% of 23S rRNA from the 50S was methylated.
Fig. 2.
Enzymatic formation of 50S subunit from the 45S precursor in vitro. (A) Partial formation of 50S subunit in the crude ribosomes. SDG profiles of ribosomal fractions from ΔrlmE, performed at 0.5 mM Mg2+ concentration after incubation at 37 °C for 2.5 h in the presence or absence of recombinant RlmE and AdoMet. (B) Partial formation of 50S subunit from the 45S precursor. SDG profiles of the mixtures at 0.5 mM Mg2+ concentration after incubation at 37 °C for 2.5 h in the presence or absence of RlmE, AdoMet, and the wash fraction from crude ribosomes. (C) Ability of the newly synthesized 50S subunit from the 45S precursor to associate with the 30S subunit. The 45S precursor was methylated by RlmE in the presence of AdoMet and the wash fraction, followed by addition of the 30S subunit and incubation at 37 °C for 30 min. SDG profiling was carried out at 10 mM Mg2+ to detect the 70S ribosome. (D) Mass-spectrometric analysis of Um2552 formation in the reconstituted 50S. The mass chromatograms represent CUmGp (m/z 987.13; black line) and CUGp (m/z 973.12; gray line) in the RNA segment G2529–C2579 of 23S rRNAs from the 45S precursor (Upper) and the reconstituted 50S subunit (Lower).
Fig. S4.
Partial enzymatic formation of the 50S subunit from the 45S precursor. (A) SDG profiles of ribosomal fractions from ΔrlmE, performed at 10 mM Mg2+ concentration after incubation at 37 °C for 2.5 h in the presence or absence of recombinant RlmE and AdoMet. (B) Mass spectrometric analysis of Um2552 formation in the 45S and 50S, which were treated with (Right) or without (Left) RlmE and AdoMet. The mass chromatograms represent CUmGp (m/z 987.13; black line) and CUGp (m/z 973.12; gray line) in the RNA segment G2529–C2579 of 23S rRNAs from the 45S precursor (Upper) and the 50S subunit (Lower). (C) Partial formation of the 50S subunit from the 45S precursor. SDG profiles of the mixtures at 0.5 mM Mg2+ concentration after incubation at 37 °C for 2.5 h in the presence or absence of RlmE, AdoMet, and the wash fraction from crude ribosomes.
Furthermore, we attempted to enzymatically reconstitute the 50S subunit in vitro from the isolated 45S particle. To this end, the isolated 45S particle was incubated with RlmE and AdoMet in the presence or absence of the wash fraction from the crude ribosomes. We could detect small amounts of 50S, which were increased by RlmE-mediated Um2552 formation in the presence of the wash fraction (Fig. 2B and Fig. S4C). About 25% of the 45S precursor was converted to 50S subunits. Moreover, the reconstituted 50S was able to associate with the 30S subunit to form the 70S ribosome (Fig. 2C). LC/MS analysis of the RNA trinucleotide C2551–G2553 in 23S rRNA clearly showed that Um2552 was concentrated in the reconstituted 50S relative to the 45S particle (Fig. 2D). Um2552 was found in ∼50% and 20% of 23S rRNAs in the 50S and 45S particles, respectively. These data clearly rule out the possibility that RlmE methylates the 50S subunit to facilitate 70S formation. Taken together, these observations demonstrate that Um2552 is indeed formed in the 45S precursor and that methylated 45S is subsequently converted into the 50S subunit.
The U2552C Mutation Causes Accumulation of the 45S Precursor.
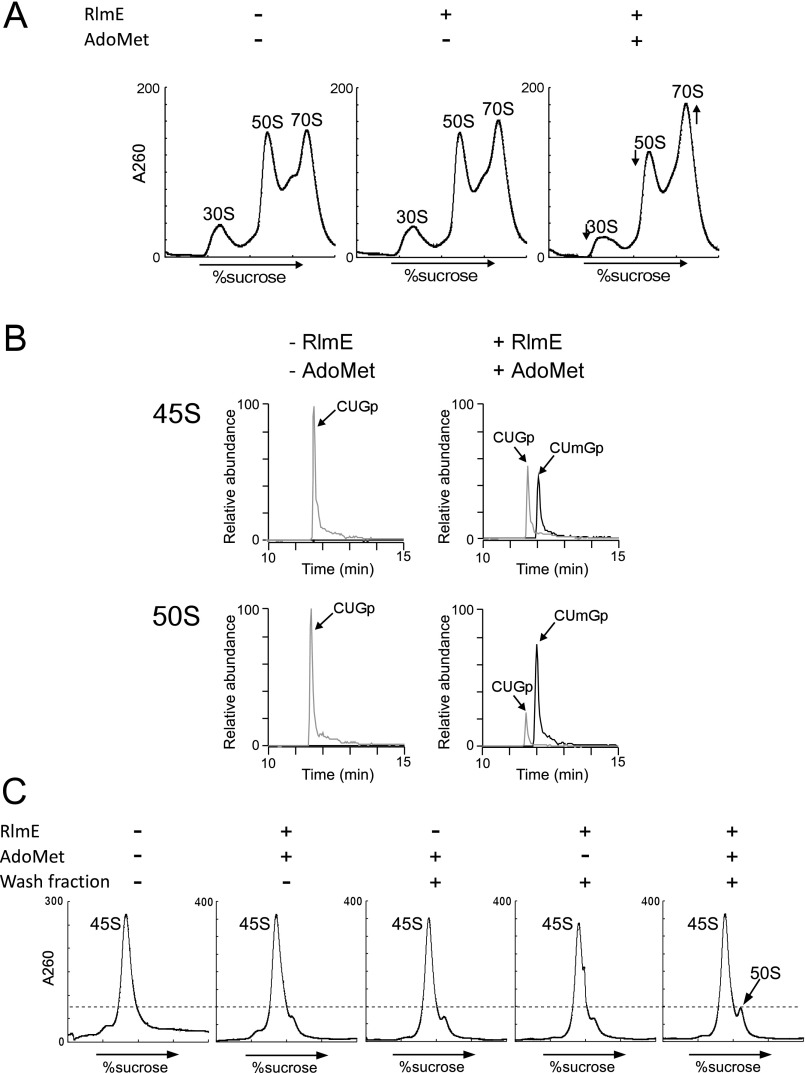
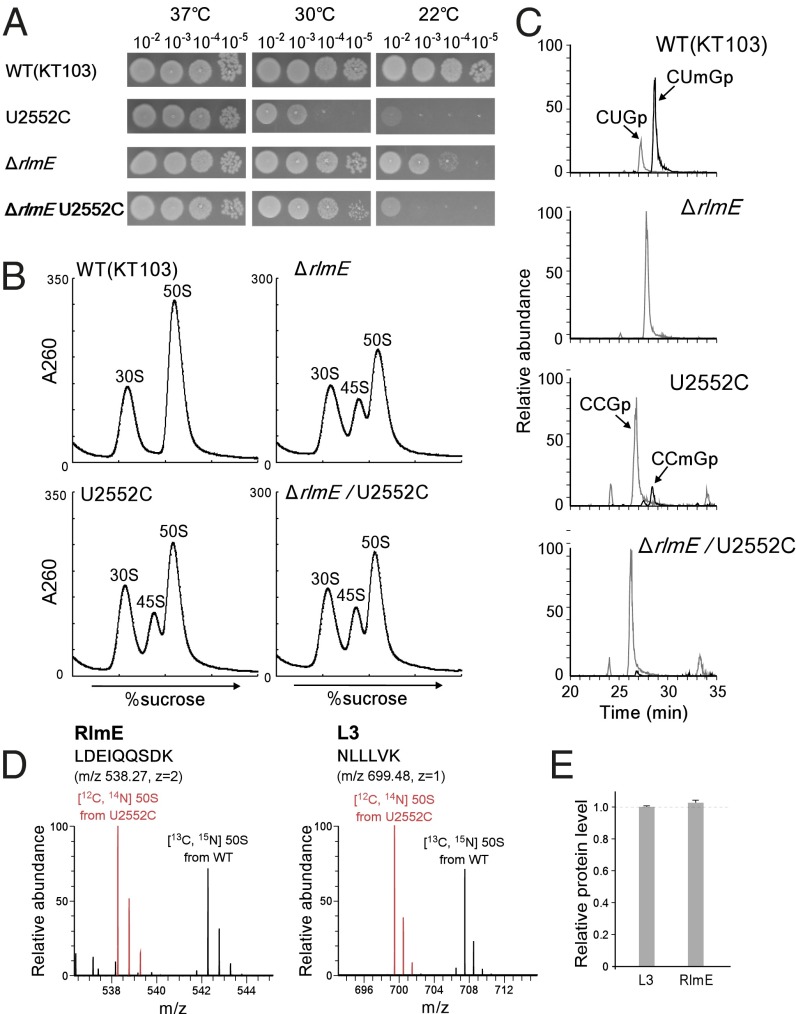
Next, we sought to clarify whether RlmE, acting as an assembly factor, transiently interacts with the 45S precursor to promote 50S assembly, or whether instead the Um2552 modification is itself required for 50S assembly. To address this issue, we generated the U2552C mutant of 23S rRNA, which is not predicted to be a substrate of RlmE. In the crystal structure of the 50S subunit, Um2552 forms a base-triple with C2556 and U1955 (Fig. 1B; and see Fig. 6A) (24–26). The C2 carbonyl group of Um2552 is involved in this interaction (see Fig. 6A); the U-to-C replacement at position 2552 does not affect this interaction because a cytosine base also has a C2 carbonyl group. In addition, the U2552C mutation does not affect peptidyl-transferase activity in vitro (18). To express the mutant rRNA, a plasmid harboring the rrnB operon with the U2552C mutation was introduced into an E. coli Δ7 prrn strain (8, 27) in which all seven chromosomal rrn operons were disrupted; consequently, all ribosomes in this strain were replaced with the U2552C mutant ribosomes. As in ΔrlmE, the U2552C mutant exhibited a cold-sensitive phenotype, but much slower growth even at 30 °C (Fig. 3A). The ribosome profile revealed that the 45S precursor accumulated in the U2552C mutant (Fig. 3B), and we confirmed that the accumulated 45S precursor increased in density and overlapped with the 50S subunit in SDG analysis performed in 10 mM Mg2+ (Fig. S5). The level of 45S accumulation in the U2552C strain is not so different from that in ΔrlmE (Fig. 3B); nevertheless, the U2552C strain grows much slower than ΔrlmE (Fig. 3A). This observation might be explained by low translational fidelity caused by the U2252C mutation that prevents 2′-O-methylation (19), in addition to the retarded 50S assembly by this mutation. We also made a double-mutant strain, ΔrlmE/U2552C, by deleting rlmE in the U2552C strain; the resultant strain was also cold-sensitive (Fig. 3A) and accumulated the 45S precursor (Fig. 3B). However, for unknown reasons, this strain grew better than the U2552C mutant (Fig. 3A). It is possible that the U2552C mutation modulates the configuration of RlmE on the ribosome, causing some adverse impact on ribosome biogenesis.
Fig. 6.
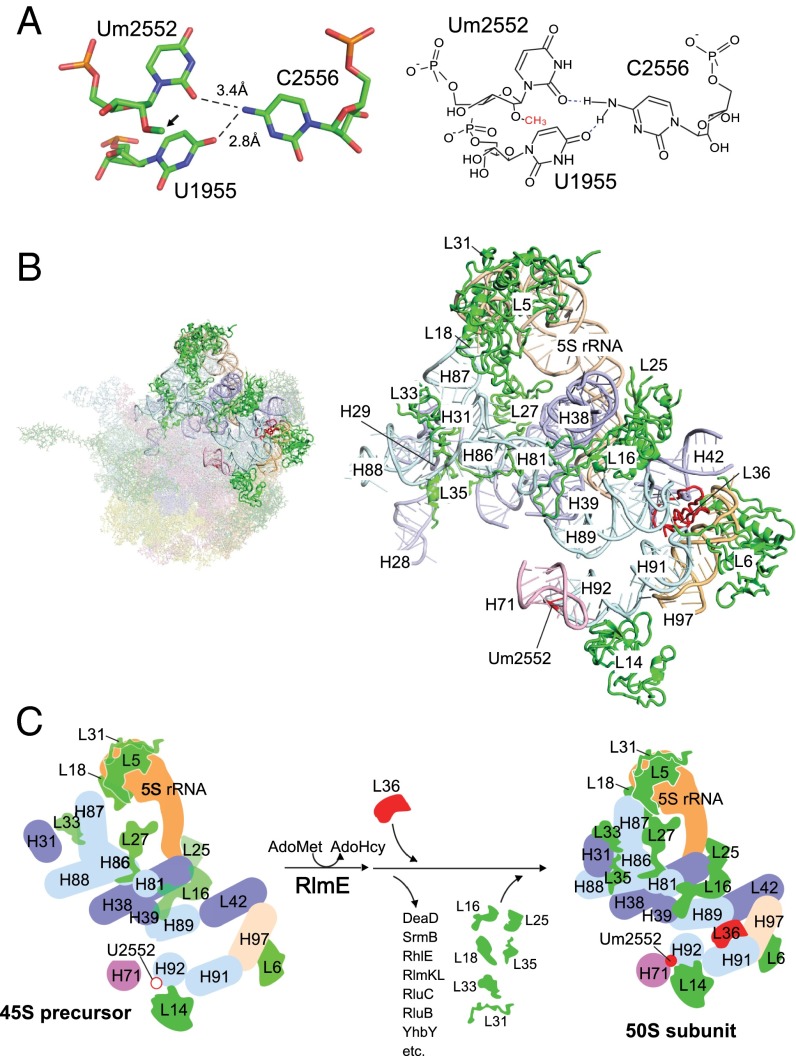
Structural insights into late steps of the 50S subunit assembly triggered by Um2552 formation. (A) Tertiary structure of the Um2552–C2556–U1955 triad (Left). Coordinates were obtained from 4YBB (26). The corresponding chemical structure is shown Right. Hydrogen bonds are shown as dashed lines. The 2′O methyl group is indicated by an arrow. (B) Tertiary structure of the area including helices 71 and 92, L36, L16, and the central protuberance. (Left) The entire structure of the 50S subunit (2AW4) (25); the bold colors indicate the area shown Right. For L31, coordinates were obtained from 2J01 (50). Um2552 and L36 are shown in red. Color codes for the domains are the same as in Fig. 5. Ribosomal proteins are shown in green. (C) Mechanistic model for late assembly of the 50S subunit from the 45S precursor triggered by Um2552 formation. RlmE-mediated formation of Um2552 (filled red circle) promotes the H92–H71 interaction, which facilitates incorporation of L36. During this process, many assembly factors attached to the 45S precursor are dissociated. In addition, many late binders, including L16, L18, L25, L31, L33, and L35, assemble to stabilize the local structure of 23S rRNA, interdomain interactions, and the central protuberance. AdoHcy, S-adenosyl-l-homocysteine.
