Significance
Epigenetic inhibitors have shown considerable promise for the treatment of malignant and inflammatory diseases. We present here the detailed characterization of a potent and highly selective inhibitor of the bromodomains of CBP (CREB binding protein)/p300. Functional preclinical data studying cells derived from patients with ankylosing spondylitis and psoriatic arthritis (two human Th17-driven diseases) show that selective inhibition of the CBP/p300 bromodomain with CBP30 strongly reduces secretion of IL-17A, without having the broader and potentially deleterious effects on cytokine production and gene transcription of the pan-BET (bromo and extraterminal domain protein family) inhibitor JQ1. CBP/p300 play a significant role in IL-17A production, and CBP/p300 inhibition is a promising therapeutic strategy in human type-17–mediated diseases such as ankylosing spondylitis and psoriatic arthritis.
Keywords: CBP/p300, bromodomain, epigenetic inhibitors, Th17, ankylosing spondylitis
Abstract
Th17 responses are critical to a variety of human autoimmune diseases, and therapeutic targeting with monoclonal antibodies against IL-17 and IL-23 has shown considerable promise. Here, we report data to support selective bromodomain blockade of the transcriptional coactivators CBP (CREB binding protein) and p300 as an alternative approach to inhibit human Th17 responses. We show that CBP30 has marked molecular specificity for the bromodomains of CBP and p300, compared with 43 other bromodomains. In unbiased cellular testing on a diverse panel of cultured primary human cells, CBP30 reduced immune cell production of IL-17A and other proinflammatory cytokines. CBP30 also inhibited IL-17A secretion by Th17 cells from healthy donors and patients with ankylosing spondylitis and psoriatic arthritis. Transcriptional profiling of human T cells after CBP30 treatment showed a much more restricted effect on gene expression than that observed with the pan-BET (bromo and extraterminal domain protein family) bromodomain inhibitor JQ1. This selective targeting of the CBP/p300 bromodomain by CBP30 will potentially lead to fewer side effects than with the broadly acting epigenetic inhibitors currently in clinical trials.
Bromodomain recognition of acetylated lysines in histones plays a key role in the epigenetic control of gene expression (1). Therapeutic targeting of bromodomains has recently been recognized as an important potential therapeutic modality in human malignant and inflammatory diseases (2–4).
The transcriptional coactivators CBP [also known as cAMP responsive element binding protein (CREB) binding protein] and the closely related p300 possess such a bromodomain (5). Strategies targeting catalytic histone acetyl transferase (HAT) activity (6), the N-terminal domain (7), or the KIX (kinase-inducible domain interacting) domain (8) have had variable efficacy. By contrast, most bromodomain acetyl-lysine binding sites have good predicted druggability (9), confirmed experimentally by the recent development of potent and selective inhibitors targeting the bromo and extraterminal domain (BET) family (10–13). The promising effects of BET inhibitors observed in cancer cell lines have stimulated the rapid optimization of these tool molecules to yield clinical candidate drugs (2). A number of weak inhibitors, such as ischemin (KD = 25 μM), have been reported to target the CBP/p300 bromodomain (14). The synthesis and binding modes of more potent CBP and BET/CBP inhibitors have been reported in the last year (15, 16). The most potent of these inhibitors, CBP30, has recently been shown to bind with low nanomolar affinity to the bromodomains of CBP/p300 (15). However, this initial study did not examine the broader specificity of CBP30 for other bromodomain targets or look at the ability of CBP30 to inhibit physiological cellular processes.
BET bromodomain inhibitors exhibit antiinflammatory activity by inhibiting expression of inflammatory genes (11). The pan-BET inhibitor JQ1 ameliorated collagen-induced arthritis and experimental autoimmune encephalomyelitis (17), both mouse models of human inflammatory disease with major Th17 components. Th17 cells, a subset of T helper cells producing IL-17A, IL-17F, IL-21, IL-22, and GM-CSF, have been identified as central effectors of several autoimmune diseases, including ankylosing spondylitis (AS), psoriasis and psoriatic arthritis (PSA), rheumatoid arthritis, Crohn’s disease, and multiple sclerosis (18–20). AS is a common chronic inflammatory disease affecting ∼0.2–0.5% of adults in the United States. AS is characterized by inflammation of the sacroiliac and spinal joints leading to bony fusion (ankylosis), pain, and disability. Treatment options have until recently been limited to nonsteroidal antiinflammatory agents and anti-TNF biologic agents (21). An anti–IL-17A antibody, secukinumab, proved efficacious in AS, further supporting the importance of Th17 cells in AS (22). This finding has intensified the search for new drug targets modulating cytokine production of Th17 cells. Cytokine-driven induction of the transcriptional Th17 profile is strongly mediated by epigenetic mechanisms (23), one facilitator being the transcriptional coactivator p300. Recent studies demonstrated that p300 binds to the Il 17a promoter in murine Th17 cells and facilitates chromatin accessibility (24).
Here, we show the potent and selective nature of inhibition of the CBP/p300 bromodomains by CBP30. We demonstrate that CBP30 inhibits IL-17A production in primary human cells and Th17 responses from patients with AS and PSA. The effect of this inhibitor is far more selective than that observed for the pan-BET bromodomain inhibitor JQ1. Our data identify a previously unidentified strategy targeting the CBP/p300 bromodomain in human inflammatory diseases with major Th17 contribution such as AS.
Results
CBP30 Preferentially Binds to the CBP/p300 Bromodomain.
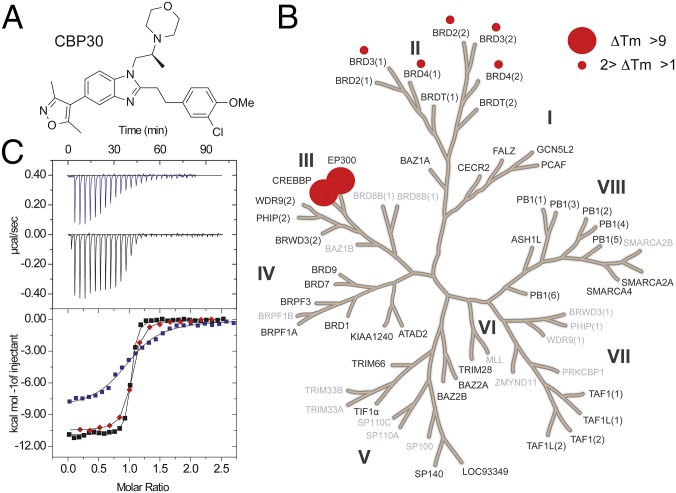
Synthetic variation on the 5-isoxazolyl-benzimidazole series of bromodomain inhibitors led to the development of CBP30 (Fig. 1A). Initial screening against 10 bromodomains suggested selectivity for CBP/p300 bromodomains (15). Here, we evaluated the selectivity of this inhibitor using temperature shift assays against a total of 45 bromodomains (Fig. 1B and Table S1), revealing only BET family bromodomains as additional targets. Notably, CBP30 did not result in significant ΔTm shifts for the testis-specific isoform BRDT (bromodomain testis-specific protein) whereas the rest of the BET family [first and second bromodomains of (bromodomain-containing protein) BRD2, BRD3, and BRD4] showed ΔTm shifts between 0.9 and 2.0 °C, suggesting low μM dissociation constants. We used isothermal titration calorimetry (ITC) to determine the dissociation constants (KDs) in solution for the bromodomains of CBP/p300 and other BET family members. ITC data revealed KDs of 26 nM and 32 nM for CBP and p300, respectively (Fig. 1C), in good agreement with affinity data already reported (15). Within the BET family, the first bromodomain of BRD4 [BRD4(1)] bound with highest affinity (KD of 885 nM), thus with 34-fold reduced selectivity compared with CBP (Table S2).
Fig. 1.
CBP30 is a selective and potent inhibitor of CBP/p300. (A) Chemical structure of CBP30. (B) Temperature shift data covering 45 human bromodomains. Screened targets are highlighted in black and targets that have not been screened in gray. Temperature shifts are represented by spheres. The gene name CREBBP corresponds to CBP protein, and EP300 to the protein p300. (C) Isothermal titration calorimetry data measuring the binding of CBP30 to BRD4 (blue), CBP (red), and p300 (black). (Upper) Raw injection heats for BRD4(1) and p300. (Lower) Normalized binding heats [BRD4(1), CBP, p300]. Nonlinear least squares fits to a single binding site model are shown as solid lines.
Table S1.
