Significance
Increasing demand for recombinant proteins leads to continuous attempts for improving the protein secretion capacity of host cells. In this study, we show that by combining high-throughput microfluidic screening with whole-genome sequencing of the selected clones from yeast libraries we can identify and map the mutations associated with significantly improved protein production. These identified mutations can be used as reverse metabolic engineering target genes in design of efficient cell factories for protein secretion. The mutations that we identified will also help in improving our understanding of the protein secretory mechanisms in yeast.
Keywords: protein secretion, yeast cell factories, droplet microfluidics, random mutagenesis, systems biology
Abstract
There is an increasing demand for biotech-based production of recombinant proteins for use as pharmaceuticals in the food and feed industry and in industrial applications. Yeast Saccharomyces cerevisiae is among preferred cell factories for recombinant protein production, and there is increasing interest in improving its protein secretion capacity. Due to the complexity of the secretory machinery in eukaryotic cells, it is difficult to apply rational engineering for construction of improved strains. Here we used high-throughput microfluidics for the screening of yeast libraries, generated by UV mutagenesis. Several screening and sorting rounds resulted in the selection of eight yeast clones with significantly improved secretion of recombinant α-amylase. Efficient secretion was genetically stable in the selected clones. We performed whole-genome sequencing of the eight clones and identified 330 mutations in total. Gene ontology analysis of mutated genes revealed many biological processes, including some that have not been identified before in the context of protein secretion. Mutated genes identified in this study can be potentially used for reverse metabolic engineering, with the objective to construct efficient cell factories for protein secretion. The combined use of microfluidics screening and whole-genome sequencing to map the mutations associated with the improved phenotype can easily be adapted for other products and cell types to identify novel engineering targets, and this approach could broadly facilitate design of novel cell factories.
The production of recombinant proteins by cell factories, including biopharmaceutical proteins and industrial enzymes, is a growing multibillion-dollar industry (1) and demands continuous improvement of the chosen cell factories. The improvements involve optimization of transcription and translation, but also of protein posttranslational modifications, folding, and trafficking. One of the remaining challenges is the rational engineering design for the optimization of the protein secretory capacity, which involves a complex secretory network (2). The complexity of the protein secretory machinery and lack of a complete understanding of its underlying mechanisms has limited the utility of rational metabolic engineering for the improvement of recombinant protein production (3). Designing an efficient cell platform for protein secretion may often require overcoming limitations at different levels, e.g., translation, protein folding, and protein trafficking, and the modification of a single metabolic engineering target may therefore be insufficient (4). Although adaptive laboratory evolution has proven a useful strategy to acquire desired phenotypes with accumulation of beneficial mutations under selective pressure (5), this approach can be more cumbersome when trying to select clones with improved protein secretion.
Alternatively, a cell library can be constructed by introducing random mutations, with the aim that some of them will give rise to clones with the improved desired phenotype. This approach requires a high-throughput screening method that allows for efficient sorting of selected clones. Conventional screening methods (e.g., screens in a 96-well plate) are low-throughput, and fluorescence-activated cell sorting is not optimal because the desired “signal” is not intracellular, but secreted, making it more difficult to optimize this approach.
Recently, droplet microfluidics-based cell sorting has emerged as a novel and powerful technology for high-throughput screening (6, 7). By generating monodisperse aqueous droplets surrounded by an immiscible oil phase, this technique allows the cell and its secreted product to be contained in isolated droplets and has been applied for enzyme direct evolution and measurement of extracellular metabolite production or consumption (8, 9). Here, we report genome-scale mapping of mutations in yeast cells selected for enhanced protein secretion by combining high-throughput droplet microfluidics screening with whole-genome sequencing of several isolated strains. The mutations identified in this study not only expand more reverse metabolic engineering targets for increasing protein secretion, but also could lead to a better understanding of the protein secretory mechanisms in yeast.
Results and Discussion
Microfluidic Screening of Yeast Mutant Libraries.
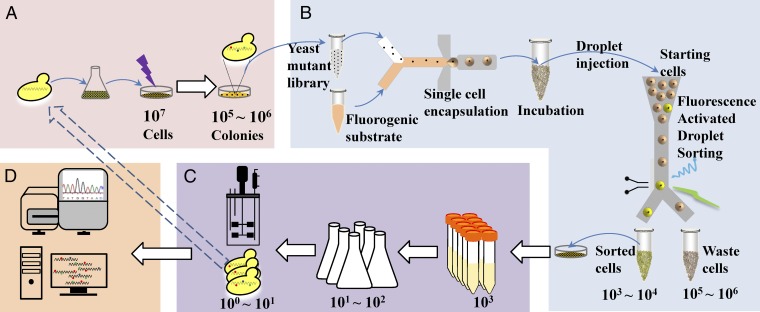
As a starting strain we used the Saccharomyces cerevisiae strain AAC where α-amylase is expressed using the POT1 plasmid system CPOTud (10). A cell culture containing 107 yeast cells was spread on a starch agar plate that was subsequently irradiated with λ = 254 nm, 4–16 mJ/cm2, giving rise to a library of cell clones (Fig. 1A). UV irradiation increased the number of random mutations, and some of the mutations could contribute to improved protein secretion. To capture the clones with increased α-amylase secretion, a droplet microfluidic system was established to encapsulate single cells and distinguish droplets based on fluorescent signals (Fig. 1B). Cells from the library were washed and resuspended in fresh medium and mixed with a fluorogenic α-amylase substrate and then encapsulated in droplets surrounded by oil. We have previously found that the fluorescence intensity of the droplets, which reflected the degradation of the fluorogenic substrate, was correlated with the α-amylase concentration (SI Appendix, Fig. S1) (11). Droplets were incubated and then injected into the sorting device. By sorting droplets with a fluorescence intensity beyond a defined threshold, higher amylase-producing mutants could be selected from a population of 105 cells within a few hours (11).
Fig. 1.