Fig. 3.
Characterization of the U2552C mutant. (A) Growth properties of a series of KT103 strains. Overnight full-growth cultures of WT (Top), U2552C mutant (Second Panels), ΔrlmE (Third Panels), and ΔrlmE/U2552C (Bottom) were serially diluted (102- to 105-fold dilutions), spotted onto LB plates, and cultivated at 37 °C for 18 h (Left), 30 °C for 24 h (Middle), or 22 °C for 50 h (Right). (B) SDG profile of ribosomes from a series of KT103-derived strains: WT KT103 (Upper Left), U2552C mutant (Lower Left), ΔrlmE (Upper Right), and ΔrlmE/U2552C (Lower Right) were cultured at 37 °C until OD600 reached ∼0.4, cultivated at 22 °C for 100 min, and then analyzed by SDG at low Mg2+ concentration. (C) Mass-spectrometric analysis of 2′-O-methylation at position 2552 in 23S rRNAs from a series of KT103-derived strains: WT (Top), ΔrlmE (Second Panel), U2552C mutant (Third Panel), and ΔrlmE/U2552C (Bottom). The mass chromatograms represent CUmGp (m/z 987.13; black line), CUGp (m/z 973.12; gray line), CCmGp (m/z 986.15; black line), and CCGp (m/z 972.13; gray line) from the RNA segment G2529–C2579 of the 23S rRNAs. (D) SILAC quantitation comparing the level of RlmE bound to U2552C 50S relative to WT 50S. Left and Right panels show mass spectra of tryptic peptides from RlmE and L3, respectively. Red and black signals represent protonated ions of unlabeled peptides from the U2552C mutant 50S and [13C6, 15N2] lysine-labeled corresponding peptides from WT 50S, respectively. Sequences, mass-to-charge ratio (m/z), and charge state (z) of the peptides are indicated. (E) Level of RlmE bound to U2552C 50S relative to WT 50S. The intensity ratio of RlmE-derived peptides between U2552C and WT 50S was normalized to that of L3-derived peptides. Average values with SD of the intensity ratios of three tryptic peptides of RlmE and L3 (Table S1) were obtained.
Fig. S5.
Accumulation of the 45S precursor in the U2552C mutant. SDG profiles of ribosomes from WT (A) and U2552C mutant (B) at low Mg2+ concentration (Left) or high Mg2+ concentration (Right).
We next analyzed 2′-O-methylation at position 2552 in the U2552C mutant. In the WT strain (KT103) (8), ∼75% of 23S rRNA was 2′-O-methylated at position 2552 (Fig. 3C). No methylation was observed in ΔrlmE or ΔrlmE/U2552C, which were used as negative controls. In the U2552C mutant, only 15% of 23S rRNA was 2′-O-methylated (Fig. 3C). Because RlmE had a weak ability to methylate C2552, the U2552C mutation efficiently abolished 2′-O-methylation at position 2552 without disruption of rlmE. It remained possible, however, that the U2552C mutation hindered RlmE binding to H92 in the 23S rRNA. Because RlmE can bind to the 50S subunit (20), we measured the relative abundance of RlmE attached to the mutant and WT 50S subunits by stable isotope labeling by amino acids in cell culture (SILAC) (28). The tryptic digests were analyzed by LC/MS to detect specific peptides containing C-terminal Lys residues from RlmE and L3 (Table S1). The unlabeled peptide from mutant 50S and the fully labeled corresponding peptide from WT 50S (with 8-Da higher mass due to the presence of [13C, 15N] Lys) were eluted simultaneously from LC and detected in the same mass spectra (Fig. 3D). The intensity ratio of these peptides was normalized against that of L3-derived peptides. The results revealed that the same amounts of RlmE were attached to the mutant and WT 50S subunits (Fig. 3E); thus, accumulation of 45S precursor in the U2552C mutant is not due to the loss of ability to bind RlmE, but instead to the lack of 2′-O-methylation at position 2552, indicating that the Um2552 modification is directly involved in 50S assembly.
Table S1.
Sequences and molecular masses of tryptic peptides from ribosomal proteins and RlmE used for SILAC analyses
| Ribosomal protein | Peptides | m/z | ||||
| Sequence | Position | Mass, Da | z | Nonlabeled | Labeled | |
| L3 | NLLLVK | 185–190 | 698.47 | 1 | 699.48 | 707.50 |
| AIQVTTGAK | 47–55 | 887.51 | 2 | 444.76 | 448.77 | |
| VDVTGTSK | 107–114 | 805.42 | 2 | 403.72 | 407.73 | |
| L5 | LHDYYK | 4–9 | 837.40 | 1 | 838.41 | 846.43 |
| SVAGFK | 73–78 | 607.33 | 1 | 608.34 | 616.36 | |
| GLSAK | 116–120 | 474.28 | 1 | 475.29 | 483.30 | |
| L6 | APVVVPAGVDVK | 7–18 | 1,149.68 | 2 | 575.85 | 579.85 |
| INGQVITIK | 19–27 | 984.60 | 2 | 493.31 | 497.31 | |
| TLNDAVEVK | 36–44 | 987.52 | 2 | 494.77 | 498.78 | |
| L7/12 | DLVESAPAALK | 86–96 | 1,112.61 | 2 | 557.31 | 561.32 |
| ALEEAGAEVEVK | 110–121 | 1,243.63 | 2 | 622.82 | 626.83 | |
| DDAEALK | 102–108 | 760.36 | 1 | 761.37 | 769.38 | |
| L9 | RAELEAK | 51–57 | 815.45 | 2 | 408.73 | 412.74 |
| NFLVPQGK | 28–35 | 901.52 | 1 | 902.51 | 910.53 | |
| VANLGSLGDQVNVK | 9–22 | 1,412.76 | 2 | 707.39 | 711.40 | |
| L10 | ALNLQDK | 2–8 | 800.44 | 2 | 401.23 | 405.23 |
| QAIVAEVSEVAK | 9–20 | 1,242.68 | 2 | 622.35 | 626.36 | |
| GVTVDK | 32–37 | 617.34 | 1 | 618.35 | 626.36 | |
| L11 | SFTFVTK | 66–72 | 828.44 | 1 | 829.45 | 837.46 |
| AQLQEIAQTK | 104–113 | 1,128.61 | 2 | 565.32 | 569.33 | |
| TPPAAVLLK | 73–81 | 908.57 | 2 | 455.29 | 459.30 | |
| L13 | DWYVVDATGK | 14–23 | 1,152.55 | 2 | 577.28 | 581.29 |
| VYYHHTGHIGGIK | 73–85 | 1,480.76 | 2 | 741.39 | 745.40 | |
| VIEIAVK | 100–106 | 770.49 | 2 | 386.25 | 390.26 | |
| L14 | YAGVGDIIK | 32–40 | 934.51 | 1 | 935.53 | 943.54 |
| GDVLK | 55–59 | 530.31 | 1 | 531.32 | 539.33 | |
| KGDVLK | 54–59 | 658.40 | 1 | 659.41 | 675.44 | |
| L15 | LNTLSPAEGSK | 3–13 | 1,115.58 | 2 | 558.80 | 562.81 |
| AAIEAAGGK | 133–141 | 786.42 | 2 | 394.22 | 398.23 | |
| GIGSGLGK | 22–29 | 687.39 | 1 | 688.40 | 696.41 | |
| L16 | GLAQGTDVSFGSFGLK | 19–34 | 1,582.80 | 2 | 792.41 | 796.41 |
| GNVEYWVALIQPGK | 87–100 | 1,572.83 | 2 | 787.42 | 791.43 | |
| TTFVTK | 128–133 | 695.39 | 1 | 696.39 | 704.41 | |
| L17 | VVEPLTITLAK | 47–57 | 1,182.72 | 2 | 541.85 | 545.85 |
| TRDNEIVAK | 70–78 | 1,044.56 | 2 | 523.78 | 527.74 | |
| DNEIVAK | 72–78 | 787.41 | 1 | 788.42 | 796.43 | |
| L18 | DAAAAVGK | 69–76 | 701.37 | 1 | 702.38 | 710.39 |
| AIAEQLK | 57–63 | 771.45 | 2 | 386.73 | 390.74 | |
| KLQELGATR | 17–25 | 1,014.58 | 2 | 508.30 | 512.31 | |
| L19 | SNIIK | 2–6 | 573.35 | 1 | 574.36 | 582.37 |
| VWVVEGSK | 30–37 | 902.49 | 1 | 903.50 | 911.51 | |
| QLEQEQMK | 7–14 | 1,032.49 | 2 | 517.25 | 521.26 | |
| L20 | FINGLK | 79–84 | 690.41 | 1 | 692.40 | 700.41 |
| VAFTALVEK | 104–112 | 976.56 | 1 | 977.57 | 985.59 | |
| QNGISYSK | 71–78 | 895.44 | 2 | 449.22 | 453.23 | |
| L21 | MYAVFQSGGK | 1–10 | 1,086.52 | 2 | 544.27 | 548.27 |
| IGVPFVDGGVIK | 49–60 | 1,199.69 | 2 | 600.85 | 604.86 | |
| QWFTDVK | 91–97 | 922.45 | 2 | 462.24 | 466.24 | |
| L22 | AAVLVK | 43–48 | 599.40 | 1 | 600.41 | 608.42 |
| IFVDEGPSMK | 74–83 | 1,121.54 | 2 | 561.78 | 565.79 | |
| KAAVLVK | 42–48 | 727.50 | 1 | 728.50 | 744.53 | |
| L23 | SNTIVLK | 27–33 | 773.46 | 2 | 387.74 | 391.75 |
| DATKAEIK | 37–44 | 874.48 | 2 | 438.25 | 446.27 | |
| APHVSEK | 13–19 | 766.40 | 2 | 384.21 | 388.21 | |
| L24 | DDEVIVLTGK | 8–17 | 1,087.58 | 2 | 544.80 | 548.80 |
| NVLSSGK | 27–33 | 703.39 | 1 | 704.40 | 712.41 | |
| VIVEGINLVK | 34–43 | 1,082.67 | 2 | 542.34 | 546.35 | |
| L25 | EAPLAIELDHDK | 35–46 | 1,349.68 | 2 | 675.85 | 679.86 |
| FPAIIYGGK | 26–34 | 964.54 | 2 | 483.27 | 487.28 | |
| VMNMQAK | 47–53 | 820.39 | 2 | 411.20 | 415.21 | |
| L27 | DHTLFAK | 56–62 | 830.43 | 2 | 416.22 | 420.23 |
| L28 | SHALNATK | 19–26 | 840.45 | 2 | 421.23 | 425.24 |
| FWVESEK | 38–44 | 923.44 | 2 | 462.73 | 466.74 | |
| ARGEKY | 73–78 | 722.37 | 2 | 362.19 | 366.20 | |
| L34 | LTVSK | 42–46 | 546.34 | 1 | 547.35 | 555.36 |
| L35 | HILTK | 31–35 | 610.38 | 1 | 611.39 | 619.40 |
| RlmE | LDEIQQSDK | 39–47 | 1,074.52 | 2 | 538.27 | 542.28 |
| DVLAPGGSFVVK | 153–164 | 1,187.66 | 2 | 594.83 | 598.84 | |
| VRKPDSSR | 187–194 | 943.52 | 2 | 472.77 | 476.78 | |
RlmE Contributes to 50S Assembly in Concert with the Incorporation of L36.
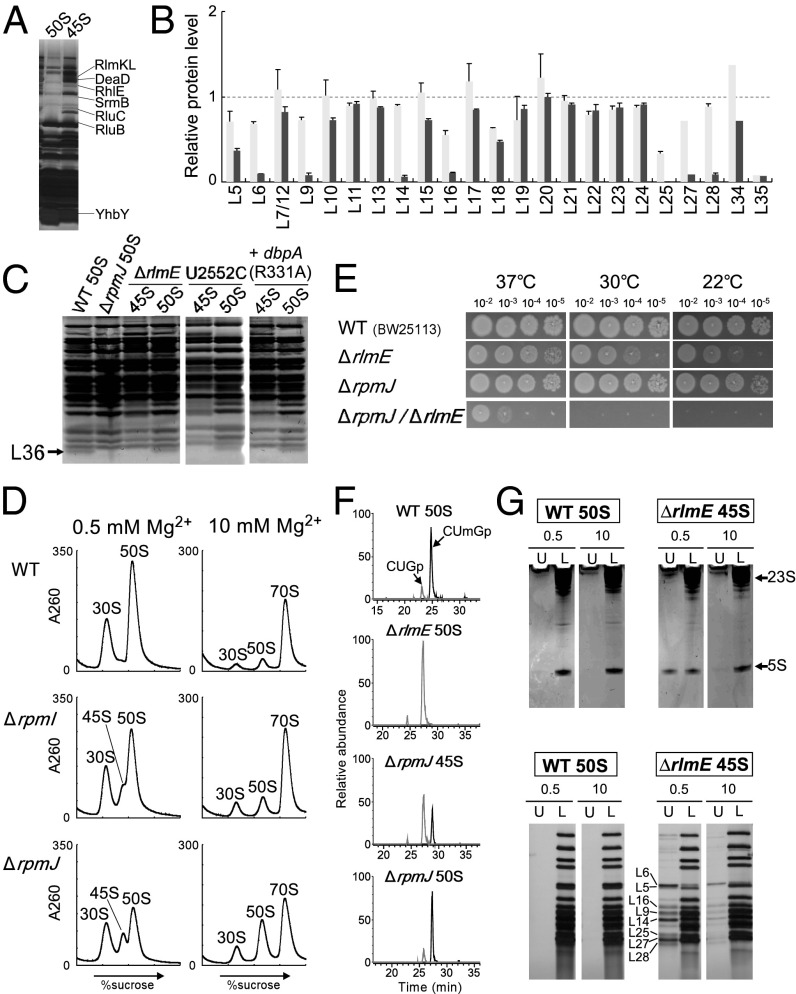
We next analyzed the protein components of the 45S precursor that accumulated in ΔrlmE cells. Comparison of the components to those in the 50S subunit revealed several additional bands in the 45S precursor, which we analyzed by peptide mass fingerprinting (Fig. 4A). The additional proteins were factors responsible for 50S assembly: three RNA helicases, DeaD, SrmB, and RhlE; three rRNA-modifying enzymes, RlmKL, RluB, and RluC; and one assembly factor, YhbY. This result also supports the idea that the 45S precursor of ΔrlmE is an authentic assembly intermediate of the 50S subunit. Possible roles of these assembly factors are discussed in SI Discussion.
