Significance
Laboratory experiments show that bacteria have surprisingly complex social lives: Like humans, they can cooperate but also cheat each other. Cooperation could benefit bacteria causing infection by coordinating attack and producing toxins in a collective effort. But can cheaters, exploiting the work of others, affect the outcome of infection? We show that populations of bacteria causing chronic lung infections in cystic fibrosis patients contain cheaters that freeload to the point where cooperation no longer pays off by not producing a compound that helps them steal iron from blood. Bad news for bacteria but good for us if we can find ways to meddle in their social lives.
Keywords: social evolution, infection, cooperation, cheating, cystic fibrosis
Abstract
Laboratory experiments show that social interactions between bacterial cells can drive evolutionary change at the population level, but significant challenges limit attempts to assess the relevance of these findings to natural populations, where selection pressures are unknown. We have increasingly sophisticated methods for monitoring phenotypic and genotypic dynamics in bacteria causing infectious disease, but in contrast, we lack evidence-based adaptive explanations for those changes. Evolutionary change during infection is often interpreted as host adaptation, but this assumption neglects to consider social dynamics shown to drive evolutionary change in vitro. We provide evidence to show that long-term behavioral dynamics observed in a pathogen are driven by selection to outcompete neighboring conspecific cells through social interactions. We find that Pseudomonas aeruginosa bacteria, causing lung infections in patients with cystic fibrosis, lose cooperative iron acquisition by siderophore production during infection. This loss could be caused by changes in iron availability in the lung, but surprisingly, we find that cells retain the ability to take up siderophores produced by conspecifics, even after they have lost the ability to synthesize siderophores. Only when cooperative producers are lost from the population is the receptor for uptake lost. This finding highlights the potential pitfalls of interpreting loss of function in pathogenic bacterial populations as evidence for trait redundancy in the host environment. More generally, we provide an example of how sequence analysis can be used to generate testable hypotheses about selection driving long-term phenotypic changes of pathogenic bacteria in situ.
Some of the most important bacterial pathogens are opportunistic in the sense that they infect a compromised human host from the surrounding environment. In cases where such infections become persistent, the evolutionary changes accompanying the transition from the environment to the human body have been the subject of intensive research, and we now have some information on what distinguishes clinical from environmental isolates (1). Surprisingly, our understanding of how this process is driven by selection often remains speculative. To study bacterial cells, we must remove them from the host environment into the laboratory, which may release them from the selection pressures that we wish to understand.
In parallel, progress has been made in understanding how bacterial populations respond to selection through in vitro experimental evolution. These studies show that phenotypic dynamics result not only in response to the environment but also, to social interactions as bacteria cooperate and compete with one another (2). Selection to outcompete neighbors can even lead to loss of traits that increase survival in the environment but are costly to produce (3–5). Such loss has been shown for a range of traits, such as extracellular enzymes, signaling molecules, and iron chelators (2). These exoproducts act as “public goods”: products that are beneficial to the group but vulnerable to exploitation by cheats that reap the benefit without paying the cost (6). Understanding selection on public goods is clinically relevant, because many are virulence factors (7, 8), and social interactions have also been shown experimentally to affect infection dynamics in vivo (9, 10).
We investigate the importance of social interactions in infectious populations of Pseudomonas aeruginosa, which is both a model organism of social evolution research and the primary cause of chronic lung infection in patients with the genetic disorder cystic fibrosis (CF). CF patients usually acquire their first P. aeruginosa infection in childhood, and these infections can persist for years, despite antibiotic treatment (11). P. aeruginosa produces an iron-scavenging molecule, pyoverdine, that acts as a cooperative public good in vitro (12). Iron is essential for growth but bound to transferrin, heme, and hemoglobin in the human host (13). P. aeruginosa circumvents this by releasing pyoverdine, which binds to iron and is taken up by a specific receptor. Detection of pyoverdine and expression of pyoverdine genes in sputum samples confirm that the pathway is active, and likely beneficial, in the CF lung environment (14, 15). However, cells that are deficient in production (i.e., potential cheaters) have also repeatedly been isolated from patients (16, 17). The pyoverdine metabolism is, therefore, an ideal system for testing whether social dynamics observed in the laboratory also occur in human hosts.
Our aim is to identify selection pressures driving any changes that we observe in pyoverdine production in the lung. Pyoverdine production may be an adaptive response to acquire a limited nutrient. It may be lost, therefore, in response to availability of other iron sources (18–20). Alternatively, production may be lost from the population even if iron is limiting as a result of cooperator–cheat dynamics. Crucially, patterns of evolution of the pyoverdine system differ depending on whether adaptation to the human lung or social interactions drive selection (Fig. 1). If pyoverdine does not provide a growth benefit in the lung, the entire system will be redundant, including receptor function. In contrast, if pyoverdine production is lost because of cheating, receptor function will remain beneficial as long as extrinsic pyoverdine is available. Only when cheating is not possible does the receptor also become redundant. We can, therefore, distinguish between the two selection pressures by determining if and when receptor function is maintained in bacteria that have lost the ability to produce pyoverdine.
Fig. 1.
The pyoverdine system. (Upper) The pyoverdine receptor FpvA spans the cell wall. In the absence of bound pyoverdine, the anti-σ factor FpvR inhibits the expression of σ-factors FpvI and PvdS. Pyoverdine acquires iron from transferrin. When ferripyoverdine binds to the receptor, FpvR releases FpvI and PvdS. Release of FpvI initiates synthesis of the receptor FpvA, and PvdS initiates synthesis of pyoverdine (illustrated by arrows). (Lower Left) If pyoverdine production is lost as an adaptation to the lung, receptor function also becomes redundant, irrespective of whether pyoverdine produced by neighbors is available. (Lower Right) However, if pyoverdine production is lost because of cheating, we expect to see retention of receptor function in the presence of pyoverdine produced by others and function only lost in the absence of pyoverdine.
