Significance
Vitamin B12 is required by humans and a variety of other organisms for diverse metabolic processes, but is produced only by a subset of microorganisms. The anaerobic biosynthesis of the “lower ligand” of B12, 5,6-dimethylbenzimidazole (DMB), is the only unknown component of the B12 biosynthetic pathway. We report the identification of the bzaABCDE genes that are necessary and sufficient for the anaerobic biosynthesis of DMB. We have characterized the role of each of the bza genes and identified three intermediates in the pathway. This finding not only completes the B12 biosynthesis pathway but also enables the sequence-based prediction of cobamides synthesized by anaerobic microorganisms.
Keywords: vitamin B12; cobalamin; cobamide; 5,6-dimethylbenzimidazole; biosynthesis
Abstract
Vitamin B12 (cobalamin) is required by humans and other organisms for diverse metabolic processes, although only a subset of prokaryotes is capable of synthesizing B12 and other cobamide cofactors. The complete aerobic and anaerobic pathways for the de novo biosynthesis of B12 are known, with the exception of the steps leading to the anaerobic biosynthesis of the lower ligand, 5,6-dimethylbenzimidazole (DMB). Here, we report the identification and characterization of the complete pathway for anaerobic DMB biosynthesis. This pathway, identified in the obligate anaerobic bacterium Eubacterium limosum, is composed of five previously uncharacterized genes, bzaABCDE, that together direct DMB production when expressed in anaerobically cultured Escherichia coli. Expression of different combinations of the bza genes revealed that 5-hydroxybenzimidazole, 5-methoxybenzimidazole, and 5-methoxy-6-methylbenzimidazole, all of which are lower ligands of cobamides produced by other organisms, are intermediates in the pathway. The bza gene content of several bacterial and archaeal genomes is consistent with experimentally determined structures of the benzimidazoles produced by these organisms, indicating that these genes can be used to predict cobamide structure. The identification of the bza genes thus represents the last remaining unknown component of the biosynthetic pathway for not only B12 itself, but also for three other cobamide lower ligands whose biosynthesis was previously unknown. Given the importance of cobamides in environmental, industrial, and human-associated microbial metabolism, the ability to predict cobamide structure may lead to an improved ability to understand and manipulate microbial metabolism.
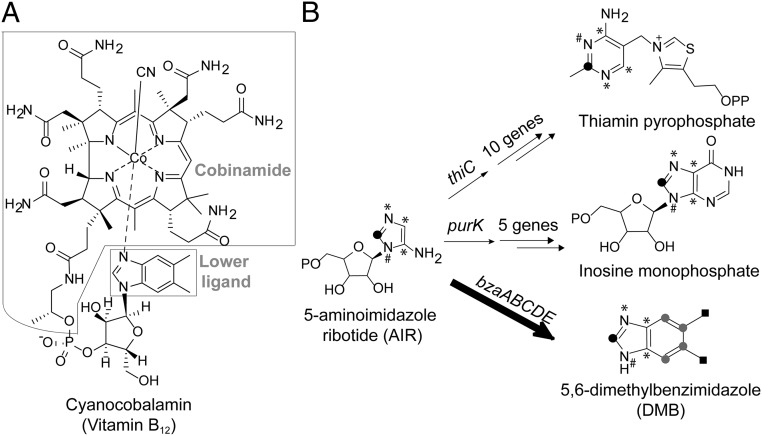
Vitamin B12 (cobalamin) is required by the majority of animals, protists, and prokaryotes, although B12 and other cobamide cofactors are synthesized exclusively by a subset of prokaryotes (1). Cobalamin is a member of the cobamide family of cofactors that are composed of a central cobalt ion coordinated by a tetrapyrrolic corrin ring, an upper ligand, and a lower ligand covalently tethered to the corrin ring by a nucleotide loop (Fig. 1A) (1). The lower ligand of cobalamin is 5,6-dimethylbenzimidazole (DMB). Purines, phenolic compounds, and other substituted benzimidazoles have also been found as cobamide lower ligands (2).
Fig. 1.
Vitamin B12 structure and the labeling pattern of DMB. (A) Structure of cyanocobalamin (vitamin B12). Cbi and the lower ligand DMB are indicated. (B) AIR is a branch point in the biosynthesis of thiamin (8), purines (27), and benzimidazoles (as shown in this study). The symbols represent the origins of atoms in each product: ●, formyltetrahydrofolate; #, glutamine; *, glycine; shaded circle, erythrose 4-phosphate or threose; ▪, methionine.
Distinct oxygen-requiring (aerobic) and oxygen-sensitive (anaerobic) cobamide biosynthesis pathways have been characterized, and each contains ∼30 genes (1). The biosynthetic pathway for DMB remained elusive until the discovery of the bluB gene, the product of which fragments reduced flavin mononucleotide (FMNH2) to produce DMB in an oxygen-dependent reaction (3–6). Currently, the only unidentified genes in the cobamide biosynthesis pathway are those required for the anaerobic biosynthesis of DMB and other benzimidazoles (7). Based on previous isotope-labeling studies, performed mainly in the anaerobic bacterium Eubacterium limosum, the biosynthesis of DMB was proposed to branch from the purine biosynthetic pathway, similar to the pyrimidine ring of the cofactor thiamin pyrophosphate (Fig. 1B) (2, 8–15). Here, we sought to identify the genes in Eubacterium limosum required for the biosynthesis of DMB.
Results
Identification of Putative DMB Biosynthesis Genes Adjacent to Cobalamin Riboswitches in E. limosum.