Fig. 4.
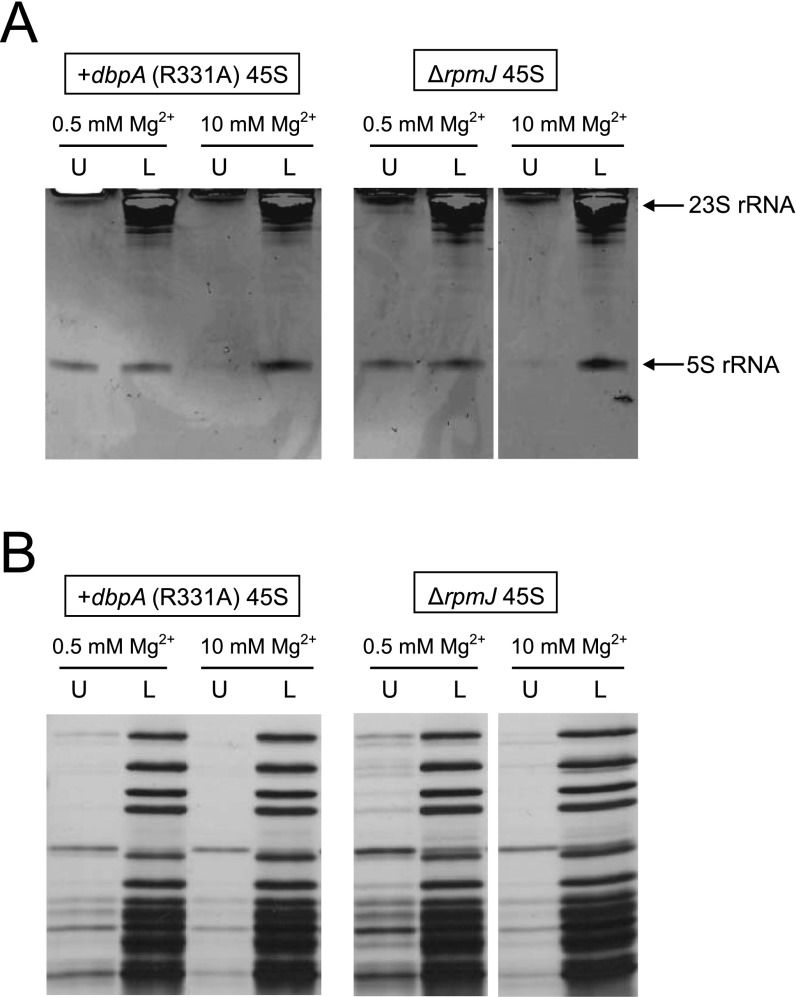
Involvement of L36 in RlmE-mediated late assembly. (A) Identification of assembly factors bound to the 45S precursor accumulated in ΔrlmE. Protein components of the 45S precursor were dissolved by SDS/PAGE. Each band was identified by peptide mass fingerprinting. (B) Relative level of ribosomal proteins in the 45S precursor versus WT 50S subunit treated (black bars) or untreated (gray bars) with high-salt solution. Relative levels of 23 ribosomal proteins were analyzed by the SILAC method. Tryptic peptides used for this analysis are listed in Table S1. The graph shows the averaged values with SD of the intensity ratios of three tryptic peptides for each protein. In the case of L27, L34, and L35 (Table S1), only one peptide was detected and available for this analysis. (C) Ribosomal proteins from the 45S precursor and the 50S subunit were resolved by urea/Tris-Tricine SDS 18% PAGE. The gel was stained with SYPRO Ruby Protein stain. Black arrow denotes L36. The 45S precursor and 50S subunit from ΔrlmE, the U2552C mutant, and a DbpA R331A-overexpressing strain were analyzed. WT and ΔrpmJ 50S subunits were used as controls. (D) SDG profiles of ribosomes from WT (Top), ΔrpmI (Middle), and ΔrpmJ (Bottom), performed at low Mg2+ concentration (Left) or high Mg2+ concentration (Right). All strains were cultured at 22 °C. (E) Severe growth defect of ΔrlmE/ΔrpmJ. Overnight saturated cultures of WT(BW25113) (Top), ΔrlmE (Second Panels), ΔrpmJ (Third Panels), and ΔrlmE/ΔrpmJ (Bottom) were serially diluted (102- to 105-fold dilutions), spotted onto LB plates, and cultivated at 37 °C for 18 h (Left), 30 °C for 24 h (Middle), or 22 °C for 51 h (Right). (F) Mass-spectrometric analysis of 2′-O-methylation at position 2552 in 23S rRNA from the indicated strains; WT 50S (Top), ΔrlmE 50S (Second), ΔrpmJ 45S (Third), and ΔrpmJ 50S (Bottom). The mass chromatograms represent CUmGp (m/z 987.13; black line) and CUGp (m/z 973.12; gray line) from the RNA segment G2529–C2579 of the 23S rRNA. (G) High-salt stripping analysis of the 45S precursor. (Upper) The result of denaturing 10% PAGE analyses of rRNAs from the 50S subunit and the 45S precursor in the upper fraction (U) and lower fraction (L), after high-salt stripping at low (0.5 mM) or high (10 mM) Mg2+ concentration. The gels were stained by ethidium bromide. (Lower) The result of 15% SDS/PAGE analyses of ribosomal proteins from the 50S subunit and 45S precursor in the upper fraction (U) and lower fraction (L), after high-salt stripping at low (0.5 mM) or high (10 mM) Mg2+ concentration. The gels were silver-stained.
Previous work demonstrated that the 45S particle of ΔrlmE lacks some ribosomal proteins, including L5, L16, L18, L25, L27, L28, L30, L33, and L35 (15, 29). We used the SILAC method to measure the abundance of ribosomal proteins in the 45S precursor relative to their abundance in the 50S subunit. Because the stoichiometries of L1, L2, L3, and L4 were unchanged (15, 29), we focused on ribosomal proteins smaller than L5. We subjected 45S and 50S to tryptic digestion and analyzed the digests by LC/MS to detect and quantitate specific peptides (Table S1) from 23 species of ribosomal proteins. We confirmed that L35 was significantly less abundant in the 45S precursor (Fig. 4B), as reported (15). In addition, the levels of L5, L6, L16, L18, L19, L25, and L27 were reduced in the 45S precursor (Fig. 4B). Although this result was basically consistent with previous reports (15, 29), we did not observe any reduction in L28. Because L28 was easily stripped off under the high-salt condition (Fig. 4B), indicating weak association of this protein, this discrepancy might be explained by differences in sample preparation. In addition, we analyzed small proteins by urea/Tris-tricine PAGE (Fig. 4C). L36, which is the smallest protein in the 50S subunit and can barely be detected by MS, was not detectable in the 45S precursor. As a control, we analyzed the 50S subunit from ΔrpmJ and confirmed that L36 was not detectable (Fig. 4C). No L36 was present in the 45S precursors accumulated in the U2552C mutant or in a strain overexpressing the DbpA R331A mutant (Fig. 4C), indicating that 45S precursors from these mutants shared similar characteristics.
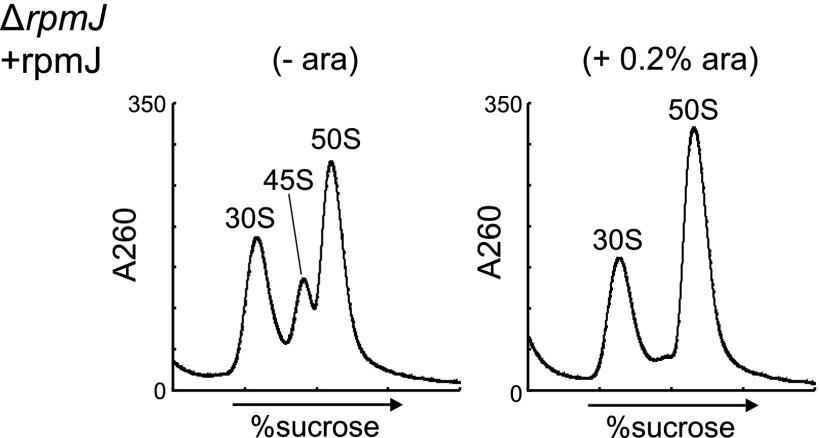
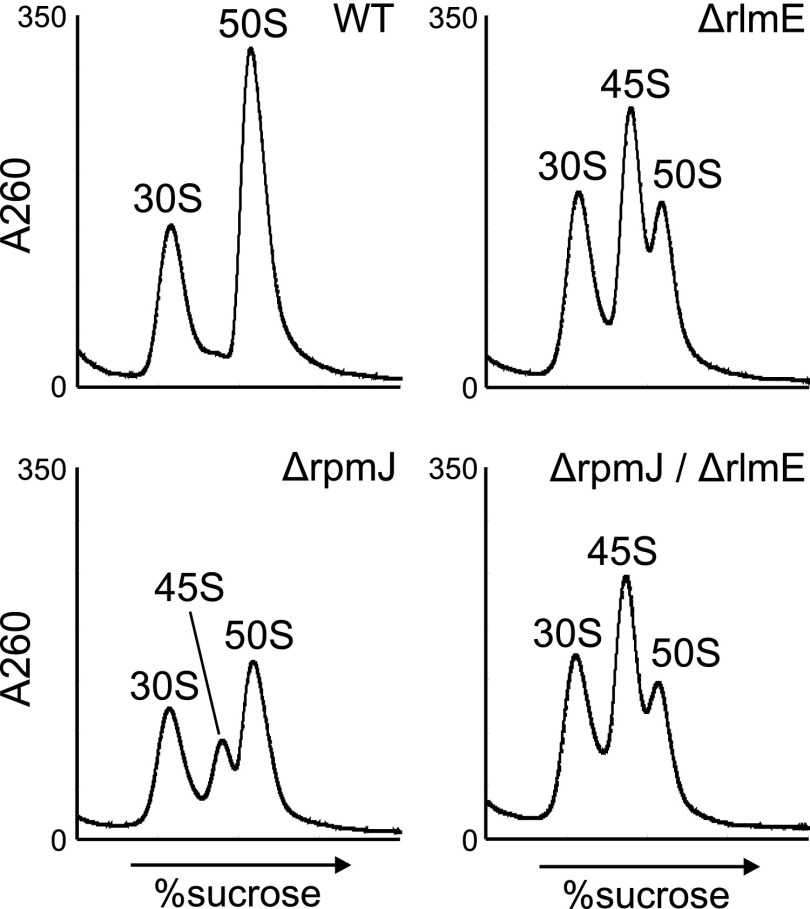
Because both L35 and L36 were absent from the 45S precursor in ΔrlmE, we investigated whether the 45S precursor also accumulated in knockout strains lacking rpmI (L35) or rpmJ (L36). In ΔrpmI, we observed a shoulder on the left side of the 50S peak at low Mg2+ concentration (Fig. 4D) that disappeared at high Mg2+ concentration (Fig. 4D), indicating marginal accumulation of the 45S precursor in ΔrpmI. Deletion of rpmJ causes accumulation of the 50S particle that is unable to productively associate with the 30S subunit (30), leading us to hypothesize that the 45S precursor would also accumulate in ΔrpmJ. Consistent with this prediction, in ΔrpmJ, we detected a 45S precursor exhibiting Mg2+-dependent alteration of its density (Fig. 4D). The level of 45S accumulation in ΔrpmJ, however, was lower than that in ΔrlmE (Fig. 1C), mirroring the more robust growth phenotype of ΔrpmJ relative to ΔrlmE (Fig. 4E). Because rpmJ is the last gene in the spc operon, we suspected that deletion of rpmJ might affect the expression of secY and other genes. To explore this possibility, we complemented ΔrpmJ with plasmid-encoded rpmJ (Fig. S6), demonstrating that 45S accumulation in ΔrpmJ is not due to a polar effect caused by rpmJ depletion; rather, L36 is required for a late assembly of the 50S subunit.
Fig. S6.
Complementation of ΔrpmJ by plasmid-encoded rpmJ. ΔrpmJ was transformed with a plasmid-encoded rpmJ under the control of the PBAD promoter (pBAD-rpmJ), and transformants were cultured in the presence (+, Right) or absence (−, Left) of l-arabinose (0.2%). Ribosome profiles were analyzed by SDG in the presence of 0.5 mM Mg2+.
To investigate the genetic interaction between L36 and RlmE, we generated a double deletion lacking both rlmE and rpmJ (ΔrlmE/ΔrpmJ). This strain exhibited a severe synthetic growth defect at 37 °C and was completely unable to grow at low temperature (Fig. 4E). Furthermore, strong accumulation of 45S precursor was also observed in the double-deletion strain (Fig. S7), indicating that incorporation of L36 acts cooperatively with modification of Um2552 by RlmE to promote late steps of 50S assembly.
Fig. S7.
SDG profiles of ribosomes from the rlmE/rpmJ double-knockout strain. SDG profiles of ribosomes from WT (Upper Left), ΔrlmE (Upper Right), ΔrpmJ (Lower Left), and ΔrlmE/ΔrpmJ (Lower Right) at low Mg2+ concentration.
We next analyzed the status of the Um2552 modification in the 45S precursor of ΔrpmJ. In the 50S subunit from WT cells, 85% of 23S rRNA was 2′-O-methylated (Fig. 4F). In ΔrpmJ, 40% of 23S rRNA was 2′-O-methylated in the 45S precursor whereas 85% was 2′-O-methylated in the 50S subunit (Fig. 4F). These data suggest that L36 incorporation is not necessary for the formation of Um2552 mediated by RlmE.
Stripping Ribosomal Components from the 45S Precursor.