Schematic workflow for microfluidics droplet screening of S. cerevisiae for higher α-amylase secretion. (A) Mutant library generation by UV irradiation. (B) High-throughput screening for improved strains by droplet microfluidic sorting. (C) Validation of α-amylase production capacity of sorted strains. (D) Genomic DNA sequencing and analysis of the selected strains.
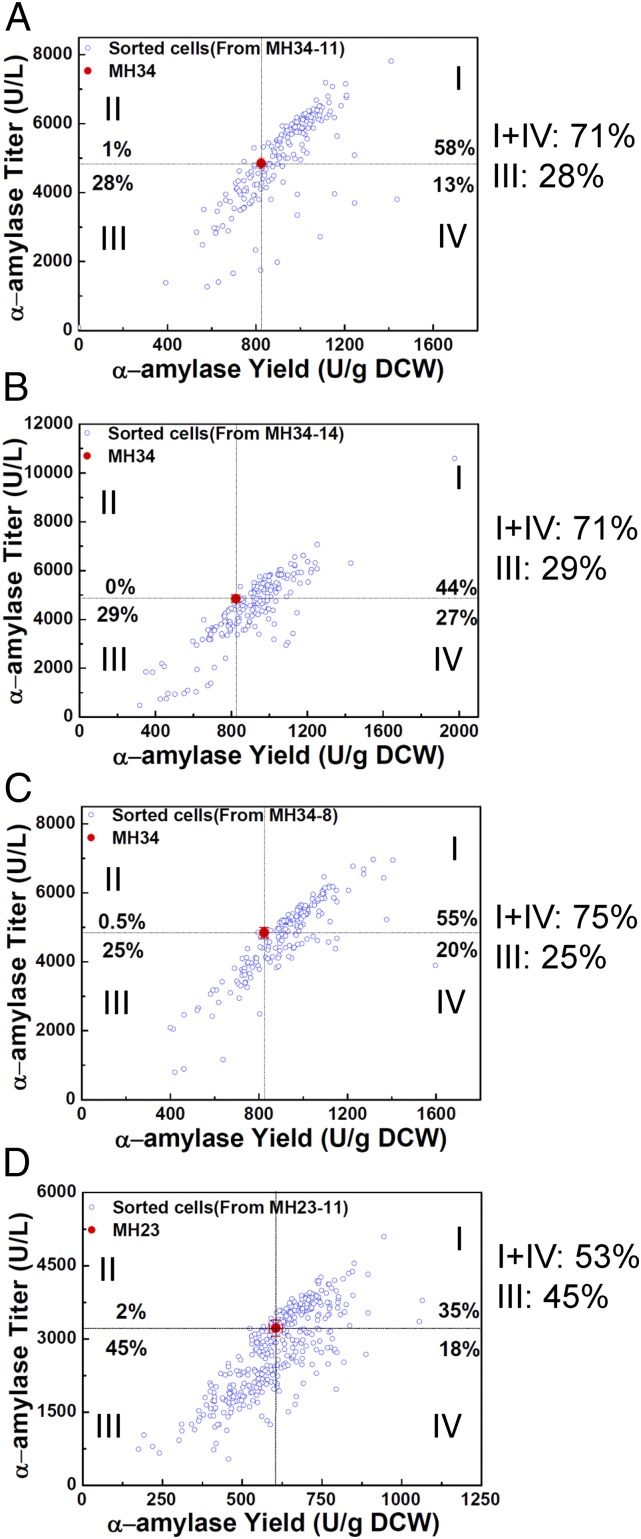
The α-amylase secretion capacity of the sorted mutants was validated by measuring α-amylase activity in the supernatant from fermentation cultures (Fig. 1C). Clones with improved α-amylase secretion selected in the first round of droplet sorting were used to generate four new mutant libraries; each new library was then used for a second round of sorting. The amylase production of 970 sorted cells from the four different libraries was validated through a secondary screen in tube fermentations. In all four libraries, droplet microfluidic screening provided an enrichment of strains with improved α-amylase production (Fig. 2). Over 70% of cells sorted from the libraries MH34-11, MH34-14, and MH34-8 achieved improved α-amylase yield compared with the reference strain MH34 (Fig. 2 A–C). However, only 53% of cells sorted from the library MH23-11 showed a yield increase comparable to the reference strain MH23, probably due to the higher fraction of sorted cells from this library compared with the other three libraries (3% vs. 1%) (SI Appendix, Fig. S2). As a control we also collected the cells discarded by microfluidic selection. The amylase secretion capacity of the discarded cell population was lower than that of the sorted cell population (SI Appendix, Fig. S3). Fifty discarded cells from the library MH34-11 were further tested in tube fermentations (SI Appendix, Fig. S4A), and a lower fraction of mutants showed α-amylase production improvement compared with the sorted cells. The average amylase titer and yield of the discarded cells was also lower than that of sorted cells from the same library (SI Appendix, Fig. S4B). This shows that the droplet microfluidic sorting system is reliable for high-throughput screening of yeast libraries for improved α-amylase secretion mutants. Of the 970 sorted cells analyzed in the secondary screening, we identified 122 mutants with at least 30% increased yield in tube fermentations (SI Appendix, Fig. S5). Twenty-nine of these mutants were selected for further verification of α-amylase secretion capacity in shake-flask fermentations, and 25 showed improved α-amylase secretion also under those conditions (SI Appendix, Fig. S6).
Fig. 2.
α-Amylase yields and titers of sorted cells tested in tube fermentations. (A) A total of 194 colonies from the library MH34-11, which was generated by UV irradiation of the mutant MH34 from the first round of sorting with a dose of 11 mJ⋅cm−2. (B) A total of 194 colonies from the library MH34-14, which was generated with a UV dose of 14 mJ⋅cm−2 on MH34. (C) A total of 194 colonies from the library MH34-8, which was generated with a UV dose of 8 mJ⋅cm−2 on MH34. (D) A total of 388 colonies from the library MH23-11, which was generated by UV irradiation of the mutant MH23 from the first round of sorting with a dose of 11 mJ⋅cm−2. I: Both titer and yield increase. II: Titer increase and yield decrease. III: Titer decrease and yield decrease. IV: Titer decrease and yield increase.
