Significance
It is widely believed that if an HIV vaccine elicits moderate in vitro titers of serum neutralizing antibodies (nAbs) against a challenge virus, it will prevent infection. This paradigm is based on studies in which passive transfer of HIV-specific nAbs protected rhesus macaques from mucosal challenge with a chimeric simian/human immunodeficiency virus. However, it is unknown whether this direct relationship can be extrapolated to protection in the setting of active immunization. Our data suggest that the relationship between serum in vitro nAb titers and protection from mucosal SIV challenge is more complex than previously recognized in the setting of active immunization, warranting further studies to understand the balance between immune activation, target cell availability, and protective antibody responses.
Keywords: SIV, vaccine, neutralization
Abstract
Although the correlates of immunological protection from human immunodeficiency virus or simian immunodeficiency virus infection remain incompletely understood, it is generally believed that medium to high titers of serum neutralizing antibodies (nAbs) against the challenge virus will prevent infection. This paradigm is based on a series of studies in which passive transfer of HIV-specific nAbs protected rhesus macaques (RMs) from subsequent mucosal challenge with a chimeric human/simian immunodeficiency virus. However, it is unknown whether nAb titers define protection in the setting of active immunization. Here we determined serum nAb titers against breakthrough transmitted/founder (T/F) SIVsmE660-derived envelope glycoprotein (Env) variants from 14 RMs immunized with SIVmac239-based DNA-prime/modified vaccinia virus Ankara-boost vaccine regimens that included GM-CSF or CD40L adjuvants and conferred significant but incomplete protection against repeated low-dose intrarectal challenge. A single Env variant established infection in all RMs except one, with no identifiable genetic signature associated with vaccination breakthrough compared with T/F Envs from four unvaccinated monkeys. Breakthrough T/F Env pseudoviruses were potently neutralized in vitro by heterologous pooled serum from chronically SIVsmE660-infected monkeys at IC50 titers exceeding 1:1,000,000. Remarkably, the T/F Env pseudoviruses from 13 of 14 monkeys were also susceptible to neutralization by autologous prechallenge serum at in vitro IC50 titers ranging from 1:742–1:10,832. These titers were similar to those observed in vaccinated RMs that remained uninfected. These data suggest that the relationship between serum nAb titers and protection from mucosal SIV challenge in the setting of active immunization is more complex than previously recognized, warranting further studies into the balance between immune activation, target cell availability, and protective antibody responses.
With the recovery and characterization of five classes of broadly neutralizing antibodies (nAbs) against HIV-1, as well as detailed information about how they develop (1, 2), and the possibility that a vaccine-elicited antibody contributed to protection in the RV144 HIV vaccine efficacy trial (3), optimism regarding antibody-mediated protection against HIV-1 has been renewed (4). Furthermore, passive administration of neutralizing monoclonal antibodies (nmAbs) to nonhuman primates (NHPs) has repeatedly been shown to provide robust protection against mucosal infection with a chimeric human/simian immunodeficiency virus, even at modest in vitro nAb IC50 titers that are readily achievable by vaccination (5–14). NHP studies have also demonstrated that protection against heterologous SIV challenge is feasible and likely mediated by antibody responses (15–18). We recently reported on the protective efficacy and immune responses elicited by SIVmac239-based DNA-prime/modified vaccinia virus Ankara-boost (DDMM) vaccination of rhesus macaques (RMs), with GM-CSF adjuvant (DgDgMM), CD40-ligand adjuvant (D40LD40LMM), or administering three MVA immunizations without DNA priming (MMM) (Fig. 1A). Each of these vaccine regimens provided statistically significant protection against a low-dose repeated heterologous SIVsmE660 intrarectal challenge, compared with the unimmunized controls (19–21). The SIVsmE660 stock (VH2000) and dosage (5,000 TCID50), immunization schedule, and challenge route were held constant during these trials, providing an opportunity to investigate in more detail the immune responses elicited across groups. Neutralizing activity was elicited by all vaccine regimens but was not associated with protection; however, this was measured indirectly using one or a few reference variants of SIVsmE660. The strongest and most consistent immune correlate that was associated with protection against acquisition in the DDMM, DgDgMM, and D40LD40LMM modalities was the presence of higher binding avidity for the SIVsmE660 envelope glycoproteins (Env). In contrast, this immune parameter was not correlated with MMM-mediated protection, perhaps reflecting that immunization with two forms of SIVmac239 Env (gp160 and gp150 by the DNA and MVA vectors, respectively) elicits a different type of antibody than gp160 alone. In outcomes like these, where protection is incomplete, the characterization of breakthrough variants can inform us about correlates of vaccine-mediated protection against acquisition (18, 22, 23). Here, we went one step further by testing the capacity of prechallenge antibodies to neutralize the exact Env variant that broke through to establish each infection. To our knowledge, this study is the first to demonstrate that vaccine-elicited serum antibodies capable of neutralizing the autologous breakthrough Env variant were present before challenge at moderate to high titers but nevertheless failed to protect against SIV infection.
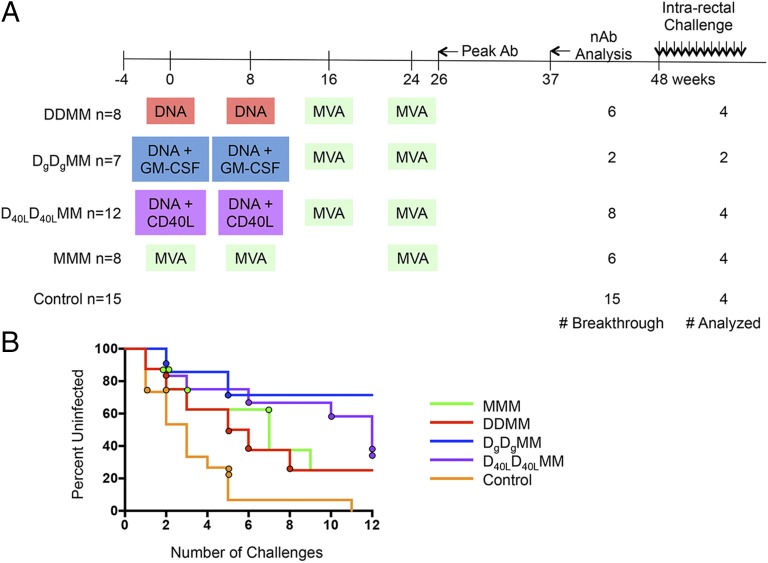
Fig. 1.
Timeline of the M11 and M12 NHP vaccine trials. (A) The immunization schedule for the M11 and M12 vaccine trials is plotted along a timeline in weeks. M11 consisted of DDMM, DgDgMM, and MMM vaccine groups, whereas the M12 trial added on the D40LD40LMM group. The agents used for priming and boosting are indicated in colored boxes, highlighting similarities and differences between vaccine groups. The total number of animals in each immunization group and the control group is shown to the left. The number of breakthrough infections and number of animals analyzed for each group is shown to the right. The time point at which the peak levels of antibody binding to SIVsmE660- and SIVmac239-derived gp140 proteins were observed is indicated by an arrow as “Peak Ab.” The time point that was analyzed for nAb activity (shown in Fig. 3B and Fig. S6) is indicated by an arrow and “nAb Analysis.” Weekly intrarectal challenges with SIVsmE660 were initiated at week 48. (B) A survival curve is shown for each of the vaccine groups, which are indicated by color in the key. The circles indicate the challenge that resulted in infection of each monkey that was included in the nAb study. For example, one control monkey (orange circles) was infected at challenge 1, one at challenge 2, and two at challenge 5.
Results
A Single Variant from the SIVsmE660 Challenge Stock Established All But One Infection.