The Mg2+-dependent alteration of its sedimentation coefficient suggests that the 45S precursor has a flexible structure in which intramolecular interactions have not yet been fully established and stabilized. We investigated this physical property by stripping ribosomal components using high concentrations of salt. Specifically, we incubated the 45S precursor from ΔrlmE and the WT 50S subunit in a solution containing 500 mM ammonium chloride in the presence of 0.5 mM or 10 mM Mg2+, and then subjected the samples to SDG to separate components [upper fraction (U) in Fig. 4G] that dissociated from the particles [lower fraction (L) in Fig. 4G]. From each fraction, rRNAs and protein components were prepared separately and then analyzed by denaturing PAGE for rRNAs (Fig. 4G, Upper) and SDS/PAGE for proteins (Fig. 4G, Lower). No components dissociated from the WT 50S under these conditions. By contrast, 5S rRNA and several protein components dissociated from the 45S precursor, especially at low Mg2+ concentration. By contrast, at high Mg2+ concentration, dissociation of these components was clearly suppressed, indicating that binding of numerous Mg2+ ions stiffens the internal structure of the 45S precursor, thereby preventing dissociation of its components. Mass-spectrometric analysis revealed the dissociated proteins in the upper fraction to be L5, L6, L9, L14, L16, L25, L27, and L28. To determine the levels of these dissociated proteins, we treated the particle with high-salt solution at low Mg2+ concentration, isolated it from the lower fraction of the SDG, and then quantitated its residual components by SILAC (Fig. 4B). We confirmed that the levels of the dissociated proteins were clearly reduced in the particle after high-salt treatment. Because L5, L18, and L25 associate with 5S rRNA, we assume that these proteins dissociate from the 45S precursor along with 5S rRNA as a result of high-salt treatment. L6, L14, and L16 are in close proximity to the region surrounding H92 and the L36-binding site, implying that the structure of this region is immature. We also carried out the same experiments on 45S precursors isolated from ΔrpmJ (Fig. S8) or from a strain that overexpressed the DbpA R331A mutant (Fig. S8); in both cases, the results were similar to those obtained with the 45S precursor from ΔrlmE.
Fig. S8.
High-salt stripping analysis of 45S particles from a DbpA R331A-overexpressing strain and ΔrpmJ. (A) Denaturing 10% PAGE analyses of rRNA from 45S particles from a DbpA R331A-overexpressing strain (Left) and ΔrpmJ (Right), in the upper (U) and lower (L) fractions, after high-salt stripping in the presence of 0.5 mM or 10 mM Mg2+. Gels were stained with ethidium bromide. (B) SDS/PAGE analyses of ribosomal proteins from 45S particles from a DbpA R331A-overexpressing strain (Left) and ΔrpmJ (Right), in the upper (U) and lower (L) fractions, after high-salt stripping in the presence of 0.5 mM or 10 mM Mg2+. The gels were silver-stained.
Structural Probing of 23S rRNA in the 45S Precursor Using Hydroxyl Radicals.
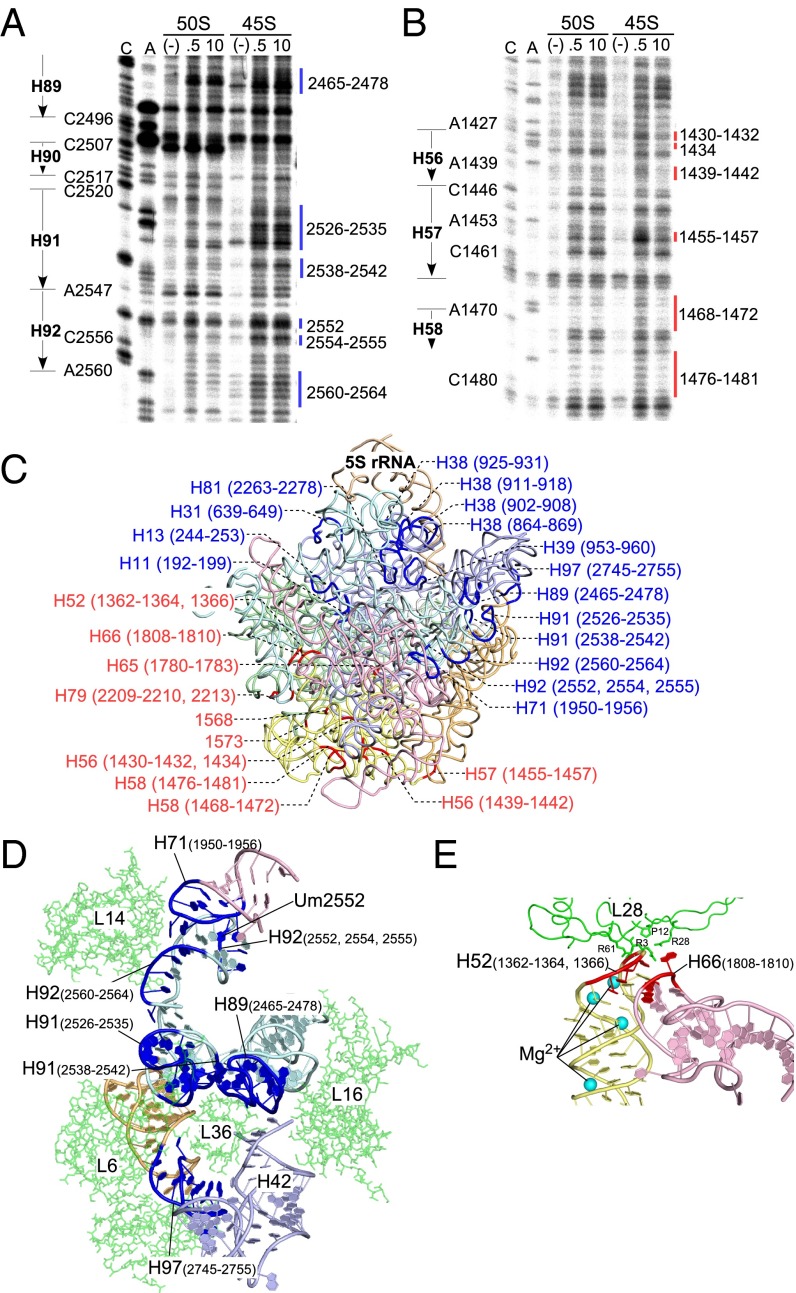
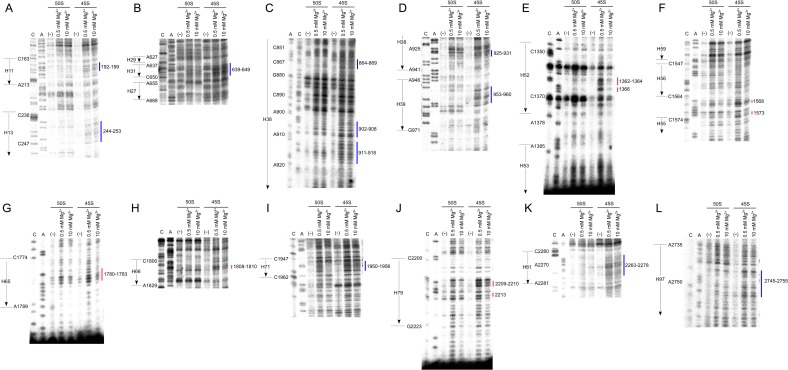
To investigate the structural differences in 23S rRNA between the 45S precursor and the 50S subunit, we performed hydroxyl-radical probing to analyze the solvent accessibility of the rRNA sugar-phosphate backbone (31). The probing was conducted at both low and high Mg2+ to identify the regions sensitive to Mg2+ concentration. Cleaved positions were determined by primer extension covering the entire sequence of the 23S rRNA (Fig. 5 A and B and Fig. S9). Compared with the 23S rRNA cleavage pattern in the 50S subunit, we found that a number of sites specifically cleaved in the 23S rRNA of the 45S precursor, which could be categorized into two types of regions. Regions of the first type were exposed to solvent specifically in the 45S precursor, regardless of Mg2+ concentration (Fig. 5A), and we designated these sites as “45S-specific cleavage” (color-coded in blue in the figures). Regions of the second type were sensitive to hydroxyl-radical cleavage only at low Mg2+ concentration (Fig. 5B), and we designated these sites as “low Mg2+-specific cleavage” (color-coded in red in the figures). We mapped both types of regions onto the crystal structure of the E. coli 50S subunit (Fig. 5C).
Fig. 5.
Structural probing of 23S rRNA in the 45S precursor. Hydroxyl-radical probing of H92 and surrounding regions in domain V (A) and regions in domain III (B) of 23S rRNA from the 50S subunit or the 45S precursor. The probing reaction was conducted at low (0.5 mM) or high (10 mM) Mg2+ concentration. The cleaved sites were detected by primer extension. Sequencing lanes with dideoxy terminators are labeled “C” and “A.” Lanes for untreated samples and samples probed at 0.5 mM and 10 mM Mg2+ are labeled “(−),” “0.5,” and “10,” respectively. The sites for the 45S-specific cleavage are shown by blue lines, with indication of their positions (A), whereas the sites for low Mg2+-specific cleavage are indicated by red lines, with indication of their positions (B). (C) Sites of 45S-specific cleavage (blue) and low-Mg2+-specific cleavage (red), with indications of their positions and helix numbers, mapped onto the tertiary structure of the 23S rRNA in the E. coli 50S subunit (PDB ID code 2AW4) (25). Color codes for domains are as follows: pale green, domain I; light blue, domain II; pale yellow, domain III; light pink, domain IV; pale cyan, domain V; light orange, domain VI; wheat, 5S rRNA. (D and E) Close-up views of the region around Um2552 and L36 (D), and the region around H52–H66 (E). Coordinates for these figures were obtained from 4YBB (26).
Fig. S9.
Hydroxyl-radical probing of 23S rRNA in the 45S precursor. Hydroxyl-radical probing of 23S rRNA from the 50S subunit or 45S precursor at low or high Mg2+ concentration. Cleaved sites were detected by primer extension. Sequencing lanes with dideoxy terminators are labeled “C” and “A.” Lanes for untreated rRNAs are labeled “(−)”. Sites of 45S-specific cleavage and low Mg2+-specific cleavage are shown by blue and red lines, respectively; numbers indicate the nucleotide positions of each site.
In the 45S precursor, the loop sequence of H92, including positions 2552, 2554, and 2555, was more sensitive to hydroxyl radicals, regardless of the Mg2+ concentration (Fig. 5 A and D). In the crystal structure, H92 interacts with H71 in domain IV via a base-triple composed of Um2552, C2556, and U1955 (Figs. 5D and 6A). We also observed 45S-specific cleavages at positions 1950–1956 in H71 (Fig. S9I). These results strongly suggest that 2′-O-methylation of Um2552 plays an important role in stabilizing the Um2552–C2556–U1955 base-triple, thereby facilitating the interdomain interaction between H92 and H71 (Figs. 1B, 5D, and 6A); therefore, the absence of 2′-O-methylation at position 2552 destabilizes this region. The weak binding of L14 in the 45S precursor (Fig. 4B) could be explained by instability of the interaction between H92 and H71 (Fig. 5D). Furthermore, we observed 45S-specific cleavages in H89, H91, and H97 (Fig. 5 A, C, and D and Fig. S9L), which, together with H42, form a binding pocket for L36 (Fig. 5D). The complete absence of L36 in the 45S precursor (Fig. 4C) is consistent with this observation. We also detected higher levels of 45S-specific cleavage in H38 (Figs. S9 C and D and S10A), which serves as a binding site for L16 and 5S rRNA. This observation explains why 5S rRNA, along with three bound proteins, dissociated from the 45S precursor after high-salt treatment (Fig. 4 B and G) and also explains the low stoichiometry and weak association of L16 in the 45S precursor (Fig. 4B). Likewise, 45S-specific cleavages were present in helices 11, 13, and 31 (Figs. S9 A and B and S10B), which forms a binding site for L35, explaining the low level of L35 in the 45S precursor (Fig. 4B). The low Mg2+-specific cleavages were apparently concentrated in domain III, including helices 52 and 55–58 (Fig. 5 B and C and Fig. S9 E and F), indicating that Mg2+-dependent alteration of the sedimentation coefficient of the 45S precursor originates from a dynamic conformational change in domain III that is mediated by Mg2+ binding. H52 in domain III and H66 in domain IV engaged in an interdomain interaction (Fig. 5E), and their contact site contained low Mg2+-specific cleavages (Fig. 5E and Fig. S9 E and H), indicating that the H52–H66 interaction in the 45S precursor is stabilized at high Mg2+ concentration.
Fig. S10.
The regions showing 45S-specific cleavage in 23S rRNA. The region around H38, L16, and 5S rRNA (A) and the region around L35 (B) in E. coli 50S subunit. All coordinates for these regions were obtained from 4YBB (26). The sites of 45S-specific cleavage are colored in blue.
SI Discussion
Assembly Factors Associated with the 45S Precursor.
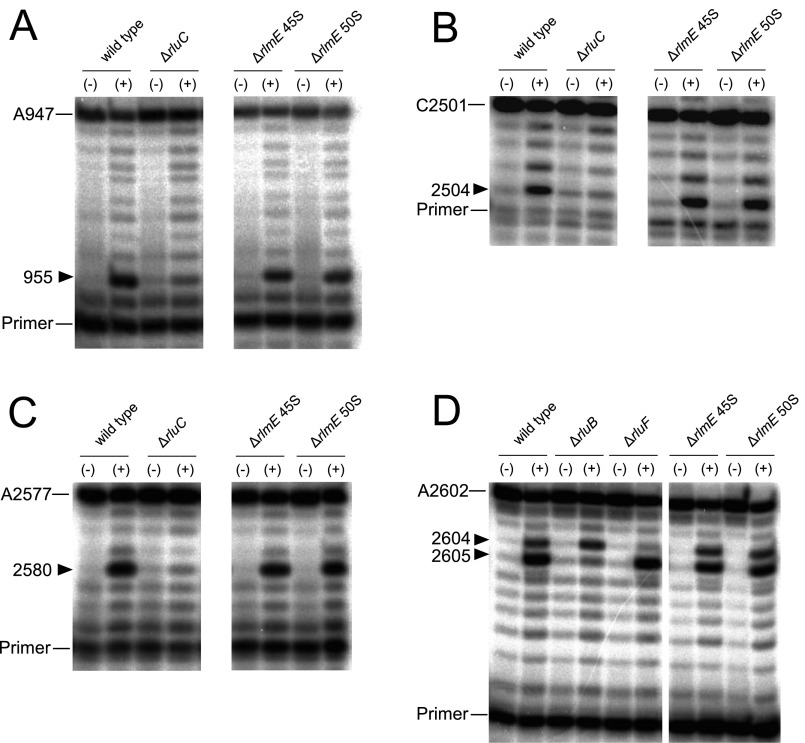
In the 45S precursor of ΔrlmE, we detected several assembly factors involved in the early stage of 50S biogenesis (Fig. 4A). SrmB and DeaD are involved in the early stage of 50S assembly (9). RhlE, which interacts genetically with srmB and deaD, is involved in the 40S accumulation (55), as is the RNA-binding protein YhbY (56). RlmKL is a fused methyltransferase responsible for forming both m7G2069 and m2G2445 in H74 of the 23S rRNA (57); both of these methylations were fully present in the 45S precursor. We reported that RlmKL has the ability to unwind H74, facilitating cooperative methylation at both positions in the early stage of 50S biogenesis (57). RluB is a pseudouridylase responsible for Ψ2605 formation (58) whereas RluC introduces Ψs at positions 955, 2504, and 2580 of the 23S rRNA (59). We analyzed the frequencies of Ψs at these positions and found that they were already introduced in the 45S precursor (Fig. S12). Considering that the target sites of these rRNA-modifying enzymes are located on the 50S subunit interface, association of these enzymes seems to prevent the 45S precursor from joining the 30S subunit; thus, these assembly factors need to dissociate from the 45S precursor during late steps of 50S assembly. Structural alteration of the 45S precursor during late assembly might be involved in this dissociation; alternatively, other assembly factors (such as GTPases) might remove these factors actively.