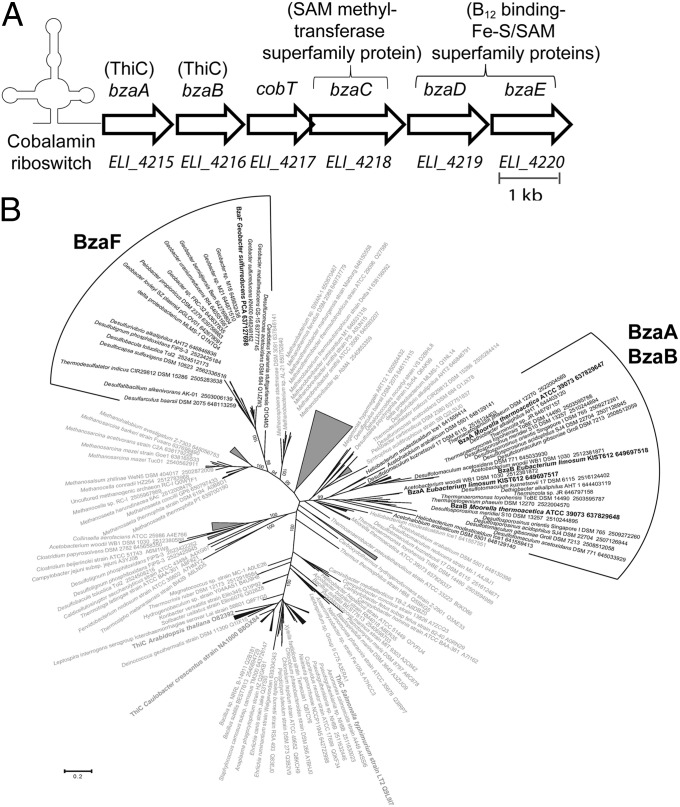
We searched the published genome sequence of E. limosum strain KIST612 (16) for genes located downstream of the regulatory elements known as cobalamin riboswitches. These regulatory sequences are commonly found adjacent to genes involved in cobamide biosynthesis and transport, including bluB (3, 17). This analysis showed that the E. limosum genome contains 13 putative cobalamin riboswitches, of which 5 are associated with homologs of known cobalamin biosynthesis genes (Fig. 2A and SI Appendix, Fig. S1). One riboswitch sequence is located at the 5′ end of a putative operon containing five genes of unknown function in addition to a homolog of cobT, which encodes the first enzyme necessary for the attachment of a lower ligand base to a cobamide precursor (Fig. 2A) (18). The first two ORFs in the operon are annotated as thiC, which encodes the enzyme 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate (HMP-P) synthase, an oxygen-sensitive radical S-adenosylmethionine (SAM) enzyme that uses the purine intermediate 5-aminoimidazole ribotide (AIR) as a substrate for the biosynthesis of the pyrimidine ring of thiamin pyrophosphate (Figs. 1B and 2A) (8). The two E. limosum thiC homologs cluster in a phylogenetic clade together with other thiC homolog pairs found associated with cobT or other genes involved in B12 biosynthesis or metabolism (Fig. 2B). These gene products share 46 and 41% identity with each other and with ThiC, respectively (SI Appendix, Table S1). We focused our subsequent functional studies on this genomic locus. Based on the results described below, we have named these genes bzaABCDE (for benzimidazole biosynthesis, Fig. 2A).
Fig. 2.
Identification of the E. limosum bza genes. (A) E. limosum bza operon. Arrows represent ORFs. Gene locus tags as given by the National Center for Biotechnology Information (NCBI) database are shown below, and new gene names are shown above. Protein annotations in the NCBI database are shown in parentheses. (B) Phylogenetic tree of BzaA, BzaB, BzaF, and ThiC. BzaA, BzaB, and BzaF homologs are in black (BzaF is discussed below); ThiC homologs are in gray. Boldface indicates experimentally verified functions. An extended version of this figure is shown in SI Appendix, Fig. S2.
Heterologous Expression of the bzaABCDE Genes in Escherichia coli Leads to DMB Biosynthesis.
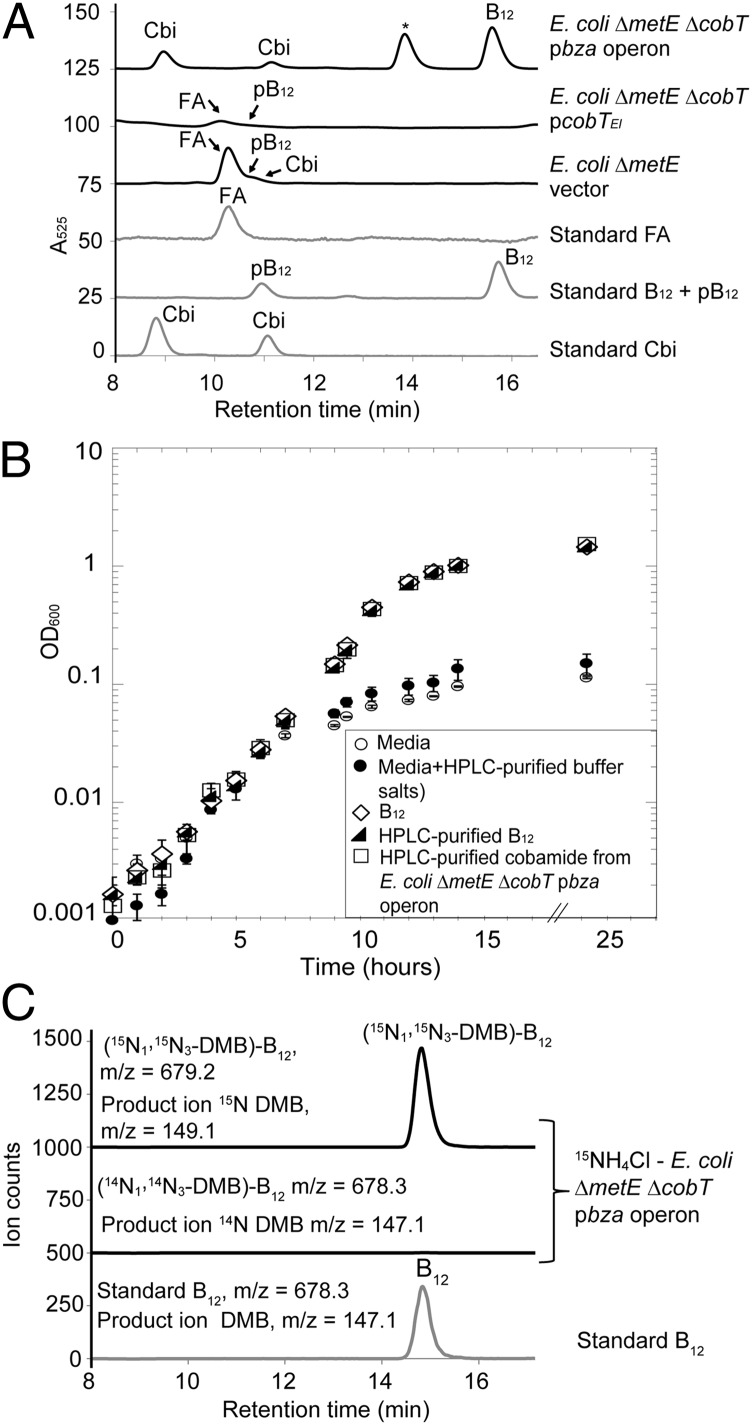
To examine the function of the E. limosum bza operon, we expressed the bza locus from E. limosum strain ATCC 10825 on a plasmid in E. coli and cultured the strain anaerobically. Although E. coli contains an incomplete cobamide biosynthesis pathway, it possesses the cobUTSC genes and therefore can form a cobamide from the advanced intermediate cobinamide (Cbi) (19, 20). In a mutant lacking the metE gene, E. coli relies on the cobamide-dependent methionine synthase encoded by metH (21). An E. coli ∆metE strain containing the empty vector attached intracellular purines to exogenously supplied Cbi to produce two purinyl cobamides, 2-methyladeninylcobamide (Factor A) and adeninylcobamide (pseudo-B12 or pB12), as revealed by HPLC analysis of corrinoid extracts (Fig. 3A). These purinyl cobamides were also detected when the E. limosum cobT gene was expressed in an E. coli ∆metE ∆cobT mutant background, indicating that the cobT gene from the bza operon is functional (Fig. 3A). Notably, when the E. limosum bza operon was expressed in the E. coli ∆metE ∆cobT strain, a cobamide was formed with a retention time and UV-visible (UV-Vis) spectrum identical to that of a standard of cyanocobalamin (vitamin B12) (Fig. 3A and SI Appendix, Fig. S3). 1H-NMR analysis of the purified cobamide confirmed its identity as cyanocobalamin (SI Appendix, Fig. S3). Furthermore, this cobamide could support the growth of Salmonella enterica provided with ethanolamine as the sole nitrogen source when grown aerobically, a condition that has been shown to require exogenous B12 (Fig. 3B) (1, 22–24).