Characterization of Selected Mutants.
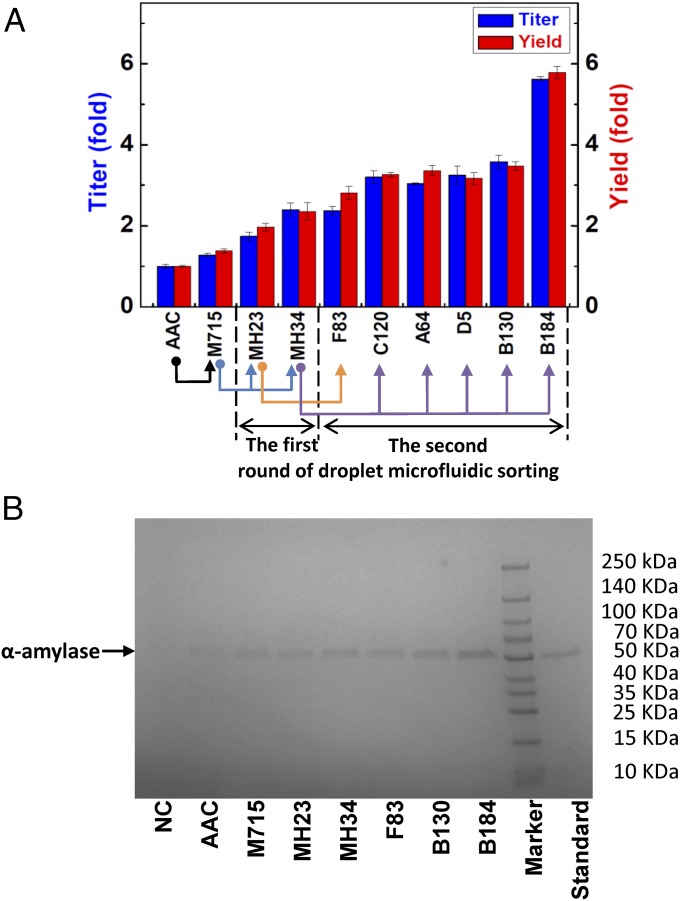
Following this final testing in shake flasks, the eight strains with the highest α-amylase production were chosen for further analysis. Of these, seven clones had more than a twofold improvement in α-amylase production compared with the initial strain AAC (Fig. 3A). The supernatants from cultures of these strains were analyzed by SDS/PAGE, which confirmed the improved secretion of amylase by the mutants (Fig. 3B). The best clone B184 had a nearly sixfold improvement in α-amylase production. In our previous work the mutant that was selected manually, M715 (12), had only about a 1.5-fold improvement in SD-2×SCAA medium (an optimized medium for protein production) compared with the wild-type strain AAC. This shows a clear potential of high-throughput screening from large libraries for selecting cell factories with improved desired phenotype.
Fig. 3.
α-Amylase production capacity of selected mutant strains. (A) α-Amylase production enhancement; the original strain (AAC) is as control. (B) SDS/PAGE analysis of the supernatant of selected mutant strains after fermentation. NC: α-amylase nonproducing strain. Standard: α-amylase standard. In all cases, the same amount (18 μL) of supernatant was loaded on the gel.
Because α-amylase was expressed using a plasmid expression system, the improvements could be associated with mutations in the plasmid. To identify whether the improvements in α-amylase production were associated with mutations in the plasmid or in the yeast genome, two additional experiments were carried out. First, plasmids were extracted from different mutant strains, and the restriction analysis did not show any mutations or differences among them (SI Appendix, Fig. S7). Second, the isolated plasmids were transformed into the wild-type strain CEN.PK 530.1C, and the production capacity of α-amylase was found to be almost identical for all strains harboring the different isolated plasmids (SI Appendix, Fig. S8), which indicates that the increase in amylase production was attributed to chromosomal mutations. Invertase is a protein endogenously secreted by S. cerevisiae, and the increased invertase activity of the best sorted mutant strains (compared with that of the original strain AAC) also indicated that the clones had improved protein secretion capacity associated with chromosomal mutations (SI Appendix, Fig. S9).
To examine the genetic stability of the isolated mutant strains, the three best mutant strains (B184, B130, and F83) were subcultured for 30 generations in a nonselective medium to eliminate the amylase expression plasmids. Loss of plasmid was confirmed by checking for inability to use starch from starch agar plates (lack of growth and lack of a halo on the agar plate). The plasmid was then transformed back into these mutant strains (SI Appendix, Fig. S10A) and fermentations were performed. The results showed that there were no differences in amylase production between the retransformed strains and the corresponding original mutant strains (SI Appendix, Fig. S10B). Thus, the mutant strains isolated for improved protein secretory capacity are genetically stable and can therefore be used for industrial applications as well as platform strains for production of other proteins.
Whole-Genome Sequencing and Data Analysis.