Melting temperatures and temperature shifts with CBP30 on bromodomain family members
| BRD target | DMSO Tm, °C | ΔTm 10 μM CBP30, °C |
| ASH1L | 53.8 ± 0.2 | −0.4 ± 0.3 |
| ATAD2 | 45.8 ± 0.7 | −0.7 ± 1.3 |
| BAZ1A | 50.8 ± 0.3 | 0.2 ± 1.0 |
| BAZ2A | 44.7 ± 0.4 | 0.3 ± 0.7 |
| BAZ2B | 53.8 ± 0.0 | 0.2 ± 0.1 |
| BRD1 | 49.1 ± 0.2 | 0.1 ± 0.1 |
| BRD2(1) | 52.5 ± 0.1 | 0.9 ± 0.4 |
| BRD2(2) | 49.5 ± 0.1 | 1.4 ± 0.0 |
| BRD3 (1) | 48.3 ± 0.1 | 1.9 ± 0.1 |
| BRD3 (2) | 49.5 ± 0.2 | 1.2 ± 0.3 |
| BRD4(1) | 49.0 ± 0.4 | 2.0 ± 0.4 |
| BRD4(2) | 50.6 ± 0.1 | 1.0 ± 0.4 |
| BRDT (1) | 48.5 ± 0.5 | 0.0 ± 0.4 |
| BRDT (2) | 51.9 ± 0.5 | 0.5 ± 0.1 |
| BRD9 | 42.4 ± 0.2 | 0.3 ± 0.2 |
| BRPF1A | 42.2 ± 0.4 | 0.3 ± 0.1 |
| BRPF3 | 46.6 ± 0.4 | 0.3 ± 0.2 |
| BRWD3 (2) | 53.3 ± 1.0 | 0.1 ± 0.7 |
| CECR2 | 50.9 ± 0.2 | 0.1 ± 0.1 |
| CBP | 45.8 ± 0.2 | 9.6 ± 0.1 |
| p300 | 41.2 ± 0.1 | 10.4 ± 0.4 |
| FALZ | 52.6 ± 0.2 | 0.4 ± 0.4 |
| GCN5L2 | 55.4 ± 0.1 | 0.2 ± 0.2 |
| KIAA1240 | 53.2 ± 0.1 | 0.3 ± 0.1 |
| LOC93349* | 56.9 ± 0.2 | 0.1 ± 0.1 |
| PB1 (1) | 58.5 ± 0.7 | 0.6 ± 0.7 |
| PB1 (2) | 53.8 ± 0.2 | 0.3 ± 0.2 |
| PB1 (3) | 45.1 ± 0.1 | 0.2 ± 0.2 |
| PB1 (4) | 46.4 ± 0.1 | 0.0 ± 0.2 |
| PB1 (5) | 48.3 ± 0.2 | 0.2 ± 0.2 |
| PB1 (6) | 61.0 ± 0.2 | −0.1 ± 0.2 |
| PCAF | 52.7 ± 0.2 | 0.3 ± 0.3 |
| PHIP(2) | 46.9 ± 0.2 | 0.2 ± 0.1 |
| SMARCA2A | 44.4 ± 0.0 | 0.1 ± 0.1 |
| SMARCA4 | 53.2 ± 0.3 | 0.2 ± 0.3 |
| SP140 | 58.2 ± 0.6 | 0.4 ± 0.8 |
| TAF1(1) | 46.3 ± 0.0 | 0.1 ± 0.0 |
| TAF1(2) | 48.1 ± 0.1 | −0.3 ± 0.1 |
| TAF1L(1) | 44.4 ± 0.1 | 0.0 ± 0.2 |
| TAF1L(2) | 46.3 ± 0.3 | 0.2 ± 0.8 |
| TIF1alpha | 39.9 ± 0.3 | 0.5 ± 0.6 |
| TIF1alpha* | 39.6 ± 0.1 | 0.6 ± 0.0 |
| TRIM28* | 59.4 ± 0.2 | 0.2 ± 0.3 |
| TRIM66* | 32.4 ± 0.9 | 0.3 ± 0.4 |
| WDR9(2) | 48.2 ± 0.4 | −0.1 ± 0.2 |
Significant changes are in bold. Mean ± SEM, n = 3.
Constructs containing the PHD-bromodomain.
Table S2.
Molecular selectivity of CBP30 for CBP and p300 compared with the BET family
| Protein | KA*, 106 M−1 | N | KD, nM | ΔHobs, cal/mol | TΔS, kcal/mol | ΔG, kcal/mol | Selectivity, fold relative to CBP |
| CBP† | 47.4 ± 4.4‡ | 0.98 ± 0.02 | 21 | −10,910 ± 48 | −0.789 | −10.1 | n/a |
| CBP | 38.5 ± 2.6 | 0.94 ± 0.02 | 26 | −10,570 ± 36 | −0.564 | −10.0 | |
| p300 | 31.4 ± 2.49 | 0.97 ± 0.03 | 32 | −10,500 ± 57 | −0.616 | −9.88 | n/a |
| BRD4(1)† | 1.17 ± 0.18 | 1.01 ± 0.03 | 854 | −8,470 ± 160 | −0.472 | −7.99 | 40.6 |
| BRD4(1) | 1.13 ± 0.07 | 0.9 ± 0.02 | 885 | −9,561 ± 107 | −1.42 | −8.14 | 34.0 |
| BRD4(2) | 0.205 ± 0.01 | 0.81 ± 0.04 | 4,878 | −3,439 ± 80 | 3.629 | −7.07 | 232 |
| BRD3(1) | 0.92 ± 0.03 | 0.93 ± 0.08 | 1,086 | −9,332 ± 85 | −1.232 | −8.1 | 52 |
| BRD3(2) | 0.456 ± 0.02 | 0.962 ± 0.02 | 2,193 | −10,630 ± 214 | 3.082 | −7.63 | 104 |
| BRD2(1) | 0.992 ± 0.03 | 0.92 ± 0.005 | 1,008 | −9,877 ± 65 | −1.801 | −8.07 | 48 |
| BRD2(2) | 0.567 ± 0.03 | 1.02 ± 0.01 | 1,763 | −4,579 ± 56 | 3.082 | −7.66 | 84 |
All experiments were carried out at 15 °C.
Data published in ref. 22 for compound 59.
Errors shown are errors of the nonlinear least square fits. n/a, not applicable.
Molecular Details of the Interaction of CBP30 with p300, BRD4, and BRD2.
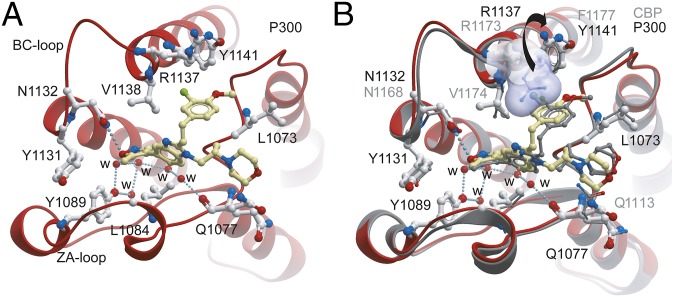
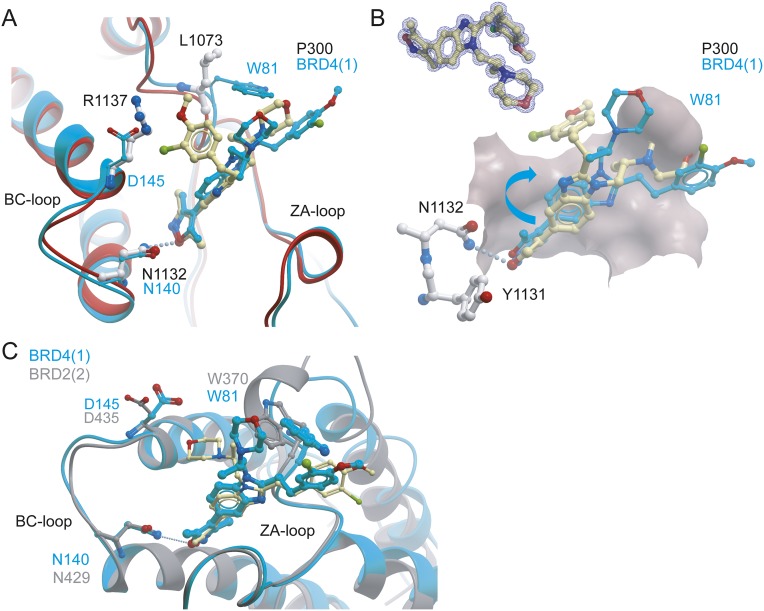
To understand the structural mechanisms for the selectivity of CBP30 for specific BET family members, we determined the cocrystal structure of CBP30 with the p300, BRD4(1), and BRD2(2) bromodomains. Crystals of the CBP30/p300 complex diffracted to 1.05 Å resolution (Table S3). As observed in the CBP bromodomain cocrystal structure (PDB ID code 4nr7), the CBP isoxazole oxygen acted as an acetyl-lysine mimetic moiety forming hydrogen bonds to the conserved asparagine (N1132) and a water-mediated interaction with the hydroxyl group of Y1089 of p300 (Fig. 2A). However, and in contrast to the CBP complex, we did not observe an induced fit binding mode of R1137, which has been reported to flip into the acetyl-lysine binding side forming a cation-π interaction. The reason for this difference in ligand coordination is due to the presence of a neighboring tyrosine residue (Y1141) that engages into a hydrogen bond interaction with R1137. In the CBP bromodomain, this position is occupied by a phenylalanine, allowing the arginine to stack with the aromatic decoration of the inhibitor. This induced fit has also been observed for other CBP bromodomain inhibitors (16). In the p300/CBP30 complex, the benzimidazole ring system was shifted, resulting in rotation of the morpholine moiety and the 3-chloro-4-methoxyphenol ring system (Fig. 2B). Additional comparison of CBP30 binding to p300 with BRD4(1) and BRD2(2) is shown in Fig. S1.
Table S3.
Crystallographic data collection and refinement statistics
| Protein ID | |||
| EP300 BRD | BRD4(1) BRD | BRD2(2) BRD | |
| Ligand | CBP30 | CBP30 | CBP30 |
| Data collection | |||
| Space group | P 61 | P 212121 | P 21212 |
| Cell dimensions | |||
| a,b,c, Å | 53.43 53.43 77.00 | 41.92 91.80 113.76 | 52.76 71.81 32.00 |
| α, β, γ, ° | 90.00 90.0 120.00 | 90.00 90.0 90.00 | 90.00 90.00 90.00 |
| Resolution, Å* | 29.60 (1.05) a | 29.87 (1.50) | 29.68 (1.40) |
| Unique observations* | 56,052 (1671) | 71,462 (3204) | 24,507 (3365) |
| Completeness (%)* | 95.9 (58.0) | 99.6 (92.8) | 99.3 (95.9) |
| Redundancy* | 10.8 (2.8) | 9.7 (8.7) | 9.2 (7.3) |
| Rsym or Rmerge* | 0.066 (0.349) | 0.098 (0.426) | 0.079 (0.165) |
| I/σI* | 22.2 (3.1) | 14.4 (4.8) | 22.3 (11.6) |
| Wavelength | 0.9795 | 0.9763 | 0.9763 |
| Phasing | MR | MR | MR |
| Refinement | |||
| Rwork/Rfree, % | 16.50/17.80 | 19.12 (21.42) | 16.44 (18.53) |
| No. of atoms | |||
| Protein/other/solvent | 976/40/189 | 3,155/116/346 | 905/30/118 |
| B-Factors, Å2 | |||
| Protein/other/solvent | 8.56/6.78/17.49 | 16.55/24.19/24.68 | 9.63/21.80/22.23 |
| rmsd bond, Å | 0.009 | 0.009 | 0.010 |
| rmsd angle, o | 1.384 | 1.422 | 1.405 |
| Ramachandran statistics | |||
| Favored,% | 100.00 | 100.00 | 100.00 |
| Outliers, % | 0.00 | 0.00 | 0.00 |
| PDB ID code | 5BT3 | 5BT4 | 5BT5 |
MR, molecular replacement.