Two Danish collections of genome-sequenced P. aeruginosa isolates provide the opportunity to study selection on pyoverdine metabolism in CF patients (Dataset S1). The first collection gives a detailed insight into changes occurring during the first 10 y of infection across 36 young CF patients with 54 different clone types (21), representing the transition from initial colonization to chronic infection. With frequent and extensive sampling from each patient (451 isolates; on average, 13 per patient), we can estimate the point of colonization of each clone type and thereby, the time period over which a given isolate has evolved. The second collection provides insight into the long-term dynamics of two clone types causing chronic infections, with samples from 24 adult patients (85 isolates) infected with the two Danish transmissible clone types DK1 and DK2 (22–24). The two transmissible clone types established and spread in the Danish CF patient group from 1973 and all of the older patients (who got chronically infected up to the beginning of the 1990s) harbor one or both of these. Afterward, segregation of patients in the clinic has largely eliminated transmission of these clone types. The DK1 and DK2 isolates, thus, typically come from now older CF patients who have each been sampled a few times, providing insight into the long-term dynamics but not on a fine scale at the early infection stage. For some of the analyses, as specified below, only the isolates from the young patients have been used.
Pyoverdine Producers and Nonproducers Co-Occur Within Patients
To detect changes in pyoverdine production during infection, we first measured pyoverdine production in 529 isolates covering more than 240 y of infection. For the first collection of isolates from regularly sampled patients, this analysis revealed a long-term decline in pyoverdine production from the point of colonization of the lung [Markov chain Monte Carlo generalized linear mixed model: b = −481 relative fluorescence units (RFUs) standardized by cell density per year since colonization, P = 0.018] (Fig. S1 and Table S1). Isolates that produced less pyoverdine than a threshold were classified as nonproducers. These nonproducers exhibited severely reduced abilities to grow in iron-limited media [Welch two-sample t test: t = −14.81, degrees of freedom (df) = 64.46, P < 0.01]; they comprised 12% of the collection (n = 54 isolates of 14 clone types) and were found in 41% (n = 15) of the young patients (Fig. S2). The nonproducers were sampled throughout the length of infection, but the proportion of nonproducers to producers increased with time (Fig. S3), resulting in a significantly later mean time of sampling for nonproducers (t = 4.14, df = 63.32, P < 0.001). Of the transmissible DK1 and DK2 isolates from the second collection, 64% (n = 50 isolates of two clone types from 20 patients) did not produce pyoverdine. Pyoverdine production varies greatly across the duration of infection, and there is frequently co-occurrence of producers and nonproducers within patients, providing the possibility for nonproducers to cheat by exploiting the supply of pyoverdine provided by other cells (Fig. S2).
Fig. S1.
Pyoverdine production measured as RFUs standardized by OD significantly decreases with length of infection in years. Isolates come from 36 different patients who were longitudinally sampled, covering 54 different clone types and three types of pyoverdine that fluoresce at different strengths. Each dot represents the pyoverdine production of one isolate (n = 451). The line represents the average decrease in pyoverdine production controlling for patient identity and pyoverdine type.
Table S1.
Pyoverdine production decreases over infection time
| Variable | Explained variation |
| Length of infection; average and lower and upper limits (RFUs standardized by OD per year) | 45%; −459, −708, and −182 |
| Pvd type: average and lower and upper limits | 30%, 4%, and 88% |
| Patient identification: average and lower and upper limits | 26%, 23%, and 50% |
| DIC | 8,934 |
Results of the Markov chain Monte Carlo generalized linear mixed model analyzing the effect on pyoverdine production of the length of infection, with pvd type and patient identification as random factors. DIC, deviance information criterion.
Fig. S2.
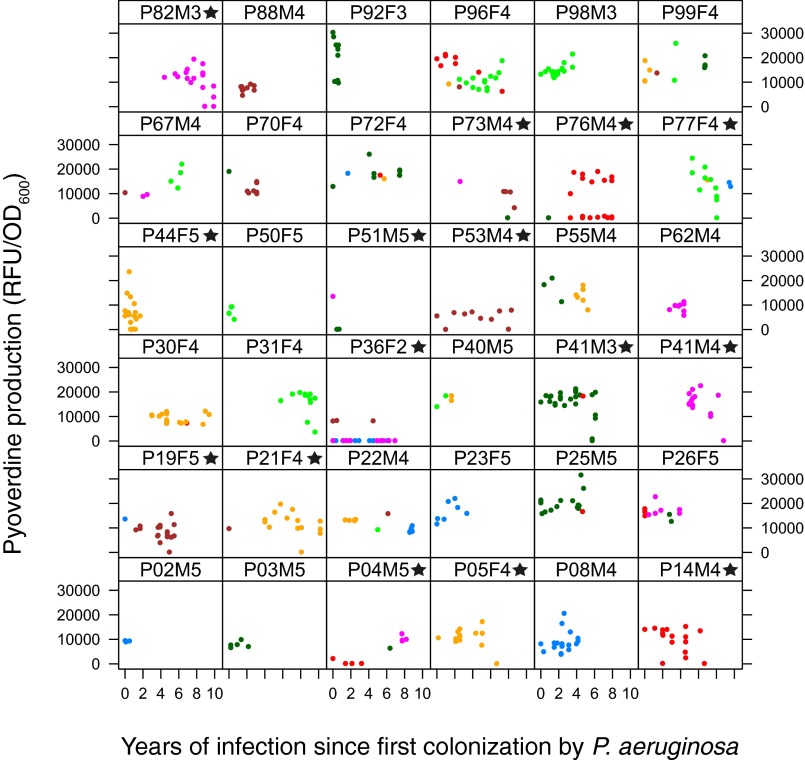
Pyoverdine production over the length of infection in years for individual patients (measured as RFUs standardized by OD). Length of infection is given as the time between sampling of an isolate and the first recording of Pseudomonas aeruginosa from the patient, irrespective of whether this isolate was included in the collection (except for patient P36F2, who, after an initial sampling of P. aeruginosa, tested negative for >10 y; therefore, only the later isolates are included). Patients who harbor isolates that do not produce pyoverdine are marked with stars. Colors represent different clone types.