We recently reported that four different SIVmac239-based DNA/MVA or MVA-only vaccination modalities were significantly but incompletely protective against repeated low-dose intrarectal challenge with heterologous viral strain SIVsmE660 in RMs (19–21). These vaccination studies, M11 and M12, were purposely designed for cross-comparison of SIVmac239-based DNA/MVA immunizations. These trials contained three groups that received two DNA primes (with no adjuvant, GM-CSF, or CD40L) at weeks 0 and 8, followed by two MVA boosts at weeks 16 and 24 (Fig. 1A). A fourth group received MMM immunizations at weeks 0, 8, and 24 without DNA priming (Fig. 1A). Intrarectal SIVsmE660 challenge with a stock designated VH2000 was initiated 20–24 wk after the last MVA immunization in all groups and continued weekly for a maximum of 12 inoculations. In the present study, transmitted/founder (T/F) Envs from breakthrough infections that occurred in the DDMM (n = 4), MMM (n = 4), DgDgMM (n = 2), and D40LD40LMM (n = 4) vaccine groups were analyzed, as well as T/F Envs from four unvaccinated control animals that became infected (Fig. 1A). These monkeys were selected based on availability of an acute infection/peak viral load plasma sample with a sufficient volume for PCR amplification and cloning and a week 37 postimmunization serum sample with a sufficient volume for neutralization assays. The monkeys were also representative of infection at various points during the challenge phase (Fig. 1B).
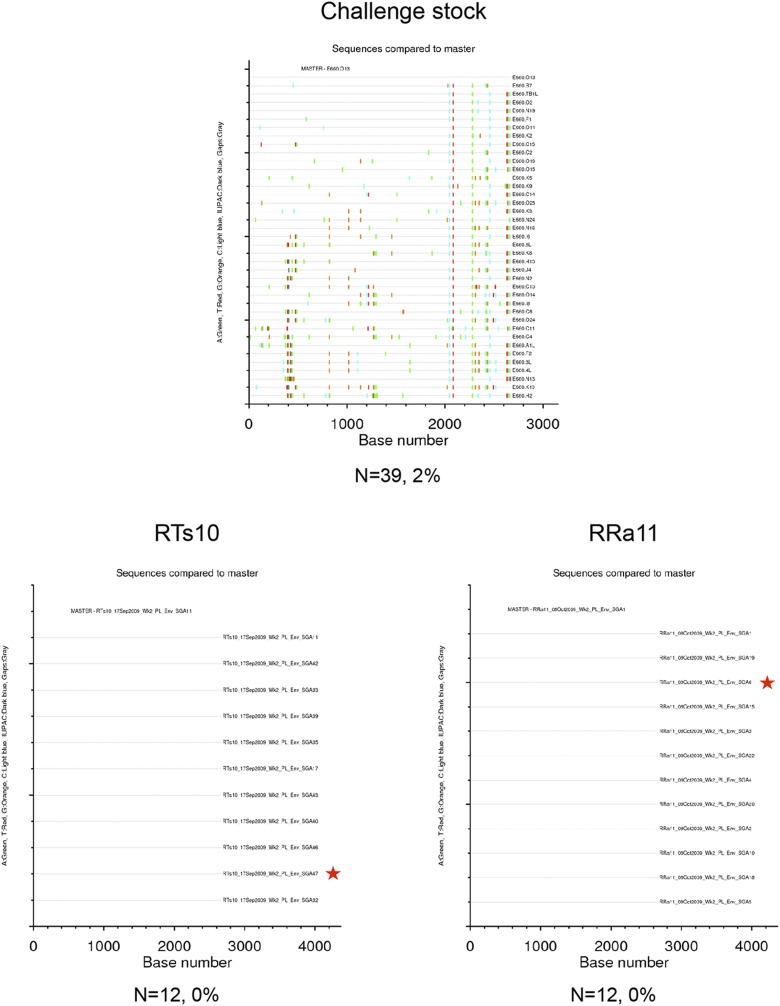
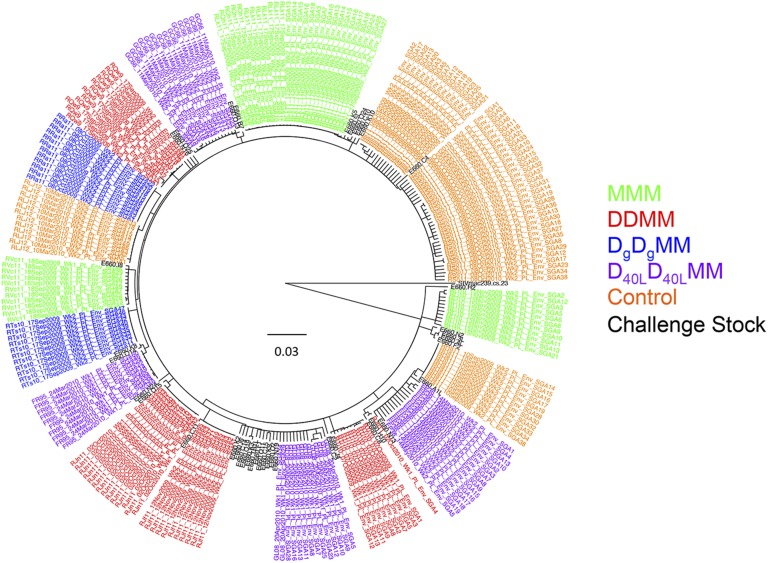
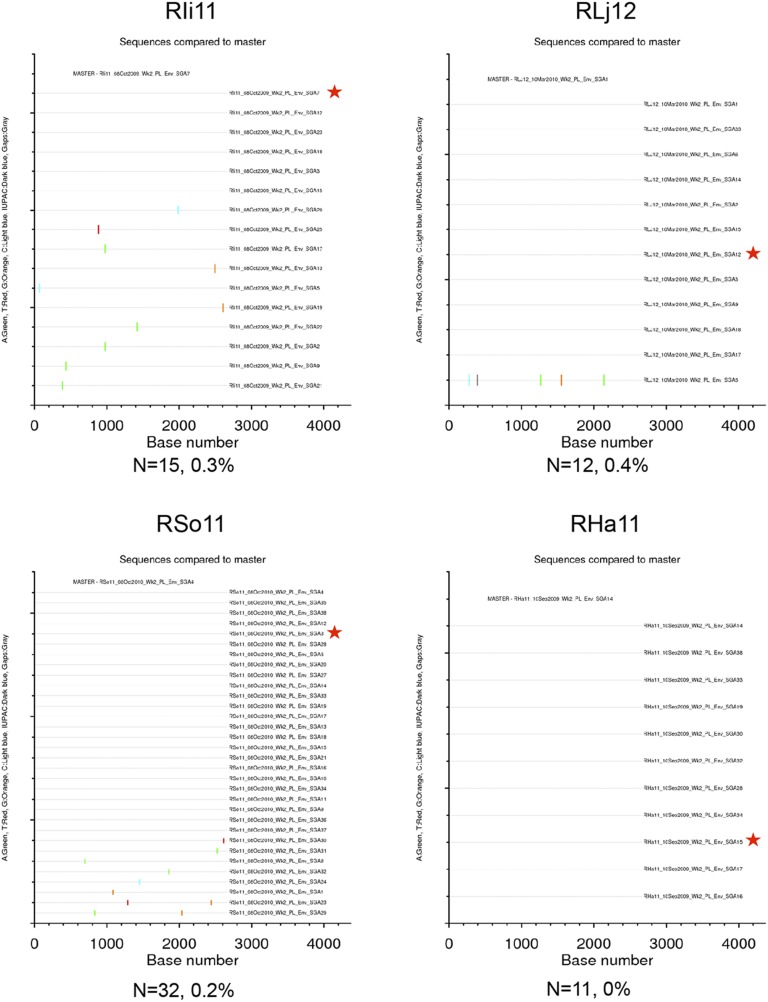
Between 10 and 32 single genome amplification (SGA) env gene sequences (which encode the Env glycoproteins) from each infected animal and 39 SGA env gene sequences from the VH2000 challenge stock were included in a genetic analysis. The VH2000 env gene sequences exhibited a maximum pairwise distance of 2% (Fig. S1), which is lower in diversity than what is generally present in an HIV-1–infected transmitting partner (24–26). Fig. 2 displays a Neighbor-joining phylogenetic tree containing 245 complete env gene nucleotide sequences obtained from the 14 vaccinated and 4 control monkeys, along with the 39 challenge stock sequences. Sequences from the vaccinated and control groups were dispersed and intermingled with variants from the challenge stock. There was no a priori evidence that vaccination had selected for transmission of a particular set of variants from within the challenge stock. The env gene sequences from each animal clustered together on the tree in all but one case (monkey RJn11), but these clusters were not supported by high bootstrap values. Further analyses revealed that the maximum pairwise distance of intra-animal env gene sequences ranged from 0 to 1.3% (Figs. S1–S5). These data therefore demonstrate that a single variant most likely established infection in 13 of the 14 monkeys, whereas RJn11 could have been initially infected by more than one variant. These data therefore suggest that the challenge virus, dose, and route of infection used in this experiment recapitulate the events occurring during transmission of HIV-1 in humans in terms of presence of a single T/F virus variant in the vast majority of individuals (24–26). In each case of breakthrough, an env gene amplicon representative of the T/F sequence, or the predominant env gene sequence in the case of RJn11, was cloned to assess neutralization sensitivity.