Fig. S12.
Analysis of pseudouridine sites introduced by RluB and RluC. rRNA extracted from the 45S precursor or 50S subunit of ΔrlmE was treated with (+) or without (−) CMC to detect pseudouridines at positions 955 (A), 2504 (B), 2580 (C), and 2605 (D) by primer extension. Total RNAs from WT, ΔrluC, ΔrluB, and ΔrluF were used as controls. Positions terminated by ddTTP or ddGTP are indicated.
Mechanistic Model for Late Assembly of the 50S Subunit Triggered by the RlmE-Mediated Um2552 Modification.
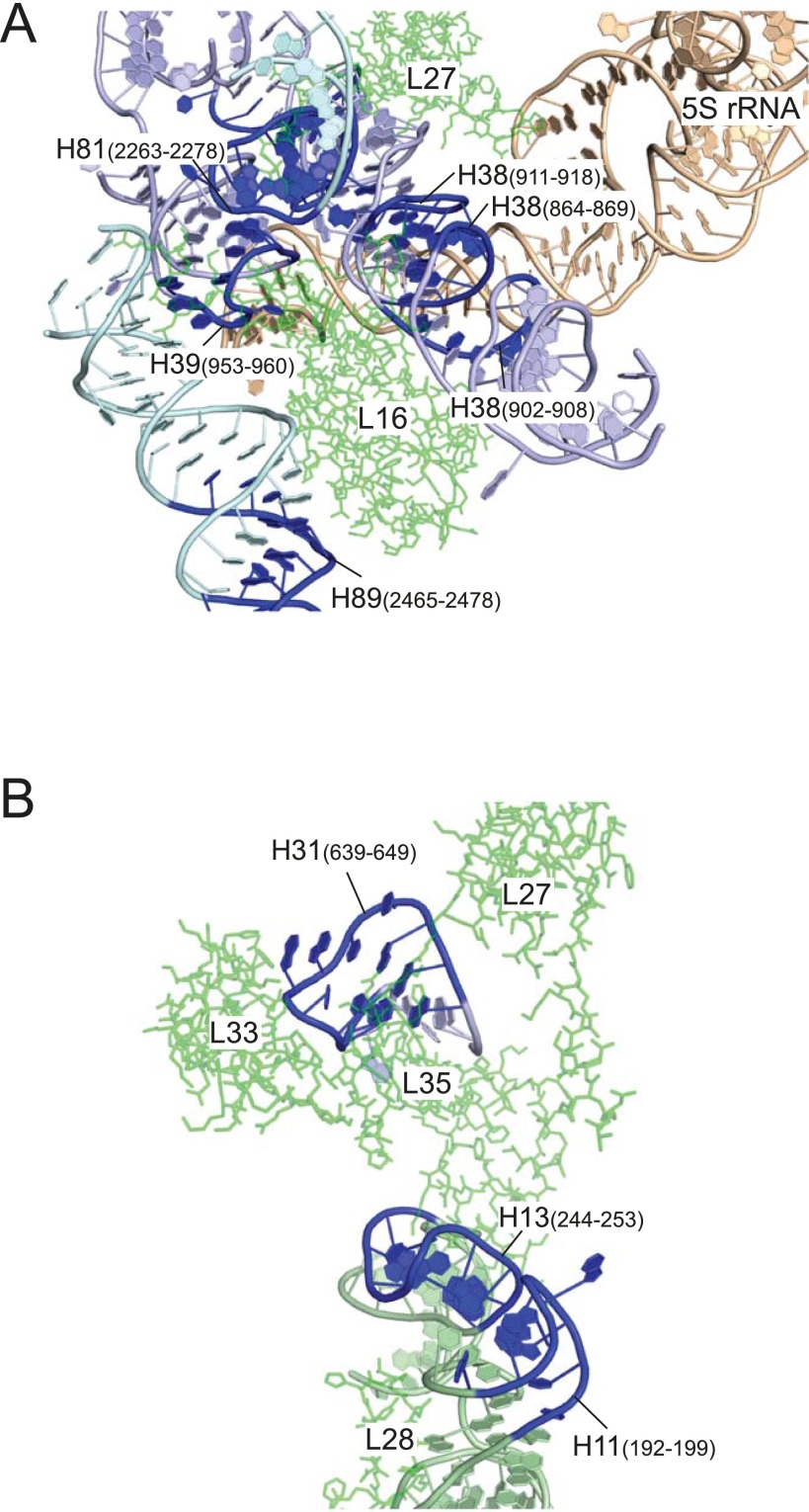
Formation of Um2552 stabilizes the A-loop structure and the Um2552–C2556–U1955 base-triple (Fig. 6A), facilitating interdomain interaction between domains V and IV via the H92–H71 association. H71 serves as a binding site for L14 (Fig. S11), which dissociates from the 45S precursor in high salt, indicating that the H92–H71 association stabilizes L14 binding (Fig. 6C). H91 serves as a major binding site for L36 (Fig. S11); upon stabilization of the H92–H71 interaction by Um2552, H91 is dragged to the correct position for recruitment of L36 (Fig. 6 B and C). L36 docks in the void space formed by the four helices H91, H89, H97, and H42 (Fig. 6B); in particular, L36 makes strong contacts with H91 via seven hydrogen bonds (25). According to our probing study of the 45S precursor, the L36-binding sites in H91, H89, and H97 exhibited the 45S-specific cleavages and were exposed to solvent (Fig. S11). H91 and H97 also serve as a binding site for L6 (Fig. S11). Approximately 70% of 45S precursors contained L6, which was completely stripped out by high-salt treatment (Fig. 4B), indicating that the association between L6 and 45S is weak. L36 incorporation facilitates assembly of domains V and VI by stabilizing the H89–H91–H97 association, thereby stabilizing L6 binding (Fig. 6C). In addition, L36 incorporation recruits and stabilizes L16 by fixing H89 in the proper position. Indeed, L16 was incorporated only inefficiently into the 45S precursor and was completely dissociated by high-salt stripping (Fig. 4B). L16 binds to H81, in addition to H89 in domain V, and it bridges H38 and H39 in domain II (Figs. S10A and S11); these binding sites were also exposed to solvent in the 45S precursor (Fig. S11). L27, which like L16 is inefficiently incorporated into the 45S precursor (Fig. 4B), binds H81, H85, and H86 in domain V and H38 in domain II (Fig. S11). Because the binding sites of L16 and L27 partially overlap, L16 incorporation affects L27 binding; thus, incorporation of L16 and L27 plays a critical role in spanning domains V and II during 50S assembly (Fig. 6 B and C). Very little L35 was present in the 45S precursor. L35 binds to helices 86 and 88 (domain V), H13 (domain I), and H31 (domain II) (Figs. S10B and S11). Association of domains V and II, as well as L27 binding to H86, might affect L35 incorporation; conversely, L35 seems to stabilize the interdomain association of domains I, II and V. Inefficient incorporation of L33 in the 45S precursor has been reported previously (15). Considering that L33 binds to H86–88 in domain V, it is reasonable to speculate that incorporation of L27 and L35 stabilizes binding of L33 (Fig. S10B). The 5S rRNA mainly interacts with H38–39 in domain II to form the central protuberance along with L5, L18, L25, and L31 (Fig. 6B and Fig. S10A). Binding sites in H38–39 for 5S rRNA were exposed to solvent in the 45S precursor (Fig. S10A), and high-salt stripping dissociated 5S rRNA along with its binders L5, L18, and L25 (Fig. 4 B and G). L31 is also incorporated inefficiently into the 45S precursor (15). These observations clearly demonstrate that the central protuberance does not form properly in the 45S precursor. Sufficient incorporation of L16 and L27 plays an important role in properly positioning H38–39 as a platform for 5S rRNA, along with the other components of the central protuberance (Fig. 6C).
Fig. S11.
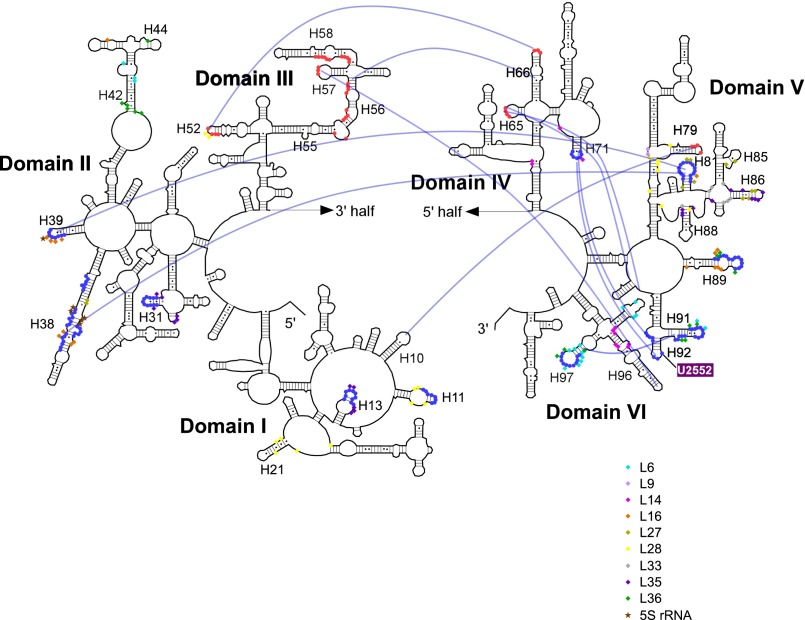
Distribution of the exposed regions in 23S rRNA in the 45S precursor and binding sites of several ribosomal proteins involved in late assembly. Sites cleaved by hydroxyl radicals detected by primer extension mapped onto the secondary structure of E. coli 23S rRNA. Sites of 45S-specific cleavage and low Mg2+-specific cleavage are indicated by blue and red closed circles, respectively. The interdomain interactions in 23S rRNA are shown as blue lines (24). Contact sites for L6, L9, L14, L16, L27, L28, L33 L35, L36, and 5S rRNA are indicated by the symbols shown in the figure, on the basis of the E. coli 50S structure (25).
Discussion
RlmE preferentially methylates the 50S subunit rather than the 45S particle that accumulates in ΔrlmE (20). On the basis of this observation, the 45S particle has been regarded as a dead-end or breakdown product of the 50S subunit (21). However, in this study, we demonstrated that the 45S particle could actually be converted to the 50S subunit in ΔrlmE (Fig. 1 E and F), demonstrating that 45S is a genuine precursor of 50S. Furthermore, we could partially reconstitute the 50S subunit from the 45S precursor in vitro by RlmE-mediated Um2552 formation in the presence of the wash fraction from crude ribosomes (Fig. 2B). This experiment is the first demonstration of the in vitro formation of 50S from its precursor, mediated by the enzymatic activity of an assembly factor. Given that most 50S subunits in cells are associated with 30S subunits at sufficient concentrations of Mg2+, it is reasonable to speculate that the physiological substrate for RlmE is not the 50S subunit, but rather the 45S precursor, because Um2552 in H92 of 23S rRNA is located on the interface side of the 50S subunit (24–26). Quantitative analyses of rRNA modifications by HPLC/MS have shown that the level of Um2552 surges during late steps of 50S assembly (32, 33), and these results are consistent with our observations.
To obtain insights into the function of an RNA modification responsible for a specific biological process, it is necessary to determine whether the RNA-modifying enzyme or the RNA modification itself plays a pivotal role in the process. In this study, we resolved this issue by examining the U2552C mutant. This mutation reduced 2′-O-methylation at position 2552 (Fig. 3C) and accumulated the 45S precursor (Fig. 3B) without hindering RlmE binding to the 50S subunit (Fig. 3 D and E). These data strongly suggest that RlmE binding to the 45S precursor does not contribute to 50S assembly; instead, Um2552 modification itself plays a critical role in triggering late steps of 50S assembly. In general, a 2′-O-methylated residue favors the C3′ endo ribose pucker conformation (34). The solution structure of H92, determined by NMR, revealed that 2′-O-methylation of Um2552 restrains the conformational dynamics of the A-loop structure by influencing Um2552–C2556 pairing (35) (Fig. 1B). In the crystal structure of the 50S subunit, Um2552 forms a base-triple along with C2556 and U1955 in H71 (Fig. 6A). If Um2552 were to adopt the C2′ endo ribose pucker, the base-triple would not be stabilized. Therefore, it is likely that the Um2552 modification stabilizes the base-triple Um2552–C2556–U1955 by adopting the C3′ endo conformation, facilitating the H92–H71 interaction and thereby promoting interdomain assembly between domains V and IV. This speculation is supported by the fact that H92 and H71 in the 45S precursor of ΔrlmE were more sensitive to hydroxyl radicals (Fig. 5D).