Fig. 3.
Characterization of the E. limosum bza operon in E. coli. (A) HPLC chromatograms of corrinoid extracts of E. coli strains expressing E. limosum genes. Peaks confirmed by LC-MS/MS are labeled; an unidentified peak is marked with an asterisk (*). Standards are shown in gray. All cobamides shown are in their cyanated forms. (B) The bza operon product supports cobalamin-dependent growth in S. enterica. Growth curves of S. enterica cultured with ethanolamine as the sole nitrogen source are shown with added cobamide product of E. coli expressing the E. limosum bza operon (Upper chromatogram in A, 15.8 min) or the indicated controls. Error bars represent SD of triplicate samples. (C) The bza operon directs de novo DMB biosynthesis. LC-MS/MS with MRM analysis is shown for a corrinoid extract of the E. coli metE cobT strain expressing the E. limosum bza operon grown in media containing 15NH4Cl.
To test whether the B12 detected above was the result of de novo biosynthesis of DMB, E. coli expressing the bza operon was grown in a medium containing 15N-labeled ammonium chloride as the sole nitrogen source. A corrinoid extract of this culture was analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) with multiple reaction monitoring (MRM). A peak that coelutes with standard cyanocobalamin and corresponding to [15N1, 15N3-DMB]-B12 was observed (top trace), whereas unlabeled B12 was not detected (middle trace), confirming that DMB is synthesized de novo in this E. coli strain (Fig. 3C). Together, these results demonstrate that expression of the E. limosum bza operon is sufficient for anaerobic DMB biosynthesis in E. coli.
Functional Analysis of the Individual bza Genes and DMB Biosynthesis Intermediates.
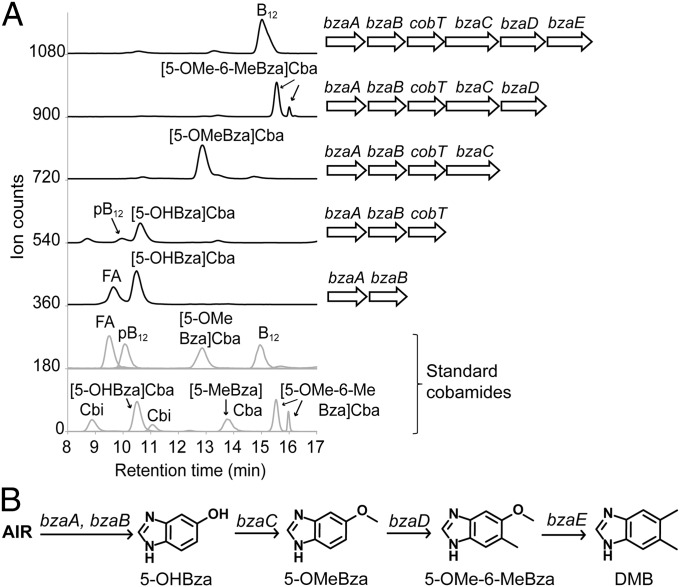
We next examined the functions of the individual bza genes by constructing a series of plasmids containing different subsets of the genes in the operon. These plasmids were introduced into the E. coli ∆metE ∆cobT or the E. coli ∆metE strain. LC-MS analysis of the corrinoids extracted from these strains showed that bzaA and bzaB were required together for the formation of the first intermediate, 5-hydroxybenzimidazole (5-OHBza), because strains expressing either gene alone were similar to a control strain containing the empty vector (Fig. 4 and SI Appendix, Fig. S4A). Expression of bzaC, which encodes a putative SAM-dependent methyltransferase, is required for methylation of this intermediate to form 5-methoxybenzimidazole (5-OMeBza) (Fig. 4 and SI Appendix, Fig. S4). The formation of the intermediate 5-methoxy-6-methylbenzimidazole (5-OMe-6-MeBza) requires bzaD, and the final step, formation of DMB, additionally requires bzaE (Fig. 4). These results demonstrate that each of the bza genes is necessary for DMB biosynthesis. When these strains were cultured under aerobic conditions, benzimidazole formation was significantly reduced, suggesting that one or more oxygen-sensitive steps are involved in this pathway (SI Appendix, Fig. S4B). However, when bzaC was expressed alone in the presence of 5-OHBza, a small amount of 5-OMeBza was formed under both anaerobic and aerobic conditions, indicating that this step is likely not oxygen-sensitive (SI Appendix, Fig. S4B). Together, these results define the roles of each bza gene and the major intermediates in the pathway leading to the formation of DMB.