Highest resolution shell (in Å) shown in parentheses.
Fig. 2.
Comparison of CBP30 binding to the p300 and CBP bromodomains. (A) Binding of CBP30 to p300. The inhibitor and most important interacting side chains are shown in ball and stick representation. Conserved water molecules are highlighted as red spheres and are labeled with “w.” Hydrogen bonds are shown as dotted lines. (B) Comparison of the CBP30 binding modes in the CBP and p300 bromodomain complex (15). The side chain of R1173 in CBP is shown in surface representation and is indicated by an arrow.
Fig. S1.
Comparison of the binding modes of CBP30 for p300 with the BRD4(1) and BRD2(2) bromodomains. (A) Comparison of the CBP30 binding modes in the p300 and BRD4(1) bromodomain complexes reveals a 170° rotation around the isoxazole–benzimidazole bond. CBP30 and key interacting residues are shown in ball and stick representation. Carbon atoms of CBP30 in the p300 complex are highlighted in yellow and in the BRD4(1) complex in cyan, respectively. (B) Alternative view of the two binding modes of CBP30 in BRD4(1) and p300 binding mode. The binding pocket of p300 is shown as a solid surface. The Inset shows a 2FoFc electron density map of the CBP30/p300 complex contoured at 2σ around CBP30. (C) Comparison of the CBP30 binding modes in BRD4(1) and BRD2(2) reveals a similar binding mode. The electron density for both benzimidazole inhibitor decorations was diffuse suggesting disorder. However, the central core of the inhibitor was well defined by electron density, allowing assignment of the binding pose. The tryptophan side chains (W81 and W370) assume a different orientation in both structures. This orientation positions the 3-chloro-4-methoxyphenol ring system next to W81 in BRD4. The aromatic interaction with the ring is probably more favorable than interactions with the morpholine moiety, which would also be sterically excluded by the bulky tryptophan side chain conserved in BET bromodomains, where the tryptophan side chain (W370) assumes a different conformation that does not allow aromatic stacking with the CBP30 3-chloro-4-methoxyphenol ring.
Kinetics of CBP30 Binding to CBP/p300.
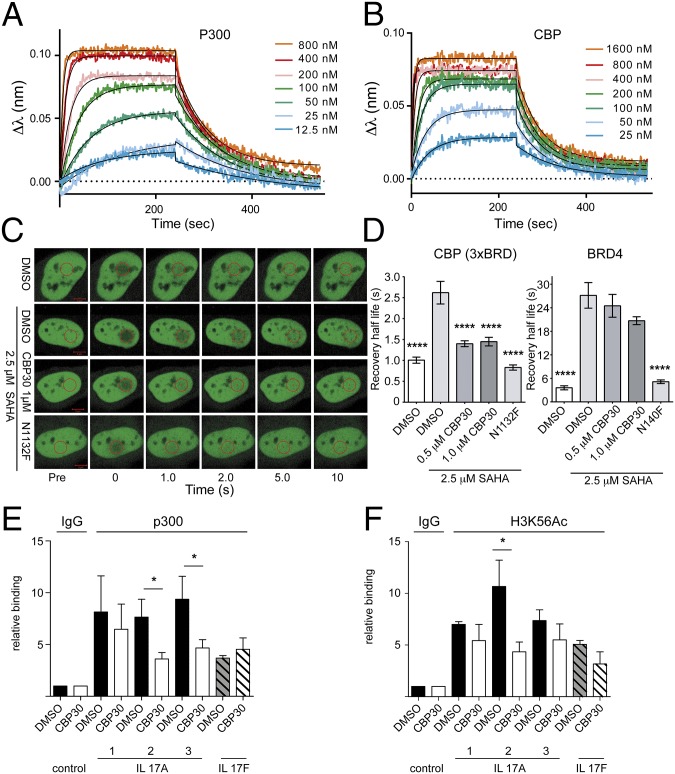
Bio-layer interference (BLI) showed similar rapid dissociation rates of CBP30 with p300 and CBP (Fig. 3 A and B). KD values calculated from the steady-state dose–response curves were 38 ± 4.7 nM and 47 ± 2.1 nM for CBP and p300, respectively, in good agreement with ITC data. Analysis of ligand-binding kinetics showed both fast and comparable dissociation on and off rates (CBP, kon, 3.21 ± 0.28 × 105 M−1⋅s−1 and kdis, 1.42 ± 0.04 × 10−2 s−1; and p300, kon 1.43 ± 0.19 × 105 M-1⋅s−1 and kdis, 9.05 ± 0.22 × 10−3 s−1). Thus, the induced fit binding mode observed in CBP bromodomain complexes does not contribute to slow binding kinetics of CBP30.
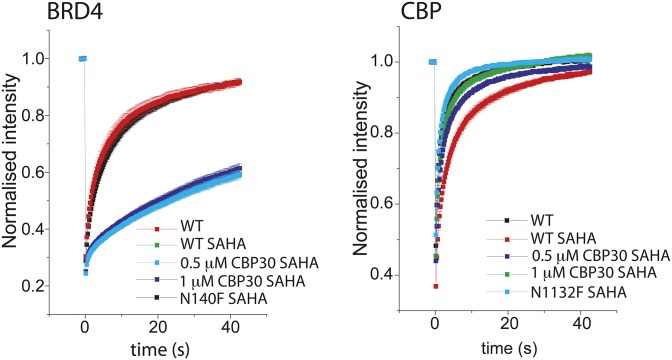
Fig. 3.
CBP30 binding kinetics and nuclear target engagement. Sensograms monitoring the kinetics of CBP30 association and dissociation with p300 (A) and with CBP (B) using BLI (bio-layer interferometry). (C) FRAP using 3× CBP BRD. Shown are nuclei of transfected cells. The bleached area is indicated by a red circle before (Pre), at bleaching (time = 0), and post-bleaching as indicated. (D) Averaged recovery half-life (n = 10) of DMSO, DMSO/SAHA (2.5 μM), and CBP30 (1 μM)/SAHA (2.5 μM) treated 3× CBP BRD or BRD4-transfected cells. Raw data traces of the fluorescent recovery are shown in Fig. S2. The pan-HDAC inhibitor SAHA was used to increase global acetylation (25). One-way ANOVA, ****P < 0.0001. CBP30 reduces p300 (E) and H3K56Ac (F) binding to the IL 17A gene locus. ChIP-q-PCR assessed in three different regions (regions 1–3) of the IL 17A and in one region of the IL 17F locus in Th17 cells treated with 2 µM CBP30 or DMSO for 24 h. Relative enrichment is expressed as mean ± SEM of four (E) and three (F) independent experiments (Student’s t test); *P < 0.05.
We used the fluorescent recovery after photobleaching (FRAP) assay to measure the displacement of the CBP bromodomain from chromatin in the presence of CBP30 (25). A GFP-labeled construct containing three CBP bromodomains flanked by a nuclear localization signal (NLS) showed significantly reduced recovery half-life in the presence of CBP30 (Fig. 3 C and D), comparable with the inactivating bromodomain mutant N1132F. By contrast, BRD4 recovery rates were only modestly affected, compared with the inactivating mutant N140F. CBP30 also inhibited binding of p300 and of acetylated histone H3K56 (a known CBP/p300 mark) to the IL 17A gene locus in chromatin immunoprecipitation experiments (Fig. 3 E and F).
Antiinflammatory Activity of CBP30 on Primary Human Cells.
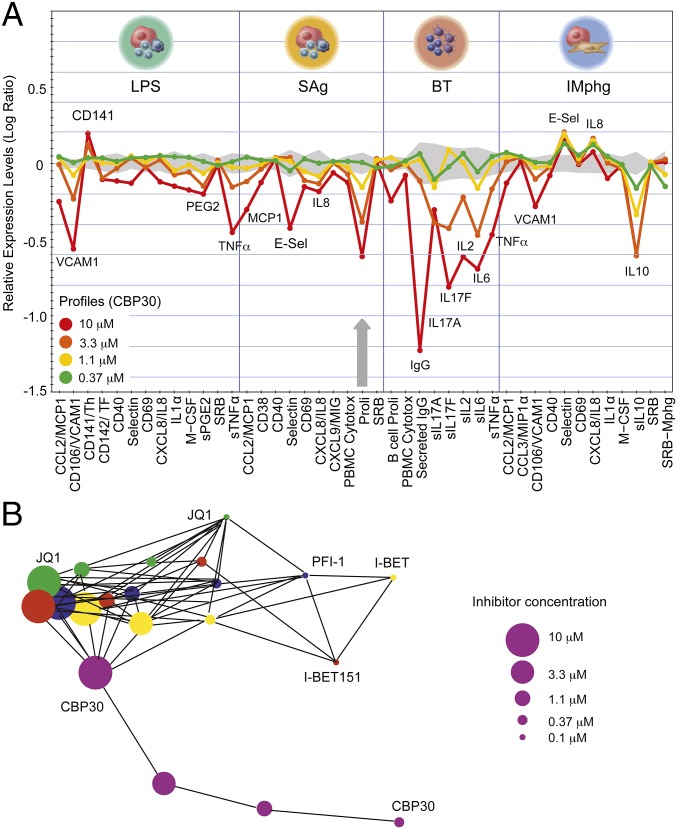
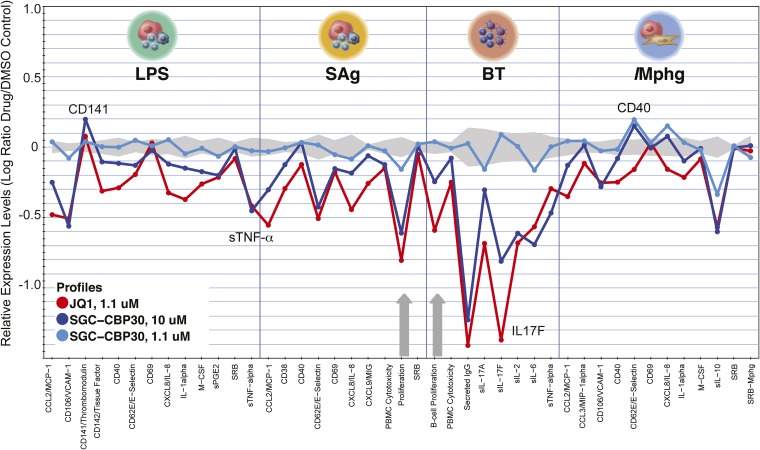
The biological activity of CBP30 was explored using a panel of 12 stimulated primary human cell types (BioMAP) (26, 27). This analysis revealed broad antiinflammatory activity but no pronounced cytotoxicity at concentrations up to 10 μM. Potential immunomodulatory effects observed included down-regulation of the cytokines IL-17, TNFα, IL-8, IL-2, IL-6, IL-1α, and IL-10, vascular cell adhesion protein 1 (VCAM-1), and cytokine monocyte chemotactic protein 1 (MCP1) (Fig. 4A and Fig. S3). In addition, inhibition of matrix metalloproteinases MMP-1 and MMP-9 and of tissue plasminogen activator (tPA) suggested potential modulation of matrix/tissue remodeling by CBP30. The BioMAP profile showed overlap with BET inhibitor profiles at high inhibitor concentrations (3.3 and 10 μM). This overlap may be due to weak BET activity of CBP30 or to coregulation of similar signaling molecules by CBP/p300 and BET (Fig. 4B and Figs. S3 and S4). Nevertheless, a distinct phenotype was detected at lower concentrations, suggesting that CBP30 does not affect BET function in cells at these concentrations (Fig. S4).