Fig. S3.
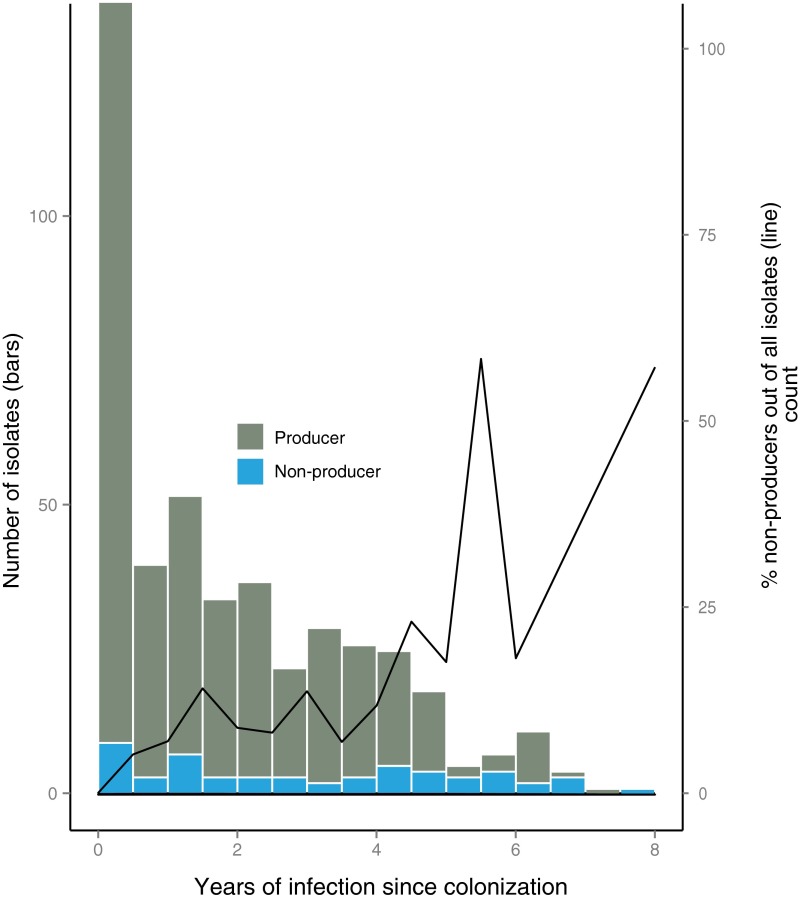
Distribution of pyoverdine-producing and -nonproducing isolates over the length of infection in years. The bars show the numbers of isolates (left y axis) cumulated for 6 mo. The distribution of pyoverdine producers (green; n = 397) is left-skewed, in part because some young patients have been followed for fewer years and in part because clone types isolated for the first time in a patient were recorded as from 0 y of infection, irrespective of other clone types harbored by the patient (Materials and Methods). Nonproducing isolates (blue; n = 54) are more evenly distributed over time. The line shows that the proportion of nonproducers to producers (right y axis), therefore, increases over time. Because of low counts for the last years (seven isolates), samples from 6 to 8 y have been cumulated.
Selection Targets Genes Involved in Both Pyoverdine Production and Uptake
Sequence analysis supports the argument that pyoverdine metabolism is a target of selection. The pyoverdine region is well-characterized (25), and the distribution of mutations across the pyoverdine genes in isolates from both collections was not random: two genes accumulated a higher number of mutations than expected by a random distribution. One is the σ-factor affecting pyoverdine biosynthesis, pvdS (pyoverdine sigma factor) [8.6× higher; P(X ≥ 14) ∼ poisson distribution (pois) (X; 1.63) < 0.001] (Fig. 2). Expression of pvdS initiates pyoverdine production (Fig. 1), and therefore, a KO of this is the most efficient way to stop production, because no costly intermediate compounds will be produced. The second target is the gene for the specific pyoverdine receptor, fpvA (ferric pyoverdine receptor A) [4.9× higher; P(X ≥ 34) ∼ pois(X; 6.92) < 0.001] (Fig. 2). Mutations in genes affecting receptor function were only observed in nonproducing isolates. These results suggest that not only pyoverdine production but also its uptake is under selection in the lung.
Fig. 2.
Distribution of mutations across the pyoverdine genes given as the ratio of observed to expected numbers of nonsynonymous SNPs and indels. Colors follow those used in Fig. 1, and other genes involved in production are light green. There were significantly more mutations than expected by random distribution in the genes pvdS and fpvA and significantly fewer mutations in the large gene pvdL (marked by an asterisk; a value of one indicates no difference). *P < 0.05.
To test whether the lung or social interactions are driving selection on the pyoverdine region, we first ensure that the underlying assumptions made by both hypotheses are met. The first hypothesis is that the observed mutations in the receptor genes compromise the ability to take up the ferripyoverdine complex, and growth assays supported this claim. We purified pyoverdine from producing isolates, and the effect of the addition of this to nonproducing isolates growing in iron-limited media was dependent on the presence of receptor mutations. Although growth in both groups was stimulated, likely because of the presence of small amounts of other iron chelators (such as pyocyanin), isolates without mutations showed a significantly higher increase than those with mutations (t = 3.13, df = 18.63, P < 0.05) (Fig. 3A).
Fig. 3.
