Significance
Aneuploidy, a condition of abnormal chromosomal content, can support adaptive mechanisms in response to environmental cues but comes at the expense of decreased proliferation and dysfunction of cellular processes. Here we show that the gain of an extra copy of chromosome XI in yeast is an adaptive mechanism to deal with oxidative stress under conditions of antioxidant deficiency. We narrowed down the effect of adaptive aneuploidy to two genes on chromosome XI, which supported increased mitochondrial abundance and respiration, which in turn provided reducing equivalents for hydroperoxide removal. Forced expression of these genes eliminated aneuploidy, improved cell growth, and was sufficient for protection against oxidative stress. Thus, aneuploidy can adaptively reprogram cellular metabolism, protecting against oxidative stress by upregulating respiration.
Keywords: thiol peroxidase, oxidative stress, aneuploidy, respiration
Abstract
Aerobic respiration is a fundamental energy-generating process; however, there is cost associated with living in an oxygen-rich environment, because partially reduced oxygen species can damage cellular components. Organisms evolved enzymes that alleviate this damage and protect the intracellular milieu, most notably thiol peroxidases, which are abundant and conserved enzymes that mediate hydrogen peroxide signaling and act as the first line of defense against oxidants in nearly all living organisms. Deletion of all eight thiol peroxidase genes in yeast (∆8 strain) is not lethal, but results in slow growth and a high mutation rate. Here we characterized mechanisms that allow yeast cells to survive under conditions of thiol peroxidase deficiency. Two independent ∆8 strains increased mitochondrial content, altered mitochondrial distribution, and became dependent on respiration for growth but they were not hypersensitive to H2O2. In addition, both strains independently acquired a second copy of chromosome XI and increased expression of genes encoded by it. Survival of ∆8 cells was dependent on mitochondrial cytochrome-c peroxidase (CCP1) and UTH1, present on chromosome XI. Coexpression of these genes in ∆8 cells led to the elimination of the extra copy of chromosome XI and improved cell growth, whereas deletion of either gene was lethal. Thus, thiol peroxidase deficiency requires dosage compensation of CCP1 and UTH1 via chromosome XI aneuploidy, wherein these proteins support hydroperoxide removal with the reducing equivalents generated by the electron transport chain. To our knowledge, this is the first evidence of adaptive aneuploidy counteracting oxidative stress.
Several fundamental biological processes such as respiration, photosynthesis, and the immune response, as well as numerous other cellular processes, lead to the production of partially reduced species of molecular oxygen known as reactive oxygen species (ROS) (1, 2). Elevated levels of ROS are often associated with oxidative stress, which manifests as damage to lipids, proteins, DNA, metabolites, and other biomolecules (3). However, ROS are not only deleterious factors but may also participate in the regulation of cellular processes. For example, hydrogen peroxide (H2O2)-mediated oxidation of cysteine residues is involved in redox signaling (4, 5). Therefore, it is important for organisms to carefully control redox homeostasis.
To deal with the dual role of ROS as deleterious or beneficial species, organisms have evolved several mechanisms. Protectors from ROS, known as antioxidants, can be categorized as nonenzymatic and enzymatic ROS molecules. The nonenzymatic antioxidants buffer the intracellular milieu against ROS toxicity. For example, ascorbate, tocopherol, flavonoids, alkaloids, and carotenoids can scavenge oxygen-derived free radicals, whereas glutathione (GSH), a highly abundant tripeptide, supports two electron reduction mechanisms (6). Enzyme antioxidants include superoxide dismutases (eliminate the superoxide radical) and catalases (eliminate H2O2), as well as many other enzymes (6).
Notable among them are peroxiredoxins (Prxs) and glutathione peroxidases (Gpxs), two families of thiol-specific peroxide scavengers (7) collectively known as thiol peroxidases. Prxs and Gpxs are highly conserved, and many organisms have multiple copies of these genes. For example, five Prxs and three Gpxs, localized to different cellular compartments, have been identified in Saccharomyces cerevisiae (8). One of the most deleterious damage forms inflicted by ROS is DNA damage (9). Many genetic disorders as well as different forms of cancer are associated with oxidative stress-induced DNA damage (10, 11). Recent studies offer direct proof that thiol peroxidases represent the first line of defense against oxidative stress-induced DNA damage (12, 13).
The observation that compromised function of thiol peroxidases causes the loss of genome stability and a broad spectrum of mutations and chromosomal rearrangements has provided further evidence that these enzymes are significant contributors to genome stability (14, 15). Aneuploidy, with its aberrant number of chromosomes, is also associated with oxidative stress (16); however, a link between thiol peroxidase status and aneuploidy has not been reported previously.
Recently, we isolated two independent strains lacking all eight thiol peroxidase genes (∆8 cells) (15). In the current study, we investigated how these cells deal with hydroperoxides. We found that both ∆8 mutants were strictly dependent on respiration and were characterized by chromosome XI (chr-XI) aneuploidy. Two genes, CCP1 and UTH1, located on this chromosome were responsible for the whole-chromosome duplication, because they allowed H2O2 elimination with the reducing equivalents of the electron transport chain (ETC). These two genes were essential for these cells, whereas their forced coexpression in ∆8 mutants allowed cells to lose the extra copy of chr-XI and increase their growth. These findings identify a mechanism of adaptive aneuploidy, which in turn points to new targets for therapeutic interventions for diseases associated with oxidative stress.
Results
Characterization of ∆8 Strains.