Fig. 4.
BioMAP profile of CBP30 on primary hematopoietic cells. (A) CBP30 profile at 0.37–10 µM. Monitored marker proteins are shown on the x axis, and relevant proteins have been highlighted. Historic variations of DMSO-treated cells are indicated by the gray shaded area, and antiproliferative effects by gray arrows. A full BioMAP including other cell types is shown in Fig. S3. Studied cell systems were as follows: peripheral blood mononuclear cells plus venular endothelial cells stimulated with LPS or SEB (SAg); B cells plus peripheral blood mononuclear cells (BT); macrophages plus venular endothelial cells (IMphg). (B) Gleason correlation (>0.85) comparing the phenotypic response (Biomap profiles across all primary tissues) of CBP30 (purple circles) with the pan-BET inhibitors (JQ1 in green, I-BET in yellow, PFI-1 in blue, and I-BET151 in red). Concentrations from 0.1 to 10 µM are indicated by the size of the circles.
Fig. S3.
Full BioMAP profile showing effects of CBP30 on primary hematopoietic cells. Response of CBP30 has been studied using four different concentrations as indicated in the figure. Monitored marker proteins are shown on the x axis, and relevant proteins have been highlighted in the figure. Historic variations of DMSO-treated cells of this platform are indicated by the gray shaded area. Antiproliferative effects are indicated by gray arrows. For details on studied cell types, see www.discoverx.com/services/drug-discovery-development-services/primary-cell-phenotypic-profiling/diversity-plus.
Fig. S4.
BioMAP comparison of the effects of CBP30 and JQ1 on primary hematopoietic cells. The effect of CBP30 was studied at 1.1 and 10 μM. Monitored marker proteins are shown on the x axis, and relevant proteins have been highlighted in the figure. Historic variations of DMSO-treated cells of this platform are indicated by the gray shaded area. Antiproliferative effects are indicated by gray arrows (JQ1). Studied cell systems were as follows: peripheral blood mononuclear cells plus venular endothelial cells stimulated with LPS; peripheral blood mononuclear cells plus venular endothelial cells (SAg); B cells plus peripheral blood mononuclear cells (BT); macrophages plus venular endothelial cells (IMphg).
Fig. S2.
Time dependence of fluorescence recovery for the BRD4 and 3xCBP fluorescence recovery after photobleaching (FRAP). Curves represent the means in the bleached area at each time point of at least 15 cells in each group, and SE bars are shown for each data point. The curves are colored as indicated in the figure.
CBP30 Inhibits Th17 Cytokine Production from Patient and Control T Cells.
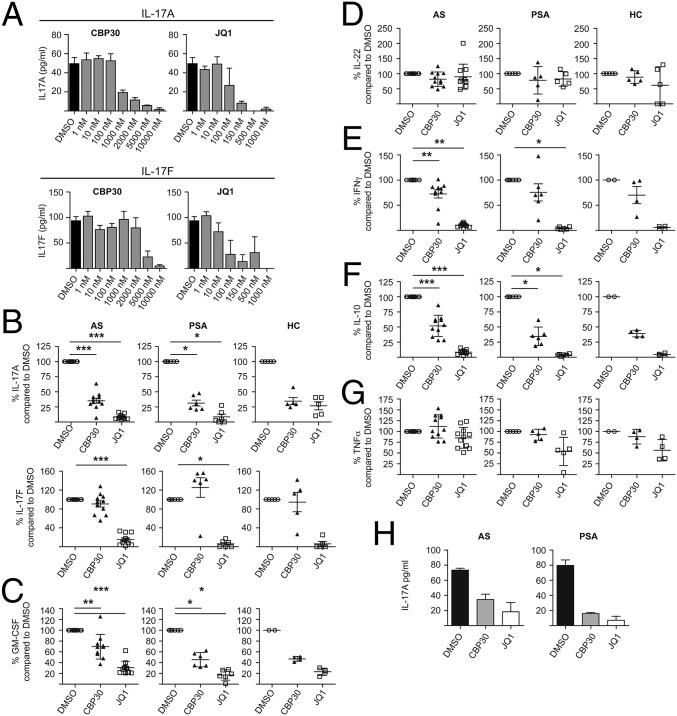
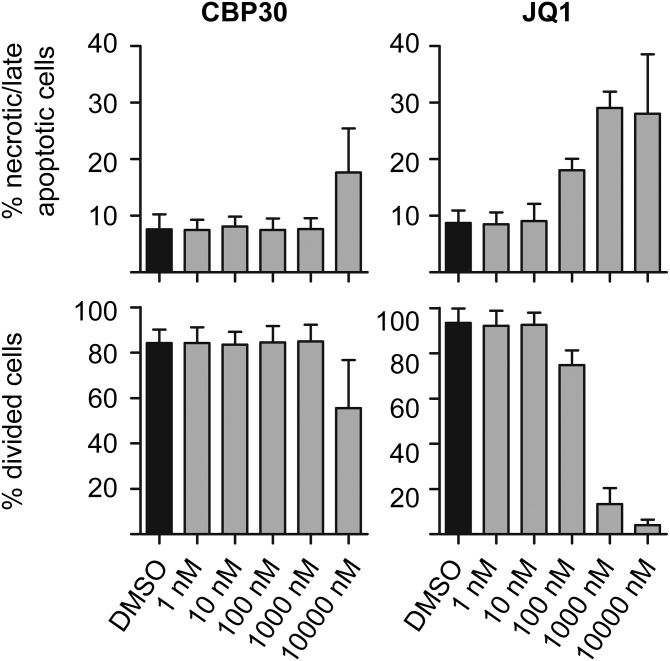
We measured cytokine secretion (ELISA) by purified CD4+ T cells from AS and PSA patients and healthy controls (HCs) cultured for 3 d under Th17-promoting conditions. An initial dose–response experiment showed that 2 µM CBP30 inhibited IL-17A production by AS CD4 T cells by 77% (Fig. 5A). This concentration did not affect cell viability or proliferation (Fig. S5). JQ1 inhibited IL-17A secretion at lower concentrations but also showed evidence of toxicity at concentrations above 100 nM (Fig. S5). In subsequent experiments, JQ1 was used at 150 nM [the concentration used by Mele et al. (17)] and CBP30 at 2 µM. CBP30 on average reduced the secretion of IL-17A by 66.3% in cells from patients with AS and PSA and from HCs (Fig. 5B).
Fig. 5.
CBP30 inhibits Th17 cytokine production by patient and control CD4 T cells. (A) IL-17A and F production from CD4+ T cells from one AS patient cultured under Th17-promoting conditions in the presence of DMSO, CBP30, or JQ1 at concentrations from 1 nM to 10 µM. ELISA of supernatant on day three. Mean ± SD of triplicates of one representative experiment (n = 4). (B) IL-17A and F production from total CD4+ T cells from 11 AS patients, 6 PSA patients, and 5 HCs cultured as in A in the presence of 2 µM CBP30 or 150 nM JQ1. Day 3 ELISA. Mean ± SEM, each triplicates. (C) GM-CSF and (D) IL-22 (LUMINEX) levels from AS patients, PSA patients, and HCs after inhibition with 2 µM CBP30 or 150 nM JQ1. Mean ± SD, n = 4–11. (E) IFNγ ELISA, mean ± SEM, (F) IL-10, and (G) TNFα (LUMINEX), mean ± SD, n = 4–11. (H) ELISA for IL-17A in the supernatant of CD4+ T cells from synovial fluid of patients with AS and PSA cultured as in A in the presence of 2 µM CBP30 and 150 nM JQ1. Data represent one of two independent experiments in triplicate, mean ± SEM. Patient demographics shown in Table S4. Wilcoxon matched-pairs signed rank test (*P < 0.05; **P < 0.01; ***P < 0.001; no asterisk, P > 0.05).
Fig. S5.
Effects of CBP30 and JQ1 on viability and proliferation of human T cells. Toxic effects of the compounds were assessed using Annexin V/7-AAD staining by flow cytometry on day 3 of culture with CBP30 or JQ1. CFSE dilution was used to evaluate % divided cells on day 6 of culture. Means ± SD are shown. Representative of n = 4 experiments.