Fig. 4.
Analysis of the functions of the individual E. limosum bza genes. (A) LC-MS/MS analysis of corrinoid extracts. E. coli strains expressing different subsets of the genes in the bza operon, as indicated on the Right, were grown as in Fig. 3A. The bzaA-bzaB plasmid was expressed in an E. coli ∆metE background and the remaining constructs in a ∆metE ∆cobT background. Standard cobamides are shown in the Lower two traces. (B) Model for the roles of the bza genes in DMB biosynthesis. The first intermediate, 5-OHBza, is synthesized from AIR (as shown below) by the bzaA and bzaB gene products. The bzaC gene product methylates 5-OHBza to form 5-OMeBza. Further methylation by the bzaD gene product converts 5-OMeBza to 5-OMe-6-MeBza, and bzaE acts next in the conversion of 5-OMe-6-MeBza to DMB.
AIR Is the Precursor of 5-OHBza, the First Intermediate in DMB Biosynthesis.
The results shown above demonstrate that 5-OHBza, the first benzimidazole intermediate, is formed from metabolites available in the E. coli cell. Given that (i) the imidazole ring of DMB shares a common metabolic origin with that of purines (2, 13), (ii) BzaA and BzaB are homologous to ThiC, and (iii) the ThiC enzyme uses the purine intermediate AIR as a substrate (8), we predicted that B12 would not be produced in an E. coli strain lacking AIR (Fig. 1B). Indeed, expression of the bza genes in an E. coli ∆metE ∆cobT strain lacking the AIR synthase encoded by the purM gene (25) did not lead to B12 production, suggesting that AIR is a required precursor of DMB (Fig. 5A). In contrast, deletion of purK, which functions in the conversion of AIR to the subsequent intermediate in purine biosynthesis, N5-carboxyaminoimidazole ribonucleotide (NCAIR), had no effect on B12 production (Fig. 5A) (26, 27). These results identify AIR as a precursor in the anaerobic B12 biosynthesis pathway. Thus, AIR is a branch point in the biosynthesis of purines, thiamin, and DMB (Fig. 1B).
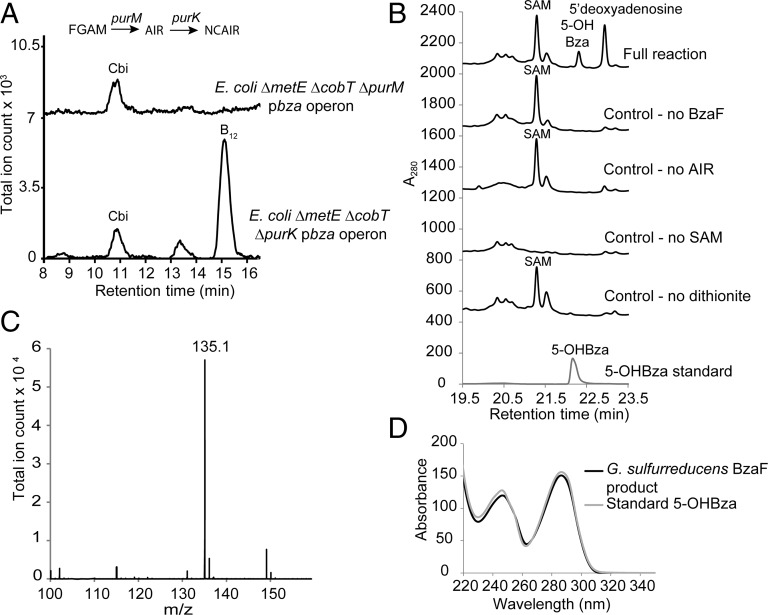
Fig. 5.
AIR is the direct precursor of 5-OHBza. (A) LC-MS/MS analysis of corrinoid extracts of purine biosynthesis mutants. The indicated E. coli strains were grown with Cbi and 15NH4Cl, and corrinoid extracts were analyzed for the presence of [15N1, 15N3-DMB]-B12 (doubly charged m/z of 679.2, product ion DMB m/z 149.1). (B) In vitro analysis of 5-OHBza biosynthesis. HPLC analysis of the in vitro reconstitution reaction of G. sulfurreducens BzaF with AIR, SAM, and dithionite shows the formation of a product that coelutes with a standard of 5-OHBza. Control reactions with one component omitted are also shown. (C) The extracted ion chromatogram of 5-OHBza (m/z 135.1) from the LC-MS analysis of the in vitro reconstitution reaction of G. sulfurreducens BzaF with AIR, SAM, and dithionite. (D) UV-Vis spectroscopy of the BzaF reaction product. The HPLC-purified G. sulfurreducens BzaF reaction product is overlaid with that of a standard sample of 5-OHBza.
In addition to AIR, threose or erythrose was proposed to be a precursor for benzimidazole biosynthesis in E. limosum and Clostridium barkeri (Fig. 1B) (12, 14). To identify the complete set of substrates required for the biosynthesis of 5-OHBza, we purified a homolog of BzaA and BzaB from Geobacter sulfurreducens and assayed its activity in vitro. In G. sulfurreducens and several other bacteria, a single thiC homolog (hereafter called bzaF) is located adjacent to cobamide biosynthesis genes (Fig. 6) and is found in a phylogenetic clade distinct from ThiC, BzaA, and BzaB (Fig. 2B). We hypothesized that this gene would also function in 5-OHBza biosynthesis. We found that, like ThiC, purified BzaF protein contains an iron–sulfur (Fe–S) cluster and uses SAM as a cofactor. Unexpectedly, when only SAM and AIR were provided to BzaF, a product with a retention time, mass, and UV-Vis spectrum identical to that of an authentic standard of 5-OHBza was observed (Fig. 5 B–D). A detailed mechanistic analysis of another 5-OHBza synthase (HBI synthase), a homolog of BzaF from Desulfuromonas acetoxidans, reveals interesting mechanistic parallels with ThiC (28). Thus, similar to thiamin pyrimidine biosynthesis, AIR is the sole substrate required for synthesizing 5-OHBza.