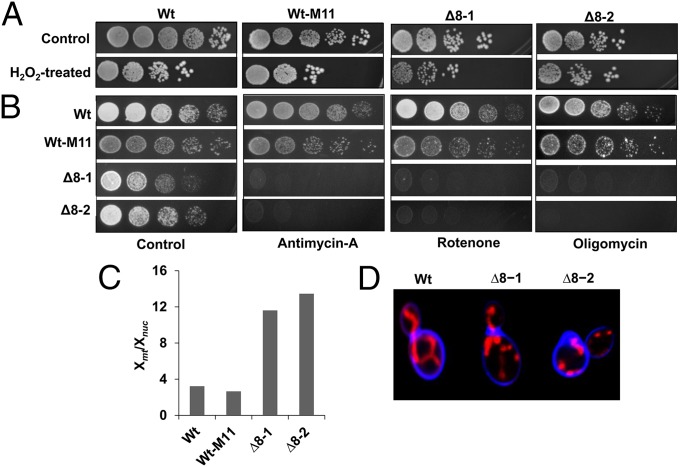
Our previous analyses of Δ8 cells revealed that thiol peroxidases are not essential; however, they surprisingly could withstand several stresses (17). Two independent Δ8 isolates also manifested phenotypes such as slow growth, short lifespan, and decreased colony size. We confirmed that both isolates grew slower than wild-type (Wt) cells but were nearly as resistant to treatment with 0.4 mM H2O2 as Wt cells (Fig. 1A). An additional observation was that Δ8 cells were dependent on respiration, as these cells were highly sensitive to antimycin-A, rotenone, and oligomycin (Fig. 1B). Consistent with the dependence on respiration, both Δ8 isolates increased their mitochondrial copy number, as measured by the ratio of mitochondrial DNA (mtDNA) to nuclear DNA (Fig. 1C). Mitochondria are known to be the major source of ROS (18); therefore, it was surprising that thiol peroxidase deficiency led to a higher, rather than lower, mitochondrial content, and forced cells to rely on respiration when grown on rich medium. We also observed structural differences in mitochondria distribution between Wt and Δ8 cells: Logarithmically grown Wt cells showed filamentous structure of mitochondria, whereas Δ8 cells exhibited punctuated mitochondria and mitochondrial clusters (Fig. 1D). Together, these data and the fact that the observed phenotypes appeared in two independent Δ8 isolates suggest that increased respiration is an adaptive feature of Δ8 cells.
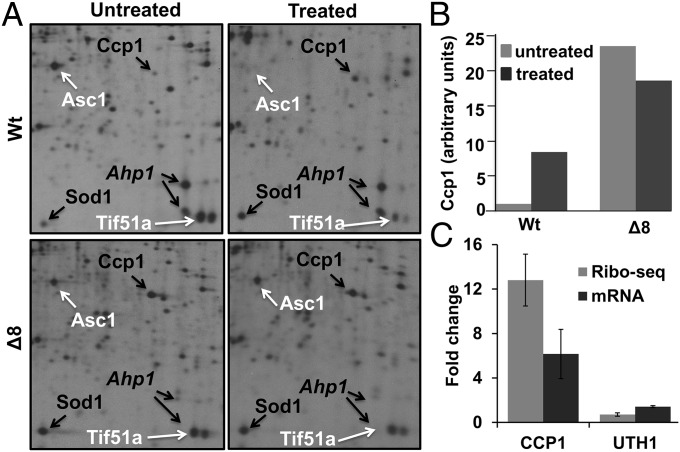
Fig. 1.
Phenotypes of ∆8 strains. (A) H2O2 tolerance. Tenfold serial dilution of Wt cells, Wt-M11 cells, and two independent ∆8 mutant isolates (∆8-1 and ∆8-2) onto YPD plates containing (H2O2-treated) or not (control) 0.4 mM H2O2. Pictures were taken after 3 d. (B) Dependence on respiration. Tenfold serial dilution assays of Wt, Wt-M11, ∆8-1, and ∆8-2 cells in the presence or absence of antimycin-A, rotenone, and oligomycin. (C) Increased mtDNA copy number in ∆8 mutant cells. The ratio of the normalized number of reads corresponding to mtDNA (Xmt) and the normalized number of reads corresponding to nuclear DNA (Xnuc) is shown for each strain. (D) Distribution of mitochondria. Mitochondria were stained with MitoTracker Red. Blue color corresponds to calcofluor, which visualizes cell membranes.
Chr-XI Aneuploidy in Δ8 Cells.
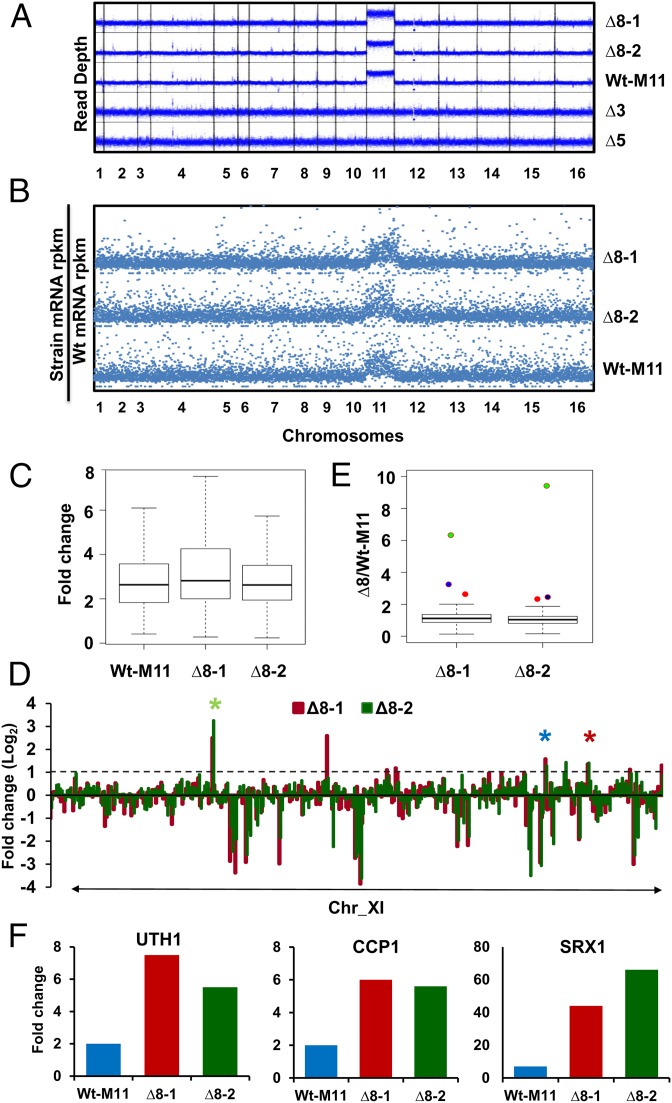
Genomic sequencing of Wt cells revealed uniform coverage of reads across the genome, as expected; however, both Δ8 strains had twice as many reads corresponding to chr-XI (Fig. 2A). As a control, we sequenced the genome and analyzed the growth and sensitivity to respiration inhibitors of the Wt-M11 strain, which is characterized by duplication of chr-XI and has a complete set of thiol peroxidase genes. This strain was not sensitive to inhibitors of respiration (Fig. 1B), but its genome coverage was identical to that of Δ8 strains (Fig. 2A). In contrast, genome sequencing of cells lacking all five Prxs (Δ5 cells) or all three Gpxs (Δ3 cells), which served as the initial strains for independent preparation of the two Δ8 isolates, did not show increased chr-XI coverage (Fig. 2A). Thus, the two Δ8 strains independently acquired a second copy of chr-XI and are characterized by aneuploidy.