We next studied the additional Th17-associated cytokines IL-17F, GM-CSF, and IL-22. CBP30 (2 µM) caused only 14.6% inhibition of IL-17F production whereas 150 nM JQ1 reduced IL-17F by up to 85% (Fig. 5A). These results were confirmed for AS and PSA patients and for HCs (Fig. 5B). Secretion of the Th17-associated proinflammatory cytokine GM-CSF was significantly lowered by both CBP30 and JQ1, in both AS and PSA patients (Fig. 5C). IL-22 was not significantly inhibited (Fig. 5D). To determine the specificity for Th17 cells, we analyzed supernatants by ELISA for IFNγ, a key cytokine produced by Th1 cells. CBP30 had less effect than JQ1 on IFNγ or IL-10 secretion (Fig. 5 E and F) and did not affect TNF α secretion (Fig. 5G). Both CBP30 and JQ1 inhibited IL-17A production by synovial fluid CD4+ T cells obtained from inflamed joints of two AS and two PSA patients (Fig. 5H). These data show that CBP30 has a more specific inhibitory effect on Th17 cytokines than JQ1.
CBP30 Has Narrower Effects on CD4 T-Cell Gene Expression than JQ1.
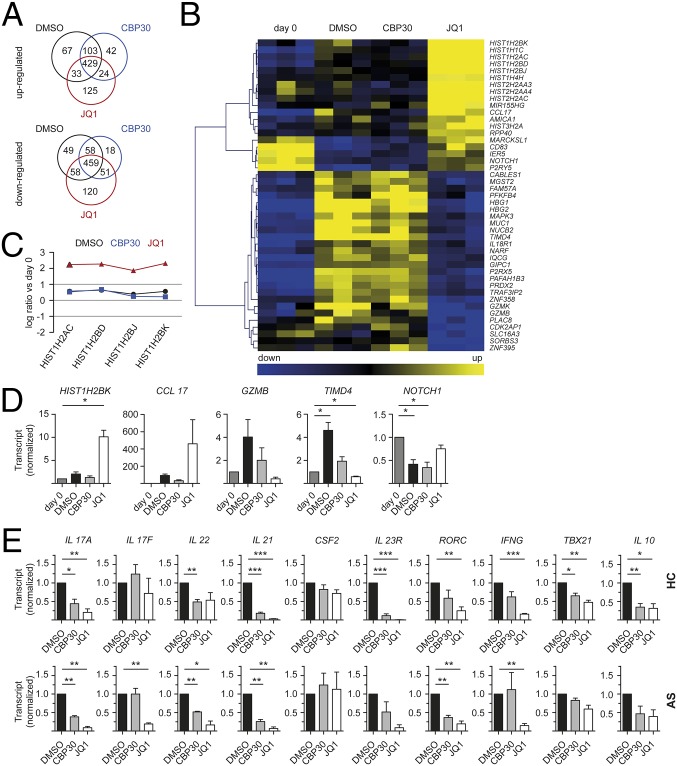
Because the observed effects of CBP30 and JQ1 on Th17-type responses lie most likely in altered transcriptional activity, we compared gene expression of unstimulated HC CD4+ T cells with cells cultured for 4 d in the presence of CBP30, JQ1, or DMSO. Although Th17-promoting conditions were used, because the starting percentages of Th17 cells were extremely low, Th17 cells made up <5% of the final lymphocyte population. During this period of stimulation, 2,087 transcripts representing 1,634 genes were significantly regulated (Fig. 6A). Fewer genes were uniquely up- or down-regulated by CBP30 than by JQ1 (42 and 18 by CBP30, respectively, vs. 125 and 120 by JQ1). Genes that were specifically up-regulated by JQ1 included genes of the histone clusters 1, 2, and 3, as well as MIR155HG and CCL17 (Fig. 6 B–D). By contrast, CBP30 had a far more limited effect on gene expression (Fig. 6 B–D).
Fig. 6.
CBP30 has more specific effects on CD4 T-cell gene expression than JQ1. (A–D) Total CD4+ T cells from three healthy controls were cultured under Th17-promoting conditions in the presence of DMSO, 2 µM CBP30, or 150 nM JQ1 for 4 d before total RNA isolation. (A) Venn diagram showing number of genes significantly up- or down-regulated greater than twofold (ANOVA < 0.05 analyzed for whole genome expression with Illumina beadchip array HT12v4.1). Pathway analysis shown in Dataset S1. (B) Hierarchical clustering heat map of selected genes showing differential regulation by CBP30 and JQ1 [genes up- or down-regulated by >1 log(2) ratio and difference >0.75 log(2) ratio between DMSO, CBP30, and JQ1]. Data from three donors are shown for each experiment. (C) Line diagram displaying up-regulation of the histone cluster genes by JQ1 but not by CBP30. (D) q-PCR on selected genes from the microarray that are differentially regulated between CBP30 and JQ1; mean ± SEM, n = 2–3, triplicates. (E) Total CD4+ T cells from four HC and three AS patients were treated as in A, and q-PCR on selected genes was performed; mean ± SEM, n = 3–4, triplicates. Paired t test (*P < 0.05, **P < 0.01, ***P < 0.001).
We further compared Th17-associated transcript expression by q-PCR in CD4+ T cells from three additional HCs and three AS patient samples cultured under Th17-favorable conditions. Both CBP30 and JQ1 significantly reduced IL 17A and IL 21 transcript levels in HC and AS patients (Fig. 6E). By contrast IL 17F transcripts were only significantly inhibited by JQ1 in AS patient samples. IL 22 transcript levels were significantly reduced by CBP30 in HC and AS patients and by JQ1 in AS patients. CSF2 levels were not significantly altered by either inhibitor. The expression of the Th17 lineage-stabilizing IL-23 Receptor (IL 23R) was significantly down-regulated by both inhibitors in HC CD4+ T cells. Transcription of the Th17 master regulator RORC was reduced by JQ1 in HC and AS patients, and by CBP30 in AS CD4+ T cells. Expression of TBX21, a Th1-associated transcription factor, was reduced significantly only in HC CD4+ T cells, and the Th1 cytokine IFNG was inhibited only by JQ1. IL 10 transcript expression was significantly reduced only in HC CD4+ T cells. Thus, CBP30 has relative selectivity for Th17-associated genes whereas JQ1 has widespread effects on many gene families.
Discussion
In this work, we show that CBP30 is a highly selective inhibitor of the bromodomains of the transcriptional coactivators CBP and p300. The ability of CBP30 to inhibit IL-17A secretion was identified using an unbiased screen of primary human cells (BioMAP). We further present preclinical efficacy in inhibiting Th17 immune responses from blood and joints of patients with two Th17-driven inflammatory diseases, ankylosing spondylitis and psoriatic arthritis. The development of epigenetic inhibitors is showing real promise in treating human disease. However, drug specificity is a key issue, with the need to avoid both predictable and unexpected off-target effects. We and others have previously described bromodomain inhibitors with broad specificity. For example, JQ1 has shown efficacy in treating both murine Th17-mediated disease and cancers in mouse models and in vitro studies (10, 17, 28). Nineteen clinical trials with BET inhibitors in oncology are underway. Nevertheless, JQ1 has very broad activity (for example, inhibiting testicular BRDT to cause testicular atrophy and reversible infertility) (29) and would not be an acceptable therapy for noncancerous diseases. Our data show that CBP30 has a far narrower specificity, both at the molecular and cellular level. We performed a detailed screen of 45 human bromodomains, greatly extending the work of Hay et al. (15), showing greater than 30-fold selectivity of CBP30 for CBP and p300 compared with other bromodomains. Of note, no binding to BRDT was detectable, and CBP30 showed only weak activity for the second bromodomains of BET proteins, a feature that may additionally contribute to its cytokine inhibitory specificity.
We show here the ability of CBP30 to profoundly inhibit secretion of the proinflammatory cytokine IL-17A by human T helper cells from healthy individuals and patients with AS and PSA. AS and PSA are common human inflammatory arthritides, both termed “seronegative,” with a combined prevalence of approximately 1% (30, 31). Both have a major IL-17–driven component, evidenced by multiple genetic, immunological, and therapeutic studies (20, 22, 32, 33). Indeed, and unlike rheumatoid arthritis, clinical studies targeting the IL-17/-23 axis with monoclonal antibodies in both AS and PSA have yielded very promising results, comparable with those seen with anti-TNF agents (22, 34). All patients studied by us had established disease, and a significant number had active disease. CBP30 also inhibited IL-17A production by Th17 cells from actively inflamed joints, showing efficacy at the site of inflammation. Overall, our data convincingly show that CBP30 effectively inhibits IL-17A production by Th17 cells from patients with AS and PSA and from HCs.
Th17 cells are heterogeneous and capable of producing multiple additional inflammatory cytokines, including GM-CSF, IL-17F, IL-21, and IL-22. We show here that CBP30 also inhibits GM-CSF cytokine secretion. Th17 cells secreting additional cytokines have been described as being particularly pathogenic, with GM-CSF in particular implicated in rheumatoid arthritis (35). The observed discrepancy between inhibitory effects of CBP30 on GM-CSF secretion and CSF2 transcription might be explained by minor differences in the experimental time course. Transcriptional analysis on a limited number of AS patients and HCs confirmed the inhibitory effects on IL 17A and also showed down-regulation of IL 21, IL 22, the transcription factor RORC (in AS patients), and the signature cytokine receptor IL 23R (in HCs). CBP30 (unlike JQ1) did not inhibit transcription or secretion of the closely related cytokine IL-17F. This observation suggests hitherto unappreciated differences in the transcriptional control of IL 17A and IL 17F. We observed no effect of CBP30 on production of TNFα (a cytokine important for protection against tuberculosis) and smaller effects of JQ1 on IL-10 and IFNγ secretion. Thus, future therapeutic use of CBP/p300 inhibitors could result in more specific inhibition of IL-17A and other Th17-type cytokines by Th cells.