Fig. S1.
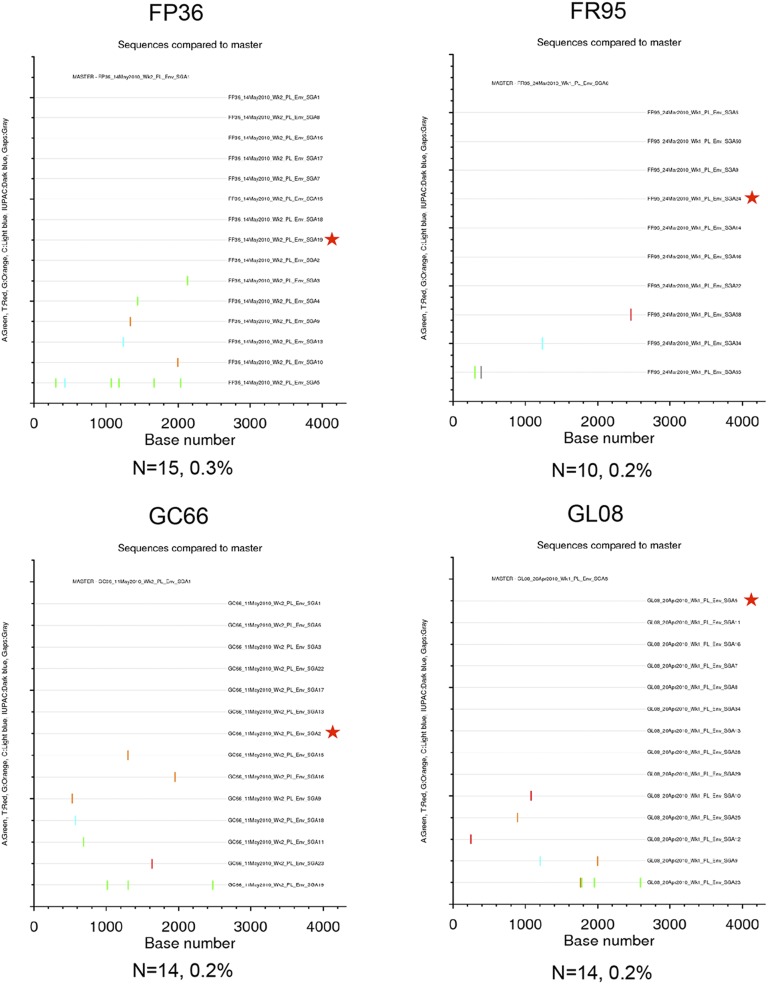
Highlighter plots of SGA env sequences derived from the VH2000 challenge stock and DgDgMM-immunized macaques. Each highlighter plot was generated using SGA-derived complete env gene nucleotide sequences, which are ∼2,700 nts in length. Aligned sequences from each dataset were exported from Geneious in the fasta format and uploaded to www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html. Colored ticks indicate mismatches from the master sequence indicated (A, green; T, red; G, orange; C, light blue; ambiguous nucleotide, dark blue; gaps, gray). A red star indicates the env gene amplicon that was cloned and used in the neutralization assays. The number of sequences and maximum pairwise distance are shown beneath each plot.
Fig. 2.
Phylogenetic tree of SGA-derived env gene nucleotide sequences from the challenge stock, vaccinated breakthrough infections, and unvaccinated control infections. A Neighbor-joining phylogenetic tree was created from complete env gene nucleotide sequences using a consensus approach in Geneious R6 v6.1.4 and annotated in Figtree v.1.4.0. The SIVmac239 env gene sequence was used as an outgroup. The horizontal bar shows the scale of genetic distance. Colors in the key indicate the vaccine or control group of the animal from which the sequences were derived. Challenge stock sequences and the SIVmac239 reference sequence are shown in black. Sequences by vaccination groups are as follows: blue, DgDgMM—RTs10, RRa11; green, MMM—RQh11, RLk11, RVc11, RLi10; orange, controls—Rha11, RIi11, RSo11, RLj12; purple, D40LD40LMM—GL08, FR95, GC66, FP36; red, DDMM—RQw9, RJn11, RTi10, RJf11.
Fig. S5.
Highlighter plots of SGA env sequences derived from unvaccinated control RMs. Each highlighter plot was generated using SGA-derived complete env nucleotide sequences, which are ∼2,700 nts in length. Aligned sequences from each dataset were exported from Geneious in the fasta format and uploaded to www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html. Colored ticks indicate mismatches from the master sequence indicated (A, green; T, red; G, orange; C, light blue; IUPAC, dark blue; gaps, gray). A red star indicates the env amplicon that was cloned and used in the neutralization assays. The number of sequences and maximum pairwise distance are shown beneath each plot.
Fig. S2.
Highlighter plots of SGA env sequences derived from D40LD40LMM-immunized RMs. Each highlighter plot was generated using SGA-derived complete env nucleotide sequences, which are ∼2,700 nts in length. Aligned sequences from each dataset were exported from Geneious in the fasta format and uploaded to www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html. Colored ticks indicate mismatches from the master sequence indicated (A, green; T, red; G, orange; C, light blue; IUPAC, dark blue; gaps, gray). A red star indicates the env amplicon that was cloned and used in the neutralization assays. The number of sequences and maximum pairwise distance are shown beneath each plot.
Fig. S3.
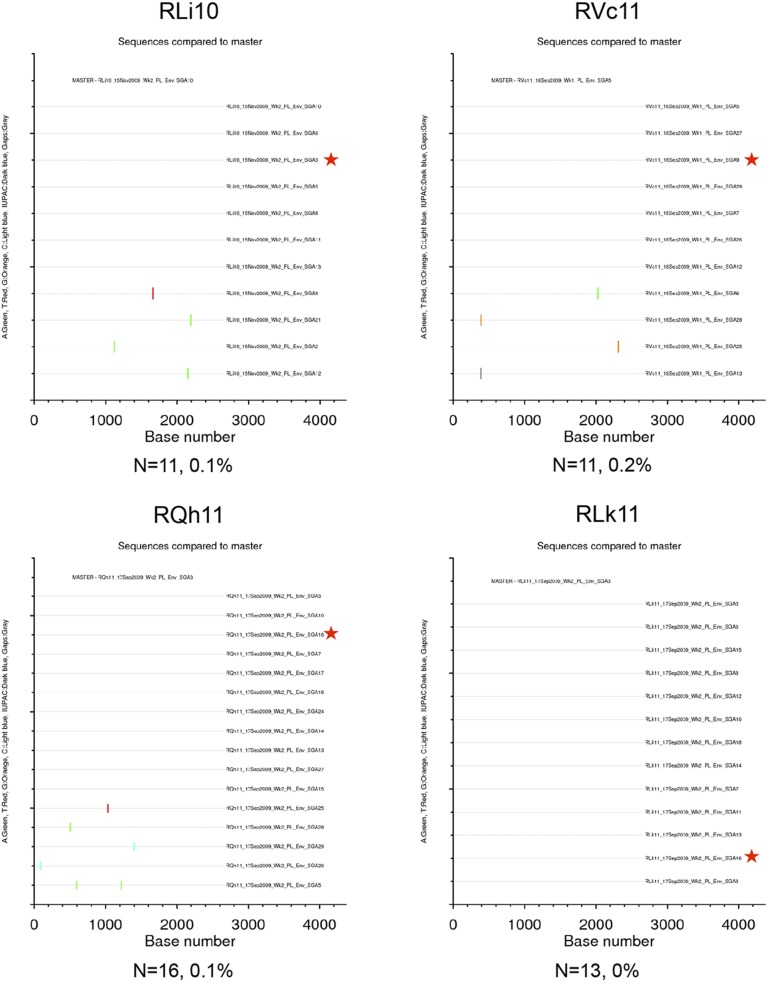
Highlighter plots of SGA env sequences derived from MMM-immunized RMs. Each highlighter plot was generated using SGA-derived complete env nucleotide sequences, which are ∼2,700 nts in length. Aligned sequences from each dataset were exported from Geneious in the fasta format and uploaded to www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html. Colored ticks indicate mismatches from the master sequence indicated (A, green; T, red; G, orange; C, light blue; IUPAC, dark blue; gaps, gray). A red star indicates the env amplicon that was cloned and used in the neutralization assays. The number of sequences and maximum pairwise distance are shown beneath each plot.