Fig. 2.
Chromosome XI aneuploidy of ∆8 cells. (A) Occurrence of chr-XI disomy in ∆8 strains identified by genome sequencing. Read depth was calculated in 100-bp windows. Genome coverage is shown for two independent ∆8 mutant isolates (∆8-1 and ∆8-2), a chr-XI aneuploid strain (Wt-M11), and strains (∆3, lacking three Gpxs; ∆5, lacking five Prxs) from which ∆8 cells were prepared. (B) Increased expression of chr-XI genes revealed by RNA-seq. Total numbers of normalized FPKM reads per strain were normalized by Wt reads, and the ratio was plotted. (C) Increased average expression of chr-XI genes in Wt-M11, ∆8-1, and ∆8-2 cells. Average expression of genes on chr-XI of Wt-M11, ∆8-1, and ∆8-2 strains was calculated as fold change compared with Wt cells. Center black bar indicates median and error bars (whiskers) indicate 95% confidence interval for C and E. (D) Expression levels of genes on chr-XI in ∆8 cells compared with those in the Wt-M11 strain. Log2 ratios of FPKM read values per gene are shown for chr-XI of ∆8-1 (red) and ∆8-2 (green) cells. Three target genes that increased expression in both ∆8 strains more than twofold are labeled with asterisks [SRX1 (green), UTH1 (blue), and CCP1 (red)]. (E) Analysis of gene expression on chr-XI. The ∆8/Wt-M11 gene expression ratios were analyzed for all chr-XI genes of ∆8-1 and ∆8-2 strains. Expression of the three genes that showed increased expression in both ∆8 strains, SRX1 (green), UTH1 (blue), and CCP1 (red), is indicated with dots. (F) Comparison of the expression of the three identified genes among strains. mRNA abundance of each gene was compared with that of Wt cells, and the ratio was plotted.
Previous studies showed that genes encoded on duplicated chromosomes tend to be more highly expressed (19, 20). Indeed, the second copy of chr-XI was fully transcriptionally active in Δ8 cells, as revealed by RNA-seq (sequencing) analyses (Fig. 2B and Dataset S1). The expression of genes encoded by chr-XI were, on average, twice as high in these cells compared with Wt cells (Fig. 2C), whereas the average gene expression derived from other chromosomes was unchanged. This observation was also confirmed by examining gene expression in Wt-M11 cells (Fig. 2B). Thus, there was a direct correspondence between chromosome copy number and derived gene expression in ∆8 strains, as shown previously for cells with other mutant backgrounds (20, 21).
Elevated Expression of Chr-XI Genes SRX1, CCP1, and UTH1 Is a Common Feature of ∆8 Strains.
Given the fact that aneuploidy can cause rapid adaptive evolution (22, 23), we investigated the possibility that chr-XI disomy, observed in ∆8 strains, was the primary genetic adaptation that accounts for the observed phenotypes of Δ8 cells and, perhaps, for their viability, resistance to oxidative stress, and dependence on respiration. First, we compared global gene expression between the ∆8 strains. The pair of ∆8 strains with identical karyotypes displayed a highly similar transcriptome profile (ρ = 0.97), whereas both showed significant changes in gene expression compared with Wt cells (15). A better reference strain to identify genes whose expression changed in response to thiol peroxidase deficiency, as opposed to changes in response to aneuploidy, is the Wt-M11 strain. With this idea in mind, we identified three genes (SRX1, CCP1, and UTH1) whose expression deviated significantly in both ∆8 strains compared with either Wt-M11 cells (P < 0.01) or Wt cells (Fig. 2 D–F). Srx1 is responsible for reducing the hyperoxidized catalytic cysteine of Tsa1 and Tsa2 (24), two of the thiol peroxidases that were deleted in ∆8 strains. Thus, the increased expression of SRX1 is likely a response to the loss of Prx functions. CCP1 encodes a heme-containing protein, which occurs in the mitochondrial intermembrane space and functions in respiring mitochondria (25, 26). It was found that Ccp1 is an efficient H2O2 scavenger that uses cytochrome c as a reductant and protects respiring yeast cells from H2O2 (27). The SUN family protein Uth1 (UTH1) is a mitochondrial inner-membrane protein that is thought to be involved in mitochondrial biogenesis, aging, cell death, and oxidative stress response (28–30). It is of interest that all three identified genes are important for mitochondrial function and protection against oxidative stress.
CCP1 and UTH1 but Not SRX1 Are Essential for ∆8 Viability.
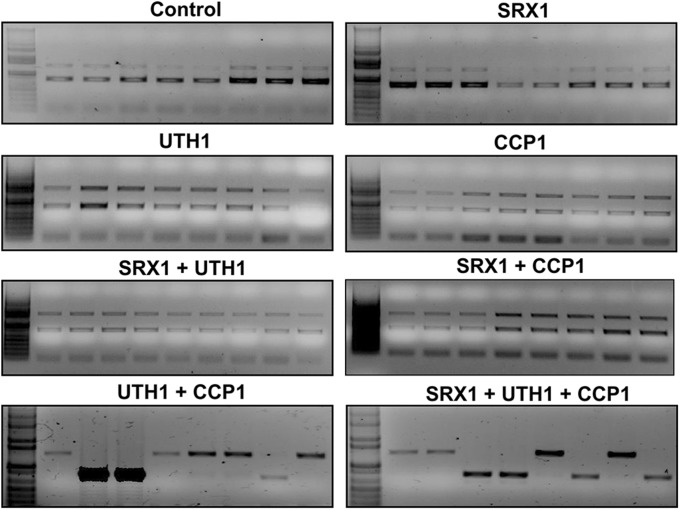
Gene expression analyses suggested that the elevated expression of the three genes might be an adaptation of Δ8 cells to thiol peroxidase deficiency. If so, Δ8 cells may require these genes for viability. To analyze the null phenotype of these genes in ∆8 cells, we constructed natamycin (NatMX) gene cassettes that contained genes as well as 1-kb regions upstream and downstream of each gene (Fig. 3A). After transformation, colonies were selected from yeast extract-peptone-dextrose (YPD) plates containing 100 µg/mL NatMX and analyzed by colony PCR. We succeeded in recovering many colonies in which SRX1 was deleted in both ∆8 and Wt cells (Fig. S1A); however, no ∆8 colonies were recovered with deletions of either CCP1 or UTH1.