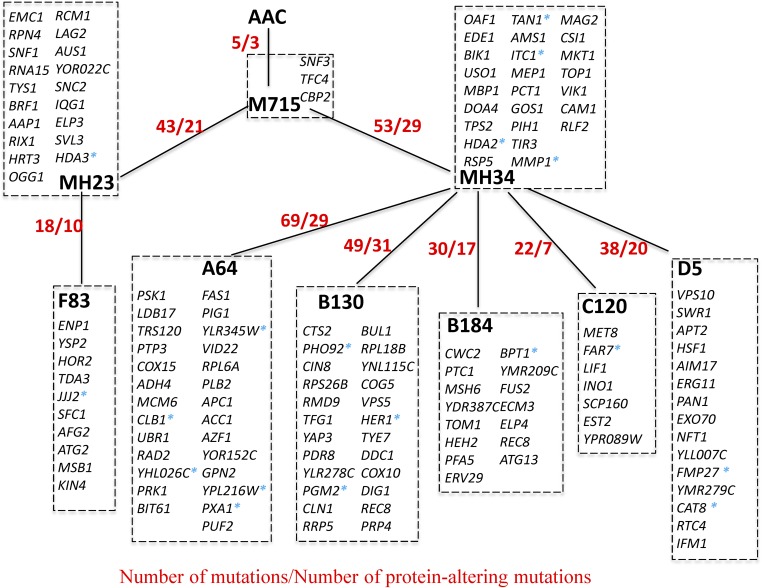
The genomes of the improved α-amylase secreting mutants together with the original parental strain AAC were sequenced to identify mutations that may contribute to the improved α-amylase secretion. Structural chromosomal variations of the mutant strains are listed in SI Appendix, Table S1. Chromosome III duplication was found in strain MH34 and its descendants; all genes on Chromosome III are listed in SI Appendix, Table S2. Chromosome duplication may cause changes in function and alter protein interaction networks and is an important process in evolution (13, 14). In addition to this large rearrangement, 330 point mutations were identified (SI Appendix, Table S3). When the single nucleotide substitutions are classified into six possible categories, the dominant mutations, accounting for 85% of all point mutations, are the C-to-T substitution and the G-to-A substitution (SI Appendix, Fig. S11). Among the 330 point mutations, 248 mutations were in coding regions, affecting 146 protein-encoding genes. Of 248 mutations in the ORFs, 79 were same-sense, 151 were missense, and 18 were nonsense mutations. Based on shared mutations, the evolutionary tree confirms the origin of strains and their relationship, which is in agreement with the experimental design (Fig. 4). Mutated genes in the strains are listed in SI Appendix, Table S4.
Fig. 4.
Evolutionary relationship between strains based on shared mutations. There are a total of 146 protein-altering mutated genes; corresponding gene names are listed. Asterisks represent nonsense mutations.
We used the Saccharomyces Genome Database (SGD) Gene Ontology (GO) Slim Mapper for the GO Slim Process analysis, where there are 101 GO categories. These categories define biological processes used in bioinformatics analyses. Individual yeast genes can be attributed, on average, to 2.23 GO Slim Processes categories. A total of 2,934 yeast genes belong to only 1 single GO category, and 1 gene belongs to 23 different GO categories, which is the maximal number. Using the GO Slim Process category analysis, we found that the 146 protein-coding genes that were selected in our screens and that carry mutations on average belong to 2.79 GO Slim Processes (SI Appendix, Fig. S12A), which is above the average (SI Appendix, Fig. S12B). This indicates that these 146 genes are possibly involved in more biological processes than average yeast genes are. This could reflect the complexity of the protein secretion pathway; i.e., proteins involved in the protein secretion pathway are involved in other cellular processes. Furthermore, of 101 biological process categories, 23 of them show a twofold or higher frequency of appearance in the mutated gene group than for the whole gene group (SI Appendix, Fig. S12C). This could point to the fact that these overrepresented processes are contributing more, or more specifically, to the higher amylase production in the sorted mutants. For example, compared with other categories, the most enriched (3.7-fold) GO process category was “Endocytosis.” This suggests that genes coding for endocytosis are potential targets for rational design for increasing protein secretion, which is consistent with a recent study reporting improvement of protein production through the blocking of endocytosis (15). Similarly, the GO process categories “organelle fusion,” “vesicle organization,” and “membrane fusion,” all related to protein trafficking, show enrichment (more than twofold) among mutated genes. We therefore speculate that the genes GOS1, SNC2, and USO1, which fall into all these three categories, could be potential engineering targets for improving protein secretion.
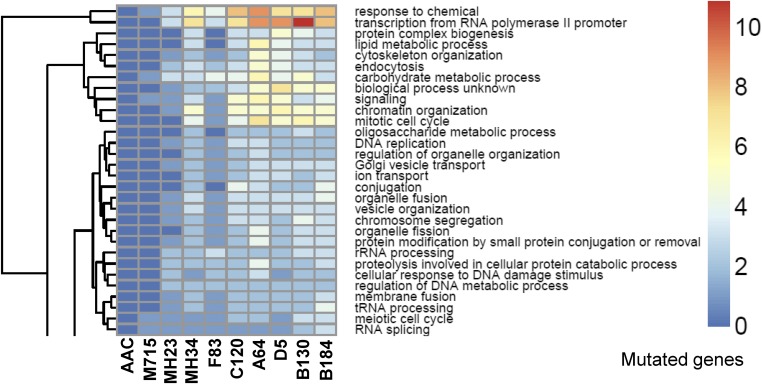
Clustering of enriched mutations is clearly shown by applying GO biological process analysis for each strain (Fig. 5). The accumulation of mutated genes from generation to generation in different GO categories may provide clues for conserved mechanisms in the evolutionary path. The clustering clearly displays that there is an accumulation of mutations down the strain pedigree. For example, more mutated genes in the GO categories “Organelle fusion,” “Membrane fusion,” and “tRNA processing” are found in the best producer B184 compared with its siblings (C120, A64, D5, and B130) (Fig. 5). Likewise, B130 has the most mutated genes in the GO category “Transcription from RNA polymerase II promoter”; and more mutated genes in the GO categories “Lipid metabolic process,” “Cytoskeleton organization,” and “Endocytosis” are found in the strains A64 and D5. The diversity leads to the different protein secretion capacities, and the analysis clearly shows that there are multiple routes that can lead to enhanced protein production.
Fig. 5.
The most significantly enriched mutation gene region in GO Slim Process categories. For the complete clustering enrichment, see SI Appendix, Fig. S13.