Transcriptional profiling of human CD4 T cells showed that CBP30 has more limited effects and far greater specificity than JQ1. Global expression arrays illustrated that JQ1 has effects on approximately fourfold more genes than CBP30. Although the global microarray showed only a limited Th17 signature (e.g., IL18R1, MIR155HG, and NOTCH1), we were nevertheless easily able to confirm the effects of CBP30 (and JQ1) on Th17-specific gene transcription on Th cells from AS patients and HCs by q-PCR. Of note, even after culture, Th17 cells made up only 2.9–8.3% of the total Th cell population (from healthy donors), explaining the lack of an obvious Th17-specific signal. Th17 cells represent <1% of healthy peripheral blood T lymphocytes (36), rising to ∼1–2% in AS, PSA, and psoriasis patients (19, 20, 36).
Transcriptional control of Th17 responses is a multistep process, involving numerous transcription factors (37). CBP/p300 is thought to be involved at an early stage, consistent with our data showing key sensitivity of human IL 17A transcription to CBP/p300 inhibition (38, 39). In murine Th17 cells, conserved noncoding sequence 2 (CNS2) is important to drive Il 17 gene transcription in coordination with RORγt and is bound by p300 (24). However, expression of Il 17f is only partially dependent on CNS2, a possible explanation for the CBP30 specificity.
We present here, to our knowledge, the first detailed characterization of CBP30, a potent and highly selective CBP/p300 bromodomain inhibitor. Functional preclinical data show effective inhibition of IL17A secretion, without the broader and potentially deleterious effects of the pan-BET inhibitor JQ1. CBP30 merits further investigation as a lead therapeutic compound for AS, PSA, and other human type-17–mediated autoimmune diseases, including psoriasis, inflammatory bowel disease, and multiple sclerosis (40–42).
Materials and Methods
Inhibitors.
SGC-CBP30 (hereafter called CBP30) and (+)-JQ1 (hereafter called JQ1) were provided by the Structural Genomics Consortium (SGC), Oxford.
ITC and Thermal Shift Assay.
Experiments were carried out on a VP-ITC microcalorimeter (MicroCal) at 15 °C in 50 mM Hepes, pH 7.5, 150 mM NaCl, using an initial injection of 2 µL, followed by 34 identical injections of 8 µL. The dilution heats, measured separately, were subtracted from the titration data. Thermodynamic parameters were calculated using ∆G = ∆H − T∆S = −RTlnKB, where ∆G, ∆H, and ∆S are the changes in free energy, enthalpy, and entropy of binding respectively. In all cases, a single binding site model was used. Thermal melting experiments were carried out using an Mx3005p Real Time PCR machine (Stratagene) as described (43).
BLI.
cDNA encoding human bromodomains was cloned, expressed, and purified as previously described (10). Kinetic ligand-binding measurements were done using an OctetRed384 instrument (ForteBio). Biotinylated protein (0.05 mg/mL) was immobilized on Super Streptavidin Biosensors. Association and dissociation measurements were done in 25 mM Hepes, pH 7.4, 100 mM NaCl, 0.01% Tween at 25 °C, with association and dissociation times of 240 s. Compounds were prepared as 1:2.5 dilutions starting from 12 μM. Binding to the reference sensors (no protein attached) was subtracted before calculations. Binding constants were calculated using the ForteBio Analysis software provided by the manufacturer.
Crystallization, Data Collection, and Structure Solution.
EP300 construct (Uniprot identifier as EP300_HUMAN Q09472-1 fragment 1048–1161), BRD4 construct (Uniprot identifier as BRD4_HUMAN O60885-1 fragment 44–168), and BRD2 construct (Uniprot identifier as BRD2_HUMAN P25440-1 fragment 344–455) were used for crystallographic studies as described in SI Materials and Methods. Data collection and refinement statistics are compiled in Table S3. The models and structure factors have PDB ID codes 5BT3 (EP300), 5BT4 [BRD4(1)], and 5BT5 [BRD2(2)].
FRAP.
FRAP studies were performed as described in SI Materials and Methods and (25).
ChIP.
ChIP-q-PCR was performed as described in SI Materials and Methods.
BioMAP Analysis of Primary Human Cells.
BioMAP assays were performed as previously described (26, 27). Data for CBP30 were compared with pan-BET inhibitors (JQ1, I-BET, PFI-1, and I-BET151) (10–13).
Patient Samples.
Heparinized venous blood (30 mL) was obtained from 14 patients with AS (modified New York criteria), 6 patients with PSA [Classification of Psoriatic Arthritis (CASPAR) criteria], and 8 HCs. Table S4 shows patient demographics and medication. Ethical permission (COREC 06/Q1606/139 and Oxfordshire Research Ethics Committee B 07/Q1605/35) and informed consent were obtained. Leukocyte cones were acquired from National Health Service Blood and Transplant.
Table S4.
Patient characteristics for samples used in ELISA/LUMINEX assays and for q-PCR
| ELISA/LUMINEX | q-PCR | ||||
| HC (n = 5) | AS (n = 11) | PSA (n = 6) | HC (n = 4) | AS (n = 3) | |
| Age, y (SD) | 52.27 (11.20) | 51.92 (10.91) | 40.0 (7.48) | 54.00 (7.00) | 44.00 (15.85) |
| Male/female, no. | 4/1 | 9/2 | 2/4 | 4/0 | 0/3 |
| HLA-B27 positive, % | na | 100 | na | na | 100 |
| BASDAI* (SD) | na | 3.38 (2.53) | na | na | 5.07 (3.55) |
| CRP,† mg/L (SD) | na | 6.47 (5.12) | 21.04 (13.99) | na | 16.53 (12.31) |
| Anti-TNF treatment, no. | na | 4/11 | 2/6 | na | 0/3 |
| DMARD treatment,‡ no. | na | 2/11 | 5/6 | na | 0/3 |
| Steroid treatment, no. | na | 1/11 | 1/6 | na | 0/3 |
Synovial fluid mononuclear cells used in Fig. 5H were from an additional AS patient with the following characteristics: 58 y, female, BASDAI 9.9, CRP 27.0 mg/L, on anti-TNF (adalimumab) and DMARD (methotrexate) treatment, no steroid treatment.
Bath ankylosing spondylitis disease activity index.
C-reactive protein.
Disease-modifying antirheumatic drug. na, not applicable. Mean and SD are shown.
Cell Purification and Cell Culture, ELISA, and LUMINEX Analysis.
CD4 T cells (5 × 104, negatively selected: 89.1% purity) were cultured in 96-well plates in conditions that promoted Th17 numbers and preserved Th1, Th2, and Treg numbers as described in SI Materials and Methods. Supernatants were analyzed with an IL-17A, IL-17F, and IFNγ ELISA kit (ebioscience) and subjected to LUMINEX analysis with a custom-made premixed Multiplex Screening Assay for IL-10, IL-22, TNFα, and GM-CSF (R&D Systems).
Transcriptional Profiling.
CD4 T cells (1 × 106) were cultured in 48-well plates, as described above, for 4 d. Microarray analysis of RNA used Illumina beadchip array HT12v4.1. Details are in SI Materials and Methods. For quantitative PCR (q-PCR), RNA was reverse transcribed with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). TaqMan probes for transcripts encoding IL 17A, IL 17F, IL 21, IL 22, CSF2, IL 10, IFNG, IL 23R, RORC, TBX21, HIST1H2BK, CCL17, GZMB, TIMD4, and NOTCH1 (see SI Materials and Methods for details) were used for q-PCR on a ViiA 7 Real-Time PCR System (Life Technologies).
Statistical Analysis.
Statistical analysis was performed using GraphPad Prism software version 5. P values less than 0.05 were considered statistically significant.
SI Materials and Methods
Thermal Shift Assay.
Thermal melting experiments were carried out using an Mx3005p Real Time PCR machine (Stratagene) as described (43). Proteins were buffered in 10 mM Hepes, pH 7.5, 500 mM NaCl and assayed in a 96-well plate at a final concentration of 2 μM in 20-μL volume. Compounds were added at a final concentration of 10 μM. SYPRO Orange (Molecular Probes) was added as a fluorescence probe at a dilution of 1:1,000. Excitation and emission filters for the SYPRO-Orange dye were set to 465 nm and 590 nm, respectively. The temperature was raised with a step of 3 °C per minute from 25 °C to 96 °C, and fluorescence readings were taken at each interval.
Protein Expression and Purification.
cDNA encoding human bromodomains was cloned, expressed, and purified as previously described (10). For purification of in vivo biotinylated protein expression, the same construct boundaries (e.g., CBP residues R1081-G1198) were subcloned into pNIC-BIO1 vector, a derivative from pNIC28-Bsa4 vector (GenBank EF198106), containing a 10 His-tag and TEV protease cleavage site at the N terminus and an in-frame biotinylation sequence inserted at the C terminus. The constructs were transformed into the BL21 (DE3)-R3-BirA cell line (BL21 derivative coexpressing BirA using a pACYC coexpression vector). Cells were grown overnight at 37 °C in 10 mL of Luria–Bertani medium with 50 μg/mL kanamycin and 34 μg/mL chloramphenicol (start-up culture). The start-up culture was diluted 1:1,000 in fresh medium, and cell growth was allowed at 37 °C to an optical density of about ∼1.0 (OD600) before the temperature was decreased to 25 °C. d-Biotine was dissolved into 10 mM bicine, pH 8.3, and added to the culture at 500 μM final concentration. The protein expression was induced for 8 h at 25 °C with 50 µM isopropyl-β-d-thiogalactopyranoside (IPTG). Proteins were purified using Ni-affinity chromatography and size exclusion chromatography.
Crystallization, Data Collection, and Structure Solution.