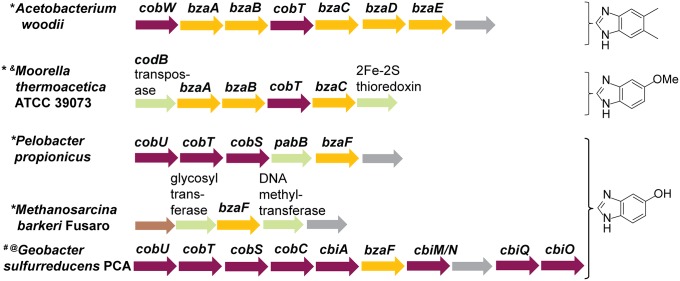
Fig. 6.
The bza genes present in an organism are predictive of the cobamide that it produces. Genetic loci of organisms identified from the JGI database that contain bza genes adjacent to cobamide biosynthesis or metabolism genes are shown (an extended list is presented in SI Appendix, Fig. S6). The benzimidazole lower ligand that these organisms are predicted to synthesize is shown on the Right. The predictions verified in previous studies are labeled with an asterisk (*). Those verified by analysis of corrinoids (#), biochemical activity (@), or heterologous expression (&) are indicated.
bza Genes Are Present in Diverse Bacteria and Archaea and Can Be Used to Predict Cobamide Structure.
Four other substituted benzimidazoles in addition to DMB have been reported as naturally occurring cobamide lower ligands in cultured anaerobic bacteria and archaea (SI Appendix, Fig. S5) (2). Our observation that three of these benzimidazoles are intermediates in the DMB biosynthetic pathway in E. limosum prompted us to examine whether homologs of the bza genes might be present in the genomes of other organisms that synthesize benzimidazolyl cobamides. Indeed, we found that the genomes of more than 25 bacteria and archaea contained putative bza operons (Fig. 6 and SI Appendix, Fig. S6). These organisms are phylogenetically diverse and are found in a variety of different environments. In the case of Acetobacterium woodii, which is reported to produce B12, homologs of all of the bza genes are present and are arranged similarly to the E. limosum bza operon, suggesting that these genes also function in DMB production (Fig. 6) (29). Several other genomes were found in which a subset of the bza genes was present. In each of the organisms previously shown to produce a benzimidazolyl cobamide, the set of bza genes present in the genome was predictive of the structure of the cobamide produced (SI Appendix, Fig. S6). We further tested the ability of genome analysis to predict cobamide production by analyzing the cobamides synthesized by G. sulfurreducens. This bacterium has been shown to produce a cobamide, but the structure has not been determined (30). Because the genome of G. sulfurreducens contains bzaF but no other bza gene homologs, we expected it to produce [5-OHBza]Cba. Indeed, LC-MS analysis of a corrinoid extract from a G. sulfurreducens culture showed the production of [5-OHBza]Cba as the major corrinoid species (SI Appendix, Fig. S7). In addition, [5-OMeBza]Cba, the cobamide previously reported to be produced by Moorella thermoacetica, was formed in an E. coli strain expressing the M. thermoacetica bza operon, which contains homologs of bzaA, bzaB, cobT, and bzaC (SI Appendix, Fig. S8) (31). Thus, we have identified not only the complete set of genes required for DMB biosynthesis in E. limosum and other B12-producing anaerobes, but also those required for the biosynthesis of other cobamide-associated benzimidazoles.
Discussion
Although the B12 biosynthetic pathway has been studied for several decades, resulting in the identification of over 40 genes involved in the anaerobic and/or aerobic biosynthesis pathways, the genes required for the anaerobic biosynthesis of the lower ligand remained elusive until now. With the present discovery, we have uncovered the last unknown component of the biosynthetic pathway for B12.
Despite the lack of phylogenetic or enzymatic commonalities between the anaerobic and aerobic routes for DMB production, some parallels exist between the two pathways. First, both the bza genes and bluB are commonly found downstream of cobalamin riboswitches and clustered with B12 biosynthesis genes in many bacterial genomes (3, 17) (SI Appendix, Fig. S6), suggesting a common mode of regulation of lower ligand biosynthesis and attachment. Second, both BzaA/B/F and BluB use the same substrate and cofactors as closely related enzymes—ThiC and nitroreductases, respectively—yet carry out a novel reaction (4, 8). It is interesting to note that, unlike the other components of the cobamide biosynthetic pathway, DMB biosynthesis has apparently evolved independently twice, underscoring the importance of DMB as a lower ligand. It remains unclear why some organisms synthesize benzimidazoles exclusively for use as cobamide lower ligands, given that lower ligands of the purine and phenolic classes are derived from other metabolic pathways.
The discovery of the anaerobic bza genes and analysis of their individual roles sheds light on several previously unresolved aspects of anaerobic benzimidazole biosynthesis. First, prior feeding studies with labeled substrates in E. limosum suggested that this pathway branches from the purine biosynthetic pathway; the present study pinpoints AIR as the direct precursor for 5-OHBza. Surprisingly, four carbon atoms from the ribose moiety of AIR are incorporated into the benzene ring component of DMB. Prior labeling studies implicated erythrose and threose as precursors of these carbon atoms, suggesting that the ribose moiety of AIR can be formed from these four-carbon sugars in E. limosum (12, 14). Second, our work agrees with feeding studies with 13C-labeled benzimidazoles that implicated 5-OHBza as the precursor of all other benzimidazoles in E. limosum and M. thermoacetica (13, 32). However, we also show that 5-OMeBza and 5-OMe-6-MeBza, which were previously suggested to be side products of the pathway, are intermediates in DMB biosynthesis (13). A third insight from those studies was that DMB, although it is a symmetric molecule, is attached to the nucleotide loop exclusively via the nitrogen atom derived from glutamine (Fig. 1B) (10–12, 14, 15). This could mean either that the bond of this nitrogen to the ribose ring was formed on an asymmetric precursor or that a close association between enzymes in the pathway prevents the release of free DMB before phosphoribosylation by CobT. Because of the proximity of the cobT and bza genes in numerous genomes (SI Appendix, Fig. S6), we speculate that the phosphoribosylation step could be coupled to the biosynthesis of DMB or one of its precursors. Curiously, both BzaD and BzaE have putative B12-binding domains, suggesting that the final two methylation steps could occur on a cobamide rather than a free base. A detailed mechanistic analysis of each of these enzymes will be required to investigate this further.