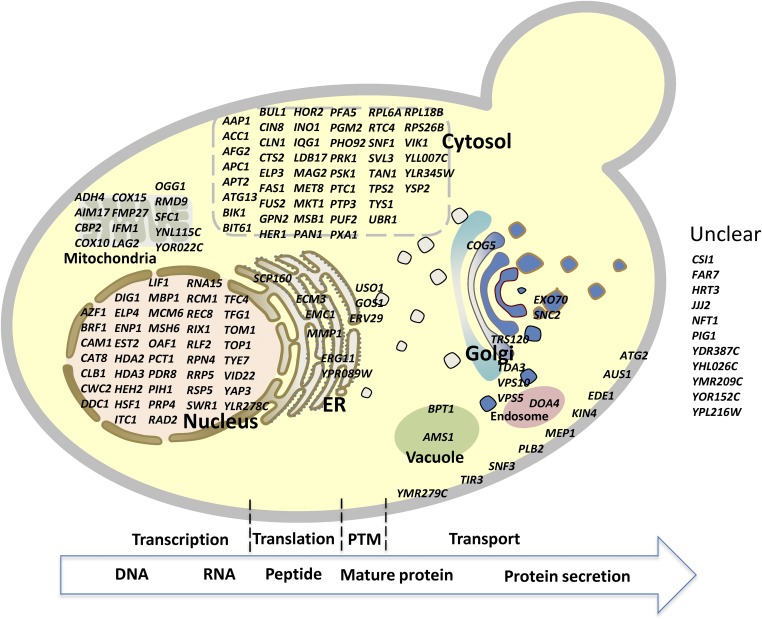
The whole protein production process can be divided into four major steps: transcription, translation, posttranslational modifications and folding to a mature protein, and protein secretion. We mapped all of the mutations on a schematic representation of the protein production process to visualize the function and location of the mutated proteins (Fig. 6). Descriptions of the mutated genes with links to the SGD are listed in SI Appendix, Table S4, which will allow for rapid access to their annotated functions. Transcription and translation, which are controlled by complex regulatory networks (16–18), are crucial for protein production, as they determine the production efficiency from DNA to peptide. Some transcriptional regulators can alter transcriptional levels in response to environmental changes; for example, Hac1p up-regulates chaperones in response an unfolded protein response (UPR), following accumulation of unfolded or mis-folded protein in the ER (19). Heat-shock response (HSR) is another universal cellular response to protect or recover cells from cellular stresses including ER stress; overexpression of the HSF1-R206S, a mutant of the major HSR transcriptional regulator heat-shock factor HSF1, can constitutively activate HSR, and heterologous protein secretion was improved by releasing mis-folded proteins causing ER stress through HSR induction of ER and cytosolic chaperones (20). It is interesting that a HSF1 mutation was also found in one of our mutant strains, D5, which may cause improved protein secretion. Nonsense mutations in HDA2 and HDA3 were found in the first round of sorted variants MH34 and MH23, respectively. Both HDA2 and HDA3 are essential regulatory subunits of the HDA1 histone deacetylase complex, and mutation in either of them will lead to the loss of catalytic activity of the HDA1 complex (21), hereby affecting relevant biological processes (22). Mutated genes in the trafficking pathway may also influence limiting steps in secretion. For example, Erv29p can serve as a cargo receptor for soluble proteins with α-factor leader packaging into COPII vesicles for secretion (23). Mutations in ERV29 may affect its activity, thereby modulating secretory efficiency in the best α-amylase–secreted strain B184. Likewise, mutations in other trafficking genes, e.g., COG5, SNC2, GOS1, USO1, etc., may alter trafficking and influence α-amylase secretion.
Fig. 6.
Genes with identified mutations mapped on the protein secretion pathway.
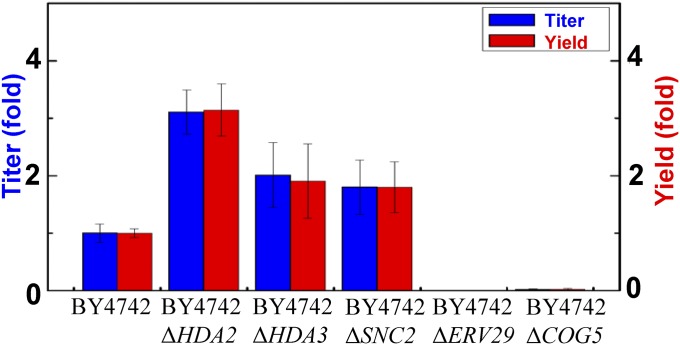
To further evaluate the relationship between protein secretion and identified mutations, we examined the α-amylase secretion capacities of the BY4742 strain with different single-gene deletions. Deletion of HDA2, HDA3, and SNC2 resulted in 3.1-, 2.0-, and 1.8-fold α-amylase secretion enhancement, respectively, compared with the parental strain BY4742. In contrast, deletion of ERV29 and COG5 resulted in defective amylase secretion (Fig. 7). These results support our findings that mutations identified in this study are associated with protein secretion and can be potentially used as reverse metabolic engineering targets. As protein secretion is generally found to depend on the properties of the protein (24), we evaluated our best performance strain for secretion of another heterologous protein, glucan 1,4-α-glucosidase. Compared with the wild-type strain CEN.PK 530.1C, B184 secreted 1.7-fold more glucan 1,4-α-glucosidase, and deletion of HDA2 in BY4742 led to a 1.3-fold increase in glucan 1,4-α-glucosidase secretion (SI Appendix, Fig. S14). These results suggest that our findings may be generally applicable for improving protein secretion, although the increased level for glucan 1,4-α-glucosidase is not as large as that for α-amylase. Careful examination of the mutated genes will be needed to unravel the underlying mechanisms and can assist in rational design and systematic engineering of protein secretion hosts.
Fig. 7.
α-Amylase secretion of yeast strains was altered by single deletion of genes, in which mutations were identified in this study.