EP300 construct (Uniprot identifier as EP300_HUMAN Q09472-1 fragment 1048–1161), BRD4 construct (Uniprot identifier as BRD4_HUMAN O60885-1 fragment 44–168), and BRD2 construct (Uniprot identifier as BRD2_HUMAN P25440-1 fragment 344–455) were used for crystallographic studies. Aliquots of the purified proteins were set up for crystallization using a mosquito crystallization robot (TTP Labtech). Coarse screens were typically setup onto Greiner three-well plates using three different drop ratios of precipitant to protein per condition (200 + 100 nL, 150 + 150 nL, and 100 + 200 nL). All crystallizations were carried out using the sitting drop vapor diffusion method at 4 °C. EP300 rod crystals with CBP30 (2.8 mM final concentration) were obtained by mixing 100 nL of the protein (13 mg/mL) and 200 nL of crystallization buffer [0.1 M trisodium citrate dehydrate, pH 5.6, 20% (vol/vol) iso-propanol, 20% (wt/vol) PEG 4000]. Bar-shaped crystals of BRD4(1) with CBP30 (5.54 mM final concentration) were obtained by mixing 200 nL of the protein (17.8 mg/mL) and 100 nL of crystallization buffer [0.1 M succinic acid, 20% (wt/vol) PEG 3350]. Plate crystals of BRD2(2) with CBP30 (8 mM final concentration) were grown by mixing equal volumes of 150 nL of protein (7 mg/mL) and crystallization buffer [0.2 M sodium/potassium tartrate, 20% (wt/vol) PEG 3350, 10% (vol/vol) ethylene glycol].
Crystals were cryoprotected using the well solution supplemented with additional 20% (vol/vol) ethylene glycol and were flash frozen in liquid nitrogen. Data were collected at Diamond Light Source beamlines I02 and I03 at a wavelength of 0.9795 Å and 0.9763 Å. Indexing and integration were carried out using XDS (44), and scaling was performed with AIMLESS (45). Initial phases were calculated by molecular replacement with PHASER using an ensemble of known bromodomain models (PDB ID codes 2OSS, 2OUO, 2GRC, 2OO1, 3DAI, 3D7C, 3DWY, and 3G0L). Unique and initial solutions were improved in a total of 50 cycles of automated protein chain tracing, starting from existing model and computed using ARP/wARP (46). Further manual building was performed with COOT (47), and refinement against maximum likelihood target using REFMAC5 (48). Thermal motions were analyzed using TLSMD (translation/libration/screw motion determination), and hydrogen atoms were included in late refinement cycles. PRODRG (49) was used to generate the CBP30 inhibitor coordinates and .cif files. All model validations were carried out using MolProbity (50). Data collection and refinement statistics are compiled in Table S3. The models and structure factors have PDB ID codes 5BT3 (EP300), 5BT4 [BRD4(1)], and 5BT5 [BRD2(2)].
FRAP.
FRAP studies were performed using a recently published protocol (25). In brief, U2OS cells were transfected (Lipofectamine 2000 transfection reagent; Life Technologies), with mammalian overexpression constructs encoding an N-terminal GFP and an NLS, followed by three tandem repeats of the extended CBP bromodomain (amino acids 868–1,341). The FRAP and imaging system consisted of a Zeiss LSM 710 laser-scanning and control system coupled to an inverted Zeiss Axio Observer.Z1 microscope equipped with a high-numerical-aperture (N.A. 1.3) 40× oil immersion objective (Zeiss). Samples were placed in an incubator chamber capable of maintaining temperature and humidity. FRAP and GFP fluorescence imaging were carried out with an argon-ion laser (488 nm) and with a PMT (photomultiplier) detector set to detect fluorescence between 500 and 550 nm. Once an initial scan had been taken, a region of interest corresponding to ∼50% of the entire GFP-positive nucleus was empirically selected for bleaching. A time-lapse series was then taken to record GFP recovery using 1% of the power used for bleaching. The image datasets and fluorescence recovery data were exported from ZEN 2009, the microscope control software. On average, 10–20 cells were imaged per group. One-way ANOVA with Tukey–Kramer correction for multiple comparisons was used to detect significant differences (P < 0.05) between treatment groups.
ChIP-q-PCR.
ChIP was performed using the Low Cell# ChIP kit (Diagenode). CD4+ T cells were negatively selected from leukocyte cones and cultured under Th17-promoting conditions as described in Cell Purification and Cell Culture. After 6 d of culture, 3 × 105 Th17 cells were purified with an IL-17 Secretion Assay (Miltenyi Biotec) for each ChIP and incubated with 2 µM CBP30 or DMSO for 24 h. Cells were cross-linked with 1% formaldehyde for 10 min, and 1.25 M Glycine was added for 5 min. Sonication was performed by three runs of 10 cycles (30 s “ON,” 30 s “OFF”) at high power setting (BioruptorPico; Diagenode). Cell lysates were immunoprecipitated with anti-p300 (C-20; Santa Cruz) and anti-H3K56Ac (C15410213; Diagenode), and DNA isolation was performed with DNA isolation buffer kit (Diagenode) according to the manufacturer’s instructions. q-PCR was performed with SYBR Green (Qiagen) on ViiA7 PCR instruments (Life Technologies), using primer pairs in the human IL 17A locus (−43, forward 5′-AGGCACAAACTCATCCATCC-3′, reverse 5′-GTCAGAACCCAGCGTTTCAT-3′; +94, forward 5′-ATGAAACGCTGGGTTCTGAC-3′, reverse 5′-CGGCTTTCAAAATCTCAAGG-3′; +4,000, forward 5′-ACCACACAACCCAGAAAGGA-3′, reverse 5′-GCAGGATGGAGTGAAGAGGA-3′) and human IL 17F locus (+29, forward 5′-GAAATCCTAGGCATGACAGTCC-3′, reverse 5′-AACACAGGCATACACAGGAAGA-3′). All values obtained were normalized to input. The relative enrichment was expressed as mean ± SEM.
Cell Purification and Cell Culture.
Mononuclear cells from peripheral blood (PBMCs) or synovial fluid (SFMCs) were isolated by Ficoll density-gradient centrifugation (Histopaque; Sigma-Aldrich). CD4+ T cells were negatively selected from PBMCs or SFMCs with a human CD4+ T-cell Isolation Kit (Miltenyi Biotec). The purity of this population was ≥89.1% by flow cytometry. CD4+ T cells were cultured in conditions that promoted Th17 numbers and preserved Th1, Th2, and Treg numbers as follows: RPMI-1640 (Sigma-Aldrich) supplemented with penicillin-streptomycin (50,000 units, 50 mg) (Sigma-Aldrich), l-glutamine (2 mM) (Gibco/Life Technologies), and 10% (vol/vol) FBS (Sigma-Aldrich) at 37 °C and 5% CO2 in a humidified atmosphere. We used recombinant human IL-2 (100 IU/mL), IL-1β (20 ng/mL), IL-6 (20 ng/mL), and IL-23 (20 ng/mL) (all Peprotech), together with anti-CD2/3/28 beads (Miltenyi Biotec) at a ratio of one bead per 20 cells in 96-well plates (5 × 104 cells per well), for 3 d for cytokine analysis and cell viability analysis, and 6 d for proliferation analysis. For transcriptional analysis, a ratio of one bead per 10 cells in 48-well plates (1 × 106 cells per well) for 4 d was used.
Cell Proliferation and Viability, ELISA, and LUMINEX Analysis.
CD4+ T cells were labeled with 5 µM CFSE (carboxyfluorescein succinimidyl ester) (Molecular Probes/Invitrogen/Life Technologies) according to the manufacturer´s instructions and cultured as above. Before flow cytometry, cells were additionally stained for surface markers (anti-CD4-APC and anti-CD8a-BV510; BioLegend) and viability (fixable viability dye eFluor780; ebioscience). Samples were analyzed on an LRSFortessa (BD Bioscience), and 2 × 104 live CD4+ T cells were recorded per sample. Data analysis was performed using FlowJo software (version X 10.0.7).
For viability assays, cells were washed twice in cold PBS and stained with anti-Annexin V-FITC and 7-AAD (both BD Bioscience) in Annexin V binding buffer (10 mM Hepes, pH 7.4, 140 mM NaCl, and 2.5 mM CaCl2) for 15 min at room temperature in the dark. Flow cytometry analysis was performed within 1 h of staining in Annexin V binding buffer.
CD4+ T cells (5 × 104 per well) were cultured in 96-well plates as described above for 3 d. Inhibitors were added at a final concentration of 1:1,000 on day 0. Supernatants were analyzed with an IL-17A, IL-17F, and IFNγ ELISA kit (ebioscience) and subjected to LUMINEX analysis with a custom-made premixed Multiplex Screening Assay for IL-10, IL-22, TNFα, and GM-CSF (R&D Systems) following the manufacturer´s instructions. Samples were read on a Bio-Plex 200 System (Bio-Rad) using Bio-Plex Manager software version 5.0.
Transcriptional Profiling.
CD4+ T cells (1 × 106) were cultured in 48-well plates as described above for 4 d. CD4+ T cells were subjected to total RNA isolation with the RNeasy Mini kit (Qiagen) on day 0 or day 4, including on-column digestion of DNA with RNase-Free DNase Set I (Qiagen). RNA quality was checked using a 2100 Bioanalyzer and a Eukaryote Total RNA Nano Assay (Agilent Technologies). Microarray analysis was performed by the Cambridge Genomic Service using Illumina beadchip array HT12v4.1. RMA (robust multiarray average) expression values were normalized by quantiles, using R (Bioconductor). Genes differentially expressed were selected using ANOVA, with a P value, based on permutations, <0.01 and a false discovery rate <0.05, using Multiple Experiment Viewer (MEV) (Dana-Farber Cancer Institute). Genes were filtered for a log ratio ≥ 1 in at least one condition. The transcripts were further organized with K-means clustering after median centering using MEV. Pathways and gene ontology overrepresentation were assessed using Ingenuity Pathways Analysis (Ingenuity Systems) (51). Data (compliant for Minimum Information About a Microarray Experiment) for the datasets used in this study have been deposited in the Gene Expression Omnibus (GEO) profiles database, www.ncbi.nlm.nih.gov/geo/ (accession no. GSE71231).