The unique physicochemical feature of the 45S precursor is the consecutive alteration of its sedimentation coefficient in response to Mg2+ concentration (Fig. 1D). This structural property of the 45S precursor can be observed only in vitro. In the cell, where the Mg2+ concentration is sufficiently high, this structural change does not occur. However, this unique characteristic provides us with useful information about the structural properties of the 45S precursor, including flexible regions and solvent-accessible regions, during ribosome biogenesis in the cell. Our structural probing of 23S rRNA from the 45S precursor revealed that helices 52, 56, 57, and 58 in domain III were more sensitive to hydroxyl-radical cleavages at low Mg2+ concentration than at high Mg2+ concentration (Fig. 5 B and C and Fig. S9), indicating that these regions undergo Mg2+-dependent structural changes. Intriguingly, H52 in domain III associates with H66 in domain IV via a “kissing-loop” interaction (Fig. 5E). Based on the observation that the interacting loops in both helices exhibited low Mg2+-specific cleavage, it is likely that the H52–H66 interaction in the 45S precursor is stabilized at high Mg2+ concentration. In the crystal structure of the 50S subunit, four Mg2+ ions are present in H52 (Fig. 5E) whereas no Mg2+ is present in H66 (26). Mg2+ ions are likely to stabilize H52 to facilitate the kissing-loop interaction with H66, allowing the association between domains III and IV (Fig. 5 C and E). Moreover, H52 interacts with L28 (Fig. 5E and Fig. S11), which dissociates from the 45S precursor after high-salt stripping (Fig. 4B). Destabilization of the H52–H66 interaction might affect the stable association of L28. Because L28 bridges domains I, III, and V of the 23S rRNA (Fig. S11), correct positioning of L28 seems to stabilize domain III by strengthening its interactions with other domains. L28 also associates with H11 (domain I), which contained 45S-specific cleavage sites (Figs. S10B and S11). H11 is situated in close proximity to H13, where L35 binds (Fig. S10B); therefore, L35 incorporation might contribute to the stable binding of L28. Taken together, these observations indicate that, within the 45S precursor, interdomain interactions within the 23S rRNA are not yet stably established. Instead, domain III has a flexible structure that can be stabilized by Mg2+ ions. The alteration of the sedimentation coefficient of the 45S precursor seems to be primarily due to stabilization of interdomain interactions and condensation of domain III in response to increasing Mg2+ concentration.
In this study, we clearly demonstrate that rlmE and rpmJ interact genetically, strongly suggesting that Um2552 and L36 play pivotal roles in late steps of the 50S subunit assembly. ΔrlmE was much more cold-sensitive (Fig. 4E), and accumulated much higher levels of the 45S precursor, than ΔrpmJ (Figs. 1C and 4D). In addition, L36 was completely absent from the 45S precursor in ΔrlmE (Fig. 4C) whereas ∼40% of 45S precursor in ΔrpmJ contained Um2552 (Fig. 4F), indicating that L36 incorporation is not required for Um2552 formation. These observations suggest that structural rearrangements of the 45S precursor induced by Um2552 are involved in incorporation of L36.
On the basis of the genetic and biochemical observations in this study, we propose a mechanistic model for late steps of the 50S subunit assembly triggered by the RlmE-mediated Um2552 modification (Fig. 6 B and C). Detailed discussion of this model is provided in SI Discussion. Um2552 stabilizes the A-loop structure and the Um2552–C2556–U1955 base-triple (Fig. 6A), which facilitates interdomain association between domains V and IV via the H92–H71 interaction, followed by recruitment of L36 via correct positioning of H91 (Fig. 6 B and C). L36 binding stabilizes four helices, H91, H89, H97, and H42 (Fig. 6B), contributing to stable binding of L6 and L16, followed by L27 incorporation. Stable binding of L16 and L27 plays a critical role in spanning domains V and II and properly positions H38–39 as a platform for 5S rRNA and the other components of the central protuberance (Fig. 6C). Association of domains V and II, as well as L27 binding to H86, affects L35 incorporation. L35 might affect stable binding of L28, which bridges domains I, III, and V. Taken together, these data indicate that all of the events in late steps of the 50S subunit assembly triggered by Um2552 formation take place on the interface side of the 45S precursor, including the central protuberance (Fig. 6B). The interface is not formed until a late step of 50S assembly; therefore, immature 50S cannot associate with the 30S subunit to create unproductive 70S ribosomes incapable of participating in translation.
The 45S particle is also found in Bacillus subtilis. RbgA/YlqF is an essential GTPase required for the proliferation of B. subtilis (36, 37). Depletion of RbgA or expression of a dominant-negative mutant of RbgA resulted in accumulation of the 45S particles (36–39). However, unlike the E. coli 45S precursor, the B. subtilis 45S particle does not alter its sedimentation coefficient, even at 10 mM Mg2+ (36, 39). In addition, there is no homolog of RbgA in E. coli (10), and no homolog of RlmE in B. subtilis; consistent with this knowledge, U2552 in 23S rRNA remains unmethylated in B. subtilis (40). Although the basic mechanisms of late assembly of the 50S subunit differ between E. coli and B. subtilis, the 45S particles share some common properties. As in the E. coli 45S precursor, substoichiometric levels of L16, L27, and L36 are present in the 45S particle that accumulates in the B. subtilis rbgA mutant (36, 37, 41). In addition, cryo-EM structures revealed that H38 and 5S rRNA are not yet oriented and that domains IV and V are not properly built up, in the B. subtilis 45S particle (41, 42). Comparisons of the structural features of the 45S particles of E. coli and B. subtilis will provide further mechanistic insights into late steps of the 50S subunit assembly.
Materials and Methods
Strains and Plasmid Constructions.
The E. coli deletion strains ΔrlmE::Kmr (JW3146) and ΔrpmJ::Kmr (JW3261) and their parental strain BW25113 were provided by the National Institute of Genetics, Japan. We disrupted rna (ribonuclease I) and rlmE in BW25113 with the cat cassette by one-step gene inactivation (43) to yield strains Δrna::Cmr and ΔrlmE::Cmr, respectively. Single-colony isolation was repeated twice for all strains constructed in this study. Knockout was confirmed by colony PCR. Δrna::Cmr was used as a recipient for construction of strains ΔrlmE::Kmr, Δrna::Cmr and ΔrpmJ::Kmr, Δrna::Cmr by P1 transduction. These strains were used for preparation of 45S precursors for a series of biochemical analyses. To construct the double-deletion strain. ΔrlmE/ΔrpmJ, ΔrlmE::Cmr was introduced into the ΔrpmJ::Kmr strain by P1 transduction. dbpA and rpmJ were PCR-amplified from the E. coli genome. The amplified products of dbpA and rpmJ were inserted into the XhoI/PstI and NcoI/PstI sites of pBAD-cMYC-HisA (Invitrogen) to yield pBAD-dbpA and pBAD-rpmJ, respectively. In pBAD-dbpA, the R331A mutation was introduced using the QuikChange site-directed mutagenesis kit (Agilent Technologies).
E.coli Δ7 prrn strain KT101 (8), which harbors pRB101 (44) encoding the rrnB operon and sacB as a counter selectable marker, was used to examine the U2552C mutation in the 23S rRNA. Disruption of rlmE in KT101 was performed by P1 transduction to generate KT101/ΔrlmE. pRB101 in WT KT101 or in KT101/ΔrlmE was completely replaced by plasmid pRB103, which harbors the rrnB operon with or without the U2552C mutation, to yield a series of KT103 strains as described (8). Sequences of primers used in these steps are listed in Table S2.
Table S2.
List of primers used in this study
| Primers/oligomers | Sequence |
| Primers used to construct vector harboring dbpA R331A | |
| pBAD-dbpA-F | tacgtacgctcgagcaccgctttttctaccctgaatgt |
| pBAD-dbpA-R | tacgtacgctgcagttattttaataaccgcacccggc |
| dbpA-R331A-F | tcatgtacatcgcatcggtgcgacagctcgtgcaggaaa |
| dbpA-R331A -R | tttcctgcacgagctgtcgcaccgatgcgatgtacatga |
| Primers used to construct vector harboring rpmJ | |
| pBAD-rpmJ-F | actgactgccatgggcaaagttcgtgcttccgtcaaga |
| pBAD-rpmJ-R | cagtcagtctgcagtcagccttggcgctgtttat |
| Primers for rna deletion | |
| rna-del-F | atgaaagcattctggcgtaacgccgcgttgctcgcggttttccggtagtcaataaaccgg |
| rna-del-R | ttaataacccgctttatcaatcacaaaggttttgccacagttggcgaaaatgagacgttg |
| Primers used to construct pRB103 harboring U2552C mutation | |
| U2552C-F | ggtcccaagggtatggccgttcgccatttaaagtg |
| U2552C-R | cactttaaatggcgaacggccatacccttgggacc |
| Oligomers used to isolate defined rRNA sequence containing position 2552 | |
| G2529-C2579-cover | gctcgcgtaccactttaaatggcgaacagccatacccttgggacctacttc |
| G2529-C2579-cover for U2552C | gctcgcgtaccactttaaatggcgaacggccatacccttgggacctacttc |
| Primers used for hydroxyl radical probing analysis | |
| 5S-102–120-R | atgcctggcagttccctac |
| 23S-2880–2903-R | aggttaagcctcacggttcattag |
| 23S-2777–2797-R | aagggtcagggagaactcatc |
| 23S-2702–2718-R | cgggcagtgccattggc |
| 23S-2582–2605-R | aactgtctcacgacgttctaaacc |
| 23S-2442–2462-R | gtatcagcctgttatccccgg |
| 23S-2339–2356-R | acgctcgcagtcaagctg |
| 23S-2234–2253-R | ccccagtcaaactacccacc |
| 23S-2127–2145-R | gcgtccacacttcaaagcc |
| 23S-2025–2046-R | cgggtacactgcatcttcacag |
| 23S-1908–1931-R | accttaggaccgttatagttacgg |
| 23S-1799–1820-R | atacgtccactttcgtgtttgc |
| 23S-1695–1714-R | atatcagcgtgccttctccc |
| 23S-1600–1618-R | tgtgtcggtttggggtacg |
| 23S-1498–1518-R | gcctcagccttgattttccgg |
| 23S-1398–1419-R | tcgcagtaacaccaagtacagg |
| 23S-1296–1314-R | gaacccttggtcttccggc |
| 23S-1197–1216-R | ccttcgcaggcttacagaac |
| 23S-1139–1158-R | gcttcggtgcatggtttagc |
| 23S-1071–1094-R | acgctttctttaaatgatggctgc |
| 23S-997–1018-R | accatgactttgggaccttagc |
| 23S-939–956-R | cacccgccgtgtgtctcc |
| 23S-834–853-R | gagatgaattcacgaggcgc |
| 23S-728–750-R | ttcaacattagtcggttcggtcc |
| 23S-611–629-R | ccttcggctcccctattcg |
| 23S-506–528-R | tgtacgtacacggtttcaggttc |
| 23S-400–420-R | gaggatggtccccccatattc |
| 23S-298–316-R | gcgcctttccagacgcttc |
| 23S-198–221-R | tcttttcctcggggtacttagatg |
| 23S_97-117_R | cggaaatcgccggttataacg |
| Primers used to construct vector harboring rlmE | |
| pET28a-rlmE-F | acgtacgtgctagcatgacaggtaagaagcgttctgc |
| pET28a-rlmE-R | acgtacgtctcgagttagggtttacgcccggtc |
| Primers used to detect pseudouridine | |
| 955 detection | cctcttcacgacggacgtta |
| 2504 detection | ccaggatgtgatgagccga |
| 2580 detection | actgtctcacgacgttctaa |
| 2604,2605 detection | cagcgcccacggcagata |
Listed are primers used in PCR, cloning, site-directed mutagenesis, gene deletion, isolation of defined rRNA sequences, probing, and detection of pseudouridines.
Analysis and Preparation of Ribosome and Subunits by SDG.
E. coli strains were cultivated in LB medium at 37 °C or 22 °C with vigorous shaking until A600 reached 0.4–0.6, quickly chilled by incubation on ice for 10 min, and harvested by centrifugation. To inhibit transcription, rifampicin was added to a final concentration of 250 µg/mL. For overexpression of the DbpA R331A mutant, 0.2% l-arabinose and 100 µg/mL ampicillin were added before inoculation of the preculture. Cell pellets from 50-mL cultures were resuspended in 1 mL of RBS buffer A [0.5 mM Mg(OAc)2, 200 mM NH4Cl, 20 mM Hepes-KOH (pH 7.6), 6 mM β-mercaptoethanol] or RBS buffer B [10 mM Mg(OAc)2, 100 mM NH4Cl, 20 mM Hepes-KOH (pH 7.6), 6 mM β-mercaptoethanol] and lysed by lysozyme treatment (1.5 mg/mL) followed by 1–3 freeze/thaw cycles. Cell lysates were clarified by centrifugation at 20,000 × g for 15 min at 4 °C. The cleared lysate (15 A260 units) was layered on top of a sucrose gradient [10–40% (wt/vol)] in RBS buffer A or B and separated by ultracentrifugation in a Beckman SW-28 Rotor at 20,000 rpm for 14 h at 4 °C. Ribosomal subunits in the gradient were fractionated on a Piston Gradient Fractionator (BIOCOMP), and the A260 was measured using a UV monitor (AC-5200; ATTO).
Ribosomal subunits used in this study were prepared from E. coli strains with the Δrna background. E. coli strains were cultivated and harvested as described above. The cell pellet was ground in a mortar and pestle for 10 min at 4 °C after addition of alumina weighing two to three times as much as the cell pellet. Ground cell pellets were then resuspended in ∼25 mL of RBS buffer C [10 mM Mg(OAc)2, 30 mM NH4Cl, 20 mM Hepes-KOH (pH 7.6), 6 mM β-mercaptoethanol] and centrifuged at 5,000 × g for 30 min at 4 °C. The supernatant was ultracentrifuged in a Beckman 70Ti rotor at 19,500 rpm for 45 min at 4 °C. After adding DNase I (5.6 U/mL), the supernatant was ultracentrifuged in a 70Ti rotor at 35,000 rpm for 4 h at 4 °C to obtain crude ribosomes. These crude ribosomes were resuspended in RBS buffer A, layered on top of a sucrose gradient [10–50% (wt/vol)] in RBS buffer A and ultracentrifuged in a Beckman SW-28 Rotor at 25,000 rpm for 15 h at 4 °C. Gradients were fractionated into more than 70 fractions on a Piston Gradient Fractionator, and absorbance at 260 nm (A260) of each fraction was measured using a SpectraMax (Molecular Devices). Fractions corresponding to 45S and 50S were pooled and recovered by ultracentrifugation in a Beckman 70Ti rotor at 40,000 rpm for 24 h at 4 °C in RBS buffer B. The pellet was resuspended in RBS buffer D [0.5 mM Mg(OAc)2, 100 mM NH4Cl, 20 mM Hepes-KOH (pH 7.6), 6 mM β-mercaptoethanol] and stored at −80 °C.
Monitoring the Ratio of Stable Isotope-Labeled rRNAs in Subunit Fractions.