In summary, we present a microfluidic droplet-sorting method for high-throughput screening of mutant yeast strain libraries for improved protein secretion. The method enables screening of large libraries as an alternative to conventional screening methods that are laborious and inefficient. The sorting method is reliable for identification of cells with improved secretion, and the improved mutants selected by sorting are genetically stable, which is beneficial for industrial application. Furthermore, we coupled this method to whole-genome sequencing of the best sorted strains and found that all of the strains have accumulated many mutations. The link between protein secretion and identified mutations was supported by single-gene deletions. Some of these mutations could be efficient targets for rational forward engineering strategies and could potentially significantly improve protein secretion. We used two rounds of high-throughput screening, but it would be possible to repeat the procedure more than twice and continuously select for even better-producing strains. This approach could easily be combined with different mutagenesis strategies that can increase the “pool” of mutations (different irradiation energies, mutagenic chemicals, etc.). In addition to the sequential mutagenesis used here, mutagenesis could also be coupled with recombination to generate even larger sets of variants for screening. Chromosomal mutations of selected strains were identified by whole genomic DNA sequencing and analyzed. GO analysis on mutated genes reveals some important process categories linked to efficient protein secretion, and we propose that more processes could be potential targets for metabolic engineering of S. cerevisiae for improved protein secretion. It is expected to release limitations of protein production and secretion of yeast cell factories at different levels by combinatorial optimization.
To facilitate future elucidation of mechanisms involved in protein secretion, we mapped the mutations by their localization and function. Detailed exploration of the different effects of mutated genes will help to better understand the protein secretory mechanisms in yeast.
Materials and Methods
Strains and Plasmids.
The yeast S. cerevisiae CEN.PK 530.1C [MATα URA3 HIS3 LEU2 TRP1 SUC2 MAL2-8c tpi1(41-707)::loxP-KanMX4-loxP] was used as the recombinant protein secretion host. The S. cerevisiae CEN.PK 530.1C was transformed with the plasmid pAlphaAmyCPOT, which contains an α-factor leader followed by an α-amylase gene from Aspergillus oryzae, named AAC (10). All mutant strains in this study are derived from the AAC strain by UV mutagenesis. S. cerevisiae BY4742 (MATα his3Δ1 leu2Δ0 lys2 Δ0 ura3 Δ0) and its single-gene deletion strains were purchased from the EUROSCARF collection (available at web.uni-frankfurt.de/fb15/mikro/euroscarf/). Plasmid p426GPD-AlphaAmy was transformed to BY4742 and its BY-single knockout strains, and α-amylase secretion was evaluated (10).
Yeast UV-Mutagenesis Library Construction.
The yeast UV-mutagenesis library was prepared as described previously (11). A single yeast colony from fresh plate was grown in the yeast extract, peptone, and dextrose (YPD) medium overnight. The cells were collected by centrifugation and washed with sterile water. The cell suspension was spread on starch agar plates and exposed to 40 W of UV light (UV cross-linker, Topac Inc.) at 254 nm for 2–8 s (corresponding to 4–16 mJ cm−2). Plates were incubated at 30 °C in the dark after UV treatment until colonies formed (∼5–7 d).
Media and Culture Conditions.
The YPD medium contained, per liter, 10 g yeast extract, 20 g peptone, and 20 g glucose. The yeast extract, peptone, and ethanol (YPE) medium contained, per liter, 10 g yeast extract, 20 g peptone, 10 g ethanol, and 0.5 g glucose. The SD-Ura medium contained 6.9 g yeast nitrogen base without amino acids, 770 mg Complete Supplement Mixture (CSM, w/o uracil), and 20 g glucose. For tube or shake flask fermentations, the SD-2×SCAA medium (26) contained, per liter, 20 g glucose, 6.9 g yeast nitrogen base without amino acids, 190 mg Arg, 400 mg Asp, 1,260 mg Glu, 130 mg Gly, 140 mg His, 290 mg Ile, 400 mg Leu, 440 mg Lys, 108 mg Met, 200 mg Phe, 220 mg Thr, 40 mg Trp, 52 mg Tyr, 380 mg Val, 1 g BSA, 5.4 g Na2HPO4, and 8.56 g NaH2PO4·H2O (pH = 6.0 by NaOH). For bioreactor fermentations, 5.4 g Na2HPO4 and 8.56 g NaH2PO4·H2O in the SD-2×SCAA were replaced by 2 g KH2PO4 (pH = 6.0 by NaOH). The starch agar plate contained, per liter, 0.04 g glucose, 10 g starch, 6.9 g yeast nitrogen base without amino acids, and 20 g agar. The starch and ethanol agar plate contained, per liter, 0.04 g glucose, 10 g starch, 10 g ethanol, 6.9 g yeast nitrogen base without amino acids, 790 mg CSM, and 20 g agar. All S. cerevisiae strains were grown at 30 °C for 96 h.
Manufacturing of Microfluidic Devices.
Microfluidic chips were manufactured with poly(dimethylsiloxane) (PDMS) and glass according to standard soft lithographic techniques (25). Briefly, masters were fabricated on 4-inch silicon wafers using SU-8 (MicroChem), an epoxy-based negative photoresist. PDMS base was mixed with a curing agent in a 10:1 ratio and poured onto the master template. After baking overnight at 65 °C, the PDMS slab was peeled off, and holes for the inlets and outlets were punched with biopsy punchers (Harris Uni-Core) and cleaned using Scotch tape. The PDMS slabs were subsequently cleaned in an ultra-sonic bath (15 min) to remove debris, followed by surface activation in oxygen plasma (Femto Scientific). The PDMS slabs and glass slides were bonded together and incubated at 65 °C for 2 h. Finally, a fluorophilic surface treatment was applied to each circuit by injecting Aquapel, flushing with pure HFE-7500 oil, and immediately purging with the filtered N2. For the sorting chips, electrodes were fabricated by heating the chip and low-melting-point solder (Indium) to 100 °C on a hot plate. The liquid solder was injected into the designed electrode channels and interfaced for connection to an off-chip voltage source using an adapter, which was fixed in electrode channel inlets as the liquid solder solidified.
Experimental Setup.