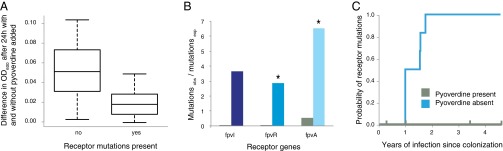
Distribution of receptor mutations supports a social adaptation scenario. (A) Presence of receptor mutations predicts function. Nonproducing isolates were grown with and without the addition of pyoverdine. Isolates without mutations (n = 16 lines of 15 clone types) showed greater induction of growth compared with those with mutations (n = 7 lines of 5 clone types; P < 0.05). Box plots of the difference in OD600 with and without pyoverdine. The middle band represents the median, the bottom and top boxes represent the 25th and 75th percentiles, respectively, and the lower and upper whiskers represent the 5th and 95th percentiles, respectively. (B) The number of observed mutations in genes affecting receptor synthesis is higher than expected in the absence of pyoverdine producers (colors follow those in Fig. 1) but not in the presence of pyoverdine producers (green bars), shown as mutations observed per mutations expected. *P < 0.05 for fpvR and fpvA. (C) Loss of receptor function is dependent on the social environment. Kaplan–Meier graph showing that the probability of acquiring receptor mutations is significantly higher when cheating on pyoverdine producers is not possible (blue line; n = 6) compared with when it is (green line; n = 4). Ticks show when the samples were censored.
The second assumption is that maintenance of the receptor is costly and that retention, therefore, indicates selection on the ability to uptake the ferripyoverdine complex. Our data also support this, because the high mutation rate of the receptor gene fpvA suggests that it is a target of selection. The cost is unlikely to be that of biosynthesis alone, because receptor expression is partly regulated by the binding of the ferripyoverdine complex, limiting expression in the absence of pyoverdine (Fig. 1). However, phage-like elements, such as some pyocins, can exploit the pyoverdine receptor and may select for modifications (26). In support of this idea, we found by protein structure prediction that mutations were significantly biased toward the extracellular region of the receptor, where interaction with and recognition of ligands occur [1.8× higher; P(X ≥ 20) ∼ pois(X; 10.88) < 0.05] (Fig. 4). Additional support for a cost of even low receptor expression comes from the transmissible DK2 clone type. In 1973, DK2 acquired a mutation in the σ-factor fpvI gene, canceling up-regulation of receptor expression in the presence of pyoverdine (Fig. 1) and also, lowering background receptor expression (18, 27). This mutation is, however, followed by no fewer than 17 unique nonsynonymous fpvA mutations in nine independent lines across patients, suggesting that even limited background expression of fpvA is costly.
Fig. 4.
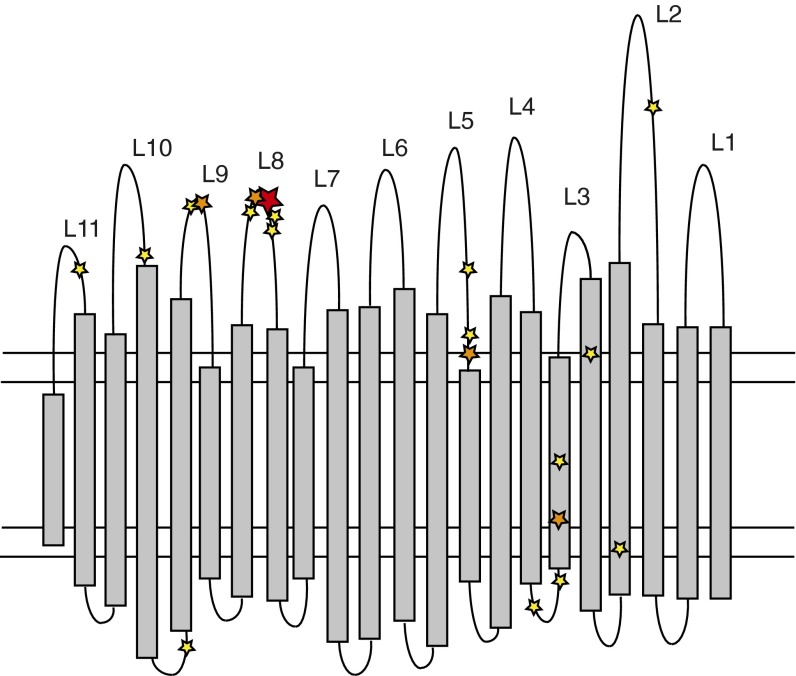
Receptor mutations found in the fpvA IIA receptor gene mapped onto the predicted transmembrane β-barrel of FpvA IIA. The β-sheets are represented by gray boxes, and the loops are represented by black lines. The extracellular loops are numbered L1–L11. Yellow stars indicate one nonsynonymous SNP or indel, orange stars represent two nonsynonymous SNPs or indels, and a red star represents four events. In L8, five adjacent amino acids were found to be altered by mutations (one of these amino acids four times, twice in both the DK1 and DK2 clonetype).
Social Interactions Drive Selection on Pyoverdine Metabolism
Given that the receptor is costly to maintain for isolates not producing pyoverdine, we then tested if the possibility of cheating selects for retention of receptor function. We found that the social environment, indeed, is key, because loss of function was dependent on loss of pyoverdine production by coinfecting isolates and not the focal isolate. In the presence of pyoverdine producers, where cheating is possible, mutations in the receptor genes [fpvA, fpvR, and fpvI (named after fecR and fecI in E. coli)] are very rare. However, in the absence of extrinsic pyoverdine, the number of mutations is significantly higher than expected for fpvA [P(X ≥ 33) ∼ pois(X; 5.06) < 0.001] and the anti-σ factor fpvR [P(X ≥ 6) ∼ pois(X; 2.1) < 0.05]. For fpvI, P(X ≥ 3) ∼ pois(X; 0.82) = 0.05 (Fig. 3B). Longitudinal sampling of nonproducing isolates from 10 patients further allowed us to measure latency to respond to changes in the social environment (Materials and Methods). Cheaters maintain receptor function for at least 4.6 y when co-occurring with pyoverdine-producers (n = 4 clone types; extent of sampling of nonproducers within a patient between 0.28 and 4.6 y). In contrast, mutations accumulate significantly faster in the absence of extrinsic pyoverdine—in less than 2 y after loss of pyoverdine (n = 6 clone types; nonproducers acquire mutations in receptor genes between 0 and 1.8 y after pyoverdine producers were last sampled from the patient, χ2 = 5.4, df = 1, P < 0.05) (Fig. 3C).