The E. coli ΔrlmE/Δrna strain was grown in LB medium at 37 °C overnight (full growth, A600 = 4). The preculture (500 μL) was inoculated to LB medium supplemented with 1 mM [ribose-13C5] adenosine (Cambridge Isotope Laboratories), and then cultured at 22 °C with vigorous shaking. When A600 reached 0.4, the cells were collected by centrifugation at 22 °C, and the pellet was washed once with LB medium containing 1 mM nonlabeled adenosine, followed by resuspension with 100 mL of LB medium (1 mM nonlabeled adenosine), and further cultured at 22 °C. Ten milliliters of the culture aliquot was collected at different time points: 0, 40, 80, and 160 min. The cell pellet was lysed by lysozyme treatment (1.5 mg/mL), followed by three freeze-thaw cycles. The resultant cell lysates were fractionated by SDG, and fractions for 30S, 45S, and 50S were collected as described above. rRNAs were extracted from each fraction by TRIzol LS and chloroform treatment, followed by isopropanol precipitation. Each rRNA (2 μg) was digested into nucleosides at 37 °C for 1 h in 60 μL of reaction mixture consisting of 25 mM ammonium acetate (pH 7.7), 1.33 microunits (μU)/μL nuclease P1 (Wako Pure Chemical Industries), and 1.33 μU/μL bacterial alkaline phosphatase A19 (Takara Bio). One-tenth of the mixture was dried in vacuo, dissolved in 20 μL of 90% acetonitrile, and subjected to Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometry (Thermo Fisher Scientific) with an Ultimate 3000 system (Thermo Fisher Scientific). The nucleosides were chromatographed on a ZIC-cHILIC column (150 mm × 2.1 mm i.d., 3.5 μm; Merck Millipore) connected to a guard column (20 mm × 2.1 mm i.d., 5 μm; Merck Millipore) at a 0.1 mL/min flow rate of a multistep linear gradient of solvent A [5 mM NH4OAc (pH 5.3) in water] and B (acetonitrile): 90–40% B for 0–30 min, 40% B for 30–40 min, 40–90% B for 40–45 min, and 90% B for 45–55 min (45). The eluent was directly conducted to an electrospray ion source of the mass spectrometer and scanned with a positive polarity mode over an m/z range of 240–300 throughout the separation. The ratio of labeled adenosine was calculated from mass chromatographic peak areas of 13C-labeled (m/z 273) and nonlabeled (m/z 268) adenosine. The experiment was repeated three times to calculate average and SD of measurements. The parameters for mass spectrometry were as follows: sheath gas flow rate, 35; aux gas flow rate, 10; spray voltage, 3.5 kV; capillary temperature, 250 °C; aux gas heater temperature, 200 °C.
Mass Spectrometry of rRNAs.
Two-hundred femtomoles of 23S rRNA isolated from the 45S precursor or 50S subunit were digested at 37 °C for 30 min in 10 µL of reaction mixture containing 10 mM NH4OAc (pH 5.3) and 25 U/µL of RNase T1. The digest was mixed with 10 µL of 0.1 M triethylamine-acetate (pH 7.0) and then analyzed by capillary liquid chromatography (LC) coupled with nano electrospray (ESI)/mass spectrometry (MS) on a linear ion trap-Orbitrap hybrid mass spectrometer (LTQ Orbitrap XL; Thermo Fisher Scientific) as described (46, 47). The m3Ψ1915-containing fragment (ΨAAC m3ΨAΨAACGp; MW 3,550.50) and the Ψ1915-containing fragment (ΨAACΨAΨAACGp; MW 3,536.49) were detected as multiply charged negative ions. Other Ψ sites were analyzed by a primer extension method (SI Materials and Methods and Fig. S12). To analyze methylation status at position 2552, a synthetic DNA (200 pmol) complementary to G2529–C2579 of E. coli 23S rRNA (Table S2) was mixed with 2.0 A260 of total RNA or 20 pmol of isolated 23S rRNA in the presence of 50 mM Hepes-KOH (pH 7.6) and 100 mM KCl. The mixture was heated at 90 °C for 5 min and then gradually cooled to 37 °C for 2.5 h, to allow the DNAs to form a heteroduplex. After adding 1 M NaOAc (pH 5.2) to a final concentration of 50 mM, the uncovered regions of 23S rRNA were removed by digestion with 500 U of RNase T1 and 0.5 μg of RNase A; digests were incubated for 60 min on ice. The heteroduplex was extracted with phenol/chloroform and precipitated with ethanol. The precipitate was resolved by 15% denaturing PAGE and visualized by staining with SYBR Gold (Molecular Probes). The heteroduplex was excised from the gel, eluted from the gel piece for 3 h at 37 °C in elution buffer [1 mM EDTA, 0.1% SDS, 0.4 M NaOAc (pH 5.0)], and precipitated with isopropanol. One-tenth of the specimen was digested with RNase T1 and subjected to MS analysis as described above.
Structural Probing of 23S rRNAs.
Hydroxyl-radical probing was performed essentially as described (48) with slight modifications. Twenty picomoles of 50S subunit or 45S precursor was dissolved in 97 µL of low Mg2+ solution [0.5 mM Mg(OAc)2, 100 mM NH4Cl, 20 mM Hepes-KOH (pH 7.6), 6 mM β-mercaptoethanol] or 97 µL of high Mg2+ solution [10 mM Mg(OAc)2, 100 mM NH4Cl, 20 mM Hepes-KOH (pH 7.6), 6 mM β-mercaptoethanol]. To generate hydroxyl radicals by the Fenton reaction, 1 µL of Fe2+-EDTA solution [100 mM Fe(SO4)2(NH4)2·6H2O and 200 mM EDTA], 1 µL of 500 mM ascorbic acid, and 1 µL of 10% H2O2 was added to each mixture. The reaction was carried out on ice for 10 min and then terminated by addition of 40 µL of 0.1 M thiourea. Samples were precipitated in ethanol and resuspended in dH2O. rRNAs were extracted by TRIzol LS followed by two chloroform treatments, precipitated with isopropanol, and then reprecipitated with ethanol. Primer extension was performed basically as described (49) with a light modification. Sequences of primers are listed in Table S2. Reaction mixture (6.5 µL) containing 2 pmol [5′-32P]-labeled primer (1–2 × 105 cpm), 0.2 pmol rRNAs, and 0.77 mM dNTPs was incubated at 65 °C for 10 min and then incubated on ice for 1 min to hybridize the primer with the rRNAs. Next, 2 µL of 5× FS buffer (Invitrogen), 0.5 µL of 0.1 M DTT, and 0.5 µL of dH2O (or 0.5 µL of 10 mM ddGTP or ddTTP for cDNA sequencing) were added, and the mixture was preincubated at 55 °C for 1 min. The reaction was started by addition of 0.5 µL (100 units) of SuperScript III (Invitrogen), incubated at 55 °C for 10 min, terminated by addition of 0.5 µL of 4 M NaOH, and boiled at 95 °C for 5 min to degrade template rRNAs. After the reaction, the cDNAs were ethanol-precipitated and subjected to 8% PAGE in the presence of 7 M urea. The gel was exposed to an imaging plate overnight, and the radiolabeled bands were visualized on an FLA-7000. The intensity of bands in each lane was quantitated using the MultiGauge software (FujiFilm). After subtracting the background intensity of nontreated lanes, sites with intensity >6000QL were selected. In the high-Mg2+ condition, if the band intensity in the 45S precursor was >1.5-fold higher than the intensity of the corresponding site in the 50S subunit, the site was defined as “45S-specific cleavage.” If the band intensity in the low-Mg2+ condition was >1.5-fold higher than the intensity of the corresponding site in the high Mg2+ condition, the site was defined as “low Mg2+-specific cleavage.” This experiment was repeated several times to confirm the reproducibility of the results.
Enzymatic Formation of the 50S Subunit from the 45S Precursor in Vitro.
A crude ribosome fraction containing the 45S precursor was prepared from ΔrlmE/Δrna, as described above. In vitro methylation of the crude ribosome fraction was performed at 37 °C for 2.5 h in a 60-µL reaction mixture containing 100 mM NH4Cl, 20 mM Hepes-KOH (pH 7.6), 6 mM Mg(OAc)2, 6 mM β-mercaptoethanol, 1 mM AdoMet, 2.5 A260 units of crude ribosomes, and 2 µM RlmE. After the reaction, the buffer was exchanged with RBS buffer A three times by an ultrafiltration using the Amicon Ultra-0.5 100K device (Merck Millipore). Then, the mixtures were resuspended in RBS buffer A, layered on top of a sucrose gradient [10–40% (wt/vol)] in RBS buffer A and ultracentrifuged in a Beckman SW-41 Rotor at 37,000 rpm for 5 h at 4 °C.
For in vitro methylation of the isolated 45S precursor, the reaction was performed at 37 °C for 2.5 h in a 200-µL reaction mixture containing 100 mM NH4Cl, 20 mM Hepes-KOH (pH 7.6), 8 mM Mg(OAc)2, 6 mM β-mercaptoethanol, 1 mM AdoMet, 1 mM ATP, 1 mM GTP, 1 A260unit of 45S precursor, and 70 µg of the wash fraction (SI Materials and Methods). After the reaction, the buffer was exchanged with RBS buffer A, and the mixture was then analyzed by SDG as described above.
To evaluate the ability of the newly synthesized 50S subunit to associate with the 30S subunit, the 30S subunit isolated from the ΔrlmE/Δrna strain was added to the reaction mixture immediately after the methylation reaction. The buffer was exchanged with RBS buffer B three times as described above, followed by incubation at 37 °C for 30 min. The mixture was then analyzed by SDG as described above.
SI Materials and Methods
Mass Spectrometry of Proteins.
Proteins resolved by SDS/PAGE were stained with Coomassie Brilliant Blue R-250, and the bands of interest were excised and subjected to in-gel digestion with trypsin. The digests were then analyzed on an ion trap mass spectrometer equipped with a nanospray ion source, and a splitless nano HPLC system (DiNa; KYA Technologies) with an octadecylsilyl (ODS) capillary column (100 × 0.15 mm, 3-μm particle size; KYA Technologies). The digests were separated in 0.1% formic acid in water, using a linear gradient from 2% to 80% of a solvent consisting of 70% (vol/vol) acetonitrile and 0.1% formic acid, for 40 min at a flow rate of 300 nL/min. Proteins were identified from the E. coli protein database by using MASCOT (Matrix Science) as a search engine for peptide mass fingerprinting.
The SILAC method (28) was used to measure the relative abundance of RlmE and ribosomal proteins. For RlmE quantification, WT KT103 was cultured in M9 medium containing 0.8 mM [13C6, 15N2] lysine (Cambridge Isotope Laboratories, Inc.) whereas the U2552C strain (KT103) was cultured in M9 medium containing 0.8 mM unlabeled lysine. The 50S subunits from WT KT103 and the U2552C strain were isolated by SDG centrifugation as described in Materials and Methods, Analysis and Preparation of Ribosome and Subunits by SDG. Equal amounts of fully labeled WT 50S and unlabeled U2552C 50S were mixed and then subjected to SDS/PAGE to resolve protein components; gels were stained with Coomassie Brilliant Blue R-250. Because RlmE is known to comigrate with L3, the protein band corresponding to L3 and RlmE was excised from the gel and digested with trypsin. The tryptic digest was analyzed by capillary LC/nano-ESI MS to detect specific peptides with C-terminal Lys residues from RlmE and L3. The unlabeled peptides from mutant 50S and the fully labeled corresponding peptides from WT 50S (with 8 Da higher mass) were eluted from LC at the same time, ionized simultaneously, and detected in the same mass spectra. The abundance ratio of RlmE in the WT and mutant 50S particles was calculated from the averaged intensity ratio of the heavy and light mass spectra of the three RlmE-derived peptides (Table S1) (51). To normalize the amount of 50S subunits from the WT and U2552C mutant, three L3-derived peptides (Table S1) were used as loading controls. The mean and SD values were calculated from the datasets of three peptides from RlmE and L3.
To determine the relative abundance of ribosomal proteins, the WT strain (BW25113 Δrna::Cmr) was cultured in M9 medium containing 0.8 mM [13C6, 15N2] lysine to label all Lys residues of ribosomal proteins. The 50S subunit from WT and the 45S precursor from ΔrlmE strain were isolated by SDG centrifugation as described in Materials and Methods, Analysis and Preparation of Ribosome and Subunits by SDG. Equal amounts of the fully labeled WT 50S and the unlabeled 45S precursor were mixed and then subjected to SDS/PAGE to resolve the protein components. Each band was excised from the gel and digested with trypsin. The digests were then subjected to capillary LC/nano-ESI MS to quantitate each ribosomal protein by SILAC, as described in the first paragraph of this section. The tryptic peptides for each protein used in this study are listed in Table S1. The proteins remaining in the 45S precursor after high-salt stripping were also quantitated by this method.
Tricine/SDS/PAGE.
Ribosomal proteins were extracted with acetic acid and precipitated in acetone, as previously described (52). The extracted proteins were subjected to Tris-tricine/urea 18% PAGE at 90 V for 13 h (53). The gel was stained with SYPRO Ruby (Molecular Probes) and visualized on an FLA-7000 imaging analyzer (FujiFilm).
High-Salt Stripping.
The 45S precursor or 50S subunit was dissolved in low Mg2+ solution [0.5 mM Mg(OAc)2, 100 mM NH4Cl, 20 mM Hepes-KOH (pH 7.6), 6 mM β-mercaptoethanol] or high Mg2+ solution [10 mM Mg(OAc)2, 100 mM NH4Cl, 20 mM Hepes-KOH (pH 7.6), 6 mM β-mercaptoethanol] and then incubated on ice for 1 h. Next, the sample was mixed with RBS buffer F [0.5 mM Mg(OAc)2, 900 mM NH4Cl, 20 mM Hepes-KOH (pH 7.6), 6 mM β-mercaptoethanol] or RBS buffer G [10 mM Mg(OAc)2, 900 mM NH4Cl, 20 mM Hepes –KOH (pH 7.6), 6 mM β-mercaptoethanol], to adjust the NH4Cl concentration to 500 mM in the presence of 0.5 mM or 10 mM Mg2+, and then incubated on ice for an additional 1 h. The mixtures were loaded on top of sucrose gradients [10–40% (wt/vol)] containing RBS buffer H [0.5 mM Mg(OAc)2, 500 mM NH4Cl, 20 mM Hepes -KOH(pH 7.6 ), 6 mM β-mercaptoethanol] or RBS buffer I [10 mM Mg(OAc)2, 500 mM NH4Cl, 20 mM Hepes-KOH (pH 7.6), 6 mM β-mercaptoethanol] and separated by ultracentrifugation in a Beckman SW-41 Rotor at 37,000 rpm for 5 h at 4 °C. The gradients were separated into an upper fraction containing ribosomal proteins and 5S rRNA and a lower fraction containing ribosomal particles. For protein analysis by SDS/PAGE, each fraction was precipitated with 20% (vol/vol) TCA, and the pellets were washed three times with acetone. For RNA analysis by denaturing PAGE, rRNAs were extracted with TRIzol LS (Invitrogen) and chloroform and then precipitated with isopropanol.