Fig. S4.
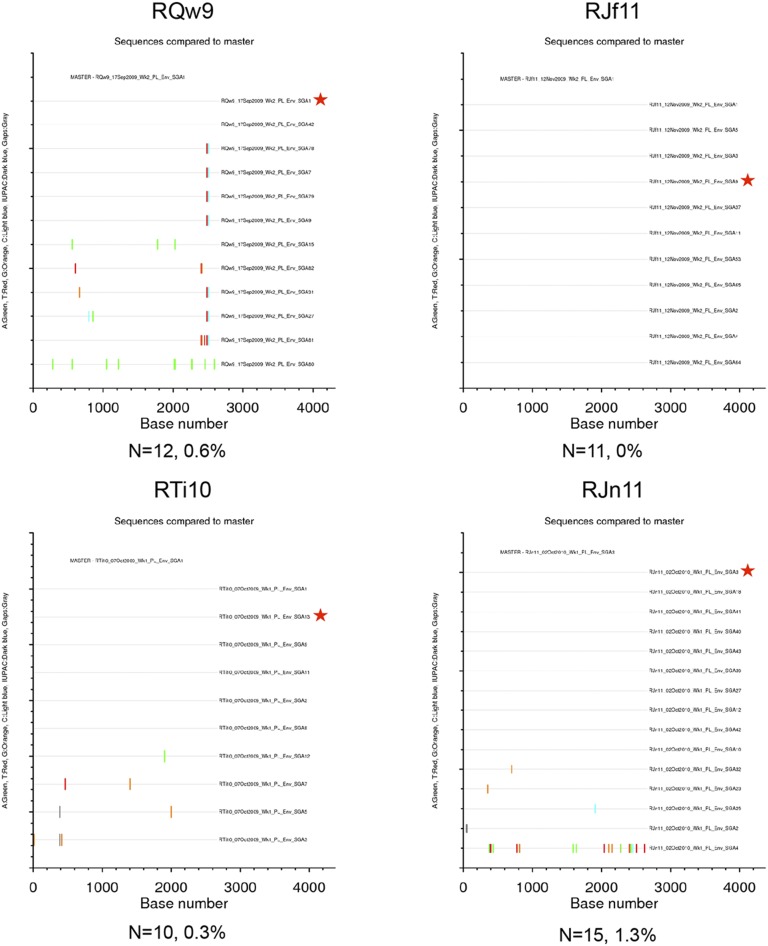
Highlighter plots of SGA env sequences derived from DDMM-immunized RMs. Each highlighter plot was generated using SGA-derived complete env nucleotide sequences, which are ∼2,700 nts in length. Aligned sequences from each dataset were exported from Geneious in the fasta format and uploaded to www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html. Colored ticks indicate mismatches from the master sequence indicated (A, green; T, red; G, orange; C, light blue; IUPAC, dark blue; gaps, gray). A red star indicates the env amplicon that was cloned and used in the neutralization assays. The number of sequences and maximum pairwise distance are shown beneath each plot.
T/F Envs from Breakthrough Infections Are Sensitive to Neutralization.
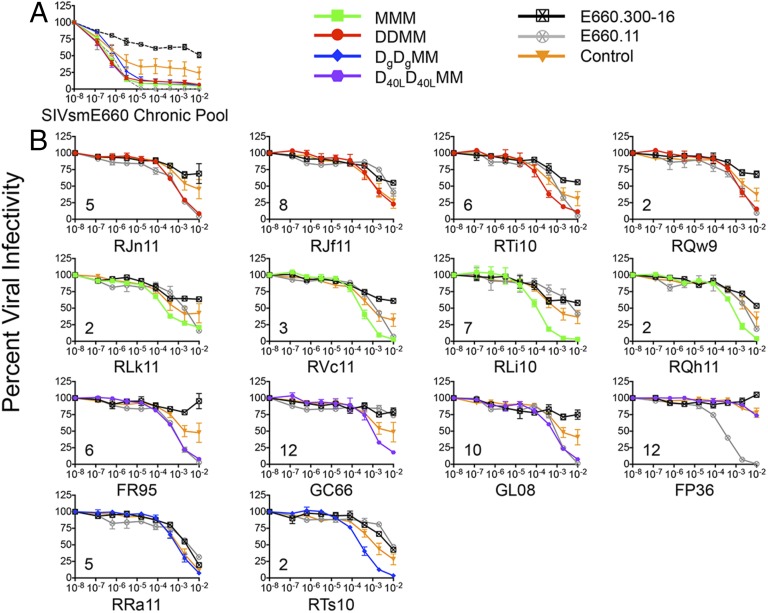
As a first step in the phenotypic characterization of the T/F Envs, we assessed their general neutralization sensitivity to pooled sera collected from nine unvaccinated RMs at 24 wk after they became infected intrarectally with the VH2000 SIVsmE660 stock. Fig. 3A demonstrates that the breakthrough T/F Env variants on the whole were highly susceptible to neutralization by antibodies in pooled serum. The majority of the T/F Envs from vaccinated animals were neutralized at an IC50 titer of more than 1:1,000,000 by the serum antibody pool, typical of the extreme neutralization sensitivity of tier 1 SIVsmE660 variants such as E660.11 (27, 28). In contrast, the T/F Envs from the control infections were less susceptible to neutralization (Fig. 3A). These data clearly demonstrate that the vaccinated breakthrough T/F Envs were not inherently neutralization resistant. We next examined whether prechallenge serum from each vaccinated animal could neutralize the autologous breakthrough T/F Env. This was accomplished by using serum samples collected 13 wk after the final MVA boost (and 11 wk before SIVsmE660 challenge) from the breakthrough infection animals, representing the midpoint between the peak binding antibody response (week 2 post-MVA) and time of challenge (week 20–24 post-MVA) (Fig. 1A).
Fig. 3.