Because DMB and other substituted benzimidazoles share a common biosynthetic pathway, our findings enable the improved prediction of cobamide biosynthesis based on analysis of genome sequence. This is especially useful considering the increasing abundance of microbial genome and metagenome sequence data, coupled with persistent challenges associated with studying most microbes in pure culture, as well as the growing recognition of the role of microbes in human health and the environment. Given the importance of cobamides in environmental, industrial, and human-associated microbes, the identification of this pathway may lead to an enhanced ability to understand and manipulate microbial metabolism.
Materials and Methods
Materials.
Cyanocobalamin (vitamin B12), dicyanocobinamide (Cbi), DMB, and 5-OMeBza were purchased from Sigma-Aldrich. 15N-ammonium chloride was purchased from Cambridge Isotopes Laboratories. 5-OHBza was synthesized as previously described (33). [5-OHBza]Cba, [5-OMeBza]Cba, [5-OMe-6-MeBza]Cba, pseudo-B12, and Factor A were purified as described (34).
Bioinformatic Analysis.
Identification of cobalamin riboswitch-regulated operons:
Nucleotide sequences of genomes of interest were searched to identify cobalamin riboswitches using Infernal v1.1 (35). A covariance model was built using the RF00174 seed alignment downloaded from Rfam (36) and Infernal’s cmbuild function. After calibrating the covariance model with the cmcalibrate function, genomes were searched with the cmsearch function. Positive hits were those satisfying the default Infernal inclusion threshold of an E-value of 0.01 or less. The chromosomal locations of the putative cobalamin riboswitches were visualized and analyzed using the Joint Genome Institute’s Integrated Microbial Genomes (IMG) web service (37).
Identification of thiC homologs:
All archaeal and bacterial genomes of “Finished” status in IMG’s system as of January 2, 2015, were searched. The function “pfam01964” was used to carry out a “Function Profile” against all finished genomes. Genomes with multiple thiC homologs were prioritized for examination of genomic context of thiC homologs. thiC homologs were screened to identify those that were contained within the same operon as cobalamin biosynthesis or transport genes.
Phylogenetic analysis of thiC homologs:
The sequences of all thiC homologs from genomes that had multiple thiC homologs were downloaded from IMG and aligned to Pfam’s PF01946 seed alignment using the “seed” option in Multiple Alignment using Fast Fourier Transform (MAFFT) v7.2 (38). The multiple sequence alignment was edited in Geneious v5.5 (39) to remove ambiguously aligned amino acids and large end gaps. After this editing step, sequences whose amino acid count was less than 60% of the length of the entire multiple sequence alignment were removed from the analysis. Seed sequences that were duplicates of sequences downloaded from IMG were also removed before phylogenetic analysis. The alignment was uploaded to the Cyberinfrastructure for Phylogenetic Research (CIPRES) web portal, and a maximum-likelihood (ML) tree was reconstructed using RAxML v8.1 (40). The amino acid substitution matrix used was the JTT matrix (41), with the Γ model of rate heterogeneity. RAxML’s rapid bootstrap was performed with 1,000 replicates, and the best-scoring ML tree was saved.
Genetic and Molecular Techniques.
E. coli strains ΔcobT::KanR (JW1969-1), ΔmetE::KnR (JW3805-1), ΔpurM::KanR (JW2484-1), and ΔpurK::KanR (JW0511-1) were obtained from the Keio collection (42). The mutations were introduced into E. coli strain MG1655 by P1 transduction (43). For construction of double and triple mutants, the KanR cassette was removed by recombination of flanking FRT sites as described (44). Double and triple mutants were constructed by P1 transduction of the KanR cassette into deletion strains.
The E. limosum and M. thermoacetica bza operons were cloned into plasmid pTH1227 (45) by Gibson assembly (46) following PCR on genomic DNA with primers P604F/P619R (E. limosum) and P615F/P616R (M. thermoacetica). Additional constructs were created similarly with the primers in SI Appendix, Table S2. Strains and plasmids are in SI Appendix, Table S3.
The nucleotide sequence of G. sulfurreducens bzaF was synthesized and cloned into plasmid THT, a pET28 vector with a tobacco etch virus protease cleavage site for removing the His-tag.
Media and Culture Conditions.
E. coli strains containing pTH1227 or its derivatives were grown at 37 °C under anaerobic conditions with 95–98% N2 and 5–2% H2 as headspace gas for 72–144 h in M9 glucose minimal medium supplemented with 100 μM isopropyl β-d-1-thiogalactopyranoside, 50 μM freshly prepared FeSO4, 10 nM Cbi, and 25 mg/L tetracycline, unless otherwise indicated. Strains containing mutations in purK or purM were grown with 50 nM Cbi, 20 μM inosine, and 20 μM thiamin (unexpectedly, both strains could grow in the absence of added thiamin, although purM strains are expected to require thiamin). When indicated, 15NH4Cl was substituted for unlabeled NH4Cl. For growth of E. coli under aerobic conditions, cultures were aerated at 250 rpm for 36–96 h. For corrinoid extractions, the strains were grown in 2-L cultures.
S. enterica serovar Typhimurium strain LT2, G. sulfurreducens PCA, E. limosum ATCC 10825, and M. thermoacetica ATCC 39073 were cultivated as described in SI Appendix, SI Methods.
Corrinoid Extraction and HPLC Analysis.
Corrinoids were extracted from cell pellets, purified, and analyzed as described in SI Appendix, SI Methods.
Chemical Analysis.