The microfluidic device was fixed on an adjustable xy-table on top of an inverted microscope (Olympus IX51). The syringes were controlled by neMESYS syringe pumps (Cetoni GmbH) except the cell suspension syringe, which was controlled by a Harvard Systems syringe pump. All syringes were connected to the circuit through polyether ether ketone tubing. A 491-nm laser was focused on the channel of the microfluidic device through the objective lens that enabled single-droplet fluorescence detection (525 ± 20 nm) using a photomultiplier tube module (Hamatsu). A high voltage amplifier unit (TREK Inc.) was connected to on-chip electrodes and amplified a purpose-built software-generated signal to create an electric field on the chip. The signal was detected and shown on an oscilloscope (GOS-522B, 20 MHz).
Encapsulation of Yeast Library in Microdroplets.
The yeast library on a starch agar plate was carefully extracted to 1 mL SD-2×SCAA medium, and the cell suspension was ultra-sonicated for 3 × 10 s at 40 W using an ultrasonic probe (Vibra-Cell, Sonics & Materials, Inc.) to avoid cell adhesion. The yeast was then washed three times by centrifuging and exchanging media, and the yeast suspension was diluted at around 1.5 × 106 cells mL−1 to achieve an average cell-to-drop ratio of 0.4. Finally, the yeast was resuspended in the fresh SD-2×SCAA medium (pH 6.0) with BSA increased to 5% (wt/wt) and incubated on ice before transfer into a syringe. The yeast suspension was injected and mixed in a 1:1 ratio with 200 µg⋅mL-1 BODIPY-starch substrate by a T-connecter just before emulsification in the droplet generation circuit. Typically, the generation chip was operated with a total aqueous flow rate of 200 µL⋅h−1 and the oil (HFE-7500 with 1% EA surfactant, a polyethylene glycol-perfluoropolyether amphiphilic block copolymer) flow rate of 1,000 µL⋅h−1, which can produce 20-pL microdroplets at 3,000 Hz for 30 min. The yeast cells were encapsulated in the droplets, and the emulsion was collected in a 1-mL plastic syringe (BD Plastipak).
Sorting of Yeast Cells Encapsulated in Microdroplets.
The emulsion generated in the generation circuit was incubated in a syringe at room temperature for 3 h and subsequently injected into the sorting chip. The sorting operation was usually performed with a flow rate of 30 µL⋅h−1 emulsion and 300 µL⋅h−1 HFE-7500 oil. Single-droplet fluorescence was acquired for each droplet approaching the sorting junction. An electric field was activated if droplet fluorescence value exceeded a predefined threshold value by supplying a voltage to the on-chip electrodes. The threshold was defined according to different cell libraries to sort a desired fraction (0.2–3%) of the droplets based on the fluorescence distribution of the droplet population. The electrodes were operated with 400- to 800-µs pulses of 800–1,000 Vp-p square waves with a frequency of 30 kHz in different experiments. Sorted droplets were collected in a syringe operated with a withdrawal rate of around 100–115 µL⋅h−1, which was fine-tuned to collect only the sorted droplets.
Extraction of Yeast Cells After Sorting.
After sorting was completed, the sorted yeast cells in the collection syringe were recovered by removing excessive oil and adding 5 µL of emulsion destabilizer. The emulsion was broken by gentle vortex, and 300 µL of fresh medium was added to extract yeast cells. The yeast cells in the fresh medium were spread on starch agar plates, which were subsequently placed at 30 °C until single colonies formed after 5–7 d. The waste material from the sorting experiment was also treated with the same procedure as a control.
Protein Measurement.
The α-amylase quantification can be calculated from enzyme activity, which was measured as described previously (10). Briefly, a Ceralpha kit was used with α-amylase from Aspergillus oryzae as a standard with an α-amylase protein conversion coefficient of 69.6 U⋅mg−1. The invertase activity was measured as described previously (26). The glucan 1,4-α-glucosidase was measured by using Amyloglucosidase Assay Reagent (Megazyme) according to the manufacturer’s instructions.
SDS/PAGE.
For SDS/PAGE analysis, strains were grown in SD-2×SCAA medium without BSA. After cultivation, the supernatant was collected by centrifugation, mixed with NuPAGE LDS Sample Buffer (4×) and NuPAGE Sample Reducing Agent (10×), and heated up to 95 °C for 5 min. Then samples were loaded onto gradient (4–20%) precast polyacrylamide gel (Mini-PROTEAN TGX gel, Bio-Rad) and stained with Coomassie blue after gel electrophoresis.
Genomic DNA Sequencing and Data Analysis.
Total genomic DNA of selected strains was extracted by using the Blood & Cell Culture DNA Kit (Qiagen). Then DNA was prepared using the Illumina DNA TruSeq protocol, according to the manufacturer’s instructions and with a fragment length of 650 bp. The samples were sequenced using the version 2 chemistry on an Illumina MiSeq, paired-end 500 cycles (2 × 250 bp). Each sample was represented by 1.1–2.0 million sequence reads. The reads were mapped to the reference genome (CEN.PK 113–7D, cenpk.tudelft.nl) using MosaikAligner version 2.1.32 (code.google.com/p/mosaik-aligner/). The alignments were postprocessed to realign potiential indels and remove likely PCR duplicates using GATK 2.3.9 (27) and Picard tools 1.100 (picard.sourceforge.net), respectively. After postprocessing, the average mapped coverage ranged from 35 to 60×.
Single nucleotide variants and small indels were detected using GATK UnifiedGenotyper and annotated using SnpEff 3.4 (snpeff.sourceforge.net). Large-scale chromosome duplications were detected by plotting the mapped coverage over all chromosomes. Smaller structural variants were detected using SVseq2 (28). All detected variants were inspected manually in a genome browser to detect and discard obvious false positives.
The GO analysis was carried out by the SGD Gene Ontology Slim Mapper on the Saccharomyces Genome Database (www.yeastgenome.org/cgi-bin/GO/goSlimMapper.pl) according to the instructions. The clustered heatmap of mutation genes of mutant strains in GO Slim Process category was made by R 3.03 with Package “pheatmap” (cran.r-project.org/web/packages/pheatmap/index.html). The information about the genes’ function and localization, etc., can be accessed from the SGD.