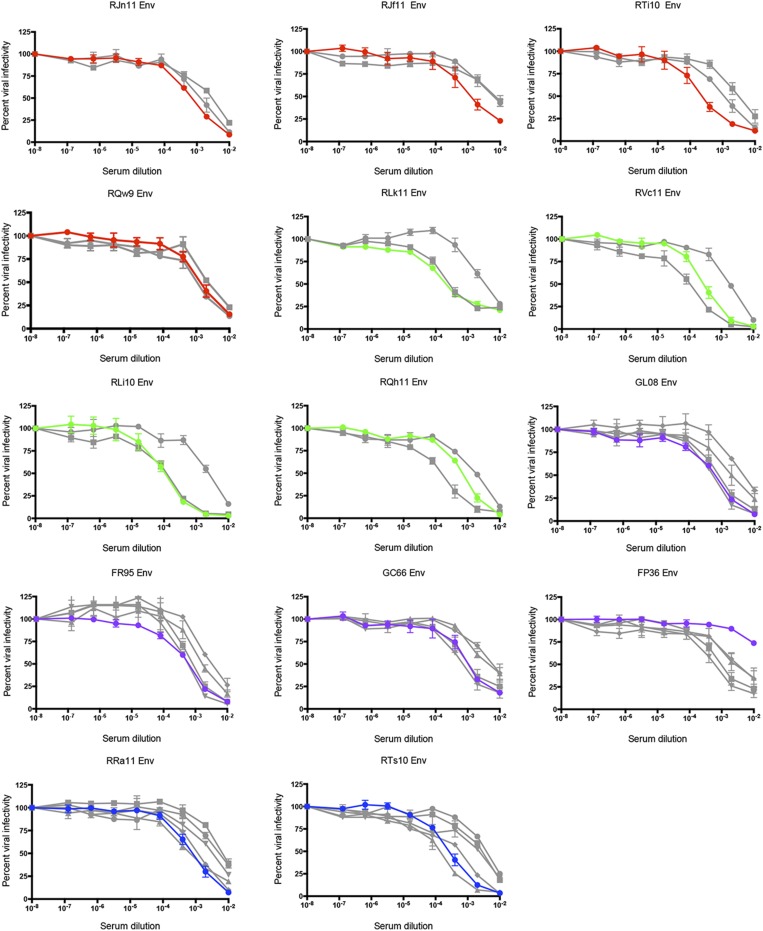
Neutralization sensitivity of breakthrough T/F Envs. T/F Env pseudoviruses from vaccinated breakthrough and unvaccinated control infections were assessed for neutralization sensitivity using the TZM-bl assay, with luciferase activity as the readout. (A) Neutralization of the control (n = 4), DDMM (n = 4), DgDgMM (n = 2), D40LD40LMM (n = 4), and MMM (n = 4) T/F Envs by a pool of serum samples collected from unvaccinated RMs at 24 wk postinfection with SIVsmE660 is shown. The data for T/F Env pseudoviruses within each vaccine or control group (percent infectivity at each serum dilution) were combined into five corresponding datasets in Prism to generate each neutralization curve (color-coded as described in the key). Pseudoviruses expressing the neutralization-sensitive Env SIVsmE660.11 and neutralization-resistant Env SIVsmE660.300–16 are included as references. (B) Neutralization of the T/F Envs by each autologous prechallenge serum sample is shown in the individual panels. The autologous breakthrough T/F Env is represented by a red (DDMM), green (MMM), purple (D40LD40LMM), or blue (DgDgMM) line in each graph; infectivity data for the four control T/F Envs were combined into a single dataset and are shown as an orange line, E660.11 as a gray line, and E660.300–16 as a black line. RLi10 (third panel, second row) is an example of a very potent autologous neutralization of the breakthrough Env; FP36 (fourth panel, third row) lacks autologous neutralization activity against the breakthrough Env. For both A and B, the reciprocal of the serum dilution from each monkey is plotted on the x axis on a log10 scale, and the percent of viral infectivity is plotted along the y axis relative to the virus only control at 100%. Each serum–Env combination was run in duplicate, and each assay was repeated independently at least twice. The variation for each point in the infectivity curve within each group is indicated by the error bars, which represent the SEM. The number of challenges at which each vaccinated monkey became infected is indicated in the bottom left corner of the graphs.
Fig. 3B demonstrates that the vaccinated breakthrough T/F Envs were susceptible to neutralization by the autologous prechallenge serum, but at IC50 titers that were several logs lower than those observed with the chronic infection pool (Fig. 3A). Individually, the IC50s of the breakthrough T/F Envs ranged from undetectable in one case (FP36) to 1:10,832 for RLi10. Although the median autologous nAb IC50 titer was 1:1,351, there were five immunized monkeys that became infected with prechallenge IC50 titers exceeding 1:3,000. Most of the breakthrough T/F Envs were completely or nearly completely neutralized at a 1:100 dilution of serum; however, some variants did retain residual infectivity at this dilution. The prechallenge serum from vaccinated breakthrough animals neutralized the sensitive reference Env SIVsmE660.11 to varying degrees, whereas neutralizing activity against the resistant reference Env SIVsmE660.300–16 was uniformly low (Fig. 3B) (27). These data collectively demonstrate that all but one vaccinated animal produced prechallenge serum antibodies capable of neutralizing the autologous breakthrough T/F Env, yet this did not protect against infection.
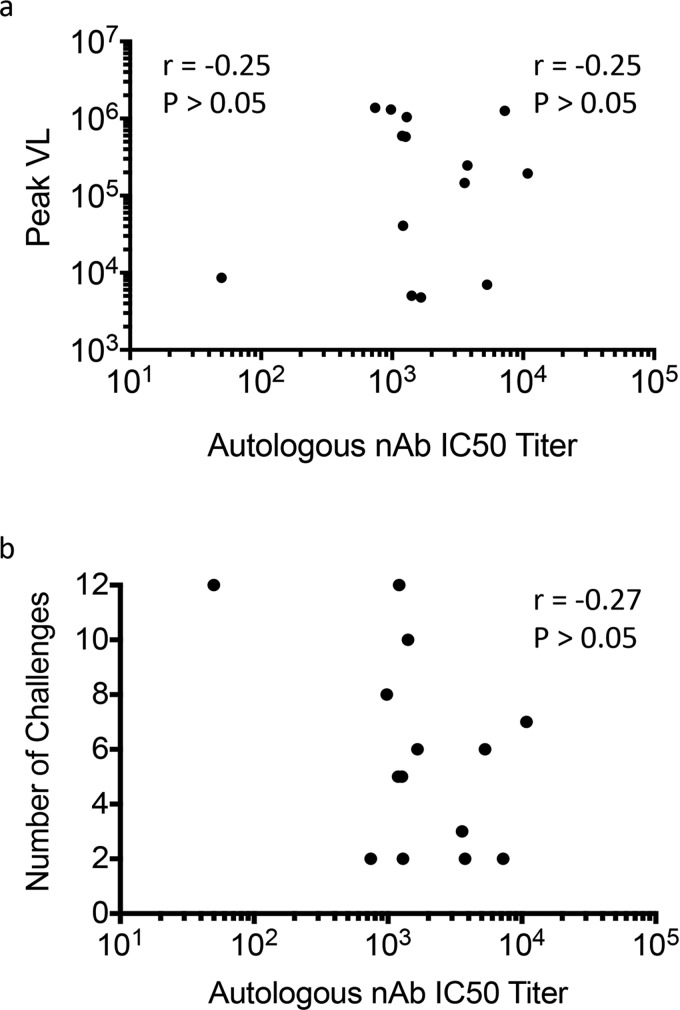
We next investigated whether vaccinated animals that were protected against SIVsmE660 challenge had superior neutralizing capacity compared with the breakthrough animals from the same vaccine group (Table S1). Prechallenge serum samples from vaccinated protected animals were tested against each breakthrough T/F Env from the same group. Neutralization potency of the protected animal serum was comparable to the autologous serum (Fig. S6), and the median IC50 titers (1:1,042 and 1:1,351, respectively) were not significantly different when analyzed by a Mann–Whitney or a paired Wilcoxon rank sum test. This result suggests that protected animals did not have substantially more potent neutralizing activity against the breakthrough T/F Env variants. This result is consistent with our previous finding that neutralizing activity against heterologous SIV Envs did not differ between infected and protected animals in the M11 and M12 trials (28). We also found no correlation between the nAb IC50 titer of the vaccinated breakthrough animals and the peak viral load or the number of SIVsmE660 challenges required for infection (Fig. S7 A and B, respectively). Thus, there was no evidence that SIV acquisition was directly linked to the individual potency of prechallenge nAb activity.
Table S1.