Fig. 3.
Aneuploidy is linked to elevated expression of CCP1 and UTH1. Representative colonies from each expression group of the laboratory evolution experiment (Fig. S1) were analyzed at 120 generations by colony PCR. Increased expression of genes CCP1 + UTH1 or CCP1 + UTH1 + SRX1 allows cells to lose the extra copy of chr-XI (two lower panels). Individual colonies expressing these genes lost one or the other copy of this chromosome, shown by the loss of the upper or lower bands. All colonies expressing individual genes or pairs of genes including SRX1 had both bands. Left lanes in each panel show size markers.
Fig. S1.
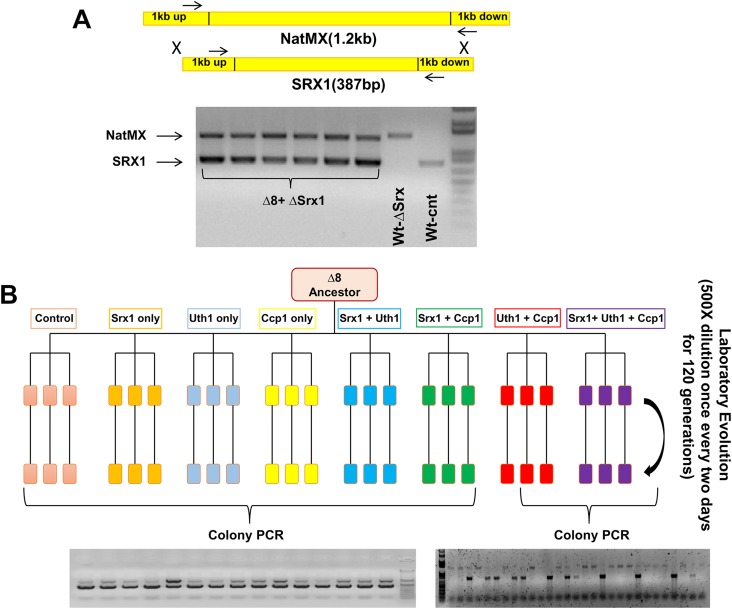
Experimental design of the laboratory evolution experiment. (A) The marker strain used in the laboratory evolution experiment. One copy of SRX1 was replaced with the NatMX marker. Primers were designed to match 100 bp upstream and downstream of SRX1. The upper (∼1.2-kb) band represents NatMX, and the lower (∼600-bp) band represents SRX1 in ∆8 cells. (B) Design of the laboratory evolution experiment. Single or combined expression of CCP1, UTH1, and SRX1 when transformed into the marker strain. Two empty vectors were also transformed and used as the control. Parallel lines splitting from the same branch represent three independent repetitions for each group. Each group was analyzed every 25th generation by assaying selected colonies by colony PCR.
Coexpression of CCP1 and UTH1 Releases the Dependence on Aneuploidy.
We designed a laboratory evolution experiment (31) to determine whether the disomy of chr-XI was selected for the increased expression of CCP1 and UTH1 in ∆8 cells. To this end, we prepared a Δ8 marker strain in which one of the two copies of SRX1 was replaced with the NatMX marker (Fig. 3A). The evolution experiment was conducted by transforming the marker strain with plasmids carrying SRX1, CCP1, and UTH1 under constitutive promoters either individually or in combination (Fig. S1B). Three independent colonies were selected from each transformation and grown on SDP medium with antibiotics for 120 generations (Fig. S1B), and populations were examined with a 25-generation increment. Strikingly, we observed spontaneous loss of chr-XI in all three lines of cells expressing CCP1 + UTH1 as well as lines expressing CCP1 + UTH1 + SRX1 (Fig. 3), whereas neither expression of individual genes nor the combinations of CCP1 + SRX1 or UTH1 + SRX1 led to this outcome. These data suggested that both CCP1 and UTH1 are required for yeast cells deficient in thiol peroxidases.
Elevated Expression of Ccp1 in ∆8 Cells.
We next used comparative 2D gel electrophoresis, because it allows a direct measure of the synthesis rate of about a third of all yeast soluble proteins (Ccp1 but not Uth1 was not detected by this approach) (Fig. 4A and Fig. S2). We found that in untreated ∆8 cells, the synthesis rate of Ccp1 was abnormally high (23.5-fold increase compared with Wt cells). The synthesis rate of Sod2 (8.6-fold), Trx2 (6.0-fold), and Sod1 (4.4-fold) was also increased, but not as much as that of Ccp1. Because these proteins are all part of the yeast H2O2-inducible response (32), their constitutive expression in Δ8 cells indicates a deregulated expression, possibly as a result of chronic oxidative stress. In comparison, Wt cell exposure to H2O2 led to an 8-fold synthesis rate increase of Ccp1, which did not match its very high expression levels seen in untreated ∆8 cells (Fig. 4B). Δ8 cell exposure to H2O2 could not further increase Ccp1 expression, suggesting that it was already maximally induced. These data further support a contribution of CCP1 gene dosage increase in ∆8 cells.
Fig. 4.
Impact of aneuploidy on protein synthesis rates. (A) Autoradiograms of 2D gel electrophoresis performed with whole extracts of 20 min [35S]Met-labeled Wt and ∆8 cells after 20 min exposure to H2O2 (0.6 mM) or left untreated, as indicated. A blown-up portion of the autoradiograms is shown. Spots of selected proteins are indicated by arrows. A few protein spots of interest are indicated by black (induced) or white (repressed) arrows. Ahp1 is shown as a control that was deleted in ∆8 cells. (B) Quantification of Ccp1 synthesis rate before (untreated) and after (treated) H2O2 treatment of Wt and ∆8 cells. (C) Transcriptional (RNA-seq) and translational (Ribo-seq) changes in CCP1 and UTH1 abundance upon 0.6 mM H2O2 treatment of Wt cells. Error bars indicate standard error of the mean.