Recombinant RlmE.
The E. coli rlmE gene was PCR-amplified from the E. coli genome and inserted between the NheI and XhoI sites of pET28a to generate pET-RlmE, which was then introduced into BL21(DE3). Recombinant RlmE with an N-terminal 6× His-tag was expressed in soluble form by induction with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and then purified on a His-trap chelating HP column (GE Healthcare Bioscience) using an ÄKTA chromatography system (GE Healthcare Bioscience). The N-terminal His-tag was removed by thrombin treatment, and then the thrombin was removed on a HiTrap Benzamidine FF (high sub) column (GE Healthcare). The pooled proteins were dialyzed against a buffer consisting of 20 mM Hepes-KOH (pH 7.6), 7 mM β-mercaptoethanol, 200 mM KCl, and 10% (vol/vol) glycerol. The protein concentration was determined using the Bio-Rad protein assay kit, with BSA as the standard. Sequences of primers used in this step are listed in Table S2.
Preparation of Wash Fraction from Crude Ribosome.
E. coli crude ribosome was obtained from 1-L culture of the ΔrlmE/Δrna strain as described above. The pellet of crude ribosome was resuspended with 10 mL of RBS buffer J [20 mM Hepes-KOH (pH 7.6), 10 mM Mg(OAc)2, 1 M NH4Cl, 6 mM β-mercaptoethanol]. The resuspended ribosomes were centrifuged in a Beckman 50Ti rotor at 45,000 rpm for 16 h at 4 °C. Ten milliliters of the supernatant was collected and precipitated with ammonium sulfate with 80% saturation concentration. The precipitate was collected by centrifugation in a Beckman 50Ti rotor at 30,000 rpm for 30 min at 4 °C. The pellet was resuspended in 100% saturated solution of ammonium sulfate and collected by centrifugation at 9,100 × g for 15 min at 4 °C. Then, the pellet was dissolved in 1 mL of RBS buffer J and dialyzed against 1 L of RBS buffer J at 4 °C overnight using Slide-A-Cassette 3500MWCO 0.5–3 mL (Thermo Fisher Scientific). Protein concentration was measured by the Bradford method using BSA as a standard.
Pseudouridine Detection by CMC Treatment and Primer Extension.
Detection of pseudouridines was performed basically as described (54). Ten micrograms of rRNA was incubated at 37 °C for 60 min in 30 μL of BEU buffer [50 mM bicine (pH 8.3), 4 mM EDTA(pH 5.0), 7 M Urea] containing 0.17 M N-cyclohexyl-N′-β-(4-methylmorpholinium)ethylcarbodiimide p-tosylate (CMC). After the reaction, the rRNAs were recovered twice by ethanol precipitation, dissolved in 40 μL of 50 mM Na2CO3 (pH 10.4), and then incubated at 37 °C for 4 h. The rRNA was recovered by ethanol precipitation, and the resultant RNA pellet was rinsed twice with 70% (vol/vol) ethanol. Primer extension and gel electrophoresis were performed basically as described in Materials and Methods for hydroxyl-radical probing. To stop the cDNA synthesis, dideoxy nucleotide triphosphate (final concentration 0.5 mM) was added to the reaction mixture: ddTTP was used for Ψ955 (Fig. S12A), Ψ2580 (Fig. S12C), and Ψ2605 (Fig. S12D), and ddGTP was used for Ψ2504 (Fig. S12B). The cDNAs were analyzed by 15% PAGE in the presence of 7 M urea. Primers used for this method are listed in Table S2.
Acknowledgments
We thank the members of the Tsutomu Suzuki laboratory for materials, technical advice, and many fruitful discussions. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan (to Tsutomu Suzuki) and by a grant from the New Energy and Industrial Technology Development Organization (to Tsutomu Suzuki).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506749112/-/DCSupplemental.
References
- 1.Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from E. coli. Nature. 1970;226(5252):1214. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- 2.Herold M, Nierhaus KH. Incorporation of six additional proteins to complete the assembly map of the 50 S subunit from Escherichia coli ribosomes. J Biol Chem. 1987;262(18):8826–8833. [PubMed] [Google Scholar]
- 3.Nierhaus KH. The assembly of prokaryotic ribosomes. Biochimie. 1991;73(6):739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- 4.Nierhaus KH, Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc Natl Acad Sci USA. 1974;71(12):4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Traub P, Nomura M. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc Natl Acad Sci USA. 1968;59(3):777–784. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 7.de Narvaez CC, Schaup HW. In vivo transcriptionally coupled assembly of Escherichia coli ribosomal subunits. J Mol Biol. 1979;134(1):1–22. doi: 10.1016/0022-2836(79)90411-x. [DOI] [PubMed] [Google Scholar]
- 8.Kitahara K, Suzuki T. The ordered transcription of RNA domains is not essential for ribosome biogenesis in Escherichia coli. Mol Cell. 2009;34(6):760–766. doi: 10.1016/j.molcel.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Iost I, Bizebard T, Dreyfus M. Functions of DEAD-box proteins in bacteria: Current knowledge and pending questions. Biochim Biophys Acta. 2013;1829(8):866–877. doi: 10.1016/j.bbagrm.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Britton RA. Role of GTPases in bacterial ribosome assembly. Annu Rev Microbiol. 2009;63:155–176. doi: 10.1146/annurev.micro.091208.073225. [DOI] [PubMed] [Google Scholar]
- 11.Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol Microbiol. 2003;48(5):1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- 12.Trubetskoy D, Proux F, Allemand F, Dreyfus M, Iost I. SrmB, a DEAD-box helicase involved in Escherichia coli ribosome assembly, is specifically targeted to 23S rRNA in vivo. Nucleic Acids Res. 2009;37(19):6540–6549. doi: 10.1093/nar/gkp685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spillmann S, Dohme F, Nierhaus KH. Assembly in vitro of the 50 S subunit from Escherichia coli ribosomes: Proteins essential for the first heat-dependent conformational change. J Mol Biol. 1977;115(3):513–523. doi: 10.1016/0022-2836(77)90168-1. [DOI] [PubMed] [Google Scholar]
- 14.Peil L, Virumäe K, Remme J. Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA. FEBS J. 2008;275(15):3772–3782. doi: 10.1111/j.1742-4658.2008.06523.x. [DOI] [PubMed] [Google Scholar]
- 15.Sharpe Elles LM, Sykes MT, Williamson JR, Uhlenbeck OC. A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit. Nucleic Acids Res. 2009;37(19):6503–6514. doi: 10.1093/nar/gkp711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang M, et al. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J Bacteriol. 2006;188(19):6757–6770. doi: 10.1128/JB.00444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caldas T, et al. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J Biol Chem. 2000;275(22):16414–16419. doi: 10.1074/jbc.M001854200. [DOI] [PubMed] [Google Scholar]
- 18.Kim DF, Green R. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol Cell. 1999;4(5):859–864. doi: 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- 19.Caldas T, Binet E, Bouloc P, Richarme G. Translational defects of Escherichia coli mutants deficient in the Um(2552) 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem Biophys Res Commun. 2000;271(3):714–718. doi: 10.1006/bbrc.2000.2702. [DOI] [PubMed] [Google Scholar]
- 20.Bügl H, et al. RNA methylation under heat shock control. Mol Cell. 2000;6(2):349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- 21.Sergiev PV, et al. Modifications of Ribosomal RNA: From Enzymes to Function. Springer; Vienna: 2011. [Google Scholar]
- 22.Tan J, Jakob U, Bardwell JC. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J Bacteriol. 2002;184(10):2692–2698. doi: 10.1128/JB.184.10.2692-2698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ero R, Peil L, Liiv A, Remme J. Identification of pseudouridine methyltransferase in Escherichia coli. RNA. 2008;14(10):2223–2233. doi: 10.1261/rna.1186608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289(5481):905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 25.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310(5749):827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 26.Noeske J, et al. High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol. 2015;22(4):336–341. doi: 10.1038/nsmb.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asai T, Zaporojets D, Squires C, Squires CL. An Escherichia coli strain with all chromosomal rRNA operons inactivated: Complete exchange of rRNA genes between bacteria. Proc Natl Acad Sci USA. 1999;96(5):1971–1976. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 29.Hager J, Staker BL, Bugl H, Jakob U. Active site in RrmJ, a heat shock-induced methyltransferase. J Biol Chem. 2002;277(44):41978–41986. doi: 10.1074/jbc.M205423200. [DOI] [PubMed] [Google Scholar]
- 30.Maeder C, Draper DE. A small protein unique to bacteria organizes rRNA tertiary structure over an extensive region of the 50 S ribosomal subunit. J Mol Biol. 2005;354(2):436–446. doi: 10.1016/j.jmb.2005.09.072. [DOI] [PubMed] [Google Scholar]
- 31.Powers T, Noller HF. Hydroxyl radical footprinting of ribosomal proteins on 16S rRNA. RNA. 1995;1(2):194–209. [PMC free article] [PubMed] [Google Scholar]
- 32.Popova AM, Williamson JR. Quantitative analysis of rRNA modifications using stable isotope labeling and mass spectrometry. J Am Chem Soc. 2014;136(5):2058–2069. doi: 10.1021/ja412084b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siibak T, Remme J. Subribosomal particle analysis reveals the stages of bacterial ribosome assembly at which rRNA nucleotides are modified. RNA. 2010;16(10):2023–2032. doi: 10.1261/rna.2160010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokoyama S, Nishimura S. 1995. Modified nucleosides and codon recognition. tRNA: Structure, Biosynthesis, and Function (ASM, Washington, DC), pp 207–223.
- 35.Blanchard SC, Puglisi JD. Solution structure of the A loop of 23S ribosomal RNA. Proc Natl Acad Sci USA. 2001;98(7):3720–3725. doi: 10.1073/pnas.051608498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uicker WC, Schaefer L, Britton RA. The essential GTPase RbgA (YlqF) is required for 50S ribosome assembly in Bacillus subtilis. Mol Microbiol. 2006;59(2):528–540. doi: 10.1111/j.1365-2958.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 37.Matsuo Y, et al. The GTP-binding protein YlqF participates in the late step of 50 S ribosomal subunit assembly in Bacillus subtilis. J Biol Chem. 2006;281(12):8110–8117. doi: 10.1074/jbc.M512556200. [DOI] [PubMed] [Google Scholar]
- 38.Schaefer L, et al. Multiple GTPases participate in the assembly of the large ribosomal subunit in Bacillus subtilis. J Bacteriol. 2006;188(23):8252–8258. doi: 10.1128/JB.01213-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuo Y, Oshima T, Loh PC, Morimoto T, Ogasawara N. Isolation and characterization of a dominant negative mutant of Bacillus subtilis GTP-binding protein, YlqF, essential for biogenesis and maintenance of the 50 S ribosomal subunit. J Biol Chem. 2007;282(35):25270–25277. doi: 10.1074/jbc.M703894200. [DOI] [PubMed] [Google Scholar]
- 40.Hansen MA, Kirpekar F, Ritterbusch W, Vester B. Posttranscriptional modifications in the A-loop of 23S rRNAs from selected archaea and eubacteria. RNA. 2002;8(2):202–213. doi: 10.1017/s1355838202013365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jomaa A, et al. Functional domains of the 50S subunit mature late in the assembly process. Nucleic Acids Res. 2014;42(5):3419–3435. doi: 10.1093/nar/gkt1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li N, et al. Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit. Nucleic Acids Res. 2013;41(14):7073–7083. doi: 10.1093/nar/gkt423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato NS, Hirabayashi N, Agmon I, Yonath A, Suzuki T. Comprehensive genetic selection revealed essential bases in the peptidyl-transferase center. Proc Natl Acad Sci USA. 2006;103(42):15386–15391. doi: 10.1073/pnas.0605970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakaguchi Y, Miyauchi K, Kang B, Suzuki T. Nucleoside analysis by hydrophilic interaction liquid chromatography coupled with mass spectrometry. Methods Enzymol. 2015;560:19–28. doi: 10.1016/bs.mie.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki T, Ikeuchi Y, Noma A, Suzuki T, Sakaguchi Y. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 2007;425:211–229. doi: 10.1016/S0076-6879(07)25009-8. [DOI] [PubMed] [Google Scholar]
- 47.Miyauchi K, Kimura S, Suzuki T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat Chem Biol. 2013;9(2):105–111. doi: 10.1038/nchembio.1137. [DOI] [PubMed] [Google Scholar]
- 48.Xu Z, O’Farrell HC, Rife JP, Culver GM. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat Struct Mol Biol. 2008;15(5):534–536. doi: 10.1038/nsmb.1408. [DOI] [PubMed] [Google Scholar]
- 49.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1(3):1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 50.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 51.Schulze WX, Mann M. A novel proteomic screen for peptide-protein interactions. J Biol Chem. 2004;279(11):10756–10764. doi: 10.1074/jbc.M309909200. [DOI] [PubMed] [Google Scholar]
- 52.Barritault D, Expert-Bezancon A, Guérin MF, Hayes D. The use of acetone precipitation in the isolation of ribosomal proteins. Eur J Biochem. 1976;63(1):131–135. doi: 10.1111/j.1432-1033.1976.tb10215.x. [DOI] [PubMed] [Google Scholar]
- 53.Schägger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1(1):16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 54.Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: Analysis by the application of a new sequencing technique. Biochemistry. 1993;32(37):9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- 55.Jain C. The E. coli RhlE RNA helicase regulates the function of related RNA helicases during ribosome assembly. RNA. 2008;14(2):381–389. doi: 10.1261/rna.800308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barkan A, et al. The CRM domain: An RNA binding module derived from an ancient ribosome-associated protein. RNA. 2007;13(1):55–64. doi: 10.1261/rna.139607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kimura S, et al. Base methylations in the double-stranded RNA by a fused methyltransferase bearing unwinding activity. Nucleic Acids Res. 2012;40(9):4071–4085. doi: 10.1093/nar/gkr1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Del Campo M, Kaya Y, Ofengand J. Identification and site of action of the remaining four putative pseudouridine synthases in Escherichia coli. RNA. 2001;7(11):1603–1615. [PMC free article] [PubMed] [Google Scholar]
- 59.Conrad J, Sun D, Englund N, Ofengand J. The rluC gene of Escherichia coli codes for a pseudouridine synthase that is solely responsible for synthesis of pseudouridine at positions 955, 2504, and 2580 in 23 S ribosomal RNA. J Biol Chem. 1998;273(29):18562–18566. doi: 10.1074/jbc.273.29.18562. [DOI] [PubMed] [Google Scholar]