Supplementary Material
Acknowledgments
We thank Yongjin Zhou for useful discussions; and the Science for Life Laboratory, the National Genomics Infrastructure (NGI), and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure. This work was funded by the Novo Nordisk Foundation and the European Research Council Grant 247013 (to J.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506460112/-/DCSupplemental.
References
- 1.Martínez JL, Liu L, Petranovic D, Nielsen J. Pharmaceutical protein production by yeast: Towards production of human blood proteins by microbial fermentation. Curr Opin Biotechnol. 2012;23(6):965–971. doi: 10.1016/j.copbio.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Feizi A, Österlund T, Petranovic D, Bordel S, Nielsen J. Genome-scale modeling of the protein secretory machinery in yeast. PLoS One. 2013;8(5):e63284. doi: 10.1371/journal.pone.0063284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hou J, Tyo KEJ, Liu Z, Petranovic D, Nielsen J. Metabolic engineering of recombinant protein secretion by Saccharomyces cerevisiae. FEMS Yeast Res. 2012;12(5):491–510. doi: 10.1111/j.1567-1364.2012.00810.x. [DOI] [PubMed] [Google Scholar]
- 4.Delic M, Göngrich R, Mattanovich D, Gasser B. Engineering of protein folding and secretion-strategies to overcome bottlenecks for efficient production of recombinant proteins. Antioxid Redox Signal. 2014;21(3):414–437. doi: 10.1089/ars.2014.5844. [DOI] [PubMed] [Google Scholar]
- 5.Caspeta L, et al. Biofuels. Altered sterol composition renders yeast thermotolerant. Science. 2014;346(6205):75–78. doi: 10.1126/science.1258137. [DOI] [PubMed] [Google Scholar]
- 6.Baret JC, et al. Fluorescence-activated droplet sorting (FADS): Efficient microfluidic cell sorting based on enzymatic activity. Lab Chip. 2009;9(13):1850–1858. doi: 10.1039/b902504a. [DOI] [PubMed] [Google Scholar]
- 7.Brouzes E, et al. Droplet microfluidic technology for single-cell high-throughput screening. Proc Natl Acad Sci USA. 2009;106(34):14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agresti JJ, et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci USA. 2010;107(9):4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang BL, et al. Microfluidic high-throughput culturing of single cells for selection based on extracellular metabolite production or consumption. Nat Biotechnol. 2014;32(5):473–478. doi: 10.1038/nbt.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Tyo KEJ, Martínez JL, Petranovic D, Nielsen J. Different expression systems for production of recombinant proteins in Saccharomyces cerevisiae. Biotechnol Bioeng. 2012;109(5):1259–1268. doi: 10.1002/bit.24409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sjostrom SL, et al. High-throughput screening for industrial enzyme production hosts by droplet microfluidics. Lab Chip. 2014;14(4):806–813. doi: 10.1039/c3lc51202a. [DOI] [PubMed] [Google Scholar]
- 12.Liu Z, et al. Improved production of a heterologous amylase in Saccharomyces cerevisiae by inverse metabolic engineering. Appl Environ Microbiol. 2014;80(17):5542–5550. doi: 10.1128/AEM.00712-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolfe KH. Yesterday’s polyploids and the mystery of diploidization. Nat Rev Genet. 2001;2(5):333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- 14.Presser A, Elowitz MB, Kellis M, Kishony R. The evolutionary dynamics of the Saccharomyces cerevisiae protein interaction network after duplication. Proc Natl Acad Sci USA. 2008;105(3):950–954. doi: 10.1073/pnas.0707293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodríguez-Limas WA, Tannenbaum V, Tyo KE. Blocking endocytotic mechanisms to improve heterologous protein titers in Saccharomyces cerevisiae. Biotechnol Bioeng. 2015;112(2):376–385. doi: 10.1002/bit.25360. [DOI] [PubMed] [Google Scholar]
- 16.Lee TI, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298(5594):799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 17.Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feist AM, Herrgård MJ, Thiele I, Reed JL, Palsson BO. Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol. 2009;7(2):129–143. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. 2011;333(6051):1891–1894. doi: 10.1126/science.1209126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou J, Osterlund T, Liu Z, Petranovic D, Nielsen J. Heat shock response improves heterologous protein secretion in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2013;97(8):3559–3568. doi: 10.1007/s00253-012-4596-9. [DOI] [PubMed] [Google Scholar]
- 21.Wu J, Carmen AA, Kobayashi R, Suka N, Grunstein M. HDA2 and HDA3 are related proteins that interact with and are essential for the activity of the yeast histone deacetylase HDA1. Proc Natl Acad Sci USA. 2001;98(8):4391–4396. doi: 10.1073/pnas.081560698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaluarachchi Duffy S, et al. Exploring the yeast acetylome using functional genomics. Cell. 2012;149(4):936–948. doi: 10.1016/j.cell.2012.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otte S, Barlowe C. Sorting signals can direct receptor-mediated export of soluble proteins into COPII vesicles. Nat Cell Biol. 2004;6(12):1189–1194. doi: 10.1038/ncb1195. [DOI] [PubMed] [Google Scholar]
- 24.Idiris A, Tohda H, Kumagai H, Takegawa K. Engineering of protein secretion in yeast: Strategies and impact on protein production. Appl Microbiol Biotechnol. 2010;86(2):403–417. doi: 10.1007/s00253-010-2447-0. [DOI] [PubMed] [Google Scholar]
- 25.McDonald JC, et al. Fabrication of microfluidic systems in poly(dimethylsiloxane) Electrophoresis. 2000;21(1):27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 26.Hou J, Tyo K, Liu Z, Petranovic D, Nielsen J. Engineering of vesicle trafficking improves heterologous protein secretion in Saccharomyces cerevisiae. Metab Eng. 2012;14(2):120–127. doi: 10.1016/j.ymben.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 27.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Wang J, Wu Y. An improved approach for accurate and efficient calling of structural variations with low-coverage sequence data. BMC Bioinformatics. 2012;13(Suppl 6):S6. doi: 10.1186/1471-2105-13-S6-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.