In conclusion, we find that the pyoverdine system of P. aeruginosa infecting lungs evolves in response to changes in the social environment, as has been found in vitro (12). Iron availability is likely to also be an important selection pressure on the pyoverdine system, which is evidenced by the persistence of this trait over years of infection in some patients. Some isolates of the DK1 clone type, for example, still produce pyoverdine, despite nearly 40 y of infection history. Crucially, however, the fitness benefits of cheating are sufficient to lead to loss of this trait from the population repeatedly. The conditions determining whether cooperation persists or breaks down are currently unknown. Subsequently, P. aeruginosa will have to acquire iron through other routes. Intriguingly, two independent studies (one on some of the isolates described here) find that the loss of pyoverdine production is followed by a shift toward private iron acquisition through increased expression of heme receptors without the use of siderophores (18, 20). This finding supports a general pattern of breakdown of cooperative behaviors late in infection (17).
This study shows that cooperator–cheat interpretations of clinical observations can be warranted, such as those in the recent study by Köhler et al. (28), which suggested that P. aeruginosa quorum sensing (QS) mutants in acute lung infections arise as cheats and not because QS is redundant in the lung. Furthermore, an intriguing experimental demonstration of the potential clinical relevance of microbial social interactions comes from the work by Koch et al. (29), which showed that intraspecific competition can select for antibiotic-resistant Staphylococcus aureus bacteria in the absence of antibiotic pressure. Together with studies on cooperator–cheat dynamics in natural populations of bacteria, such as iron acquisition of Vibrio in seawater (30) and toxin production in Bacillus infecting moth larvae (31), this finding calls for a reevaluation of how we interpret evolutionary change of natural microbial populations.
Materials and Methods
Sampling.
P. aeruginosa samples were collected by bronchoalveolar lavage and endolaryngal suction and from expectorates or acquired during endoscopic sinus surgery at the Copenhagen Cystic Fibrosis Center, Rigshospitalet as described previously (32). From a culture plate of the samples, one to four isolates were selected as representative of the dominant microbiota. For the study of early evolution of the pyoverdine system, 451 isolates of 54 different clone types from 36 young CF patients were included. The patient age at first infection varied from 1.4 to 25.7 y (mean ± SD = 9.38 ± 6.15 y), and the samples were collected over a range of 0.5–10.17 y for each patient (21). To address the long-term adaptations, 85 isolates of the two Danish transmissible clone types DK1 and DK2 from 24 patients were added (seven samples were available as genome sequences only) that were sampled between 1973 and 2012 (22–24).
Measurement of Pyoverdine Production.
Pyoverdine production was measured following the work by Kümmerli et al. (33). Isolates were cultured from frozen stocks in 2 mL King's B (KB) medium in 24-well plates and incubated overnight at 37 °C. The cultures were read at optical density 600 nm (OD600), standardized to an OD600 of 0.1 by dilution with M9 minimal media, and inoculated into 96-well plates with iron-limited casamino acid (CAA) medium (200 μL media, 2 μL culture in six replicates). After incubation for 48 h at 37 °C, the fluorescence was measured at 400/460 nm excitation/emission with a 475-nm cutoff in addition to OD600. Pyoverdine production per cell was calculated as RFUs corrected for OD600. No reliable measurements were obtained from 45 isolates, because they failed to grow in the iron-limited media (OD600 > 0.03 after 48 h of incubation). These isolates were scored as nonproducers and assigned a fluorescence measure of 150 RFUs standardized by OD, comparable with that of other nonproducing isolates when grown in iron-limited CAA medium. For the low-producing isolates, there was a distinct gap between isolates producing <880 RFUs standardized by OD (54 isolates; lower 11.5th quantile of the distribution) and the remainder producing >2,020 RFUs standardized by OD. The former were classified as nonproducers.
The effect of the length of infection on pyoverdine production was tested in R (www.R-project.org) with a Markov chain Monte Carlo generalized linear mixed model using the MCMCglmm package (34). The length of infection at sampling time was estimated as the number of years since the first recording of the sampled clone type in the patient. The pvd type and patient identification were included as random effects. The random effects were assigned uninformative priors, and the model was run with 3 million iterations, of which the first 500,000 iterations were discarded as burn in. Sample distributions were visualized in R with the ggplot2 package (35). The three pvd types previously characterized for P. aeruginosa were all present in young patients. The pvd type II (214 isolates and 25 clone types) was found to dominate in 16 patients; the pvd type I (175 isolates and 22 clone types) was found to dominate in 13 patients, 1 patient had two isolates of the pvd type I and two isolates of the pvd type II, and 5 patients predominantly had pvd type III (62 isolates and 7 clone types). In 19 patients, more than one P. aeruginosa clone type was found, and in 12 of these, multiple pvd types were present. The mean RFUs standardized by OD were significantly higher for pvd type I than for pvd types II and III and significantly lower for pvd type II compared with pvd type III (nonproducers were excluded; one-way ANOVA, F = 69.98, df = 2, P > 0.001).
Test of Receptor Function.