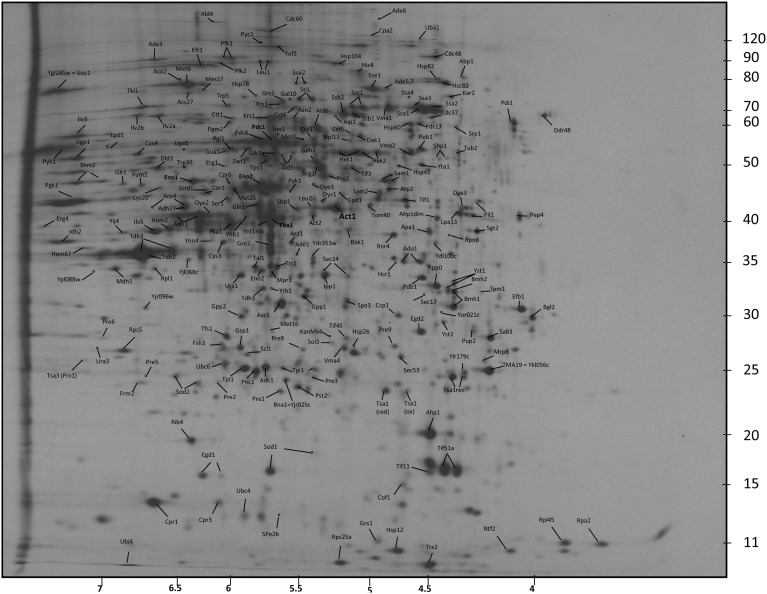
Fig. S2.
Master gel of Wt cells treated with H2O2. The gel represents an example of 2D gels analyzed in the study. The location of protein spots identified through mass spectrometry analysis is shown. The isoelectric point is shown on the horizontal axis, and molecular mass (in kDa) is on the vertical axis.
Loss of the Extra Chromosome Improved Growth Rates.
We further examined the impact of losing the extra chr-XI on cell growth. All three lines of ∆8 cells expressing CCP1 and UTH1 as well as all three lines of ∆8 cells expressing CCP1, UTH1, and SRX1 showed an increased growth rate compared with the original ∆8 cells (Fig. 5A). To further examine how these genes and aneuploidy influence cell growth, we analyzed the growth of disomic ∆8 cells in which UTH1 and CCP1 were expressed individually or in combination. The data revealed that UTH1 and CCP1 did rescue the growth phenotype, whereas the loss of the extra chr-XI improved cell growth (Fig. S3). Overall, the data suggest that thiol peroxidase deficiency led to adaptive aneuploidy, wherein the extra chr-XI supported increased expression of CCP1 and UTH1. Although aneuploidy also affected cell fitness, for example it reduced growth rate and lifespan, this adaptation allowed the removal of H2O2 by Ccp1 with cytochrome c as a reductant, with Uth1 assisting in this process. This explains the resistance of ∆8 cells to oxidative stress and their dependence on respiration, because it provided a source of reducing equivalents for H2O2 clearance.
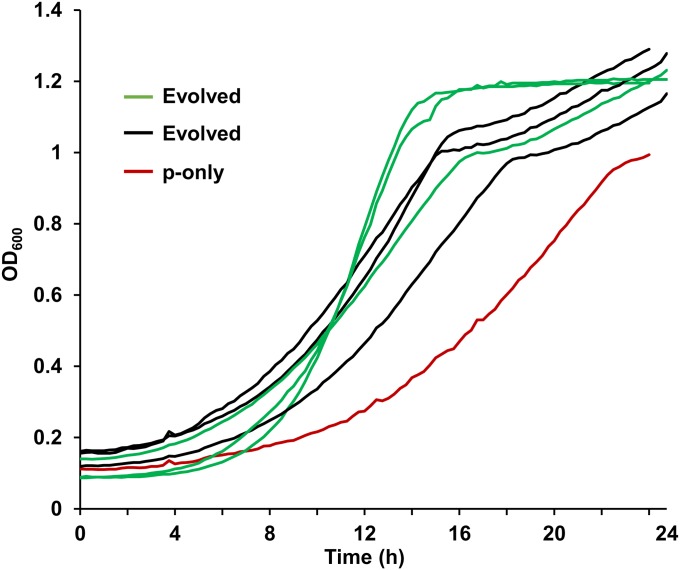
Fig. 5.
Improved growth of evolved ∆8 cell populations that lost the extra chr-XI. At the end of the laboratory evolution experiment, each population was analyzed for their cell-growth properties. The color scheme is as follows: initial ∆8 cells (red) harboring plasmid only as control (p-only), three independent populations that express CCP1 + UTH1 (black), and three independent populations expressing CCP1 + UTH1 + SRX1 (green).
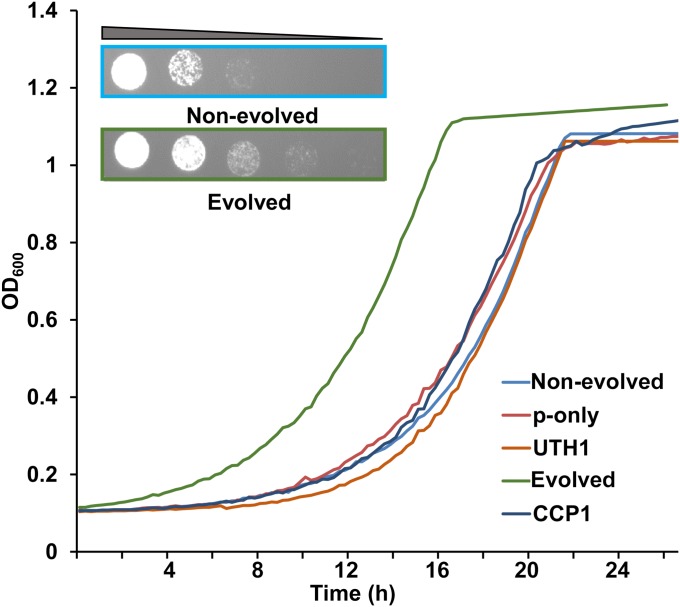
Fig. S3.
Growth of disomic ∆8 cells in which UTH1 and CCP1 were expressed individually or in combination. The nonevolved strain is the strain in which the two proteins are expressed but the extra chr-XI is present. The evolved strain lacks the extra chr-XI, following long-term expression of the two proteins. (Inset) Growth of nonevolved (Upper; blue box) and evolved (Lower; green box) populations by spot assays. The strains expressing individual genes (CCP1 or UTH1) during long-term culture are shown for comparison, and exhibit slower growth compared with the evolved strain.