Summary of RMs included in the study
| Monkey ID | Trial | Vaccine group | Challenge status | T/F Env cloned |
| Rzp10 | M11 | DDMM | Uninfected | N/A |
| RNi11 | M11 | DDMM | Uninfected | N/A |
| RQw9 | M11 | DDMM | Infected | Yes |
| RJn11 | M11 | DDMM | Infected | Yes |
| RTi10 | M11 | DDMM | Infected | Yes |
| RJf11 | M11 | DDMM | Infected | Yes |
| RFm11 | M11 | DgDgMM | Uninfected | N/A |
| RKk11 | M11 | DgDgMM | Uninfected | N/A |
| RPv10 | M11 | DgDgMM | Uninfected | N/A |
| RWg11 | M11 | DgDgMM | Uninfected | N/A |
| RLe11 | M11 | DgDgMM | Uninfected | N/A |
| RTs10 | M11 | DgDgMM | Infected | Yes |
| RRa11 | M11 | DgDgMM | Infected | Yes |
| RUh11 | M11 | MMM | Uninfected | N/A |
| RKk10 | M11 | MMM | Uninfected | N/A |
| RQh11 | M11 | MMM | Infected | Yes |
| RLk11 | M11 | MMM | Infected | Yes |
| RVc11 | M11 | MMM | Infected | Yes |
| RLi10 | M11 | MMM | Infected | Yes |
| GM06 | M12 | D40LD40LMM | Uninfected | N/A |
| GM99 | M12 | D40LD40LMM | Uninfected | N/A |
| RKb12 | M12 | D40LD40LMM | Uninfected | N/A |
| RQk11 | M12 | D40LD40LMM | Uninfected | N/A |
| GL08 | M12 | D40LD40LMM | Infected | Yes |
| FR95 | M12 | D40LD40LMM | Infected | Yes |
| GC66 | M12 | D40LD40LMM | Infected | Yes |
| FP36 | M12 | D40LD40LMM | Infected | Yes |
| RHa11 | M11 | Control | Infected | Yes |
| RIi11 | M11 | Control | Infected | Yes |
| RSo11 | M11 | Control | Infected | Yes |
| RJj12 | M12 | Control | Infected | Yes |
Fig. S6.
Neutralization of breakthrough T/F Envs by prechallenge serum from vaccinated protected monkeys compared with the autologous prechallenge serum. Neutralization of each breakthrough T/F Env by prechallenge serum samples from monkeys that were immunized with the same vaccine but remained uninfected after 12 challenges is shown in the individual panels. The autologous serum sample is represented by a red (DDMM), green (MMM), purple (D40LD40LMM), or blue (DgDgMM) line in each graph; the vaccination group-matched serum samples from uninfected animals against the same T/F Env are shown in gray. The reciprocal of the serum dilution is plotted on the x axis on a log10 scale, and the percent of viral infectivity is plotted along the y axis relative to the virus-only control at 100%. Each serum–Env combination was run in duplicate, and each assay was repeated independently at least twice. The variation for each point in the infectivity curve within each group is indicated by the error bars, which represent the SEM.
Fig. S7.
Lack of correlation between the prechallenge autologous nAb IC50 titer and peak viral load or number of challenges required for infection. A Spearman analysis was performed using Prism 6.0 to determine whether the autologous nAb IC50 titer for each vaccinated breakthrough monkey (n = 14) was correlated with (A) the peak plasma viral load (copies per mL) or (B) the number of SIVsmE660 intrarectal challenges required for infection. The nAb IC50 titer is plotted along the x axis in both plots. In both cases, the correlation between the two parameters was not significant.
Discussion
Identifying the immune correlates of protection against HIV/SIV continues to be a central pursuit for the vaccine research community (29–31). Multiple vaccine studies in NHPs and humans have implied that functional antibody responses against Env are necessary for protective immunity (3, 18, 32). Furthermore, passive administration of nmAbs to RMs has unambiguously been shown to provide protection against mucosal SHIV infection (5–13), with in vitro serum nAb IC50 titers in the range of ∼1:200 emerging as predictive of protection (7, 12, 14). However, it was not known whether these low to moderate nAb titers would extend to vaccine-elicited antibody protection, as previous studies have reported a disconnect between in vitro nAb and a protective effect (33–35). Here, we directly demonstrate that prechallenge serum nAb titers against the autologous breakthrough T/F Env variants were not predictive of protection. Remarkably, these breakthrough variants were surprisingly sensitive to autologous neutralization, with in vitro nAb titers frequently exceeding 1:1,000, and were similarly sensitive to neutralization by prechallenge serum from vaccinated protected animals.
These results highlight an important and well-recognized difference between passively transferred and vaccine-elicited antibody protection; the latter occurs in the presence of ongoing host immune responses. Thus, it is important to consider that vaccination itself may alter the mucosal environment such that transmission is favored over protection (31, 36, 37). This could result from the generation and homing of vaccine-elicited CD4+ T cells to the portal of entry, which in turn mitigates the beneficial effects of the vaccine-elicited humoral immune response (38–40). Indeed, the efficiency of horizontal and vertical SIV transmission in NHP hosts in the absence of vaccination is linked to the availability of susceptible target cells in the mucosa (41–43). Furthermore, there is strong evidence that genital inflammation reduces the stringency of HIV-1 transmission (25, 44, 45) and increased target cell availability increases the risk of HIV-1 superinfection (46).
Additional caveats may also contribute to the marked discrepancy between in vitro neutralization and in vivo protection that we observed here. It is possible that vaccine-elicited antibodies capable of potent neutralization in the TZM-bl assay lack certain biological properties that are essential for preventing acquisition in vivo (14, 47). Cell-to-cell transmission could play a more prominent role in intrarectal SIV infection than is currently recognized (48–51), and higher concentrations of antibodies with certain specificities could then be necessary to prevent establishment of infection (52, 53). It is similarly possible that the atypically high neutralization sensitivity of SIVsmE660 Env variants involves antibody specificities that are greatly enhanced in the TZM-bl assay and have limited relevance to those that neutralize HIV-1 (27). Finally, we were not able to directly assess the neutralizing activity in rectal secretions or in serum on the day of challenge. However, Env-specific serum IgG titers induced by DNA/MVA immunization were stable in RMs and humans during a similar time frame as was analyzed in refs. 54 and 55.
Despite these caveats, our results collectively demonstrate that the DNA/MVA vaccination regimens administered in the M11 and M12 trials consistently elicited prechallenge antibodies that potently neutralized the autologous breakthrough T/F variants in vitro, but these antibodies were not protective in vivo. The lack of overt selection for more neutralization-resistant breakthrough variants supports the concept that the serum in vitro nAb titer was not the defining feature of vaccine-mediated protection. The findings are consistent with the idea that desirable vaccine-elicited immune responses such as nAbs can be negated by a concurrent increase in the availability and/or susceptibility of CD4+ T cells and suggest that the interplay between the two parameters needs to be better elucidated. These data further suggest that the relationship between serum nAb titers and protection from mucosal SIV challenge in the setting of active immunization is more complex than previously recognized and that additional studies of this interaction are warranted to better inform the design and evaluation of Env-based vaccinations.
Materials and Methods
Ethics Statement.
The Emory University Institutional Animal Care and Use Committee (AWA A3180-01) approved these studies of NHPs under protocols 215–2007Y and 139–2008Y. This study was conducted in strict accordance with US Department of Agriculture regulations and the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. SIV-infected Indian RMs (Macaca mulatta) were housed in standard NHP primate cages and received standard primate feed, as well as fresh fruit and enrichment daily, and had continuous access to water. Animals had continuous access to enrichment resources, including objects for perching and other manipulanda. Animal welfare was monitored daily. Appropriate procedures were performed to ensure that potential distress, pain, or discomfort was alleviated. The sedatives Ketamine (10 mg/kg) and Telazol (4 mg/kg) were used for blood draws. Euthanasia using Pentobarbital (100 mg/kg) under anesthesia was performed only when deemed clinically necessary by veterinary medical staff and according to Institutional Animal Care and Use Committee endpoint guidelines.
SGA and Sequencing of SIVsmE660 env Genes.