For quantitative PCR, RNA was reverse transcribed with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). TaqMan Fast Universal PCR Master Mix and TaqMan probes for transcripts encoding IL 17A (Assay ID Hs00174383_m1), IL 17F (Hs00369400_m1), IL 21 (Hs00222327_m1), IL 22 (Hs01574154_m1), CSF2 (Hs00929873_m1), IL 10 (Hs00961622_m1), IFNG (Hs00989291_m1), IL 23R (Hs00332759_m1), RORC (Hs01076122_m1), TBX21 (Hs00203436_m1), HIST1H2BK (Hs00955067_g1), CCL17 (Hs00171074_m1), GZMB (Hs01554355_m1), TIMD4 (Hs00293316_m1), and NOTCH1 (Hs01062014_m1) were used for q-PCR on a ViiA 7 Real-Time PCR System (Life Technologies). Assays were performed in triplicate, and gene expression levels were normalized to ACTB (β-actin) (Hs01060665_g1) or RPL13A (Hs04194366_g1), and internal controls (no cDNA) with ΔΔCt calculations.
Supplementary Material
Acknowledgments
A.H. was funded by Deutsche Forschungsgemeinschaft Grant HA 7021/1-1. This work was funded by the Oxford National Institute for Health Research Biomedical Research Centre/Unit and by the Structural Genomics Consortium, a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, the Canada Foundation for Innovation, Genome Canada, GlaxoSmithKline, Janssen, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development and Innovation, Pfizer, Takeda, and Wellcome Trust Grant 092809/Z/10/Z. P.B. is in part funded by the Innovative Medicines Initiative (IMI)-funded project Unrestricted Leveraging of Targets for Research Advancement and Drug Discovery (ULTRA-DD).
Footnotes
Conflict of interest statement: P.B. declares research support from Merck Research Laboratories.
This article is a PNAS Direct Submission. S.K. is a guest editor invited by the Editorial Board.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE71231). The models and structure factors have been deposited in the Protein Data Bank, www.pdb.org {PDB ID codes 5BT3 [EP300], 5BT4 [BRD4(1)], and 5BT5 [BRD2(2)]}.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501956112/-/DCSupplemental.
References
- 1.Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat Struct Mol Biol. 2013;20(9):1040–1046. doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- 2.Filippakopoulos P, Knapp S. Targeting bromodomains: Epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13(5):337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 3.Muller S, Filippakopoulos P, Knapp S. Bromodomains as therapeutic targets. Expert Rev Mol Med. 2011;13:e29. doi: 10.1017/S1462399411001992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belkina AC, Nikolajczyk BS, Denis GV. BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J Immunol. 2013;190(7):3670–3678. doi: 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276(17):13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 6.Arif M, et al. Mechanism of p300 specific histone acetyltransferase inhibition by small molecules. J Med Chem. 2009;52(2):267–277. doi: 10.1021/jm800657z. [DOI] [PubMed] [Google Scholar]
- 7.Emami KH, et al. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected] Proc Natl Acad Sci USA. 2004;101(34):12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Best JL, et al. Identification of small-molecule antagonists that inhibit an activator: Coactivator interaction. Proc Natl Acad Sci USA. 2004;101(51):17622–17627. doi: 10.1073/pnas.0406374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vidler LR, Brown N, Knapp S, Hoelder S. Druggability analysis and structural classification of bromodomain acetyl-lysine binding sites. J Med Chem. 2012;55(17):7346–7359. doi: 10.1021/jm300346w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468(7327):1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Picaud S, et al. PFI-1, a highly selective protein interaction inhibitor, targeting BET Bromodomains. Cancer Res. 2013;73(11):3336–3346. doi: 10.1158/0008-5472.CAN-12-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hewings DS, et al. 3,5-dimethylisoxazoles act as acetyl-lysine-mimetic bromodomain ligands. J Med Chem. 2011;54(19):6761–6770. doi: 10.1021/jm200640v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hay DA, et al. Discovery and optimization of small-molecule ligands for the CBP/p300 bromodomains. J Am Chem Soc. 2014;136(26):9308–9319. doi: 10.1021/ja412434f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rooney TP, et al. A series of potent CREBBP bromodomain ligands reveals an induced-fit pocket stabilized by a cation-π interaction. Angew Chem Int Ed Engl. 2014;53(24):6126–6130. doi: 10.1002/anie.201402750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mele DA, et al. BET bromodomain inhibition suppresses TH17-mediated pathology. J Exp Med. 2013;210(11):2181–2190. doi: 10.1084/jem.20130376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 19.Shen H, Goodall JC, Hill Gaston JS. Frequency and phenotype of peripheral blood Th17 cells in ankylosing spondylitis and rheumatoid arthritis. Arthritis Rheum. 2009;60(6):1647–1656. doi: 10.1002/art.24568. [DOI] [PubMed] [Google Scholar]
- 20.Bowness P, et al. Th17 cells expressing KIR3DL2+ and responsive to HLA-B27 homodimers are increased in ankylosing spondylitis. J Immunol. 2011;186(4):2672–2680. doi: 10.4049/jimmunol.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sieper J. Treatment challenges in axial spondylarthritis and future directions. Curr Rheumatol Rep. 2013;15(9):356. doi: 10.1007/s11926-013-0356-9. [DOI] [PubMed] [Google Scholar]
- 22.Baeten D, et al. Anti-interleukin-17A monoclonal antibody secukinumab in treatment of ankylosing spondylitis: A randomised, double-blind, placebo-controlled trial. Lancet. 2013;382(9906):1705–1713. doi: 10.1016/S0140-6736(13)61134-4. [DOI] [PubMed] [Google Scholar]
- 23.Wei G, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30(1):155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, et al. Transcription of Il17 and Il17f is controlled by conserved noncoding sequence 2. Immunity. 2012;36(1):23–31. doi: 10.1016/j.immuni.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philpott M, et al. Assessing cellular efficacy of bromodomain inhibitors using fluorescence recovery after photobleaching. Epigenetics Chromatin. 2014;7:14. doi: 10.1186/1756-8935-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciceri P, et al. Dual kinase-bromodomain inhibitors for rationally designed polypharmacology. Nat Chem Biol. 2014;10(4):305–312. doi: 10.1038/nchembio.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berg EL, et al. Chemical target and pathway toxicity mechanisms defined in primary human cell systems. J Pharmacol Toxicol Methods. 2010;61(1):3–15. doi: 10.1016/j.vascn.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Roderick JE, et al. c-Myc inhibition prevents leukemia initiation in mice and impairs the growth of relapsed and induction failure pediatric T-ALL cells. Blood. 2014;123(7):1040–1050. doi: 10.1182/blood-2013-08-522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matzuk MM, et al. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150(4):673–684. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reveille JD, Weisman MH. The epidemiology of back pain, axial spondyloarthritis and HLA-B27 in the United States. Am J Med Sci. 2013;345(6):431–436. doi: 10.1097/maj.0b013e318294457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dean LE, et al. Global prevalence of ankylosing spondylitis. Rheumatology (Oxford) 2014;53(4):650–657. doi: 10.1093/rheumatology/ket387. [DOI] [PubMed] [Google Scholar]
- 32.Cortes A, et al. International Genetics of Ankylosing Spondylitis Consortium (IGAS); Australo-Anglo-American Spondyloarthritis Consortium (TASC); Groupe Française d’Etude Génétique des Spondylarthrites (GFEGS); Nord-Trøndelag Health Study (HUNT); Spondyloarthritis Research Consortium of Canada (SPARCC); Wellcome Trust Case Control Consortium 2 (WTCCC2) Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 2013;45(7):730–738. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans DM, et al. Spondyloarthritis Research Consortium of Canada (SPARCC); Australo-Anglo-American Spondyloarthritis Consortium (TASC); Wellcome Trust Case Control Consortium 2 (WTCCC2) Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43(8):761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McInnes IB, et al. PSUMMIT 1 Study Group Efficacy and safety of ustekinumab in patients with active psoriatic arthritis: 1 year results of the phase 3, multicentre, double-blind, placebo-controlled PSUMMIT 1 trial. Lancet. 2013;382(9894):780–789. doi: 10.1016/S0140-6736(13)60594-2. [DOI] [PubMed] [Google Scholar]
- 35.Burmester GR, et al. EARTH Study Group Efficacy and safety of mavrilimumab in subjects with rheumatoid arthritis. Ann Rheum Dis. 2013;72(9):1445–1452. doi: 10.1136/annrheumdis-2012-202450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jandus C, et al. Increased numbers of circulating polyfunctional Th17 memory cells in patients with seronegative spondylarthritides. Arthritis Rheum. 2008;58(8):2307–2317. doi: 10.1002/art.23655. [DOI] [PubMed] [Google Scholar]
- 37.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151(2):289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 immune axis: From mechanisms to therapeutic testing. Nat Rev Immunol. 2014;14(9):585–600. doi: 10.1038/nri3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Byun JS, et al. Dynamic bookmarking of primary response genes by p300 and RNA polymerase II complexes. Proc Natl Acad Sci USA. 2009;106(46):19286–19291. doi: 10.1073/pnas.0905469106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Annunziato F, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204(8):1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pène J, et al. Chronically inflamed human tissues are infiltrated by highly differentiated Th17 lymphocytes. J Immunol. 2008;180(11):7423–7430. doi: 10.4049/jimmunol.180.11.7423. [DOI] [PubMed] [Google Scholar]
- 42.Matusevicius D, et al. Interleukin-17 mRNA expression in blood and CSF mononuclear cells is augmented in multiple sclerosis. Mult Scler. 1999;5(2):101–104. doi: 10.1177/135245859900500206. [DOI] [PubMed] [Google Scholar]
- 43.Fedorov O, Niesen FH, Knapp S. Kinase inhibitor selectivity profiling using differential scanning fluorimetry. Methods Mol Biol. 2012;795:109–118. doi: 10.1007/978-1-61779-337-0_7. [DOI] [PubMed] [Google Scholar]
- 44.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evans PR. An introduction to data reduction: Space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3(7):1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Schüttelkopf AW, van Aalten DM. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 50.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez FO. 2012. Analysis of gene expression and gene silencing in human macrophages. Curr Protoc Immunol, Suppl 96, Unit 14.28, pp 14.28.1–14.28.23.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.