Discussion
Aneuploidy, an abnormal number of chromosomes, is a type of genome alteration often associated with disorders such as cancer (33) and Down syndrome (34). Although the effects of aneuploidy on cell physiology remain incompletely understood, it is known that it can interfere with cell proliferation and create imbalances in protein composition irrespective of the specific karyotypes (20). However, aneuploidy can also be beneficial; namely, emergence of aneuploidy can create adaptive phenotypes under selective pressure (21, 35). Studies in yeast have shown that deletion of even a single gene may cause aneuploidy and that up to 8% of S. cerevisiae ORF knockout library strains are aneuploid. In addition, deleting a single paralog may be sufficient to gain an extra chromosome, as is the case of the ribosomal RPS24A and RPS24B paralogous pair (19).
In this study, we discovered that two independently prepared strains lacking all eight thiol peroxidases are characterized by chr-XI aneuploidy. The absence of thiol peroxidases created a selective condition (presumably as a result of intrinsic oxidative stress), so we examined what possible fitness advantage is created by the extra copy of chr-XI. Phenotypic adaptations often proceed through point mutations, resulting in changes in protein activities or expression levels (31). However, our previous analysis showed that no common mutations, including gross genome insertions or deletions, occurred independently in both ∆8 strains (15). We analyzed the gene expression profile of the duplicated chromosome by RNA-seq. Of the 306 genes encoded by chr-XI, two genes (CCP1 and UTH1) were found to be responsible for the observed adaptive whole-chromosome duplication. Ccp1 and Uth1 are both nuclear-encoded mitochondria-localized proteins required for mitochondrial biogenesis and function (36, 37). Although many reports have been published describing phenotypes associated with deficiency in these genes, their molecular functions remain unclear. Studies on Ccp1, a heme-containing protein, suggest that it functions as a mitochondrial ROS scavenger (26, 27). This would be consistent with the observed essentiality of mitochondrial respiration, which is required for the function of Ccp1. Alleviation of oxidative stress by Ccp1 may then support the growth of ∆8 cells. Our proteomic data revealed that Ccp1 protein expression was very high in ∆8 cells, even in the absence of H2O2 treatment, which normally induces Ccp1 expression in Wt cells. This finding may explain the observation that ∆8 cells do not suffer from severe oxidative stress, as revealed by the analyses of ROS and sensitivity of cells to various stresses (17). Our data suggest that Uth1 also plays a critical role in protecting cells from oxidative stress. One possibility is its role in mitochondrial biogenesis, because ∆8 cells showed increased mitochondrial abundance. Increased expression of UTH1 may be required for maintaining functional mitochondria. Analysis of RNA-seq and ribosome profiling data following a 30-min incubation of cells with 0.2 mM H2O2 (38) supports this possibility. In agreement with the proteomics data, H2O2 treatment led to a 6-fold increase in transcription and 12-fold increase in translation of Ccp1, but there was no change in Uth1 levels (Fig. 4B). Even though aneuploidy can introduce adaptive traits under certain conditions, organismal fitness occurs at the expense of slow proliferation. We tested this observation in our model and showed that losing the extra chromosome leads to a positive effect on ∆8 cells: The evolved ∆8 populations grew faster compared with the original mutant cells.
Overall, we have identified an example of adaptive dosage compensation where whole-chromosome duplication promotes increased expression of two genes and supports the growth of thiol peroxidase null strains. Our results clearly show that aneuploidy can rapidly compensate for the absence of major protectors against oxidative stress. It would be of interest to examine how this finding applies to multicellular organisms, considering that aneuploidy is highly detrimental for them. Oxidative stress and aneuploidy contribute to many pathological conditions, including different types of cancers. In this regard, experimental approaches addressing this relationship may bring fundamental insights into these processes and improve our ability to manipulate these systems for research and therapeutic purposes.
Materials and Methods
Yeast Strains and Plasmids.
Isolation of mutant strains lacking all eight thiol peroxidases was reported previously (15, 17). Details regarding all strains used in the current study can be found in Table S1. Yeast strains were grown on YPD medium. The strain used in the laboratory evolution experiment was created by replacing one copy of SRX1 (i.e., on one of the two chr-XIs) with a NatMX resistance (100 µg/mL) cassette in a ∆8 strain. For expression of genes of interest, two plasmids were used. SRX1 was cloned into the pCEV plasmid (39) containing Zeocin (ZeoMX) resistance, whose expression was controlled by the constitutive TEF1 promoter. For expression of CCP1 and/or UTH1, we modified a recently constructed triple-expression plasmid, pATP425 (40), by inserting a bialaphos (PatMX) cassette along with a TEF1 promoter and terminator at the SacI-PacI restriction sites. We inserted CCP1 and/or UTH1 sequences at the SacI-NotI and PmeI-FseI restriction sites, wherein their expression was controlled under the TDH3 and ADH1 promoters, respectively. PatMX (300 µg/mL) and ZeoMX (100 µg/mL) selection was performed on SDP medium (1 g/L proline, 6.7 g/L yeast nitrogen base, 20 g/L glucose). Cells were transformed with plasmids to express individual genes. After colonies were selected on both plates, combinatory transformations were performed and the colonies were selected on SDP media containing ZeoMX and PatMX for coexpression.
Table S1.
Yeast strains used in this study
| Strain | Designation of Wt and mutant cells | Genotype | Source |
| BY4741 | Wt | MATa, his3 leu2 met15 ura3 | American Type Culture Collection |
| A13771 | Wt-M11 | MATa, ade2-1, leu2-3, ura3, trp1-1, his3-11,15, can1-100, GAL, [phi+], intergenic region (430900-431000) between YKL006C-A and YKL006W::HIS3, intergenic region (430900-431000) between YKL006C-A and YKL006W::KanMX6 | (22) |
| GY25 | ∆3Gpx | MATa, his3 leu2 met15 ura3 ∆gpx1:URA3, ∆gpx2:HIS3, ∆gpx3: KAN | (17) |
| GY14 | ∆5Prx | MATa, his3 leu2 met15 ura3 ∆tsa1:KAN, ∆tsa2:LEU2, ∆dot5:MET15, ∆ahp1:HIS3, ∆prx1:URA3 | (17) |
| GY100 | ∆8 (all ∆Prx + all ∆Gpx) | MATa, his3 leu2 met15 ura3 ∆tsa1:KAN, ∆tsa2:LEU2, ∆dot5:MET15, ∆ahp1:HIS3, ∆prx1:URA3, ∆gpx2:HYG, ∆gpx1:URA3, Δgpx3:KAN | (17) |
| GY151 | ∆8 (all ∆Prx + all ∆Gpx) | MATa, his3 leu2 met15 ura3 ∆tsa1:KAN, ∆tsa2:LEU2, ∆dot5:MET15, ∆ahp1:HIS3, ∆prx1:URA3, ∆gpx2:HYG, ∆gpx1:NAT, Δgpx3:KAN | (15) |
| GY152 | ∆8 (all ∆Prx + all ∆Gpx) + ∆Srx1 | MATa, his3 leu2 met15 ura3 ∆tsa1:KAN, ∆tsa2:LEU2, ∆dot5:MET15, ∆ahp1:HIS3, ∆prx1:URA3, ∆gpx2:HYG, ∆gpx1:URA3, Δgpx3:KAN, ∆srx1:NAT | This study |
H2O2 Sensitivity and Analyses of Cell Growth.