Plasma collected at 7 or 14 d postinfection was used for 96-well SGA of SIVsmE660 env genes. Plasma viral loads in these samples ranged from 6.74 × 102 to 3.62 × 106 copies per mL. Viral RNA was extracted from plasma using the QIAmp viral RNA kit according to the manufacturer’s instructions (Qiagen). Reverse transcription of viral RNA into cDNA was performed using the SuperScript III kit and reverse primer SM-ER1 CTATCACTGTAATAAATCCCTTCCAGTCCC, following the manufacturer’s instructions (Invitrogen). For SGA, the dilution of a cDNA template that resulted in one-fourth to one-third of the PCR reactions being positive was used, as described previously in refs. 25, 56. The first round of PCR was performed in a 15 μL volume using the Phusion Hotstart II High Fidelity DNA Polymerase (Thermo Scientific) and the following primers: forward SIVsm/macEnvF1, CCTCCCCCTCCAGGACTAGC, and reverse SIVsm/macEnvR1, TGTAATAAATCCCTTCCAGTCCCCCC. The cycling conditions were 98 °C for 2 min; 10 cycles of 98 °C for 10 s, 55 °C for 45 s, and 68 °C for 2 min; 25 cycles of 98 °C for 10 s, 55 °C for 45 s, and 68 °C for 2 min, adding 5 s to the extension per cycle; 72 °C for 30 min; and 4 °C hold. The second round of PCR was performed with the same enzyme in a 10 μL volume using 1 μL of the first round of PCR as a template and the following primers: forward SIVsmEnvF2M, CACCTATGATAGACATGGAGACACCCTTGAAGGAGC, and reverse SIVsmEnvR2, ATGAGACATRTCTATTGCCAATTTGA. The cycling conditions were 98 °C for 2 min; 45 cycles of 98 °C for 10 s, 55 °C for 45 s, and 68 °C for 2.5 min; 72 °C for 30 min; and 4 °C hold. PCR amplicons were purified using the Roche High Pure PCR Product Purification Kit following the manufacturer’s instructions. Between 10 and 32 SGA amplicons were sequenced per animal using BigDye Terminator sequencing (Applied Biosystems). The following primers were used: forward F1, ATCCATTTCAGAAGTGGATGTGCCCACTCC; F2, AGGGTTAAAAAGGGACAAAAGGATAGAATA; F3, TTGTAGAGGAGAATTCTTATACTGCAAAAT; F4N, AAACATTGGTGCCTAATTGGAGCAATATGA; and reverse, R1 GCAAAGCATAACCTGGCGGTGCACAATATC; R2, CTCCTTCCCTAGGAGGCAAATATACATTTT; R3N, TTGCAATTCATACATATTCTTTTCTTGCTG; and R4, TCATCTTCATCATCCACATCATCCATGTTT. Sequencher v5 was used to generate contigs, and sequences with evidence of mixed peaks were omitted from the analysis. Geneious v6.1.7 was used to translate nucleotide sequences, create alignments, calculate pairwise diversity, and generate a Neighbor-joining phylogenetic tree. Highlighter plots were generated to identify the consensus sequence for the T/F Env variant (www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html). GenBank accession numbers are as follows: FJ579014–FJ579055 for the VH2000 challenge stock sequences and KP734847–KP735110 for the SIVsmE660 SGA and cloned env sequences.
Neutralization Assay.
An SGA-derived env gene amplicon that was representative of the T/F consensus sequence for each monkey was purified using the Qiagen PCR Purification Kit (Qiagen) and was cloned into pCDNA3.1dV5His TOPO-TA (Invitrogen) using approaches described previously (57–59). An env gene clone whose sequence was identical to the matching SGA amplicon was used to generate a pseudovirus by transfecting the Env-expressing plasmid DNA alongside the HIV-1 SG3ΔEnv proviral backbone DNA into 293T cells, using the Fugene HD reagent as recommended (Promega). Pseudovirus stocks were collected from the 293T cell supernatants at 48–72 h after transfection, clarified by centrifugation, divided into small volumes, and frozen at –80 °C. Fivefold serial dilutions of heat-inactivated serum samples were assayed for their inhibitory potential against the Env pseudoviruses using the TZM-bl indicator cell line, with luciferase as the readout, as described previously (57, 59–63). TZM-bl cells were plated and cultured overnight in flat-bottomed 96-well plates. A pseudovirus (2,000 IU per well) in DMEM with ∼3.5% (vol/vol) FBS (HyClone) and 40 μg/mL DEAE-dextran was mixed with serial dilutions of plasma or serum and subsequently added to the plated TZM-bl cells. At 48 h postinfection, the cells were lysed and luciferase activity was measured using a BioTek Synergy HT multimode microplate reader with Gen 5, v2.0 software. The average background luminescence from a series of uninfected wells was subtracted from each experimental well, and infectivity curves were generated using GraphPad Prism v6.0d, where values from experimental wells were compared against a well containing a virus without a test reagent (100% infectivity). Neutralization IC50 titer values were calculated in Graphpad Prism v6.0d using the dose–response inhibition analysis function with variable slope, log-transformed x values, and normalized y values.
Acknowledgments
We gratefully acknowledge Dr. Brandon Keele for providing SGA sequences from the SIVsmE660 VH2000 challenge stock; Samuel R. DeVictor, Geovonni P. Bell, Lena A. Sheorey, Andrew D. Chang, and Mariam Dvalishvili for technical assistance; Drs. George Shaw and Katharine Bar for providing the SIVsmE660 reference Env plasmids E660.11 and E660.300-16 and their sequences; and the National Institute of Allergy and Infectious Diseases/National Institutes of Health for Grants R01-AI58706 (to C.A.D.), U19-AI096187 (to E.H. and R.R.A.), and P51-RR000165 and P51-OD011132 (to the Yerkes National Primate Research Center).
Footnotes
Conflict of interest statement: R.R.A. and H.L.R. are coinventors of DNA/MVA vaccine technology that has been licensed to GeoVax.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FJ579014–FJ579055 and KP734847–KP735110).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509731112/-/DCSupplemental.
References
- 1.Doria-Rose NA, et al. NISC Comparative Sequencing Program Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509(7498):55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liao HX, et al. NISC Comparative Sequencing Program Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496(7446):469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366(14):1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derdeyn CA, Moore PL, Morris L. Development of broadly neutralizing antibodies from autologous neutralizing antibody responses in HIV infection. Curr Opin HIV AIDS. 2014;9(3):210–216. doi: 10.1097/COH.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shibata R, et al. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat Med. 1999;5(2):204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 6.Mascola JR, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6(2):207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 7.Hessell AJ, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15(8):951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parren PW, et al. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J Virol. 2001;75(17):8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baba TW, et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000;6(2):200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 10.Pegu A, et al. Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor. Sci Transl Med. 2014;6(243):243ra88. doi: 10.1126/scitranslmed.3008992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moldt B, et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci USA. 2012;109(46):18921–18925. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shingai M, et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med. 2014;211(10):2061–2074. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudicell RS, et al. NISC Comparative Sequencing Program Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J Virol. 2014;88(21):12669–12682. doi: 10.1128/JVI.02213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hessell AJ, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5(5):e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Letvin NL, et al. Immune and genetic correlates of vaccine protection against mucosal infection by SIV in monkeys. Sci Transl Med. 2011;3(81):81ra36. doi: 10.1126/scitranslmed.3002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flatz L, et al. Gene-based vaccination with a mismatched envelope protects against simian immunodeficiency virus infection in nonhuman primates. J Virol. 2012;86(15):7760–7770. doi: 10.1128/JVI.00599-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel V, et al. DNA and virus particle vaccination protects against acquisition and confers control of viremia upon heterologous simian immunodeficiency virus challenge. Proc Natl Acad Sci USA. 2013;110(8):2975–2980. doi: 10.1073/pnas.1215393110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roederer M, et al. Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature. 2014;505(7484):502–508. doi: 10.1038/nature12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwa S, et al. CD40L-adjuvanted DNA/modified vaccinia virus Ankara simian immunodeficiency virus SIV239 vaccine enhances SIV-specific humoral and cellular immunity and improves protection against a heterologous SIVE660 mucosal challenge. J Virol. 2014;88(17):9579–9589. doi: 10.1128/JVI.00975-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai L, et al. Prevention of infection by a granulocyte-macrophage colony-stimulating factor co-expressing DNA/modified vaccinia Ankara simian immunodeficiency virus vaccine. J Infect Dis. 2011;204(1):164–173. doi: 10.1093/infdis/jir199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai L, et al. SIVmac239 MVA vaccine with and without a DNA prime, similar prevention of infection by a repeated dose SIVsmE660 challenge despite different immune responses. Vaccine. 2012;30(9):1737–1745. doi: 10.1016/j.vaccine.