The 54 isolates from young patients and the 50 isolates from older patients who were found to not produce pyoverdine were tested for their ability to take up pyoverdine, because this uptake is a prerequisite for them to act as cheats. A purified sterile solution of pyoverdine was obtained following the work by Meyer et al. (36). In short, a producing strain was grown in 5 mL CAA overnight at 37 °C and 9 × g, transferred to 250 mL CAA, and grown overnight at 37 °C and 14 × g. The culture was centrifuged at 9,400 × g for 15 min, and the supernatant was passed through an XAD-4 Amberlite Column. The column was washed with ddH2O, and the pyoverdine was eluted with methanol and distilled deionized water (ddH20) [50%/50% (vol/vol)], dried, dissolved in ddH2O, filter-sterilized, and standardized so that 2 μL inoculated in 200 μL CAA was equivalent to the RFUs of a WT-producing isolate after 24 h of culture. The nonproducing isolates were grown in KB medium overnight; OD600 standardized to 0.1 and 2 μL inoculated into a 96-well plate with iron-limited CAA media in six replicates with and without purified pyoverdine were added to the media. Wells with purified pyoverdine without cells served as negative controls. OD600 was measured after 24 h of incubation at 37 °C as described above. The growth induction was calculated as the difference in OD600 between cells grown with and without pyoverdine after 24 h of growth. Nonproducers were tested with pyoverdine of their own pvd type. The distribution of clone types in the collection of nonproducing isolates was heavily skewed toward the transmissible DK1 and DK2 isolates. To account for this skew, the average growth increase was calculated for each clone type for each independent line of mutations, resulting in 16 lines of 15 different clone types without receptor mutations and 7 different clone types with receptor mutations, of which 3 were DK1 lines. Bacterial fitness in iron-limited CAA is strongly correlated with the production of pyoverdine (37). To confirm that an observed fitness benefit by the addition of supernatant was caused by pyoverdine, the experiment was repeated in CAA not iron-limited by apotransferrin for nine isolates, representing all three pvd types. These controls showed no significant effect of added pyoverdine when iron is not bound by transferrin (paired t test: t = 0.74, df = 8, P = 0.48).
Identification of Mutations in Pyoverdine Genes.
Mutations in genes known to be important for the production of pyoverdine were identified by sequencing isolates [Illumina HiSeq for isolates from the young patients as described previously (21) and Illumina’s GAIIx or HiSeq2000 for the transmissible DK1 and DK2 clone types as described previously (22–24)]. Previously unpublished sequences are described in Dataset S2. The reads were mapped to reference sequences with the Burrows–Wheeler alignment tool (bio-bwa.sourceforge.net), and alignments were generated with the paired-end reads setting or single-end reads setting. Alignments were filtered to remove unmapped reads, sorted, and indexed, and each isolate was assigned to a read group using Picard Tools (broadinstitute.github.io/picard/). Differences between isolates of the same clone type were identified using Samtools (samtools.sourceforge.net), and mutations were manually checked using IGV (www.broadinstitute.org/igv). Dataset S5 shows the full pipeline used. Whether SNPs were synonymous or nonsynonymous was determined manually in ExPASy translate (web.expasy.org/translate). Reads from all isolates were mapped to four reference regions: the pvdS/pvdG/pvdL region containing the σ-factor controlling the main pyoverdine operon; the main pyoverdine operon containing 17 genes, which is highly diverse with three different variants characterized (25); the pyoverdine chromophore (pvc) region coding the chromophore biosynthesis gene cluster; and the gene for the alternative pyoverdine receptor fpvB (Datasets S3 and S4). For one patient with 18 isolates, the sequencing depth was only of sufficient quality to determine the clone type and not SNPs and indels between isolates. Larger deletions were called conservatively, and therefore, varying sequencing depth of, in particular, some of the single-end reads may have caused some of such events to not have been identified. Isolates of one of the transmissible strains of the Copenhagen CF community, DK1, were observed in one patient as a result of transmission from an older patient (21). Mutations in these and the isolates from the second collection were identified by comparison with the WT-like isolates of both from 1973.
Mutations were identified in 56 of the isolates from the young patients, 12 clone types from 13 patients, and 68 isolates from of the DK1 and DK2 clone types from 19 patients (including 11 isolates of DK1 from a young patient). We identified 171 nonsynonymous SNPs, 35 deletions, 3 insertions, 93 synonymous SNPs, and 8 intergenic mutations from the complete isolate collection (Datasets S3 and S4). For each gene, we calculated the expected number of mutations given a random distribution. This expected value was found as the total number of independent nonsynonymous SNPs and indels (209) divided by the average size of the pyoverdine region (72.228 kb) times the gene size (0.39–13.02 kb). Two genes (pvdJ and pvdI) differ in size between the pyoverdine types, and an average was used. The pvdD gene is only found in types I and III, and the expected number of mutations was weighted by the distribution of pvd- types (22% of samples with mutations were of types I and III). The probability of finding the observed distribution of mutations was calculated per gene as P(X ≥ mutationsobserved) ∼ pois(X; mutationsexpected) < 0.05. All mutations were classified as having occurred in the presence (27% of all mutations) or absence of pyoverdine production (73% of all mutations; sampling of a producing isolate from the same patient less than 1 y before the mutation occurred). For the receptor genes (fpvI, fpvA, and fpvR), the expected number of mutations in the presence and absence of pyoverdine was compared with the observed as described above. The older patients harboring DK1 and DK2 isolates had been sampled less frequently than the young patients, and therefore, the order of mutations accumulating could not always be inferred. When an isolate had acquired mutations affecting both production and receptor, we expected that production had been lost first, but in these cases, all mutations were characterized as having occurred in the absence of production.