Cells were grown at 30 °C, and OD600 measurements were taken at 15-min intervals using a Bioscreen C MBR instrument until cells reached early stationary phase. Resistance of Wt and mutant strains to H2O2 was determined with spot assays. For each strain, overnight cultures were adjusted to OD600 0.6, and 5 μL of serial dilutions (10-fold each) was spotted on solid medium that contained H2O2 at the indicated concentrations. Cells were grown for 3 d at 30 °C and pictures were taken.
Dependence on Respiration.
Sensitivity of yeast cells to antimycin-A, rotenone, and oligomycin was assessed with spot assays. Cultures were collected at OD600 0.6, and 5 μL of 10-fold serial dilutions was spotted on solid medium containing 10 µg/mL antimycin A, 20 µg/mL rotenone, or 7.5 µg/mL oligomycin. For visualization of mitochondria under the microscope, logarithmically grown cells were washed three times with 1× saline buffer and incubated with calcofluor-white (Sigma) and MitoTracker Red (Invitrogen) in the same buffer for 30 min. Cells were washed and visualized under a fluorescence microscope. To assess a relative abundance of mtDNA in yeast strains, we calculated the read coverage of mitochondrial and nuclear genomes, and the abundance of mtDNA (Xmt) was plotted as the ratio of the coverage of Xmt and the nuclear genome (Xnuc) coverage for each strain.
Genome Sequencing and RNA-Seq Analysis.
Genome sequencing and RNA-seq analyses were performed on various yeast strains as described previously (15, 38). We further used the Genome Analysis Toolkit to the calculate the depth of coverage for each chromosome (41). The “DepthOfCoverage” tool was applied to sorted *.bam files to generate a genome-wide table of sequencing depth. A custom Perl script modified this table by omitting highly abundant mitochondrial sequences (>100× coverage than the nuclear genome) and ribosomal DNA (>50× than the rest of the nuclear genome). We also normalized the coverage by the total number of reads per sample. The graph was plotted with the ggplot2 package within the R statistical environment (42). Each dot is an average of the sequencing depths of 100 nt. There are 100,000 dots per graph. RNA-seq (FASTQ) files were aligned to the yeast reference genome; Ensembl build R64-1-1 was downloaded from the Illumina iGenome resource at support.illumina.com/sequencing/sequencing_software/igenome.html using Bowtie 2 v2.1.0 (43) and the spliced-read mapper TopHat (v2.0.9) (44), with reference gene annotations to guide the alignment (-G argument in TopHat). Fragments per kilobase of transcript per million mapped (FPKM) reads were calculated using Cufflinks v2.1.1 (45), with default parameters.
Laboratory Evolution Experiment.
The laboratory evolution experiment was carried out by serial dilution. Cells were grown until reaching stationary phase at 30 °C with moderate shaking and then diluted by a factor of 1:500 with fresh SDP medium (∼4.5 generations per dilution) containing ZeoMX and/or PatMX antibiotics. This procedure was repeated until loss of the extra chromosome was detected by colony PCR. Briefly, 10 µL each culture was spread onto SDP plates containing ZeoMX and/or PatMX. After 2 d, several colonies were selected and incubated at 30 °C in 2.5 µL water containing 20 U/mL zymolase-20T solution for 30 min and for 5 min at 95 °C in PCR tubes. A drop of mineral oil was added on top to prevent evaporation. After cooling on ice, 50 µL water was added to each tube and 1.25 µL was used as a template for PCR.
Analysis of Protein Expression.
Strains were grown at 30 °C in YNB defined medium containing (wt/vol) 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose, supplemented with appropriate amino acids and 0.2 mM GSH. When OD600 reached 0.35, H2O2 (0.6 mM) was added or not (t0); at t20 (20 min), cells were pulse-labeled with [35S]methionine (150 µCi), and at t40 (40 min) they were collected. Preparation of whole-cell protein extracts and 2D gel electrophoresis were performed as described (46). Dried gels were analyzed on a Storm 860 Molecular Imager, and spot intensity quantification was performed with ImageQuant software (GE Healthcare), as described (47). For select proteins, a synthesis rate index was calculated as the value of spot intensity signal normalized to total gel intensity signal.
Supplementary Material
Acknowledgments
We thank Dr. Angelika Amon for a chr-XI aneuploid strain (Wt-M11), Dr. Gilles Lagniel for the proteome analysis, and Dr. Jun Ishii for providing the pATP425 vector. This work was supported by NIH Grant GM065603 (to V.N.G.) and Grant PLBIO INCA_5869 from Agence Nationale pour la Recherche (ANR) ERRed and from Institut National du Cancer (InCA) (to M.B.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genome reads reported in this paper have been deposited in the BioProject database, www.ncbi.nlm.nih.gov/bioproject (accession no. PRJNA287442). Transcriptome (RNA-sequencing) reads and fragments per kilobase of transcript per million mapped read values have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE70036). Ribosome profiling (Ribo-sequencing) data have also been deposited in the GEO database (accession no. GSE59573).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1505315112/-/DCSupplemental.