The UV-Vis spectra were collected during HPLC analysis using Agilent ChemStation software. Samples were further characterized on an Agilent 6410 liquid chromatograph-triple quadrupole mass spectrometer using an Agilent Zorbax SB-Aq column to identify cobamides based on their characteristic transitions and retention times (34). For corrinoid extracts from strains grown in 15NH4Cl, the MS method was modified to detect the following precursor and product ion transitions, respectively: 679.3, 149.1 (B12); 673.3, 137.1 ([5-OHBza]Cba); 680.3, 151.1 ([5-OMeBza]Cba); 687.3, 165.1 ([5-OMe-6-MeBza]Cba); 675.3, 141.1 (pseudo-B12); 682.3, 155.1 (Factor A). For NMR analysis, cobamides were purified by HPLC using an Agilent 1200 series fraction collector. A standard sample of cyanocobalamin and a blank buffer sample were also collected under the same HPLC purification conditions. These samples were analyzed using a Bruker Biospin Avance II 900 MHz NMR spectrometer equipped with a TXI cryoprobe accessory at the California Institute for Quantitative Biosciences Berkeley core facility.
Biochemical Characterization of G. sulfurreducens BzaF.
His-tagged G. sulfurreducens PCA BzaF was expressed and purified as described in SI Appendix, SI Methods.
Enzymatic Assays for LC-MS and HPLC Purification of 5-OHBza.
Purified G. sulfurreducens BzaF (120 µM) was incubated with dithionite (10 mM), AIR (3 mM), and SAM (5 mM) at room temperature for 60 min. Controls included reactions lacking either AIR, SAM, dithionite, or the enzyme. The protein was removed with 10-kDa cutoff filters, and the filtrate was analyzed by HPLC and LC-MS.
Supplementary Material
Acknowledgments
We thank S. Yi and J. Pelton for assistance with chemical analysis; M. Mehta for assistance with culturing G. sulfurreducens; J. Roth, S. Ragsdale, and D. Lovley for bacterial strains; and members of the Taga laboratory for critical reading of the manuscript. This work was supported by National Science Foundation Grant MCB1122046 and a Hellman Family Faculty Fund award (to M.E.T.) and by Robert A. Welch Foundation Grant A-0034 (to T.P.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. KT347435 (E. limosum ATCC 10825 bza operon) and KT347436 (M. thermoacetica ATCC 39073 bza operon)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509132112/-/DCSupplemental.
References
- 1.Roth JR, Lawrence JG, Bobik TA. Cobalamin (coenzyme B12): Synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 2.Renz P. Biosynthesis of the 5,6-dimethylbenzimidazole moiety of cobalamin and of the other bases found in natural corrinoids. In: Banerjee R, editor. Chemistry and Biochemistry of B12. John Wiley & Sons, Inc.; New York: 1999. pp. 557–575. [Google Scholar]
- 3.Campbell GRO, et al. Sinorhizobium meliloti bluB is necessary for production of 5,6-dimethylbenzimidazole, the lower ligand of B12. Proc Natl Acad Sci USA. 2006;103(12):4634–4639. doi: 10.1073/pnas.0509384103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taga ME, Larsen NA, Howard-Jones AR, Walsh CT, Walker GC. BluB cannibalizes flavin to form the lower ligand of vitamin B12. Nature. 2007;446(7134):449–453. doi: 10.1038/nature05611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins HF, et al. Bacillus megaterium has both a functional BluB protein required for DMB synthesis and a related flavoprotein that forms a stable radical species. PLoS One. 2013;8(2):e55708. doi: 10.1371/journal.pone.0055708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray MJ, Escalante-Semerena JC. Single-enzyme conversion of FMNH2 to 5,6-dimethylbenzimidazole, the lower ligand of B12. Proc Natl Acad Sci USA. 2007;104(8):2921–2926. doi: 10.1073/pnas.0609270104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warren MJ. Finding the final pieces of the vitamin B12 biosynthetic jigsaw. Proc Natl Acad Sci USA. 2006;103(13):4799–4800. doi: 10.1073/pnas.0601030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee A, et al. Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily. Nat Chem Biol. 2008;4(12):758–765. doi: 10.1038/nchembio.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Höllriegl V, Lamm L, Rowold J, Hörig J, Renz P. Biosynthesis of vitamin B12. Different pathways in some aerobic and anaerobic microorganisms. Arch Microbiol. 1982;132(2):155–158. doi: 10.1007/BF00508722. [DOI] [PubMed] [Google Scholar]
- 10.Lamm L, Heckmann G, Renz P. Biosynthesis of vitamin B12 in anaerobic bacteria. Mode of incorporation of glycine into the 5,6-dimethylbenzimidazole moiety in Eubacterium limosum. Eur J Biochem. 1982;122(3):569–571. [PubMed] [Google Scholar]
- 11.Lamm L, Hörig JA, Renz P, Heckmann G. Biosynthesis of vitamin B12. Experiments with the anaerobe Eubacterium limosum and some labelled substrates. Eur J Biochem. 1980;109(1):115–118. doi: 10.1111/j.1432-1033.1980.tb04775.x. [DOI] [PubMed] [Google Scholar]
- 12.Munder M, Vogt JR, Vogler B, Renz P. Biosynthesis of vitamin B12 in anaerobic bacteria. Experiments with Eubacterium limosum on the incorporation of D-[1-13C]erythrose and [13C]formate into the 5,6-dimethylbenzimidazole moiety. Eur J Biochem. 1992;204(2):679–683. doi: 10.1111/j.1432-1033.1992.tb16681.x. [DOI] [PubMed] [Google Scholar]
- 13.Renz P, Endres B, Kurz B, Marquart J. Biosynthesis of vitamin B12 in anaerobic bacteria. Transformation of 5-hydroxybenzimidazole and 5-hydroxy-6-methylbenzimidazole into 5,6-dimethylbenzimidazole in Eubacterium limosum. Eur J Biochem. 1993;217(3):1117–1121. doi: 10.1111/j.1432-1033.1993.tb18344.x. [DOI] [PubMed] [Google Scholar]
- 14.Vogt JR, Lamm-Kolonko L, Renz P. Biosynthesis of vitamin B-12 in anaerobic bacteria. Experiments with Eubacterium limosum and D-erythrose 14C-labeled in different positions. Eur J Biochem. 1988;174(4):637–640. doi: 10.1111/j.1432-1033.1988.tb14145.x. [DOI] [PubMed] [Google Scholar]