Mutations in the fpvA receptor gene were further localized to functional regions. The crystal structure of the pvd type I receptor has been resolved (38), but this protein has only 28% similarity at the amino acid level, with types II and IIA receptors harboring 94% of the mutations. With the PRED-TMBB tool (bioinformatics.biol.uoa.gr/PRED-TMBB), the strands and loops of the β-barrel of FpvA I, FpvA II, FpvA IIA, and FpvA III were predicted, and for FpvA I, a good congruence with the known crystal structure was found. Of 34 mutations, 31 were found in the β-barrel of the receptor (Fig. 4). The extracellular loops of the β-barrel make up 32% of the protein, but 20 mutations (58%) were found in the coding regions for these loops. Two mutations were in the plug part of the protein, and the remainder was in the β-sheets (5), the cytosolic loops (3), or the N terminus of the receptor (3).
For nonproducing isolates, we tested whether mutations in receptor genes fpvI, fpvA, and fpvR were more likely to occur in the absence than in the presence of pyoverdine producers and at the timescale at which mutations occur. For the majority of patients, we only had one nonproducing sample of a given clone type (27 samples; one had an fpvA mutation), but for four patients, we had two or more longitudinal samples of nonproducing isolates (the same clone type with the same mutations) where pyoverdine-producing isolates had been sampled <1 y before the nonproducers. The extent of sampling of nonproducers from each of these patients varied between 0.28 and 4.6 y. None of these carried mutations in the receptor genes. For six patients, we had nonproducing isolates where no other producers had been sampled >1 y before. All of these lines acquired receptor mutations over the sampling period. Three lines had been sampled repeatedly <2 y apart (after 1.58, 1.6, and 1.8 y), and mutations were assumed to have occurred in this short timeframe. The third was the transmissible DK2 line first sampled as a WT pyoverdine-producing strain in 1973 from one patient and then later the same year, from another patient with a mutation in both σ-factor genes, pvdS and fpvI. Both were assumed to have been mutated within 1 y. The last two were DK1 transmissible clone types, where the first isolate sampled from the patient was negative for both production and receptor and thus, scored as having lost receptor function within the first year of production loss, although the exact timing was unclear. For one of these patients, P28F1, producing isolates were also sampled, but whole-genome phylogenetic analysis following the work by Markussen et al. (22) suggested that the first receptor mutation had occurred in patient P46F0 in the absence of pyoverdine producers and subsequently, had been transmitted to P28F1. The analysis was run first with the latter two cases scored as receptor lost within 1 y of loss of pyoverdine production and subsequently, more conservatively without these two lines. Using the survival package in R (39), we compared the probability of acquiring no receptor mutations in the presence or absence of producing isolates. In both cases, we found a significant difference, because mutations only accumulated in the absence of pyoverdine [χ2 = 5.4 (all samples)/4.2 (two samples excluded), df = 1, P < 0.05].
Supplementary Material
Acknowledgments
We thank Melanie Ghoul for guidance on the pyoverdine measurements, Rolf Kümmerli for advice on pyoverdine purification, and Macarena Toll-Riera for sharing her expertise in bioinformatics. We also thank Melanie Ghoul, Kevin Foster, Paul Harvey, and Stuart West for providing feedback on the manuscript. S.B.A. was funded by Villum Fonden Grant 95-300-13894 and Lundbeck Fonden Grant R108-2012-10094. H.K.J. was funded by a clinical research stipend from The Novo Nordisk Foundation and Rigshospitalet Rammebevilling 2015-17 and Lundbeckfonden Grant R167-2013-15229. A.S.G. was funded by the Royal Society, a L'Oreal/UNESCO For Women In Science award, and a European Research Council grant (SESE).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper are in Dataset S2.
See Commentary on page 10577.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508324112/-/DCSupplemental.
References
- 1.Sokurenko EV, Hasty DL, Dykhuizen DE. Pathoadaptive mutations: Gene loss and variation in bacterial pathogens. Trends Microbiol. 1999;7(5):191–195. doi: 10.1016/s0966-842x(99)01493-6. [DOI] [PubMed] [Google Scholar]
- 2.Xavier JB. Social interaction in synthetic and natural microbial communities. Mol Syst Biol. 2011;7:483. doi: 10.1038/msb.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Velicer GJ, Kroos L, Lenski RE. Developmental cheating in the social bacterium Myxococcus xanthus. Nature. 2000;404(6778):598–601. doi: 10.1038/35007066. [DOI] [PubMed] [Google Scholar]
- 4.Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450(7168):411–414. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 5.Sandoz KM, Mitzimberg SM, Schuster M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci USA. 2007;104(40):15876–15881. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4(8):597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 7.Popat R, Crusz SA, Diggle SP. The social behaviours of bacterial pathogens. Br Med Bull. 2008;87(1):63–75. doi: 10.1093/bmb/ldn030. [DOI] [PubMed] [Google Scholar]
- 8.Leggett HC, Brown SP, Reece SE. War and peace: Social interactions in infections. Philos Trans R Soc Lond B Biol Sci. 2014;369(1642):20130365. doi: 10.1098/rstb.2013.0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rumbaugh KP, et al. Quorum sensing and the social evolution of bacterial virulence. Curr Biol. 2009;19(4):341–345. doi: 10.1016/j.cub.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 10.Czechowska K, McKeithen-Mead S, Al Moussawi K, Kazmierczak BI. Cheating by type 3 secretion system-negative Pseudomonas aeruginosa during pulmonary infection. Proc Natl Acad Sci USA. 2014;111(21):7801–7806. doi: 10.1073/pnas.1400782111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen SK, et al. Evolution and diversification of Pseudomonas aeruginosa in the paranasal sinuses of cystic fibrosis children have implications for chronic lung infection. ISME J. 2012;6(1):31–45. doi: 10.1038/ismej.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin AS, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430(7003):1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 13.Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010;6(8):e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunt TA, Peng W-T, Loubens I, Storey DG. The Pseudomonas aeruginosa alternative sigma factor PvdS controls exotoxin A expression and is expressed in lung infections associated with cystic fibrosis. Microbiology. 2002;148(Pt 10):3183–3193. doi: 10.1099/00221287-148-10-3183. [DOI] [PubMed] [Google Scholar]