References
- 1.Apel K, Hirt H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 2.Geiszt M, Kopp JB, Várnai P, Leto TL. Identification of renox, an NAD(P)H oxidase in kidney. Proc Natl Acad Sci USA. 2000;97(14):8010–8014. doi: 10.1073/pnas.130135897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cross CE, et al. Oxygen radicals and human disease. Ann Intern Med. 1987;107(4):526–545. doi: 10.7326/0003-4819-107-4-526. [DOI] [PubMed] [Google Scholar]
- 4.Rhee SG, Chae HZ, Kim K. Peroxiredoxins: A historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38(12):1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 5.Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194(1):7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408(6809):239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 7.Wood ZA, Schröder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28(1):32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 8.Park SG, Cha MK, Jeong W, Kim IH. Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. J Biol Chem. 2000;275(8):5723–5732. doi: 10.1074/jbc.275.8.5723. [DOI] [PubMed] [Google Scholar]
- 9.Huang ME, Kolodner RD. A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Mol Cell. 2005;17(5):709–720. doi: 10.1016/j.molcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 10.D’Errico M, Parlanti E, Dogliotti E. Mechanism of oxidative DNA damage repair and relevance to human pathology. Mutat Res. 2008;659(1-2):4–14. doi: 10.1016/j.mrrev.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30(1):2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neumann CA, et al. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature. 2003;424(6948):561–565. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- 13.van Loon B, Markkanen E, Hübscher U. Oxygen as a friend and enemy: How to combat the mutational potential of 8-oxo-guanine. DNA Repair (Amst) 2010;9(6):604–616. doi: 10.1016/j.dnarep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Iraqui I, et al. Peroxiredoxin Tsa1 is the key peroxidase suppressing genome instability and protecting against cell death in Saccharomyces cerevisiae. PLoS Genet. 2009;5(6):e1000524. doi: 10.1371/journal.pgen.1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaya A, et al. Thiol peroxidase deficiency leads to increased mutational load and decreased fitness in Saccharomyces cerevisiae. Genetics. 2014;198(3):905–917. doi: 10.1534/genetics.114.169243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorla GR, Malhi H, Gupta S. Polyploidy associated with oxidative injury attenuates proliferative potential of cells. J Cell Sci. 2001;114(Pt 16):2943–2951. doi: 10.1242/jcs.114.16.2943. [DOI] [PubMed] [Google Scholar]
- 17.Fomenko DE, et al. Thiol peroxidases mediate specific genome-wide regulation of gene expression in response to hydrogen peroxide. Proc Natl Acad Sci USA. 2011;108(7):2729–2734. doi: 10.1073/pnas.1010721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nemoto S, Takeda K, Yu ZX, Ferrans VJ, Finkel T. Role for mitochondrial oxidants as regulators of cellular metabolism. Mol Cell Biol. 2000;20(19):7311–7318. doi: 10.1128/mcb.20.19.7311-7318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes TR, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25(3):333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 20.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317(5840):916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 21.Rancati G, et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135(5):879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheltzer JM, Amon A. The aneuploidy paradox: Costs and benefits of an incorrect karyotype. Trends Genet. 2011;27(11):446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G, Rubinstein B, Li R. Whole chromosome aneuploidy: Big mutations drive adaptation by phenotypic leap. BioEssays. 2012;34(10):893–900. doi: 10.1002/bies.201200069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425(6961):980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 25.Sels AA, Cocriamont C. Induced conversion of a protein precursor into cytochrome C peroxidase during adaptation of yeast to oxygen. Biochem Biophys Res Commun. 1968;32(2):192–198. doi: 10.1016/0006-291x(68)90368-9. [DOI] [PubMed] [Google Scholar]
- 26.Kwon M, Chong S, Han S, Kim K. Oxidative stresses elevate the expression of cytochrome c peroxidase in Saccharomyces cerevisiae. Biochim Biophys Acta. 2003;1623(1):1–5. doi: 10.1016/s0304-4165(03)00151-x. [DOI] [PubMed] [Google Scholar]
- 27.Martins D, Kathiresan M, English AM. Cytochrome c peroxidase is a mitochondrial heme-based H2O2 sensor that modulates antioxidant defense. Free Radic Biol Med. 2013;65:541–551. doi: 10.1016/j.freeradbiomed.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 28.Kissová I, Deffieu M, Manon S, Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem. 2004;279(37):39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- 29.Kennedy BK, Austriaco NR, Jr, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80(3):485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- 30.Bandara PD, Flattery-O’Brien JA, Grant CM, Dawes IW. Involvement of the Saccharomyces cerevisiae UTH1 gene in the oxidative-stress response. Curr Genet. 1998;34(4):259–268. doi: 10.1007/s002940050395. [DOI] [PubMed] [Google Scholar]
- 31.Elena SF, Lenski RE. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4(6):457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 32.Godon C, et al. The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem. 1998;273(35):22480–22489. doi: 10.1074/jbc.273.35.22480. [DOI] [PubMed] [Google Scholar]
- 33.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396(6712):643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 34.Hassold TJ, Jacobs PA. Trisomy in man. Annu Rev Genet. 1984;18:69–97. doi: 10.1146/annurev.ge.18.120184.000441. [DOI] [PubMed] [Google Scholar]
- 35.Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313(5785):367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daum G, Böhni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257(21):13028–13033. [PubMed] [Google Scholar]
- 37.Camougrand NM, Mouassite M, Velours GM, Guérin MG. The “SUN” family: UTH1, an ageing gene, is also involved in the regulation of mitochondria biogenesis in Saccharomyces cerevisiae. Arch Biochem Biophys. 2000;375(1):154–160. doi: 10.1006/abbi.1999.1655. [DOI] [PubMed] [Google Scholar]
- 38.Gerashchenko MV, Lobanov AV, Gladyshev VN. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci USA. 2012;109(43):17394–17399. doi: 10.1073/pnas.1120799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vickers CE, Bydder SF, Zhou Y, Nielsen LK. Dual gene expression cassette vectors with antibiotic selection markers for engineering in Saccharomyces cerevisiae. Microb Cell Fact. 2013;12:96. doi: 10.1186/1475-2859-12-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishii J, et al. Three gene expression vector sets for concurrently expressing multiple genes in Saccharomyces cerevisiae. FEMS Yeast Res. 2014;14(3):399–411. doi: 10.1111/1567-1364.12138. [DOI] [PubMed] [Google Scholar]
- 41.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. R Core Development Team (2015) R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna). Available at https://www.r-project.org. Accessed March 1, 2015.
- 43.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J, et al. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J Biol Chem. 1999;274(23):16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- 47.Hasan R, et al. The control of the yeast H2O2 response by the Msn2/4 transcription factors. Mol Microbiol. 2002;45(1):233–241. doi: 10.1046/j.1365-2958.2002.03011.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.