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rolland M, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490(7420):417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rolland M, et al. Genetic impact of vaccination on breakthrough HIV-1 sequences from the STEP trial. Nat Med. 2011;17(3):366–371. doi: 10.1038/nm.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derdeyn CA, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303(5666):2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 25.Haaland RE, et al. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 2009;5(1):e1000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boeras DI, et al. Role of donor genital tract HIV-1 diversity in the transmission bottleneck. Proc Natl Acad Sci USA. 2011;108(46):E1156–E1163. doi: 10.1073/pnas.1103764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopker M, et al. Heterogeneity in neutralization sensitivity of viruses comprising the SIVsmE660 isolate and vaccine challenge stock. J Virol. 2013;87(10):5477–5492. doi: 10.1128/JVI.03419-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kilgore KM, et al. Characterization and implementation of a diverse SIVsm envelope panel in the assessment of neutralizing antibody breadth elicited in rhesus macaques by multi-modal vaccines expressing the SIVmac239 ENVELOPE. J Virol. 2015;89(16):8130–8151. doi: 10.1128/JVI.01221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JH, Excler JL, Michael NL. Lessons from the RV144 Thai phase III HIV-1 vaccine trial and the search for correlates of protection. Annu Rev Med. 2015;66:423–437. doi: 10.1146/annurev-med-052912-123749. [DOI] [PubMed] [Google Scholar]
- 30.Tomaras GD, Haynes BF. Advancing toward HIV-1 vaccine efficacy through the intersections of immune correlates. Vaccines (Basel) 2014;2(1):15–35. doi: 10.3390/vaccines2010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis GK, DeVico AL, Gallo RC. Antibody persistence and T-cell balance: Two key factors confronting HIV vaccine development. Proc Natl Acad Sci USA. 2014;111(44):15614–15621. doi: 10.1073/pnas.1413550111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barouch DH, et al. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature. 2012;482(7383):89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeVico A, et al. Antibodies to CD4-induced sites in HIV gp120 correlate with the control of SHIV challenge in macaques vaccinated with subunit immunogens. Proc Natl Acad Sci USA. 2007;104(44):17477–17482. doi: 10.1073/pnas.0707399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eugene HS, Pierce-Paul BR, Cragio JK, Ross TM. Rhesus macaques vaccinated with consensus envelopes elicit partially protective immune responses against SHIV SF162p4 challenge. Virol J. 2013;10:102. doi: 10.1186/1743-422X-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas MA, et al. HIV-1 CD4-induced (CD4i) gp120 epitope vaccines promote B and T-cell responses that contribute to reduced viral loads in rhesus macaques. Virology. 2014;471-473:81–92. doi: 10.1016/j.virol.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fouts TR, et al. Balance of cellular and humoral immunity determines the level of protection by HIV vaccines in rhesus macaque models of HIV infection. Proc Natl Acad Sci USA. 2015;112(9):E992–E999. doi: 10.1073/pnas.1423669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fauci AS, Marovich MA, Dieffenbach CW, Hunter E, Buchbinder SP. Immunology. Immune activation with HIV vaccines. Science. 2014;344(6179):49–51. doi: 10.1126/science.1250672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staprans SI, et al. Enhanced SIV replication and accelerated progression to AIDS in macaques primed to mount a CD4 T cell response to the SIV envelope protein. Proc Natl Acad Sci USA. 2004;101(35):13026–13031. doi: 10.1073/pnas.0404739101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carnathan DG, et al. Activated CD4+CCR5+ T cells in the rectum predict increased SIV acquisition in SIVGag/Tat-vaccinated rhesus macaques. Proc Natl Acad Sci USA. 2015;112(2):518–523. doi: 10.1073/pnas.1407466112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tenbusch M, et al. Risk of immunodeficiency virus infection may increase with vaccine-induced immune response. J Virol. 2012;86(19):10533–10539. doi: 10.1128/JVI.00796-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma D, et al. International Vervet Research Consortium SIVagm infection in wild African green monkeys from South Africa: Epidemiology, natural history, and evolutionary considerations. PLoS Pathog. 2013;9(1):e1003011. doi: 10.1371/journal.ppat.1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chahroudi A, et al. Target cell availability, rather than breast milk factors, dictates mother-to-infant transmission of SIV in sooty mangabeys and rhesus macaques. PLoS Pathog. 2014;10(3):e1003958. doi: 10.1371/journal.ppat.1003958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandrea I, et al. Paucity of CD4+ CCR5+ T cells may prevent transmission of simian immunodeficiency virus in natural nonhuman primate hosts by breast-feeding. J Virol. 2008;82(11):5501–5509. doi: 10.1128/JVI.02555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carlson JM, et al. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 2014;345(6193):1254031. doi: 10.1126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sagar M, et al. Identification of modifiable factors that affect the genetic diversity of the transmitted HIV-1 population. AIDS. 2004;18(4):615–619. doi: 10.1097/00002030-200403050-00005. [DOI] [PubMed] [Google Scholar]
- 46.Blish CA, et al. Association between cellular immune activation, target cell frequency, and risk of human immunodeficiency virus type 1 superinfection. J Virol. 2014;88(10):5894–5899. doi: 10.1128/JVI.00187-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hessell AJ, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449(7158):101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 48.Bernard-Stoecklin S, Gommet C, Cavarelli M, Le Grand R. Nonhuman primate models for cell-associated simian immunodeficiency virus transmission: The need to better understand the complexity of HIV mucosal transmission. J Infect Dis. 2014;210(Suppl 3):S660–S666. doi: 10.1093/infdis/jiu536. [DOI] [PubMed] [Google Scholar]
- 49.Sagar M. Origin of the transmitted virus in HIV infection: Infected cells versus cell-free virus. J Infect Dis. 2014;210(Suppl 3):S667–S673. doi: 10.1093/infdis/jiu369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whaley KJ, Mayer KH. Strategies for preventing mucosal cell-associated HIV transmission. J Infect Dis. 2014;210(Suppl 3):S674–S680. doi: 10.1093/infdis/jiu398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sallé B, et al. Infection of macaques after vaginal exposure to cell-associated simian immunodeficiency virus. J Infect Dis. 2010;202(3):337–344. doi: 10.1086/653619. [DOI] [PubMed] [Google Scholar]
- 52.Malbec M, et al. Broadly neutralizing antibodies that inhibit HIV-1 cell to cell transmission. J Exp Med. 2013;210(13):2813–2821. doi: 10.1084/jem.20131244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abela IA, et al. Cell-cell transmission enables HIV-1 to evade inhibition by potent CD4bs directed antibodies. PLoS Pathog. 2012;8(4):e1002634. doi: 10.1371/journal.ppat.1002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goepfert PA, et al. HVTN 205 Study Group National Institutes of Allergy and Infectious Diseases HIV Vaccines Trials Network Specificity and 6-month durability of immune responses induced by DNA and recombinant modified vaccinia Ankara vaccines expressing HIV-1 virus-like particles. J Infect Dis. 2014;210(1):99–110. doi: 10.1093/infdis/jiu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iyer SS, et al. Codelivery of envelope protein in alum with MVA vaccine induces CXCR3-biased CXCR5+ and CXCR5- CD4 T cell responses in rhesus macaques. J Immunol. 2015;195(3):994–1005. doi: 10.4049/jimmunol.1500083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salazar-Gonzalez JF, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82(8):3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li B, et al. Nonpathogenic simian immunodeficiency virus infection of sooty mangabeys is not associated with high levels of autologous neutralizing antibodies. J Virol. 2010;84(12):6248–6253. doi: 10.1128/JVI.00295-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rong R, et al. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 2009;5(9):e1000594. doi: 10.1371/journal.ppat.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li B, et al. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J Virol. 2006;80(11):5211–5218. doi: 10.1128/JVI.00201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murphy MK, et al. Viral escape from neutralizing antibodies in early subtype A HIV-1 infection drives an increase in autologous neutralization breadth. PLoS Pathog. 2013;9(2):e1003173. doi: 10.1371/journal.ppat.1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lynch RM, et al. The B cell response is redundant and highly focused on V1V2 during early subtype C infection in a Zambian seroconverter. J Virol. 2011;85(2):905–915. doi: 10.1128/JVI.02006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rong R, et al. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J Virol. 2007;81(3):1350–1359. doi: 10.1128/JVI.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rong R, et al. Unique mutational patterns in the envelope alpha 2 amphipathic helix and acquisition of length in gp120 hypervariable domains are associated with resistance to autologous neutralization of subtype C human immunodeficiency virus type 1. J Virol. 2007;81(11):5658–5668. doi: 10.1128/JVI.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]