- 15.Haas B, Kraut J, Marks J, Zanker SC, Castignetti D. Siderophore presence in sputa of cystic fibrosis patients. Infect Immun. 1991;59(11):3997–4000. doi: 10.1128/iai.59.11.3997-4000.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Vos D, et al. Study of pyoverdine type and production by Pseudomonas aeruginosa isolated from cystic fibrosis patients: Prevalence of type II pyoverdine isolates and accumulation of pyoverdine-negative mutations. Arch Microbiol. 2001;175(5):384–388. doi: 10.1007/s002030100278. [DOI] [PubMed] [Google Scholar]
- 17.Jiricny N, et al. Loss of social behaviours in populations of Pseudomonas aeruginosa infecting lungs of patients with cystic fibrosis. PLoS One. 2014;9(1):e83124. doi: 10.1371/journal.pone.0083124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marvig RL, et al. Within-host evolution of Pseudomonas aeruginosa reveals adaptation toward iron acquisition from hemoglobin. MBio. 2014;5(3):e00966-14. doi: 10.1128/mBio.00966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter RC, et al. Ferrous iron is a significant component of bioavailable iron in cystic fibrosis airways. MBio. 2013;4(4):e00557-13. doi: 10.1128/mBio.00557-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen AT, et al. Adaptation of iron homeostasis pathways by a Pseudomonas aeruginosa pyoverdine mutant in the cystic fibrosis lung. J Bacteriol. 2014;196(12):2265–2276. doi: 10.1128/JB.01491-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marvig RL, Sommer LM, Molin S, Johansen HK. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet. 2015;47(1):57–64. doi: 10.1038/ng.3148. [DOI] [PubMed] [Google Scholar]
- 22.Markussen T, et al. Environmental heterogeneity drives within-host diversification and evolution of Pseudomonas aeruginosa. MBio. 2014;5(5):e01592-14. doi: 10.1128/mBio.01592-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jelsbak L, et al. Molecular epidemiology and dynamics of Pseudomonas aeruginosa populations in lungs of cystic fibrosis patients. Infect Immun. 2007;75(5):2214–2224. doi: 10.1128/IAI.01282-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L, et al. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci USA. 2011;108(18):7481–7486. doi: 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith EE, Sims EH, Spencer DH, Kaul R, Olson MV. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. J Bacteriol. 2005;187(6):2138–2147. doi: 10.1128/JB.187.6.2138-2147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tümmler B, Cornelis P. Pyoverdine receptor: A case of positive Darwinian selection in Pseudomonas aeruginosa. J Bacteriol. 2005;187(10):3289–3292. doi: 10.1128/JB.187.10.3289-3292.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rédly GA, Poole K. FpvIR control of fpvA ferric pyoverdine receptor gene expression in Pseudomonas aeruginosa: Demonstration of an interaction between FpvI and FpvR and identification of mutations in each compromising this interaction. J Bacteriol. 2005;187(16):5648–5657. doi: 10.1128/JB.187.16.5648-5657.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Köhler T, Buckling A, van Delden C. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc Natl Acad Sci USA. 2009;106(15):6339–6344. doi: 10.1073/pnas.0811741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koch G, et al. Evolution of resistance to a last-resort antibiotic in Staphylococcus aureus via bacterial competition. Cell. 2014;158(5):1060–1071. doi: 10.1016/j.cell.2014.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cordero OX, Ventouras L-A, DeLong EF, Polz MF. Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc Natl Acad Sci USA. 2012;109(49):20059–20064. doi: 10.1073/pnas.1213344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raymond B, West SA, Griffin AS, Bonsall MB. The dynamics of cooperative bacterial virulence in the field. Science. 2012;337(6090):85–88. doi: 10.1126/science.1218196. [DOI] [PubMed] [Google Scholar]
- 32.Johansen HK, et al. Colonisation and infection of the paranasal sinuses in cystic fibrosis patients is accompanied by a reduced PMN response. J Cyst Fibros. 2012;11(6):525–531. doi: 10.1016/j.jcf.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Kümmerli R, Jiricny N, Clarke LS, West SA, Griffin AS. Phenotypic plasticity of a cooperative behaviour in bacteria. J Evol Biol. 2009;22(3):589–598. doi: 10.1111/j.1420-9101.2008.01666.x. [DOI] [PubMed] [Google Scholar]
- 34.Hadfield JD. MCMC methods for multi-response generalised linear mixed models: The MCMCglmm R package. J Stat Softw. 2010;33:1–22. [Google Scholar]
- 35.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; New York: 2009. [Google Scholar]
- 36.Meyer JM, et al. Siderotyping of fluorescent Pseudomonas: Molecular mass determination by mass spectrometry as a powerful pyoverdine siderotyping method. Biometals. 2008;21(3):259–271. doi: 10.1007/s10534-007-9115-6. [DOI] [PubMed] [Google Scholar]
- 37.Jiricny N, et al. Fitness correlates with the extent of cheating in a bacterium. J Evol Biol. 2010;23(4):738–747. doi: 10.1111/j.1420-9101.2010.01939.x. [DOI] [PubMed] [Google Scholar]
- 38.Cobessi D, et al. The crystal structure of the pyoverdine outer membrane receptor FpvA from Pseudomonas aeruginosa at 3.6 angstroms resolution. J Mol Biol. 2005;347(1):121–134. doi: 10.1016/j.jmb.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 39.Therneau T. 2014. A package for survival analysis in S. R package version 2.37-4. Available at https://cran.r-project.org/web/packages/survival/index.html. Accessed March 18, 2014.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.