- 15.Vogt JR, Renz P. Biosynthesis of vitamin B-12 in anaerobic bacteria. Experiments with Eubacterium limosum on the origin of the amide groups of the corrin ring and of N-3 of the 5,6-dimethylbenzimidazole part. Eur J Biochem. 1988;171(3):655–659. doi: 10.1111/j.1432-1033.1988.tb13836.x. [DOI] [PubMed] [Google Scholar]
- 16.Roh H, et al. Complete genome sequence of a carbon monoxide-utilizing acetogen, Eubacterium limosum KIST612. J Bacteriol. 2011;193(1):307–308. doi: 10.1128/JB.01217-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem. 2003;278(42):41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- 18.Trzebiatowski JR, Escalante-Semerena JC. Purification and characterization of CobT, the nicotinate-mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase enzyme from Salmonella typhimurium LT2. J Biol Chem. 1997;272(28):17662–17667. doi: 10.1074/jbc.272.28.17662. [DOI] [PubMed] [Google Scholar]
- 19.Escalante-Semerena JC. Conversion of cobinamide into adenosylcobamide in bacteria and archaea. J Bacteriol. 2007;189(13):4555–4560. doi: 10.1128/JB.00503-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence JG, Roth JR. The cobalamin (coenzyme B12) biosynthetic genes of Escherichia coli. J Bacteriol. 1995;177(22):6371–6380. doi: 10.1128/jb.177.22.6371-6380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerjee RV, Johnston NL, Sobeski JK, Datta P, Matthews RG. Cloning and sequence analysis of the Escherichia coli metH gene encoding cobalamin-dependent methionine synthase and isolation of a tryptic fragment containing the cobalamin-binding domain. J Biol Chem. 1989;264(23):13888–13895. [PubMed] [Google Scholar]
- 22.Anderson PJ, et al. One pathway can incorporate either adenine or dimethylbenzimidazole as an alpha-axial ligand of B12 cofactors in Salmonella enterica. J Bacteriol. 2008;190(4):1160–1171. doi: 10.1128/JB.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeter RM. Cobalamin-dependent 1,2-propanediol utilization by Salmonella typhimurium. J Gen Microbiol. 1990;136(5):887–896. doi: 10.1099/00221287-136-5-887. [DOI] [PubMed] [Google Scholar]
- 24.Roof DM, Roth JR. Ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1988;170(9):3855–3863. doi: 10.1128/jb.170.9.3855-3863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith JM, Daum HA., 3rd Nucleotide sequence of the purM gene encoding 5′-phosphoribosyl-5-aminoimidazole synthetase of Escherichia coli K12. J Biol Chem. 1986;261(23):10632–10636. [PubMed] [Google Scholar]
- 26.Tiedeman AA, et al. Nucleotide sequence analysis of the purEK operon encoding 5′-phosphoribosyl-5-aminoimidazole carboxylase of Escherichia coli K-12. J Bacteriol. 1989;171(1):205–212. doi: 10.1128/jb.171.1.205-212.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Morar M, Ealick SE. Structural biology of the purine biosynthetic pathway. Cell Mol Life Sci. 2008;65(23):3699–3724. doi: 10.1007/s00018-008-8295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehta AP, et al. (in press) Anaerobic 5-hydroxylbenzimidazole formation from aminoimidazole ribotide: An unanticipated intersection of thiamin and vitamin B12 biosynthesis. [DOI] [PMC free article] [PubMed]
- 29.Stupperich E, Eisinger HJ, Kräutler B. Diversity of corrinoids in acetogenic bacteria. P-cresolylcobamide from Sporomusa ovata, 5-methoxy-6-methylbenzimidazolylcobamide from Clostridium formicoaceticum and vitamin B12 from Acetobacterium woodii. Eur J Biochem. 1988;172(2):459–464. doi: 10.1111/j.1432-1033.1988.tb13910.x. [DOI] [PubMed] [Google Scholar]
- 30.Yan J, Ritalahti KM, Wagner DD, Löffler FE. Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to organohalide-respiring Dehalococcoides mccartyi strains. Appl Environ Microbiol. 2012;78(18):6630–6636. doi: 10.1128/AEM.01535-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irion E, Ljungdahl L. Isolation of factor 3m coenzyme and cobyric acid coenzyme plus other B12 factors from Clostridium thermoaceticum. Biochemistry. 1965;4(12):2780–2790. doi: 10.1021/bi00888a031. [DOI] [PubMed] [Google Scholar]
- 32.Wurm R, Weyhenmeyer R, Renz P. On the biosynthesis of 5-methoxybenzimidazole. Precursor-function of 5-hydroxybenzimidazole, benzimidazole and riboflavin. Eur J Biochem. 1975;56(2):427–432. doi: 10.1111/j.1432-1033.1975.tb02249.x. [DOI] [PubMed] [Google Scholar]
- 33.Crofts TS, Seth EC, Hazra AB, Taga ME. Cobamide structure depends on both lower ligand availability and CobT substrate specificity. Chem Biol. 2013;20(10):1265–1274. doi: 10.1016/j.chembiol.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Men Y, et al. Identification of specific corrinoids reveals corrinoid modification in dechlorinating microbial communities. Environ Microbiol. 2014 doi: 10.1111/1462-2920.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29(22):2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nawrocki EP, et al. Rfam 12.0: Updates to the RNA families database. Nucleic Acids Res. 2015;43(Database issue):D130–D137. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markowitz VM, et al. IMG: The Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 2012;40(Database issue):D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 42.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silhavy TJ, Berman ML, Enquist LW. Experiments with Gene Fusions. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1984. [Google Scholar]
- 44.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng J, Sibley CD, Zaheer R, Finan TM. A Sinorhizobium meliloti minE mutant has an altered morphology and exhibits defects in legume symbiosis. Microbiology. 2007;153(Pt 2):375–387. doi: 10.1099/mic.0.2006/001362-0. [DOI] [PubMed] [Google Scholar]
- 46.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.